Abstract
Talin is a large dimeric protein that couples integrins to cytoskeletal actin. Here, we report the structure of the C-terminal actin-binding domain of talin, the core of which is a five-helix bundle linked to a C-terminal helix responsible for dimerisation. The NMR structure of the bundle reveals a conserved surface-exposed hydrophobic patch surrounded by positively charged groups. We have mapped the actin-binding site to this surface and shown that helix 1 on the opposite side of the bundle negatively regulates actin binding. The crystal structure of the dimerisation helix reveals an antiparallel coiled-coil with conserved residues clustered on the solvent-exposed face. Mutagenesis shows that dimerisation is essential for filamentous actin (F-actin) binding and indicates that the dimerisation helix itself contributes to binding. We have used these structures together with small angle X-ray scattering to derive a model of the entire domain. Electron microscopy provides direct evidence for binding of the dimer to F-actin and indicates that it binds to three monomers along the long-pitch helix of the actin filament.
Keywords: actin, electron microscopy, structure, talin, THATCH domain
Introduction
Talin is one of a number of cytoskeletal proteins, including α-actinin, filamin, tensin and ILK, implicated in linking members of the integrin family of αβ-heterodimeric cell adhesion molecules to filamentous actin (F-actin). Talin (2541 amino acids) is composed of a globular head (residues 1–400), containing a FERM domain, connected to a flexible rod (residues 482–2541) by a short linker sequence containing a calpain-II cleavage site (Critchley, 2004). The FERM F3 subdomain contains a binding site for the β-integrin cytoplasmic domain, and recent structural studies have provided a detailed understanding of how F3 recognises both the NPxY motif (Garcia-Alvarez et al, 2003) and membrane proximal sequences within the β-integrin cytodomain (Wegener et al, 2007). The talin rod is made up of a series of amphipathic helical bundles, a number of which contain binding sites for the cytoskeletal protein vinculin (Papagrigoriou et al, 2004; Gingras et al, 2005), which is thought to stabilise focal adhesions (Saunders et al, 2006), possibly by crosslinking talin to F-actin. Vinculin recognises a series of hydrophobic residues on one side of a helix (Gingras et al, 2005); both X-ray and NMR structures show that these residues are buried in the core of the helical bundle, which must unfold to allow vinculin binding. Indeed, intact talin binds vinculin with low affinity (Patel et al, 2006); the mechanisms for activating the vinculin-binding sites in the talin rod remain to be determined. The talin rod also contains a second integrin-binding site (residues 1984–2113) that appears to be essential for focal adhesion assembly (Moes et al, 2007), although it is unclear how β-integrin interacts with this site. Finally, the C-terminal region of talin (residues 2300–2541) contains a binding site for F-actin (Hemmings et al, 1996) that is homologous to that in the yeast protein Sla2p, the huntingtin-interacting protein HIP1, and the related protein HIP1R. This highly conserved domain has been referred to as an I/LWEQ motif (McCann and Craig, 1997) or more recently the THATCH (talin/HIP1R/Sla2p actin tethering C-terminal homology) domain (Brett et al, 2006). This region of talin is predicted to contain six helices; the N-terminal is the least conserved and appears to negatively regulate actin binding, while the C-terminal helix is required for talin dimerisation (Senetar et al, 2004).
Here, we report the NMR structure of talin residues 2300–2482, a five-helix bundle, which has a similar structure to the HIP1R THATCH core domain, and the crystal structure of helix 6 that forms an antiparallel dimer. Small-angle X-ray scattering (SAXS) together with these two structures allows us to propose a model for the entire talin 2300–2541 dimer, which is elongated in solution. We have mapped the residues involved in actin binding in the five-helix bundle to one face of the domain. Binding requires dimerisation, and is negatively regulated by helix 1, which is on the opposite face to the actin-binding site. Electron microscopy, interpreted with the help of the overall shape of the dimer determined by SAXS, establishes how the dimer interacts with actin filaments and positions the N termini accessible and on opposite sides of the filaments.
Results
Structure of the C-terminal actin-binding domain of talin
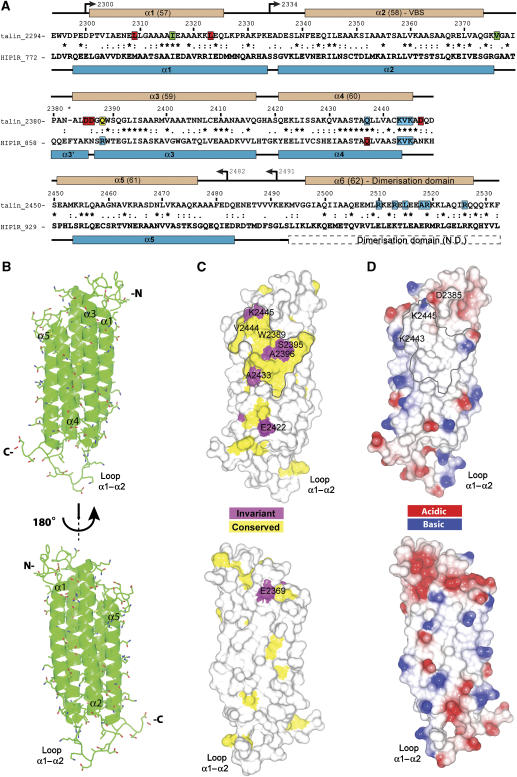
Initially, we used secondary structure prediction and NMR spectroscopy of a range of constructs to identify a talin polypeptide containing the C-terminal actin-binding site suitable for NMR structure determination (see Supplementary Results and Supplementary Figure S1). These studies demonstrated the presence of a stable globular domain (residues 2300–2482) connected by a flexible linker to a helical dimerisation domain (residues 2496–2529). The structure of talin 2300–2482 comprises five antiparallel α-helices (Figure 1A and B; see also Supplementary Results and Supplementary Figure S2D), as described for the homologous HIP1R actin-binding domain (referred to as the THATCH core) (Brett et al, 2006). The helical bundle is stabilised by hydrophobic interactions. There are hydrophobic cores at each end of the bundle separated by a set of small hydrophilic side chains (Thr 2356, 2404, 2435 and Ser 2467) reminiscent of the ‘threonine belt' observed in the structure of talin 782–889 (Fillingham et al, 2005). The hydrophobic core at the N-terminal end of the bundle is arranged around the aromatic ring of Trp 2389 (Supplementary Figure S2C) and incorporates the side chain of the conserved Gln 2367, which points into the bundle. The core at the C-terminal end is made up of the hydrophobic side chains of Leu, Ile and Val residues, and is capped by the aromatic ring of Phe 2341. Helices 2 and 3 are the longest (32 residues), while the other three are approximately two turns shorter (22–27 residues). Helix 2 is connected to the neighbouring helices by long loops (14 and 9 residues), while the other two loops are relatively short (4–5 residues). The long loop between helices 1 and 2 is unstructured and highly dynamic, as indicated by sharp NMR resonances and the lack of NOEs (Figure 1B and Supplementary Figure S2A). The loop between helices 2 and 3 has restricted mobility due to the hydrophobic contacts made by the Val 2376 side chain and the presence of Pro 2380.
