Abstract
To ensure survival in the face of genomic insult, cells have evolved complex mechanisms to respond to DNA damage, termed the DNA damage checkpoint. The serine/threonine kinases ataxia telangiectasia-mutated (ATM) and ATM and Rad3-related (ATR) activate checkpoint signaling by phosphorylating substrate proteins at SQ/TQ motifs. Although some ATM/ATR substrates (Chk1, p53) have been identified, the lack of a more complete list of substrates limits current understanding of checkpoint pathways. Here, we use immunoaffinity phosphopeptide isolation coupled with mass spectrometry to identify 570 sites phosphorylated in UV-damaged cells, 498 of which are previously undescribed. Semiquantitative analysis yielded 24 known and 192 previously uncharacterized sites differentially phosphorylated upon UV damage, some of which were confirmed by SILAC, Western blotting, and immunoprecipitation/Western blotting. ATR-specific phosphorylation was investigated by using a Seckel syndrome (ATR mutant) cell line. Together, these results provide a rich resource for further deciphering ATM/ATR signaling and the pathways mediating the DNA damage response.
Keywords: DNA damage, mass spectrometry, phosphorylation
Maintaining the integrity of the genome is of utmost importance for cellular survival. For this reason, cells have evolved complex mechanisms to inhibit cell cycle progression in response to genomic insult, termed the DNA damage checkpoint (1). Activating checkpoint mechanisms gives cells time to repair or bypass the damage using specialized DNA polymerases or, in cases of high levels of damage, to activate apoptotic pathways (2). Elucidating pathways involved in checkpoint activation and maintenance continues to be an active area of research.
A family of phosphoinositol-3-phosphate kinase-like kinases are critical to the proper function of the DNA damage checkpoint. The two central kinases involved are ataxia telangiectasia-mutated (ATM) and ATM and Rad3-related (ATR). This kinase family also includes DNA-dependent protein kinase (DNA-PK) and a more recently discovered member of the family, SMG1 (3). These kinases are activated in response to DNA damage and subsequently phosphorylate targets responsible for such diverse activities as blocking cell cycle progression, coordinating DNA repair activities, and affecting transcription of DNA damage response genes. ATR is activated in response to a variety of damaging agents: UV light, alkylating agents such as methyl methanesulfonate (MMS), and chemical inhibitors of DNA replication such as aphidicolin and hydroxyurea (4, 5). ATM, however, is primarily involved in the response to double-strand breaks, such as those caused by gamma irradiation (IR) (6). Deficiency in ATM/R, as well as other components of the DNA damage checkpoint, has been found to cause debilitating diseases such as ataxia telangiectasia (ATM mutants), Fanconi's anemia, Seckel syndrome (ATR mutants), and the avoidance of checkpoint activation to allow cancer progression.
In response to DNA damage, ATM/R phosphorylate checkpoint kinases Chk1 and Chk2, as well as p53, which block cell cycle progression by inhibiting cyclin-dependent kinase activity (2, 7–10). In addition to the phosphorylation of Chk1, Chk2, and p53, other substrates of ATM/R have been identified, including BRCA1, NBS1, MDM2, CtIP, SMC1, MDC1, FANCD2, and 53BP1, among others (11). ATM/R phosphorylate serine or threonine residues with glutamine (Q) at the +1 position (the so-called SQ/TQ motif) but nearby positively charged residues inhibit phosphorylation of peptides by ATM/R (12).
Until recently, substrates for ATM/R have been found only one or a few at a time. To identify large numbers of novel ATM/R substrates (sites containing the minimum ATM/R phosphorylation motif SQ or TQ not previously experimentally shown to be phosphorylated in human cells), we have extended our immunoaffinity technology for isolating phosphotyrosine-containing peptides (13) to the ATM/R substrate motif SQ/TQ, similar to a recent study investigating phosphorylation in response to IR damage of DNA (14).
Here, we describe the identification of 570 SQ/TQ sites on 464 proteins from UV light- (UV) damaged cells, most of which (498, or 87%) are previously undescribed. We show that >200 of these sites are at least 2-fold more abundant in UV-damaged than undamaged cells. Finally, ATR-specific phosphorylation in response to UV damage was investigated by using an ATR-deficient cell line. Among the many previously uncharacterized sites we found, those that were both more abundant after UV damage and less abundant in the ATR mutant cell line are highly likely to be bona fide UV-inducible ATR substrates, suitable for future hypothesis-driven functional studies. This study provides many potential substrates of ATM/R phosphorylated in response to UV damage of DNA, demonstrates the versatility and power of this method in elucidating previously undescribed substrates, as predicted (15), and more clearly defines signaling pathways involved in the DNA damage response.