Figure 1.
Solution structure of the C-terminal actin-binding domain of talin (residues 2300–2482). (A) Sequence alignment of mouse talin1 with human HIP1R THATCH domain. Symbols denote the degree of conservation: (*) identical, (:) conservative substitution and (.) semi-conservative substitutions. Secondary structures of mouse talin and human HIP1R THATCH core are shown above and below the alignment, respectively—the position of the putative dimerisation domain is indicated. N.D.—structure not determined. Numbering is from mouse talin (P26039). The talin residues mutated are highlighted depending on their effects on F-actin binding: red—increased binding; green—binding similar to wild type; blue—decreased binding. Residue Q2388 is highlighted in yellow. The residues mutated in HIP1R that are equivalent to those analysed in talin are also highlighted for comparison. (B) Ribbon drawing of a representative low-energy structure showing the overall topology of the five-helix bundle of the C-terminal actin-binding domain of talin. (C) Map of conserved surface residues. Magenta—invariant residues; yellow—residues that are highly conserved. (D) Map of surface charge.
As predicted from sequence homology (Figure 1A), the structure most similar to the C-terminal actin-binding domain of talin is that of the HIP1R THATCH core domain (Brett et al, 2006), which shows 35% sequence identity. For the whole structure, a relatively high r.m.s.d. was obtained (main-chain atoms 4.77 Å; all heavy atoms 5.12 Å), but the r.m.s.d. was much lower for the helical regions (main-chain atoms 2.92 Å; all heavy atoms 3.37 Å) (Supplementary Figure S2B). Many structural features are conserved between the two helical bundles, and the majority of the conserved residues occupy similar positions. In particular, the side chain of Trp 2389 has the same orientation in both structures (Supplementary Figure S2C), highlighting its role as a key residue in the hydrophobic core. However, there are also significant differences between the two structures. Helices 1, 2 and 5 are substantially shorter at the C-terminal end of the bundle than the equivalent helices in HIP1R, leading to a less elongated structure and the formation of the large unstructured loop between helices 1 and 2. The shortness of helix 1 can be attributed to the presence of Pro 2328 at the end of the helix, followed by a group of charged residues (Figure 1A). The proline side chain makes contacts with the hydrophobic core and is positioned level with the ends of the other helices. The main difference at the N-terminal end of the bundle is the absence of the short capping helix α3′ that is present in the HIP1R structure (Figure 1A and Supplementary Figure S2C). Instead, there is an extra helical turn in helix 3, followed by a largely immobilised loop. It appears that the key residues for the formation and packing of the HIP1R capping helix are Phe 861 and Tyr 862, which are absent in talin. In addition, Pro 2380 occupies a position corresponding to the middle of helix α3′ in HIP1R. The absence of the α3′ helix in talin affects the position of the N-terminal end of the helices. In the HIP1R structure, the α3′ helix is wedged between the ends of helices 1 and 2, pushing helix 1 out. In talin, the absence of the capping helix allows helix 1 to move closer to helix 2, resulting in a structure that looks more closed at the N-terminal end of the bundle than in HIP1R.
Structure of the talin dimerisation domain
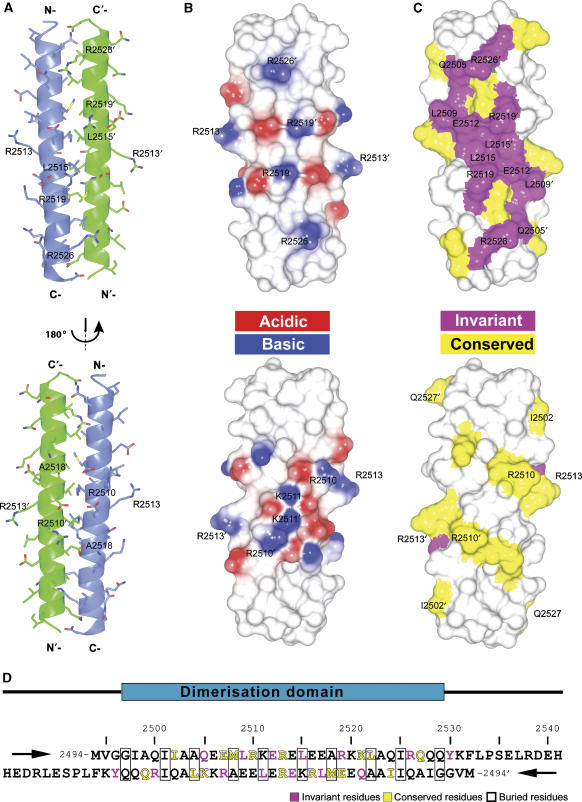
Using secondary structure prediction and NMR, the optimal domain boundaries of the talin dimerisation domain were shown to be residues 2494–2541 (see Supplementary Results and Supplementary Figure S3). Large crystals that diffracted X-rays to 2.2-Å resolution were readily obtained using sparse matrix screening, and the crystal structure was determined using single-wavelength anomalous diffraction from a selenomethionine derivative (Supplementary Figure S4A, Supplementary Table SII and Supplementary Results). The two monomers in the asymmetric unit superimpose well onto one another (average r.m.s.d. for main-chain atoms 0.34 Å, and for all heavy atoms 1.08 Å), the main differences between the monomers being the orientations of long side chains of solvent-exposed residues. Each monomer is composed of a long straight helix approximately 48 Å in length (Figure 2A). The helices form an antiparallel coiled-coil dimer with a small angle between the helices. The formation of the dimer buries approximately 30% (1539 Å2) of the total surface area. Both ends of the dimer are nonpolar with a highly charged belt in the middle (Figure 2B). There is a salt bridge cluster in the centre of the dimer formed by the side chains of K2511 and E2514 from each monomer, with the four side chains making intra- and intermolecular contacts within the cluster (Supplementary Figure S4B). Additionally, intramolecular salt bridges are observed for E2516/R2519 in both monomers, stabilising the helix (Supplementary Figure S4C). Interestingly, E2507 from one of the monomers forms an intermolecular salt bridge with K2521, while in the other monomer this residue forms an intramolecular salt bridge with R2510 (Supplementary Figure S4B). In all other cases, the charged groups are too far apart to make a salt bridge and some of the side chains are hydrogen-bonded to nearby solvent molecules.
Figure 2.
Structure of the talin dimerisation domain. (A) Cartoon representation of the crystal structure of the dimerisation helix (2496–2529) showing the antiparallel coiled-coil dimer. (B) Surface electrostatic potential of the dimer. (C) Map of conserved residues: magenta—invariant residues; yellow—highly conserved residues. (D) Sequence of residues 2494–2541, which includes the dimerisation helix—two antiparallel peptide sequences are shown.