Results and Discussion
Immunoaffinity Isolation and Mass Spectrometry Identification of Phosphopeptides.
To identify novel ATM/R substrates, we developed two antibodies that broadly react with the ATM/R substrate consensus motif. One ATM/R substrate motif antibody was generated and characterized as described (16). The second antibody was intended to be specific for phospho-Chk2 (T26/S28) but was found to detect both phosphorylated Chk2 and other ATM/R-dependent phosphorylation sites (17). Further characterization of this antibody as a bona fide ATM/R substrate motif antibody is shown in supporting information (SI) Fig. 4.
We used the ATM/R substrate antibodies combined with liquid chromatography–tandem mass spectrometry (LC-MS/MS) to isolate and identify phosphopeptides from UV-treated M059K glioblastoma cells. Sequences were assigned to MS/MS spectra with Sequest (18), allowing for phosphorylation at serine and threonine residues. Phosphorylation sites found at SQ/TQ motifs were considered highly likely to be correct, because the phosphorylated motif was not considered in the assignment process yet was expected based on antibody recognition specificity. A complete table of identified phosphorylation sites is provided in SI Table 3 and in the PhosphoSite bioinformatics resource (Cell Signaling Technology). SI Table 3 lists only phosphopeptides found to contain at least one SQ/TQ motif. These sites are likely to be direct ATM/R substrates or substrates of other SQ/TQ-directed kinases. In some cases, nonmotif phosphorylation sites are present on the same peptide as a motif site. These sites have also been reported, although because of lack of motif, their assignment is more ambiguous. Frequency maps derived by aligning all phosphopeptides found in the analysis (not just SQ/TQ phosphopeptides) for the two motif antibodies are shown in SI Fig. 5. The analysis yielded 570 SQ/TQ phosphorylation sites from 464 proteins, and 368 were localized with at least 95% certainty (SI Table 4) based on probability-based scoring of phosphorylation site localization (19). Nearly all (498, or 87%) of these sites were previously undescribed human phosphorylation sites, based on information contained in the PhosphoSite bioinformatics resource. One hundred seventy-eight of these 498 sites were also found in another recent study investigating IR damage-induced SQ/TQ phosphorylation (14). Before that study, only 304 human SQ/TQ sites were known, and only 90 of these sites were known substrates of ATM/ATR/DNA-PK (from the PhosphoSite bioinformatics resource). Thus, this study has increased the number of potential substrates of ATM/R phosphorylated in response to UV damage of DNA >2-fold from all previously known SQ/TQ sites and >5-fold from all previously known DNA damage-inducible SQ/TQ sites.
Phosphorylation of Diverse Protein Classes Induced upon UV Damage of DNA.
We next measured the changes in the abundance of these SQ/TQ phosphorylation sites in response to UV damage as a way of pinpointing sites associated with ATM/R signaling. For each phosphorylation site identified in M059K glioblastoma cells with or without UV damage, the intensity of the precursor peptide ion was measured at the apex of its chromatographic peak, using a semiquantitative analytical approach (see Table 1 for a partial list and SI Table 5 for the complete list). Peptide ions whose intensities were increased at least 2-fold by UV damage were included in the tables. This analysis yielded 216 UV-inducible SQ/TQ sites on 180 proteins. Of the 216 sites, 192 (88%) were previously undescribed. Approximately half (99 of 192) of these sites were also found by Matsuoka et al. (14). The 216 UV-inducible SQ/TQ sites represent a significant increase in the number of potential UV damage-dependent ATM/R substrates from the 90 damage-inducible sites previously known.
Table 1.
Selected peptides found by semiquantitative analysis to be at least 2-fold more highly phosphorylated in UV-damaged M059K cells than undamaged M059K cells
| Name | Protein type | Phospho-Site accession no. | Site(s) | Sequence | Normalized peak height |
UV: control ratio | ATM/R 1 | ATM/R 2 | |
|---|---|---|---|---|---|---|---|---|---|
| Control | UV damaged | ||||||||
| p400 | Apoptosis | Q96L91 | 2,050 | AEEFVVLS*QEPSVTETIAPK | 6.2 | 14.7 | 2.4‡ | ● | |
| Claspin | Cell cycle regulation | Q9HAW4 | 950 | FTS*QDASTPASSELNK | 1.2 | 3.7 | 3.1‡ | ● | |
| MDC1 | Cell cycle regulation | Q14676 | 659 | ENLTDLVVDTDTLGEST*QPQR | 76.0 | 457.6 | 6.0‡ | ● | |
| MDC1 | Cell cycle regulation | Q14676 | 1,086, 1,095 | QDGS*QEAPEAPLS*SELEPFHPKPK | 4.1 | 49.6 | 12.2‡ | ● | |
| MDC1 | Cell cycle regulation | Q14676 | 513 | S*QASTTVDINTQVEK | 31.5 | 319.9 | 10.2‡ | ● | |
| NASP | Cell cycle regulation | P49321 | 464 | VQIAANEET*QER | 1.4 | 3.0 | 2.1‡ | ● | |
| NuMA-1 | Cell cycle regulation | Q14980 | 1,744, 1,757† | LPRT*QPDGTSVPGEPAS*PISQR | 3.7 | 20.6 | 5.6‡ | ● | |
| NuMA-1 | Cell cycle regulation | Q14980 | 395 | LSQLEEHLS*QLQDNPPQEK | 8.3 | 99.3 | 11.9‡ | ● | |
| NuMA-1 | Cell cycle regulation | Q14980 | 1,744, 1,757† | T*QPDGTSVPGEPAS*PISQR | 0.8 | 5.9 | 7.3‡ | ● | |
| VCP | Cell cycle regulation | P55072 | 783† | FPSGNQGGAGPS*QGSGGGTGGSVYTEDNDDDLYG | 1,181.3 | 7,354.2 | 6.2‡ | ● | |
| FANCD2 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q9BXW9 | 592†, 596† | SES*PSLT*QER | 0.2 | 1.9 | 10.7‡ | ● | |
| MCM2 | Chromatin, DNA-binding, DNA repair or DNA replication protein | P49736 | 108† | AIPELDAYEAEGLALDDEDVEELTAS*QR | 6.6 | 18.9 | 2.9‡ | ● | |
| MRE11A | Chromatin, DNA-binding, DNA repair or DNA replication protein | P49959 | 676 | IMS*QSQVSK | 1.6 | 6.7 | 4.1‡ | ● | |
| NBS1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | O60934 | 58 | NHAVLTANFSVTNLS*QTDEIPVLTLK | 3.9 | 12.8 | 3.3‡ | ● | |
| NBS1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | O60934 | 343† | TTTPGPSLS*QGVSVDEK | 22.1 | 57.0 | 2.6‡ | ● | ● |
| Rad50 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q92878 | 635† | KEEQLSSYEDKLFDVCGS*QDFESDLDR | 7.1 | 21.0 | 3.0‡ | ● | |
| RFC1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | P35251 | 190† | RKELS*QNTDESGLNDEAIAK | 3.1 | 42.7 | 13.8‡ | ● | |
| Rif1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q5UIP0 | 1098 | CDIPAMYNNLDVSQDTLFTQYS*QEEPMEIPTLTR | 4.1 | 14.5 | 3.5‡ | ● | |
| Rif1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q5UIP0 | 1518 | KADPENIKSEGDGT*QDIVDK | 2.9 | 17.6 | 6.1 | ● | |
| RPA1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | P27694 | 180 | AAGPSLSHTSGGT*QSK | 1.5 | 7.9 | 5.2‡ | ● | |
| Smc1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q14683 | 956, 966† | GTMDDISQEEGS*SQGEDSVSGS*QR | 3.6 | 117.6 | 32.7‡ | ● | |
| Smc1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q14683 | 957†, 966† | GTMDDISQEEGSS*QGEDSVSGS*QR | 3.5 | 113.6 | 32.5 | ● | |