No density was observed for the first three residues at the N terminus of both monomers where a pair of glycines (G2496/G2497) destabilise the helix. The last 12 residues at the C-terminus (2530–2541) are also disordered and no density was observed in the crystal structure—this is consistent with [1H,15N]HSQC NMR spectra where a dozen sharp peaks are observed (Supplementary Figure S3B). The dimer is stabilised by nonpolar interactions involving side chains of I2501, A2504, M2508, L2515, A2518, L2522 and I2525 located on the same face of the helix (Figure 2D). The aliphatic part of the side chain of K2511 also contributes to the hydrophobic core and the amino group is stabilised away from the core by a salt bridge with E2514 (Supplementary Figure S4B). Most of these residues are conserved in the THATCH domains identified to date (Brett et al, 2006). Interestingly, the solvent-exposed residues Q2505, L2509, E2512, L2515, R2519, R2526 and Y2530 are even more conserved (Figure 2C and D) and are clustered on one face of the dimer, resulting in a conserved surface made up of both charged and hydrophobic residues. These characteristics suggest that this surface may be involved in actin binding.
Identification of residues involved in actin binding
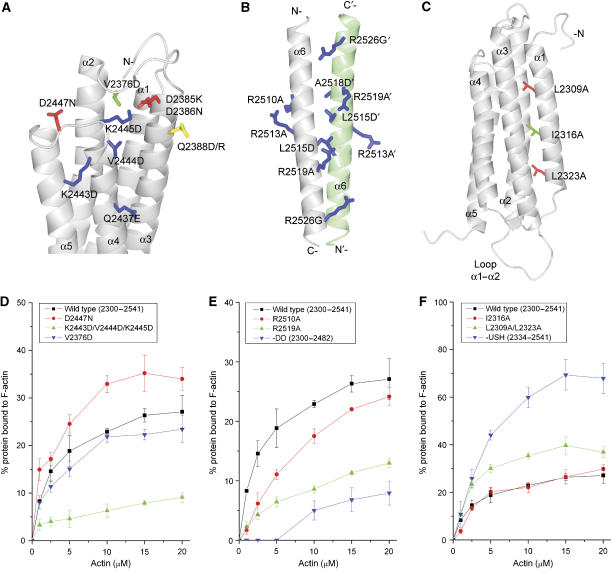
The five-helix bundle that comprises the core of the C-terminal actin-binding domain of talin contains a number of conserved surface-exposed residues that are predominantly clustered on the face made up of helices 3 and 4 (Figure 1C). This part of the surface consists of an extensive hydrophobic patch surrounded by positively charged groups (Figure 1D), characteristics that make it a good candidate for a region involved in F-actin binding. Indeed, the actin-binding site in the HIP1R THATCH domain has been mapped to the equivalent surface (Brett et al, 2006). However, this surface is on the face of the domain opposite helix 1, which has been shown to negatively regulate actin binding (Senetar et al, 2004). In addition, actin binding depends on the presence of the C-terminal dimerisation helix (see below). To map the residues in talin 2300–2541 directly or indirectly involved in actin binding, we tested the effects of a series of mutations on its affinity for F-actin. In the presence of a six-fold molar excess of F-actin, ∼30% of wild-type talin 2300–2541 co-sedimented with F-actin (Figure 3D), a result similar to that previously observed for talin and HIP1R (Senetar et al, 2004). The single mutations Q2388D, Q2437E, K2443D, V2444D and K2445D (equivalent to those analysed in the HIP1R THATCH domain; Brett et al, 2006) all caused a significant reduction in F-actin binding (Table I), and the triple mutant K2443D/V2444D/K2445D reduced binding to 27% of wild type. Interestingly, although the Q2437E mutant reduced binding affinity, the equivalent mutation in HIP1R (Q916E) increased binding. While this difference is puzzling, our results clearly demonstrate that incorporating acidic amino acids into the actin-binding surface of talin reduces binding. Both the Q2388D mutation in talin (Supplementary Figure S5D and Table I) and the equivalent R867D mutation in HIP1R reduce binding to F-actin; interestingly, substituting talin Q2388 with arginine had no effect on binding, indicating that either a basic or an uncharged residue at this position can be tolerated. Talin D2447, located close to the conserved basic residues K2443 and K2445, is homologous to N926 in HIP1R and a talin D2447N mutant increased F-actin binding to 144% that of wild type (Figure 3D and Table I). Thus, reducing the acidic charge in the proximity of the conserved basic residues (Figure 1C and D) increases the ability of talin to bind F-actin.
Figure 3.
Identification of residues in the C-terminal actin-binding site of talin, which contributes to binding. (A–C) Ribbon diagrams highlighting the mutations introduced in talin 2300–2541. (A) F-actin-binding surface on the core five-helix bundle, (B) the dimerisation domain and (C) the USH. Residues are colour coded according to the effects of the mutation on F-actin binding compared to wild type: red—increase in binding; green—no change; blue—decrease in binding. Residue Q2388 is shown in yellow. (D–F) Quantitative analysis of the effects of talin mutations on F-actin binding (means of three independent experiments) as determined using the actin-co-sedimentation assay described in Materials and methods. Bars represent standard deviations. The data for all the mutants analysed are shown in Supplementary Figures S5, S7 and S8.
Table 1. Effects of mutations in talin 2300–2541 on F-actin binding.
| Helices | Residue mutated | Position mutated | Co-sedimentation pellet (% of WT) | Dimer |
|---|---|---|---|---|
| 1–6 | Wild type (2300–2541) | — | 100±3 | Yes |
| 1–6 | V2376D | ABS | 95±5 | Yes |
| 1–6 | Q2388D | ABS | 59±5 | Yes |
| 1–6 | Q2388R | ABS | 102±9 | Yes |
| 1–6 | Q2437E | ABS | 62±13 | Yes |
| 1–6 | K2443D | ABS | 43±5 | Yes |
| 1–6 | V2444D | ABS | 54±10 | Yes |
| 1–6 | K2445D | ABS | 48±10 | Yes |
| 1–6 | D2447N | ABS | 144±8 | Yes |
| 1–6 | K2443D/V2444D/K2445D | ABS | 27±6 | Yes |
| 1–6 | D2385K/D2386N | ABS | 113±13 | Yes |
| 1–6 | D2385K/D2386N/D2447N | ABS | 138±16 | Yes |
| 1–6 | R2510A | DD | 77±5 | Yes |
| 1–6 | R2513A | DD | 66±14 | Yes |
| 1–6 | L2515D | DD | 36±6 | No |
| 1–6 | A2518D | DD | 43±10 | No |
| 1–6 | L2515D/A2518D | DD | 42±7 | No |
| 1–6 | R2519A | DD | 37±3 | No |
| 1–6 | R2526G | DD | 33±5 | No |
| 1–6 | R2526A | DD | 37±9 | No |
| 1–5 | DD deletion (2300–2482) | DD | 22±7 | No |
| 1–6 | L2309A | USH | 113±3 | Yes |
| 1–6 | I2316A | USH | 96±9 | Yes |
| 1–6 | L2323A | USH | 127±3 | Yes |
| 1–6 | I2316A/L2323A | USH | 135±13 | Yes |
| 1–6 | L2309A/L2323A | USH | 154±7 | Yes |
| 2–6 | USH deletion (2334–2541) | USH | 262±18 | Yes |
| ABS, actin-binding surface; DD, dimerisation domain; USH, upstream helix. | ||||
| Summary of the results from F-actin co-sedimentation assays performed at 10 μM F-actin and 4 μM talin. | ||||
One of the key structural differences between the THATCH cores of talin and HIP1R is in the region between helices 2 and 3, where a loop is observed in talin and a short α3′ helix in HIP1R (Figure 1A). Valine 2376 in the loop contacts W2389 in the centre of the actin-binding site (Figures 1C and 3A), and we therefore made a V2376D mutant to study the effects on binding of destabilising this loop. The [1H,15N]HSQC NMR spectrum of this mutant showed few changes in comparison with the wild-type protein, indicating that the protein adopted the correct fold, and its ability to bind F-actin was not affected (Figure 3D). We conclude that this poorly conserved loop, which is located on the opposite side of the bundle, is probably not involved in F-actin binding, and that V2376 is not important for maintaining the hydrophobic core near W2389.