| Smc1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q14683 | 957† | GTMDDISQEEGSS*QGEDSVSGSQR | 33.7 | 137.0 | 4.1‡ | ● | |
| Smc1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q14683 | 358, 360† | MEEES*QS*QGRDLTLEENQVK | 130.4 | 3,023.6 | 23.2‡ | ● | |
| Smc1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q14683 | 360† | MEEESQS*QGRDLTLEENQVK | 108.9 | 1,164.1 | 10.7‡ | ● | |
| Lamin A/C | Nuclear Envelope | P02545 | 390†, 392†, 395 | LRLS*PS*PTS*QR | 1.2 | 3.5 | 2.9‡ | ● | |
| Lamin A/C | Nuclear Envelope | P02545 | 390†, 395 | LRLS*PSPTS*QR | 7.9 | 28.7 | 3.7 | ● | |
| Pnk1 | Phosphatase | Q9UNF8 | 111, 118, 126 | T*PESQPDT*PPGTPLVS*QDEKR | 1.5 | 46.8 | 31.0‡ | ● | |
| Pnk1 | Phosphatase | Q9UNF8 | 114, 118, 126 | TPES*QPDT*PPGTPLVS*QDEKR | 1.5 | 45.7 | 30.3‡ | ● | |
| Pnk1 | Phosphatase | Q9UNF8 | 118, 122, 126 | TPESQPDT*PPGT*PLVS*QDEKR | 1.9 | 39.8 | 20.4‡ | ● | |
| DNA-PK | Protein kinase, Ser/Thr | P78527 | 2,638†, 2,645†, 2,647† | AT*QQQHDFT*LT*QTADGR | 0.7 | 8.3 | 11.3‡ | ● | |
| DNA-PK | Protein kinase, Ser/Thr | P78527 | 2,645†, 2,647† | ATQQQHDFT*LT*QTADGR | 4.1 | 15.3 | 3.8‡ | ● | |
| DNA-PK | Protein kinase, Ser/Thr | P78527 | 2,612† | STVLTPM#FVETQAS*QGTLQTR | 14.4 | 6,008.4 | 417.2‡ | ● | |
| 53BP1 | Transcriptional regulator | Q12888 | 855 | ADDPLRLDQELQQPQT*QEK | 70.5 | 238.4 | 3.4‡ | ● | |
| 53BP1 | Transcriptional regulator | Q12888 | 580 | FVPAENDSILMNPAQDGEVQLS*QNDDKTK | 82.4 | 170.0 | 2.1‡ | ● | |
| 53BP1 | Transcriptional regulator | Q12888 | 1,068 | GNLLHFPSS*QGEEEKEKLEGDHTIR | 414.0 | 2,661.8 | 6.4‡ | ● | |
| 53BP1 | Transcriptional regulator | Q12888 | 543, 552† | IDEDGENT*QIEDTEPM#S*PVLNSK | 0.9 | 7.2 | 7.8‡ | ● | |
| 53BP1 | Transcriptional regulator | Q12888 | 855 | LDQELQQPQT*QEK | 6.8 | 136.3 | 20.1‡ | ● | |
| 53BP1 | Transcriptional regulator | Q12888 | 1,094, 1,101, 1,104 | QSQQPMKPIS*PVKDPVS*PAS*QK | 64.7 | 192.5 | 3.0‡ | ● | ● |
| 53BP1 | Transcriptional regulator | Q12888 | 105 | VADPVDSSNLDTCGSIS*QVIEQLPQPNR | 6.1 | 29.5 | 4.8‡ | ● | |
| NFI-A | Transcriptional regulator | Q12857 | 300, 305 | S*PGSGS*QSSGWHEVEPGMPSPTTLK | 22.7 | 434.1 | 19.1‡ | ● | |
| BAP1 | Ubiquitin system | Q92560 | 592, 596 | GSSPSIRPIQGS*QGSS*SPVEK | 206.5 | 1,542.5 | 7.5‡ | ● | |
| MUF1 | Ubiquitin system | Q15345 | 305 | RST*QESLTAGGTDLKR | 15.6 | 87.2 | 5.6‡ | ● | |
| RNF40 | Ubiquitin system | O75150 | 127 | CHESQGELSSAPEAPGT*QEGPTCDGTPLPEPGTSELR | 2.7 | 30.2 | 11.3 | ● | |
| UBLE1A | Ubiquitin system | Q9UBE0 | 185 | VAKVS*QGVEDGPDTKR | 1.7 | 4.4 | 2.5 | ● | |
| UREB1 | Ubiquitin system | Q7Z6Z7 | 3,377 | ACSPCSS*QSSSSGICTDFWDLLVK | 66.4 | 133.9 | 2.0 | ● | |
| UREB1 | Ubiquitin system | Q7Z6Z7 | 2,377, 2,391 | SGEDES*QEDVLMDEAPSNLS*QASTLQANR | 5.6 | 27.8 | 4.9 | ● | |
| UREB1 | Ubiquitin system | Q7Z6Z7 | 2,391 | SGEDESQEDVLM#DEAPSNLS*QASTLQANR | 105.9 | 424.6 | 4.0 | ● | |
†Sites known to be phosphorylated, although not necessarily by ATM/R.
‡Fold changes measured manually.