At the N terminus of helix 3, there are two aspartate residues (D2385 and D2386) that are near the conserved basic residues within the actin-binding site, and we therefore mutated these to the equivalent residues in HIP1R, that is, D2385K and D2386N (Figure 3A). Interestingly, the double mutation produced only a small increase in binding (Table I). Since we had already shown that a D2447N mutation increased binding significantly (144%), we made a triple mutant (D2385K/D2386N/D2447N) but this bound no better than the D2447N mutant alone. The sequence conservation around D2385 and D2386 is low and the structural features of talin and HIP1R are very different in this region, suggesting that it is not critical for F-actin binding. As described above, Q2388 on the adjacent helical turn of helix 3 could be mutated to arginine without any effect on F-actin binding, but a mutation to an acidic residue did reduce binding to 59% of wild type (Table I). Overall, the mutagenesis data show that exposed residues on two adjacent helical turns have very different effects on F-actin binding, indicating that the talin–actin contact site is a well-defined hydrophobic patch surrounded by positively charged residues.
The talin dimerisation domain is important for F-actin binding
Complete removal of the dimerisation domain reduced F-actin binding to 22% of the wild type (Figure 3E and Table I). To check whether this effect is due to conversion of the dimer to the monomer, or to loss of additional contacts between the dimerisation domain itself and F-actin, we made a series of mutants designed to inhibit dimer formation with minimum effect on the properties of the helices. Three point mutants (R2526G, L2515D and A2518D) were all monomeric as determined by gel filtration (Supplementary Figure S6) and NMR, as was the double mutant L2515D/A2518D (Table I). For these mutants, we observed a large increase in the number of sharp intense signals in the [1H,15N]HSQC spectra and an overall reduction in the resonance line-widths. The positions of the majority of the resonances corresponding to the five-helix bundle remain unchanged. We conclude that the above mutations lead to the complete unfolding of the dimerisation domain, while the five-helix bundle remains unperturbed. These mutants showed much reduced actin binding, although they did bind to actin significantly better than the polypeptide lacking the entire dimerisation domain (Table I), suggesting that the dimerisation domain may also interact with actin.
Analysis of the sequence of the dimerisation domain shows that all the invariant residues cluster on one face of the domain (Figure 2C and D). We have mutated two invariant basic residues, R2519 and R2526, to alanine to reduce the overall positive charge of this surface (Figure 3B). Surprisingly, these two mutants are monomeric (Table I) and as a result their F-actin-binding capacity was substantially reduced (Figure 3E and Table I). The core of this antiparallel dimer is not a typical leucine zipper and is not highly hydrophobic. Indeed, the crystal structure shows many salt bridges between the two helices, and our mutational analysis suggests that they are critical to maintain a stable fold. We therefore mutated two conserved though not invariant residues (R2510 and R2513) to alanine (Figure 2C and D). R2510 is on the opposite face of the dimer and makes salt bridges to neighbouring residues (Figure 2A), while R2513 is at the edge of the dimer and is not involved in any salt bridges (Figure 3B). Gel filtration and NMR data show that both mutants are dimeric (Table I), but both showed a significant decrease in F-actin binding (Figure 3E, Table I and Supplementary S7C). Taken together, the data show that the dimerisation of the actin-binding domain is essential for F-actin binding, in agreement with the recent work of Smith and McCann (2007), and also suggest that the dimerisation domain itself might contribute to binding through electrostatic interactions.
Effects of mutating the upstream helix on actin binding
The N-terminal helix 1 of the actin-binding domain, also termed the upstream helix (USH), was identified as a conserved structural element that decreases the actin-binding capacity of this family of proteins (Senetar et al, 2004). Removal of this helix in talin led to a 2.6-fold increase relative to the full domain in the fraction of talin (2334–2541) bound to actin (Figure 3F), in agreement with published data (Senetar et al, 2004). However, there was also a slight increase in the amount of this polypeptide that pelleted in the absence of F-actin; NMR shows that removal of the USH alters the conformation of the domain, which may decrease stability and lead to a tendency to aggregate with time. The USH is anchored to the rest of the domain through nonpolar interactions involving L2309, I2316 and L2323 (Figure 3C), and the effect of mutating these residues on F-actin binding is shown in Figures 3F, Supplementary Figure S8 and Table I. The L2309A or L2323A mutations led to a small but reproducible increase in binding, while the double mutant (L2309A/L2323A) showed a larger increase in binding (154% that of wild type; Figure 3F). An increase in binding was also observed for the double mutant I2316A/L2323A (135%), while no change was observed for the I2316A single mutant. Although none of the mutations increased binding to the same degree as removing the entire USH (Table I), they had little effect on the solubility of the protein compared to removal of the entire USH. These results support previous conclusions that the USH negatively regulates actin binding, through a conformational mechanism, details of which remain unclear.
SAXS analysis of the C-terminal actin-binding domain of talin shows an extended dimeric protein
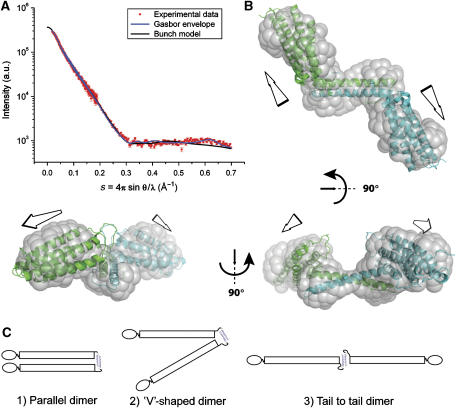
To investigate the overall shape of the talin 2300–2541 dimer, we carried out SAXS experiments. From the scattering profile (Figure 4A), the maximum linear dimension (Dmax) for the talin dimer is 124 (±6) Å, suggesting that the dimer is elongated. Similarly, the distribution of scattering mass of the dimer (as indicated by the radius of gyration, Rg) gives Rg=37.0 (±0.3) Å, a high value for a 52-kDa dimer; one would expect an Rg value of ∼25 Å for a spherical particle of similar mass. These observations clearly demonstrate that the dimer is characterised by an extended non-globular arrangement of the THATCH domain, consistent with the results from analytical gel filtration (see Supplementary Results).