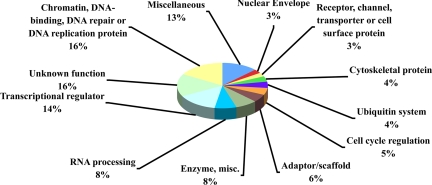
Semiquantitative analysis showed peptides differentially phosphorylated from a wide range of protein classes (Fig. 1). Interestingly, some protein classes expected to be found in the analysis, such as cell cycle and apoptosis proteins, have relatively low numbers of peptides, whereas other classes, such as adaptor/scaffold or ubiquitin system proteins, were found in higher numbers than expected. Overall, 21 different protein classes were found in the analysis, with an additional 16% of peptides having unknown functions. The diversity of protein classes found to be differentially phosphorylated in response to UV damage suggests that ATM/R-dependent phosphorylation affects cellular processes not previously thought to be involved in the DNA damage response. Our results clearly show that UV-inducible phosphorylation at SQ/TQ motifs is widespread among protein classes, as was also seen in response to IR damage of DNA (14).
Fig. 1.
Semiquantitative analysis of control vs. UV-damaged M059K cells reveals diversity of known/previously undescribed substrates. Pie chart shows protein classes identified in the semiquantitative analysis. All classes not shown have been collapsed into the “miscellaneous” category. The percentage of the total for each protein class is shown.
Confirmation of Differential Phosphorylation.
Confirmation of differential phosphorylation between undamaged and UV-damaged M059K cells was first attempted through quantitative stable isotope labeling by amino acids in cell culture (SILAC) analysis using a unit-resolution ion trap mass spectrometer (SI Table 6) (20). A number of peptides that appear in the semiquantitative analysis were confirmed by SILAC analysis. In fact, of the 54 SQ/TQ phosphorylation sites in the SILAC analysis that increased by at least 2-fold upon UV damage, 45 were found in the semiquantitative analysis (marked with a √ symbol in SI Table 6). Of those same 54 phosphorylation sites, 42 sites are previously uncharacterized. Ideally, SILAC analysis is performed with a high-resolution mass spectrometer, but the lower-resolution ion trap instrument we used was sufficient to confirm many of the semiquantitative analysis measurements. SILAC analysis resulted in fewer phosphopeptide identifications than semiquantitative analysis, because part of the instrument duty cycle was spent collecting MS/MS spectra for both the light- and heavy-isotope forms of the same phosphopeptide. The SILAC ratios in SI Table 6 are minimum fold changes, because baseline noise limits ratio measurement when signal intensities are relatively low.
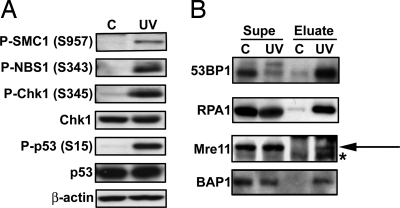
Further confirmation of sites seen in the semiquantitative analysis was performed through Western blotting (Fig. 2A). First, known UV damage-inducible phosphorylation sites seen were confirmed, including S957 of SMC1 and S343 of NBS1. Additionally, phosphorylation of Chk1 and p53 was confirmed, although these peptides were not detected in our system. This may have been due to the specificity of the antibodies used (see SI Fig. 5). An exception is S15 of p53, which has the exact motif preferred by ATM/R substrate motif antibody-1 (LSQE). Because of the high density of N-terminal p53 phosphorylation sites, the tryptic peptide containing p53 S15 may have contained more modifications than the peptide identification software allowed.
Fig. 2.
Western blotting and IP/Western blotting confirm phosphorylation of known/previously undescribed substrates of ATM/R seen in immunoaffinity purification/LC-MS/MS analysis. (A) Untreated (C) or UV-treated (UV) M059K lysates were subjected to Western blotting with the antibody indicated. (B) Proteins were immunoprecipitated from untreated or UV-treated M059K cell lysates with ATM/R substrate motif antibody-1 (53BP1, RPA1, and Mre11), or antibody-2 (BAP1) and blotted with the indicated antibody. “Supe” denotes post-IP supernatants; “Eluate” denotes IP elutions. For clarity, an arrow is shown to denote the band of interest in the Mre11 blot, whereas the asterisk (*) denotes a contaminating band.
To confirm some of the previously undescribed sites described in this study, proteins were immunoprecipitated with the same motif antibodies used for the site discovery studies described above, then blotted with the respective total protein antibody (instead of a phosphorylation site-specific antibody). Fig. 2B shows Western blots for 53BP1, RPA1, Mre11, and BAP1. As seen in Fig. 2B, more of each protein was immunoprecipitated in the UV sample than the control, providing further evidence that these are, in fact, previously undescribed UV-inducible substrates of ATM/R. We cannot exclude the possibility that the motif antibodies immunoprecipitated another phosphorylated protein complexed with one of the proteins shown. Ideally, a reciprocal immunoprecipitation (IP with total antibody, blot with motif antibody) would be performed, but this was not feasible because of poor performance of the motif antibodies as Western blot reagents. That the reported phosphorylation sites on all four proteins are consensus ATM/R substrate sites does give more confidence that these sites are correctly assigned. Differential phosphorylation of two of the proteins shown (53BP1 and BAP1) has also been independently confirmed by quantitative SILAC analysis (see SI Table 6). 53BP1 is a known ATM/R substrate, but the sites we describe here are distinct from the known sites of phosphorylation. We have therefore confirmed differential phosphorylation of both known and previously uncharacterized substrates of ATM/R in response to UV damage of DNA.
ATR-Specific Phosphorylation.
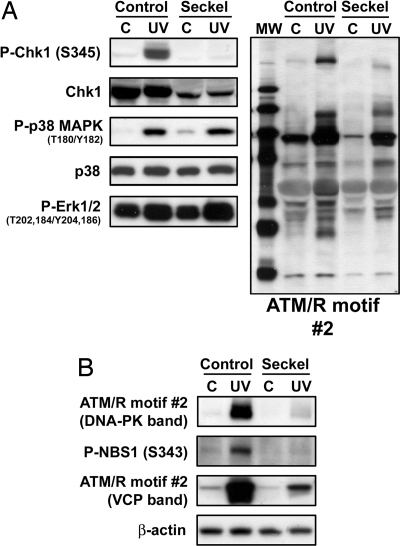
UV damage primarily activates the ATR branch of the DNA damage checkpoint. Seckel syndrome is an autosomal recessive disorder causing dwarfism and mental retardation that has been mapped to mutations in ATR. To further delineate the phosphorylation sites regulated specifically by ATR, we compared the Seckel syndrome fibroblast cell line GM18366 (ATR 2101A>G mutant, http://ccr.coriell.org/nigms) to an age- and race-matched control fibroblast cell line, GM00200. This ATR mutation both decreases ATR protein levels and causes defects in the response to UV damage of DNA (21). Western blots were performed to probe for UV-induced activation of ATM/R-dependent and -independent pathways in this particular cell line. As seen in Fig. 3A, S345 of Chk1 is phosphorylated in response to UV damage in the control but not the Seckel cell line. Total Chk1 levels are lower in the Seckel cell line but not low enough to account for the complete absence of Chk1 phosphorylation. Phosphorylation of p38 MAPK was induced by UV damage in both cell lines. Thus, although this Seckel cell line is deficient in an ATR-dependent response to UV damage (Chk1 S345 phosphorylation), the cells remain proficient in other ATR-independent DNA damage responses (such as p38 MAPK T180/Y182 phosphorylation).
Fig. 3.
Characterization of the DNA damage response in a Seckel cell line compared with a matched control/confirmation of immunoaffinity purification/LC-MS/MS results. (A) GM00200-matched control cells (Control) or GM18366 Seckel syndrome cells (Seckel) were untreated (C) or UV-treated (UV) and blotted with the indicated antibody. (B) Control or Seckel cells were untreated (C) or UV-treated (UV) and blotted with the indicated antibody against sites found in the semiquantitative analysis. ATM/R motif antibody-2 was shown to detect phosphorylated DNA-PK and valosin-containing protein (VCP) (17).
Next, both control and Seckel cell lines were grown, damaged with UV light, and subjected to immunoaffinity purification/LC-MS/MS analysis. SI Table 7 shows a semiquantitative comparison between the control and Seckel syndrome cell lines, listing peptides with intensities at least 2-fold higher in the control than in the Seckel cell line (more abundant when ATR is present and activated). The differentially phosphorylated sites included known ATM/R substrates such as DNA-PK S2612, NBS1 S343, and SMC1 S957. Differential phosphorylation of DNA-PK, NBS1, and VCP was confirmed by Western blotting (Fig. 3B). Twenty-nine of the 39 SQ/TQ phosphorylation sites seen in the semiquantitative analysis are previously uncharacterized. Because two different cell lines were used, we cannot exclude the possibility that some differences seen were due to differences in protein levels between the cell lines. It was, therefore, of interest to find sites for which there was higher confidence in differences seen, such as those differentially phosphorylated in both semiquantitative studies.