Figure 4.
SAXS data for the dimeric talin polypeptide 2300–2541. (A) Experimental scattering profile of the talin dimer (red) compared with the theoretical scattering curves from the shape reconstructed ab initio with GASBOR (blue line), and the structural model of the dimer obtained with the rigid body modelling program BUNCH (black line). The goodness of fit of GASBOR and BUNCH profiles versus experimental data is indicated by their χ2 values (χ2=2.5 and 2.2, respectively). (B) Three orthogonal views of the talin dimer model (monomers in cyan and green) deduced using BUNCH fitted within the shape envelope provided by GASBOR and derived from experimental scattering data alone (transparent grey surface). (C) The talin C-terminal dimerisation domain suggests that full-length talin may adopt a number of conformations, for example, (1) a parallel dimer (2) a V-shaped dimer or (3) an extended dimer.
Ab initio shape reconstructions from the experimental SAXS profile alone using GASBOR (Svergun et al, 2001) highlight the elongated shape of the dimeric talin 2300–2541 polypeptide; the calculated molecular envelope is shown in Figure 4B. The high-resolution structures of the dimerisation helix and the five-helix bundle were used as rigid bodies to model the dimeric talin polypeptide using BUNCH (Petoukhov and Svergun, 2005). The BUNCH model is shown in Figure 4B superimposed on the model-independent shape reconstruction (see also Supplementary movie). The ab initio shape and the rigid body model reconstructions were evaluated against the experimental scattering profiles assuming a two-fold symmetry constraint; both reproduce the features of the experimental scattering profiles well (Figure 4A).
The arrangement of the two five-helix bundles in the dimer seems to be conserved, since all rigid body models yielded an angle of approximately 130° between their long axes. Interestingly, the angle between the two antiparallel helices in the dimerisation domain is approximately 120°. This may indicate an interaction between the bottom of the five-helix bundle and one face of the dimerisation domain (possibly the conserved face described in Figure 2C); the twist in the dimerisation domain would determine the arrangement of the two five-helix bundles. A number of residues might be involved in stabilising the observed domain arrangement within the dimer. These include a loop of 15 amino-acid residues between helices 1 and 2 that could contact the dimerisation domain, a segment of ∼20 amino-acid residues between helix 5 and the dimerisation domain and a stretch of 14 residues at the C terminus of the dimerisation domain (Figures 1A and 2D), totalling 49 residues (98 residues in the dimer).
Comparison of the [1H,15N]HSQC spectra of (i) talin 2294–2541, containing both the core five-helix bundle and the dimerisation domain, (ii) the five-helix bundle alone and (iii) two different polypeptides spanning the dimerisation domain (residues 2494–2541 and 2481–2541) confirms that the two domains interact. For example, G2496 and G2497 are mobile in the isolated dimerisation domain but become immobilised in talin 2294–2541, as shown by severe broadening of the resonances. Similarly, residues 2532–2538, which are C-terminal to the structured part of the dimerisation domain are highly dynamic in the isolated domain, but become immobilised in talin 2294–2541. The changes in dynamic properties indicate a direct interaction between the THATCH core domain and the N- and C-terminal ends of the dimerisation domain. However, the core domain retains a significant degree of independent mobility within talin 2294–2541, as indicated by the relatively modest increase in line widths of the NMR resonances relative to the isolated five-helix bundle. This suggests that the area of contact with the core domain is relatively small, and is probably at the C-terminal end of the domain.
Electron microscopy of the C-terminal domain of talin bound to actin filaments
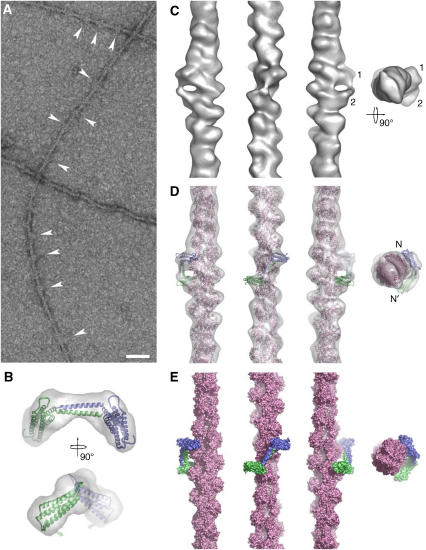
To determine where the C-terminal domain of talin binds on the actin filament, we used the talin 2334–2541 construct lacking the USH, since it has a higher affinity for F-actin. Differential scanning calorimetry (DSC) showed that this construct is properly folded and stable under the conditions used, and formation of a complex with F-actin was verified by co-sedimentation and DSC (Supplementary Figure S9). Using electron microscopy, extra density was visible decorating the actin filaments at both pH 7.0 and 7.5 in the presence of the talin construct (Figure 5A arrows), clearly indicating binding. Two complementary image-reconstruction approaches for helically symmetric structures were applied to two independent data sets. The analysis of the resulting three-dimensional (3D) reconstructions shows that the helical symmetry is consistent with the values reported for decorated actin filaments by us and others. However, difference mapping of reconstructions of F-actin with and without talin 2334–2541 did not provide any clear difference peaks that might correspond to the talin domain.
Figure 5.
The C-terminal actin-binding site in talin binds to the sides of actin filaments. (A) The talin fragment binds to the side of actin filaments at specific sites (arrowheads). This binding does not follow helical symmetry. The scale bar represents 50 nm. (B) Two orthogonal views of the dimer model (monomers in blue and green cartoon representation) and the envelope derived by SAXS (transparent grey). The small grey arrows indicate the direction of the twist that can be used to improve the fit of the SAXS model into the 3D reconstruction. (C) Surface representation of the 3D reconstruction of F-actin decorated with the talin C-terminal domain. The three views perpendicular to the filament axis are related by successive 90° anticlockwise rotations around the axis. The pointed end of the filament is to the top of the figure for these views. The rightmost view is along the filament axis from the pointed end towards the barbed end. The two connected densities are indicated (1 and 2) (D) Docked atomic models of F-actin (pink) and a dimer of the talin C-terminal domain (monomers in blue and green) inside the 3D reconstruction (transparent grey). Views as in (C). (E) Molecular surface of the docked models. Views and colours as in (D).
Consequently, we used an image analysis approach that does not rely on helical symmetry. The resulting 3D reconstruction shows two distinct but connected densities attached to the filament (Figure 5C). These densities are fully consistent in shape and size with the helical bundle of the talin C-terminal domain. In fact, the SAXS-based dimer model fits nicely into the extra density (Figure 5D), indicating that the conformation of actin-bound dimers and dimers in solution are similar. A minor twist (20°) around the dimerisation domain (Figure 5B) improves the fit dramatically and also increases contact area with the actin filament. The docking indicates that the talin domain binds to three monomers along the filament. Thus, it cannot support helical symmetry, explaining why the helical reconstructions did not show additional density that can be attributed to talin.