The results of this study on phosphorylation in Seckel cells and of the study on UV-damage-responsive phosphorylation in M059K glioblastoma cells provide an overlap that can be used to associate these sites with activation of the ATR protein kinase: These two studies have 19 SQ/TQ sites in common (shown in Table 2). These phosphorylation sites are the most likely to be bona fide ATR substrates phosphorylated in response to UV damage for three reasons: They contain the minimum SQ/TQ motif, they were more highly phosphorylated in UV-damaged than undamaged samples, and their phosphorylation in response to UV damage was decreased in an ATR mutant cell line. These phosphorylation sites are therefore excellent candidates for follow-up hypothesis-driven studies aimed at verifying the kinase–substrate relationships revealed in this work and for determining functional consequences of phosphorylation.
Table 2.
Peptides found by semiquantitative analysis to be at least 2-fold more highly phosphorylated in 1.GM00200 control cells + UV damage than GM18366 Seckel cells + UV damage and 2.UV-damaged M059K cells than undamaged M059K cells
| Name | Protein type | PhosphoSite accession no. | Site(s) | Sequence | C/UV ratio | C/Seckel ratio |
|---|---|---|---|---|---|---|
| DBC-1 | Apoptosis | Q8N163 | 454† | AAEAAPPT*QEAQGETEPTEQAPDALEQAADTSR | 19.6‡ | 2.5‡ |
| MDC1 | Cell cycle regulation | Q14676 | 513 | S*QASTTVDINTQVEK | 10.2‡ | 8.1‡ |
| NuMA-1 | Cell cycle regulation | Q14980 | 1744, 1,757† | LPRT*QPDGTSVPGEPAS*PISQR | 5.6‡ | 2.3‡ |
| VCP | Cell cycle regulation | P55072 | 783† | FPSGNQGGAGPS*QGSGGGTGGSVYTEDNDDDLYG | 6.2‡ | 4.2‡ |
| CROP | Chromatin, DNA-binding, DNA repair or DNA replication protein | O95232 | 425, 429, 431 | ESDTKNEVNGTSEDIKS*EGDT*QS*N | 4.7 | 6.4‡ |
| Smc1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q14683 | 957† | GTMDDISQEEGSS*QGEDSVSGSQR | 4.1 | 3.6 |
| Smc1 | Chromatin, DNA-binding, DNA repair or DNA replication protein | Q14683 | 358, 360† | MEEES*QS*QGRDLTLEENQVK | 23.2‡ | 4.0‡ |
| EYA3 | Phosphatase | Q99504 | 262, 266 | LSSGDPSTS*PSLS*QTTPSKDTDDQSR | 18.1‡ | 2.4‡ |
| Pnk1 | Phosphatase | Q9UNF8 | 118, 122, 126 | TPESQPDT*PPGT*PLVS*QDEKR | 20.4‡ | 8.8‡ |
| DNA-PK | Protein kinase, Ser/Thr | P78527 | 2,612† | STVLTPM#FVETQAS*QGTLQTR | 417.2‡ | 3.8‡ |
| RBM21 | RNA processing | Q9H6E5 | 750 | GHEAAQEWS*QGEAGK | 8.1‡ | 7.0‡ |
| SFRS14 | RNA processing | Q8IX01 | 7 | RIT*QETFDAVLQEK | 3.5‡ | 3.4‡ |
| 53BP1 | Transcriptional regulator | Q12888 | 1,094, 1,101, 1,104 | QSQQPMKPIS*PVKDPVS*PAS*QK | 3.0‡ | 5.5‡ |
| CUTL1 | Transcriptional regulator | P39880 | 1,357 | AAPSSEGDSCDGVEATEGPGSADTEEPKS*QGEAER | 29.9‡ | 5.3‡ |
| Daxx | Transcriptional regulator | Q9UER7 | 424 | LQGTSSHSADTPEASLDSGEGPSGM#AS*QGCPSASR | 9.1‡ | 9.2‡ |
| MCEF | Transcriptional regulator | Q9UHB7 | 487, 499 | VNPHKVS*PASSVDSNIPSS*QGYKK | 2.5‡ | 3.5‡ |
| ZNF318 | Transcriptional regulator | Q5VUA4 | 27, 34 | YIS*QEEGPLS*PFLGQLDEDYR | 18.4‡ | 2.9‡ |
| UREB1 | Ubiquitin system | Q7Z6Z7 | 2,391 | SGEDESQEDVLMDEAPSNLS*QASTLQANR | 4.0‡ | 3.3‡ |
†Sites known to be phosphorylated, although not necessarily by ATM/R.
‡Fold changes measured manually.