The model places the two helical bundles towards two opposite sides of the filament. The upper bundle is placed close to subdomain 2 of the central actin monomer and is also close to the back (C-terminal region) of subdomain 1 of the upper actin monomer and the top of subdomain 1 of the central actin monomer. The lower talin helical bundle is close to the lowest actin monomer of the three-monomer interaction site. The dimerisation domain is close to the N terminus (subdomain 1) of the central actin monomer. In this configuration, residues implicated in actin binding by the mutagenesis experiments are placed close to the surface of F-actin.
Discussion
We have determined the solution structure of the talin C-terminal actin-binding domain, a five-helix bundle that has a similar fold to the HIP1R THATCH core domain (Brett et al, 2006). Using mutagenesis, we have defined the actin-binding surface on the bundle as a region of highly conserved residues forming a hydrophobic patch lined by basic residues. Binding to F-actin is negatively regulated by helix 1 (the USH), on the opposite face of the bundle to this site, although the physiological significance of this remains to be determined. In the full-length protein, this domain is C-terminal to a series of helical bundles that make up the talin rod, and the mechanical force exerted on talin might weaken the interaction between the USH and the core of the bundle, thereby increasing its affinity for F-actin.
An intriguing feature of proteins containing a THATCH domain is the C-terminal helix that seems to enhance actin binding by supporting dimer formation. Here, we describe for the first time the structure of one of these domains, the talin dimerisation domain, which forms an antiparallel dimer. NMR indicates that additional intramolecular interactions between the unstructured regions N- and C-terminal to the dimerisation helix may also help to stabilise the antiparallel dimer. The structure suggests that full-length talin might adopt a wide variety of conformations, including an extended tail-to-tail dimer (Figure 4C). This notion is supported by data showing that HIP1R is a rod-shaped dimer with globular heads at either end (Engqvist-Goldstein et al, 2001). Talin has previously been reported to form an antiparallel dimer (Goldmann et al, 1994), which is difficult to reconcile with our results on the C-terminal domain. Mutagenesis of the dimerisation domain clearly demonstrates the importance of this domain in supporting high-affinity actin binding. Intriguingly, most mutations in the dimerisation helix rendered the domain monomeric, and F-actin binding was markedly reduced (Table I). However, we were able to identify two mutants (R2510A and R2513A) that retained the ability to form dimers while showing a reduction in F-actin binding, suggesting that the dimerisation domain itself might contribute to actin binding.
Electron microscopy and image analysis studies together with DSC and co-sedimentation assays provide direct evidence for binding of the dimeric C-terminal domain of talin to filamentous actin. The 3D reconstruction indicates that the dimeric talin construct binds to three actin monomers along the long-pitch helix of the filament (Figure 5). This is surprising because most F-actin-binding proteins tend to bind two monomers (McGough, 1998), often involving a prominent hydrophobic pocket in the filament primarily composed of subdomain 1 (Dominguez, 2004) with some contributions from subdomain 2 of the long-pitch neighbour below (Volkmann et al, 2000). In our model, the upper helical bundle of the talin dimer is indeed located close to this consensus binding site on F-actin, contacting two actin monomers along the long-pitch helix. This is consistent with the hydrophobic nature of the binding site determined by mutagenesis. The second talin helical bundle is mainly bound to the front of a single actin monomer right below these two, and the dimerisation domain is close enough to make contact with the negatively charged N-terminal region of subdomain 1 in the central actin monomer of the three.
One consequence of this mode of binding and the intrinsic symmetry of the dimer is that there must be two non-equivalent modes of actin binding. This is consistent with a study on an isolated monomeric THATCH core domain (Galkin et al, 2005) where two different modes of binding were identified. However, in contrast to that study, we were not able to observe F-actin binding by the monomeric talin five-helix bundle (equivalent to the HIP1R THATCH core domain), indicating that both sets of contacts seen in our model are necessary to produce a stable complex. The model assumes that there are no major rearrangements between the domains in solution (as determined by SAXS) and the actin-bound form of the dimer. This assumption is fully consistent with the shape of the additional density, which can be accounted for well by docking the SAXS dimer structure into the density. A slight twist of the dimer bringing the helical bundles in a more parallel position results in an even better fit. The model for the actin-bound dimer places the residues implicated in actin binding close to the filament surface and the N termini on two opposite sides of the actin filament. However, this assignment is not unique and shows only that there is no contradiction between the dimer placement in the model and the other data.
The finding that the dimeric C-terminal actin-binding site of talin binds to a single actin filament explains why this domain fails to bundle actin filaments under the conditions used in this study. In contrast, Smith and McCann (2007) report that this domain has bundling activity, as determined by low-speed centrifugation and by negative stain electron microscopy. All our experiments using the construct lacking the USH were carried out within 24–48 h of purification, and the integrity of the fold was confirmed by DSC. We found that the protein has a tendency to aggregate with time as detected by NMR, and this may explain the discrepancy between the two studies. It is well established that full-length talin has actin-bundling activity, as does the talin rod (Schmidt et al, 1999). However, it is important to note that talin contains at least two other regions that bind F-actin, namely the talin FERM domain (Lee et al, 2004) and residues 951–1327 in the centre of the talin rod (Hemmings et al, 1996). It will be important to establish the role of each of these actin-binding sites in a cellular context. Initial studies using talin1 knockout cells have shown that the C-terminal region of talin is required to support coupling of surface-associated fibronectin to the actin cytoskeleton (Jiang et al, 2003), but further studies are required to establish whether this was due to loss of the C-terminal actin-binding site or to the fact that the protein was monomeric. The results reported here pave the way for such studies.
Materials and methods
Protein expression and purification
Mouse talin1 cDNAs were amplified by PCR and cloned into either pET-151/D-TOPO (Invitrogen) or pETM-20 (EMBL, Heidelberg, Germany). Proteins were purified from Escherichia coli BL21 Star (DE3) as previously described (Gingras et al, 2006). Recombinant talin 2494–2541 was expressed in the B834 strain of E. coli and cultured in minimal media containing selenomethionine. Protein concentrations were determined using their respective extinction coefficient at 280 nm.
NMR spectroscopy
NMR experiments for resonance assignment and structure determination of the talin 2300–2482 fragment were performed with 1 mM protein, 20 mM sodium phosphate, 50 mM NaCl, 2 mM DTT, pH 6.0, containing 10% (v/v) 2H2O. NMR spectra of talin 2300–2482 were obtained at 45°C using Bruker AVANCE DRX 600 and AVANCE DRX 800 spectrometers equipped with CryoProbes. Spectra were processed using TopSpin (Bruker) or NMRPipe (Delaglio et al, 1995) and analysed using Analysis (Vranken et al, 2005). Backbone and side-chain assignments were obtained using standard triple- and double-resonance experiments. NMR structure calculations were performed as described previously (Gingras et al, 2006); details are given in Supplementary Materials and methods, with structural statistics in Supplementary Table SI. The set of 20 lowest energy structures has been submitted to the Protein Data Bank under PDB code 2JSW.
X-Ray crystallography
Crystals of talin 2494–2541 were obtained at 19°C by vapour diffusion using 100 mM citrate, 11% (w/v) PEG 3000, 200 mM NaCl, pH 4.2. Protein at 8.0 mg/ml in 20 mM Tris–HCl, 0.2 M NaCl, 2 mM DTT, pH 8.0 was mixed with an equal volume of precipitant. Crystals belong to the space group P4132 with a, b, c=98.8 Å, α, β, γ=90°. The crystals contained one dimer per asymmetric unit with a solvent content of 66.4%. Data sets were collected for native crystals at ESRF beamline ID23-1, using an ADSC Q315R CCD detector, and for the selenomethionine derivative at ESRF, beamline 14–3, using an ADSC Q4R CCD detector. The model converged to an Rwork of 25.7% and Rfree of 31.5% for all data between 30 and 2.2 Å. The structure has been deposited in the Protein Data Bank under PDB code 2QDQ. Figures were generated with PYMOL (http://www.pymol.org) and ccp4mg (Potterton et al, 2002).
Actin co-sedimentation assays
Muscle G-actin was purified from rabbit skeletal muscle (Pardee and Spudich, 1982) and polymerised in 10 mM Tris, 50 mM NaCl, 100 μM ATP, 1 mM DTT, 1 mM MgCl2, pH 7.0. Assays were performed using 4 μM talin and concentrations of F-actin ranging from 0 to 25 μM. The mixture was incubated for 60 min at room temperature and centrifuged at 100 000 r.p.m. for 30 min at 22°C using a Beckman Optima TM ultracentrifuge. Supernatants and pellets were analysed on 12% SDS–PAGE gels, stained using Coomassie blue and scanned. Protein levels in the pellet were determined using Image J software (http://rsb.info.nih.gov/ij/) and normalised using talin loading controls (2 μM). Each mutant was analysed in triplicate.
Solution X-ray scattering data collection and analysis
SAXS experiments were carried out at station 2.1 of the Synchrotron Radiation Source at Daresbury using a multiwire gas detector covering a momentum transfer range of 0.02 Å−1 <q<0.70 Å−1 with q=4π sin Θ/λ (where 2Θ is the scattering angle and λ the X-ray wavelength of 1.54 Å). Measurements on talin 2300–2541 were performed at 4°C, at concentrations of 2 and 10 mg/ml in 20 mM sodium phosphate, 50 mM NaCl, 2 mM DTT, pH 6.5. Data were accumulated in 60-s frames and before averaging, frames were inspected for X-ray-induced damage or aggregation. No protein aggregation was detected and the linearity of the Guinier plot (Supplementary Figure S10) indicates that the protein solutions were homogeneous. The background was subtracted using the scattering from the buffer solution alone. Data reduction was carried out with software provided at SRS Daresbury and subsequent analysis was carried out with programs from the ATSAS package (Konarev et al, 2006). Particle shapes were reconstructed ab initio with the bead modelling program GASBOR (Svergun et al, 2001), which represents the protein as a chain of dummy residues centred at the Cα positions. In addition, the program BUNCH (Petoukhov and Svergun, 2005) was applied using the atomic coordinates of individual domains reported here. Rigid body modelling allows exploration of possible positions and arrangements of domains and likely conformations of flexible polypeptide segments consistent with the experimental scattering profile.
Electron microscopy
Rabbit skeletal muscle actin was prepared and stored as described (Volkmann et al, 2000). F-actin was used within 2–3 weeks of preparation. Talin 2234–2541 was used within 1 or 2 days of preparation. The sample was diluted to 0.02 mg/ml just before the application to glow-discharged 400-mesh copper grids coated with holey carbon film. The filaments were washed twice with 50 mM NaCl, 20 mM Pipes, 1 mM MgCl2 at pH 7.0 or 7.5. Talin 2234–2451 (5 μl of 0.06 mg/ml) was applied to the grids. Following 30 s incubation in a humidified chamber, a fresh drop of 5 μl protein was added, incubated for another minute, blotted and stained with 2% aqueous uranyl acetate and air dried or plunge frozen in liquid N2-cooled ethane. Images were recorded at a dose of ∼10 e−/Å2 with a Tecnai 12 electron microscope (FEI Electron Optics, Hillsboro, OR) at 120 keV with a nominal magnification of 52 000 and 1.5 μm defocus. A total of 53 images were digitised using a SCAI scanner (Z/I Imaging Corporation, Englewood, CO) with pixel size of 0.4 nm on the sample.
Image processing of EM data
To generate 3D reconstructions, we first applied standard (Owen and DeRosier, 1993) and hybrid approaches (Egelman, 2000; Volkmann et al, 2005) for structures with helical symmetry. We selected only filaments that clearly showed decoration by visual inspection. Control reconstructions of F-actin alone were also calculated. After optimal alignment (Hanein and DeRosier, 1999), difference maps between the decorated filaments and the F-actin controls were calculated. None of the difference maps showed statistically significant additional density attributable to the attached talin domain. Attempts at sorting into subgroups did not improve the results. Next, we used a reconstruction strategy that does not rely on helical symmetry. Briefly, we selected 374 motifs that showed a clear density attached to the side of the filament. These were processed using standard single-particle analysis procedures. We built a starting model from F-actin with a talin dimer attached in arbitrary orientation close to the centre of the image. The extra density in the model was necessary to ensure that the extra mass in the experimental data aligned properly. The reconstruction procedure was repeated with a different starting model that converged to a similar reconstruction. An atomic model of F-actin was docked into the density using CoAn (Volkmann and Hanein, 1999). The SAXS-based dimer models were docked into the remaining density. The Gasbor-based model (see below) fits significantly better than the BUNCH model.
Docking of atomic models into SAXS envelopes and variance analysis
The top-scoring nine Gasbor models were converted into electron density (see Supplementary Materials and methods). These densities were optimally aligned using the CoAn algorithm (Volkmann and Hanein, 1999), filtered to a resolution of 15 Å, and averaged. The variance distribution was relatively uniform within the envelope. Docking of the atomic models of the five-helix bundle indicated that neither the orientation of the domain around its long axis nor its direction can be fixed based on the data. The orientation of the long axis itself can be fixed within ±15°. There is also a translational uncertainty of ∼6 Å. Discrepancy maps (Volkmann and Hanein, 2003) were calculated to remove the contribution of the five-helix bundle from the density and allow docking of the dimerisation domain. Again, the orientation around the long axis is not well determined by the data. The orientation of the long axis is fixed by the symmetry but there is an ∼8 Å uncertainty in translation.
Supplementary Material
Supplementary data
supplementary movie
Acknowledgments
We thank Peter Moody for substantial help with the crystallography and Andrey Bobkov for the DSC data. The work in Leicester was supported by the Wellcome Trust, Cancer Research UK and the NIH Cell Migration Consortium Grant U54 GM64346 from the National Institute of General Medical Sciences (NIGMS). The electron microscopy and image analysis studies performed at the Burnham Institute for Medical Research were supported by the NIH Cell Migration Consortium Grant U54 GM64346 from the NIGMS to DH and NV.
References
- Brett TJ, Legendre-Guillemin V, McPherson PS, Fremont DH (2006) Structural definition of the F-actin-binding THATCH domain from HIP1R. Nat Struct Mol Biol 13: 121–130 [DOI] [PubMed] [Google Scholar]
- Critchley DR (2004) Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem Soc Trans 32: 831–836 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Dominguez R (2004) Actin-binding proteins—a unifying hypothesis. Trends Biochem Sci 29: 572–578 [DOI] [PubMed] [Google Scholar]
- Egelman EH (2000) A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85: 225–234 [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Warren RA, Kessels MM, Keen JH, Heuser J, Drubin DG (2001) The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J Cell Biol 154: 1209–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham I, Gingras AR, Papagrigoriou E, Patel B, Emsley J, Critchley DR, Roberts GC, Barsukov IL (2005) A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure 13: 65–74 [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Koleske AJ, Egelman EH (2005) The Arg non-receptor tyrosine kinase modifies F-actin structure. J Mol Biol 346: 565–575 [DOI] [PubMed] [Google Scholar]
- Garcia-Alvarez B, de Pereda JM, Calderwood DA, Ulmer TS, Critchley D, Campbell ID, Ginsberg MH, Liddington RC (2003) Structural determinants of integrin recognition by talin. Mol Cell 11: 49–58 [DOI] [PubMed] [Google Scholar]
- Gingras AR, Vogel KP, Steinhoff HJ, Ziegler WH, Patel B, Emsley J, Critchley DR, Roberts GC, Barsukov IL (2006) Structural and dynamic characterization of a vinculin binding site in the talin rod. Biochemistry 45: 1805–1817 [DOI] [PubMed] [Google Scholar]
- Gingras AR, Ziegler WH, Frank R, Barsukov IL, Roberts GC, Critchley DR, Emsley J (2005) Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J Biol Chem 280: 37217–37224 [DOI] [PubMed] [Google Scholar]
- Goldmann WH, Bremer A, Haner M, Aebi U, Isenberg G (1994) Native talin is a dumbbell-shaped homodimer when it interacts with actin. J Struct Biol 112: 3–10 [DOI] [PubMed] [Google Scholar]
- Hanein D, DeRosier D (1999) A new algorithm to align three-dimensional maps of helical structures. Ultramicroscopy 76: 233–238 [DOI] [PubMed] [Google Scholar]
- Hemmings L, Rees DJ, Ohanian V, Bolton SJ, Gilmore AP, Patel B, Priddle H, Trevithick JE, Hynes RO, Critchley DR (1996) Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J Cell Sci 109 (Pt 11): 2715–2726 [DOI] [PubMed] [Google Scholar]
- Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP (2003) Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424: 334–337 [DOI] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, Svergun DI (2006) ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Cryst 39: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Bellin RM, Walker DL, Patel B, Powers P, Liu H, Garcia-Alvarez B, de Pereda JM, Liddington RC, Volkmann N, Hanein D, Critchley DR, Robson RM (2004) Characterization of an actin-binding site within the talin FERM domain. J Mol Biol 343: 771–784 [DOI] [PubMed] [Google Scholar]
- McCann RO, Craig SW (1997) The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA 94: 5679–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A (1998) F-actin-binding proteins. Curr Opin Struct Biol 8: 166–176 [DOI] [PubMed] [Google Scholar]
- Moes M, Rodius S, Coleman SJ, Monkley SJ, Goormaghtigh E, Tremuth L, Kox C, van der Holst PP, Critchley DR, Kieffer N (2007) The integrin binding site 2 (IBS2) in the talin rod domain is essential for linking integrin beta subunits to the cytoskeleton. J Biol Chem 282: 17280–17288 [DOI] [PubMed] [Google Scholar]
- Owen C, DeRosier D (1993) A 13-Å map of the actin–scruin filament from the limulus acrosomal process. J Cell Biol 123: 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagrigoriou E, Gingras AR, Barsukov IL, Bate N, Fillingham IJ, Patel B, Frank R, Ziegler WH, Roberts GC, Critchley DR, Emsley J (2004) Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J 23: 2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA (1982) Purification of muscle actin. Methods Enzymol 85 (Pt B): 164–181 [DOI] [PubMed] [Google Scholar]
- Patel B, Gingras AR, Bobkov AA, Fujimoto LM, Zhang M, Liddington RC, Mazzeo D, Emsley J, Roberts GC, Barsukov IL, Critchley DR (2006) The activity of the vinculin binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod. J Biol Chem 281: 7458–7467 [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J 89: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M (2002) The CCP4 molecular-graphics project. Acta Crystallogr D 58: 1955–1957 [DOI] [PubMed] [Google Scholar]
- Saunders RM, Holt MR, Jennings L, Sutton DH, Barsukov IL, Bobkov A, Liddington RC, Adamson EA, Dunn GA, Critchley DR (2006) Role of vinculin in regulating focal adhesion turnover. Eur J Cell Biol 85: 487–500 [DOI] [PubMed] [Google Scholar]
- Schmidt JM, Zhang J, Lee HS, Stromer MH, Robson RM (1999) Interaction of talin with actin: sensitive modulation of filament crosslinking activity. Arch Biochem Biophys 366: 139–150 [DOI] [PubMed] [Google Scholar]
- Senetar MA, Foster SJ, McCann RO (2004) Intrasteric inhibition mediates the interaction of the I/LWEQ module proteins Talin1, Talin2, Hip1, and Hip12 with actin. Biochemistry 43: 15418–15428 [DOI] [PubMed] [Google Scholar]
- Smith SJ, McCann RO (2007) A C-terminal dimerization motif is required for focal adhesion targeting of talin1 and the interaction of the talin1 I/LWEQ module with F-actin. Biochemistry 46: 10886–10898 [DOI] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N, Hanein D (1999) Quantitative fitting of atomic models into observed densities derived by electron microscopy. J Struct Biol 125: 176–184 [DOI] [PubMed] [Google Scholar]
- Volkmann N, Hanein D (2003) Docking of atomic models into reconstructions from electron microscopy. Methods Enzymol 374: 204–225 [DOI] [PubMed] [Google Scholar]
- Volkmann N, Hanein D, Ouyang G, Trybus KM, DeRosier DJ, Lowey S (2000) Evidence for cleft closure in actomyosin upon ADP release. Nat Struct Biol 7: 1147–1155 [DOI] [PubMed] [Google Scholar]
- Volkmann N, Liu H, Hazelwood L, Krementsova EB, Lowey S, Trybus KM, Hanein D (2005) The structural basis of myosin V processive movement as revealed by electron cryomicroscopy. Mol Cell 19: 595–605 [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59: 687–696 [DOI] [PubMed] [Google Scholar]
- Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID (2007) Structural basis of integrin activation by talin. Cell 128: 171–182 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
supplementary movie