Conclusions
Here, we extend our previously described technology for immunoaffinity isolation of phosphotyrosine peptides (13) to the analysis of phosphorylated serine/threonine motifs such as pSQ and pTQ. Immunoprecipitation from UV-damaged cell lysates using ATM/R substrate motif antibodies resulted in the identification of 570 SQ/TQ phosphorylation sites from 464 proteins. Many of these sites were found to be differentially phosphorylated after UV damage, significantly increasing the number of UV-inducible potential ATM/R substrates from the 90 sites currently known. To date, the only known damage-inducible SQ/TQ-directed kinases are ATM, ATR, DNA-PK, and SMG1. However, we cannot rule out the existence of other as-yet-unidentified SQ/TQ kinases that may contribute to phosphorylation at the sites described here. Quantitative SILAC, Western blotting, and IP/Western blotting were used to confirm differential phosphorylation of many peptides found in the semiquantitative analysis. Finally, the use of an ATR-deficient (Seckel syndrome) cell line yielded a group of phosphorylation sites both induced by damage and absent without wild-type ATR. These sites represent the most likely targets of ATR phosphorylation in response to UV damage of DNA.
The proteins differentially phosphorylated upon UV damage comprise a surprisingly wide range of protein classes. Some, such as cell cycle proteins or DNA-interacting proteins, were expected to be found in the analysis in high numbers. Within these expected classes, important findings were made, such as the confirmed phosphorylation of RPA1 and Mre11. These discoveries may shed light on established DNA damage pathways, providing a potential mechanistic basis for previously seen genetic and physical interactions. This work supports the finding from Matsuoka et al. (14) that the DNA damage response leads to phosphorylation of a much larger number of substrates than was previously thought. That study primarily examined the phosphorylation profile induced in response to IR damage of DNA, known to activate ATM-dependent signaling. Our study investigates phosphorylation in response to UV damage of DNA, which induces ATR-dependent signaling. Our study both supports and extends this recent work in two ways. First, we report not only a number of sites that overlap with the other study but also many potential substrates specific to our work; and second, we examine phosphorylation in response to activation of the ATR branch of the DNA damage checkpoint as opposed to the ATM branch. That many of the substrates were found in both studies suggests the presence of generalized mechanisms to deal with genetic insult regardless of the branch of the checkpoint activated or the way in which the damage is repaired. Differences seen between the two data sets may reflect differences in substrate preference between ATM (IR damage) and ATR (UV damage), differences in cell type, or differences in the immunoprecipitating antibodies used.
In dissecting protein kinase signaling networks, a critical step is identifying substrates that mediate downstream biological responses. For protein kinases where a consensus phosphorylation motif is known, it is now possible to produce antibodies directed against phosphorylated motifs that broadly recognize many potential substrates. When combined with mass spectrometry, these antibodies can be used to probe and extend specific signaling networks as described here for ATM/R and the UV damage response. Identified downstream components of the ATM/R signaling network provide a necessary starting point for functional studies and will stimulate insights into diseases such as ataxia telangiectasia, Fanconi's anemia, Seckel syndrome, and the avoidance of cell cycle checkpoints in cancer, a critical early event in cancer progression (22).
Materials and Methods
Cell Lines and Reagents.
M059K glioblastoma cells were from the American Type Culture Collection (ATCC). GM18366 Seckel syndrome cells and GM00200-matched control cells were from the Coriell Cell Repository. For culture conditions/UV damage conditions, see SI Text.
Western Blotting.
Detection of total and phosphorylated proteins was performed by using standard methods (see Cell Signaling Technology). Antibodies used were: anti-phospho-ATM/R substrate motif, anti-phospho-Chk2 (T26/S28) (ATM/R substrate motif-2), anti-Chk1, anti-phospho-Chk1 (S345), anti-p38 MAPK, anti-phospho-p38 MAPK (T180/Y182), anti-phospho-SMC1 (S957), anti-phospho-NBS1 (S343), anti-p53, anti-phospho-p53 (S15), anti-beta actin, anti-53BP1, anti-RPA1, and anti-Mre11 (Cell Signaling Technology), and anti-BAP1 (Santa Cruz Biotechnology).
Immunoprecipitation/LC-MS/MS Mass Spectrometry.
Phosphopeptides were immunoprecipitated by using the PhosphoScan Kit (Cell Signaling Technology), with the modifications outlined in SI Text.
LC-MS/MS was performed as described (23), with the modifications outlined in SI Text.
Generation of Frequency Maps.
Redundant peptide lists for M059K cells treated with UV from all experiments were entered into pattern explorer from Scansite (www.scansite.mit.edu) (24). Resulting frequency maps were exported to Excel. Z values (deviation from expected frequency of an amino acid based on amino acid composition of all proteins in the SwissProt database) from frequency map data were input into TIGR MeV V.3.1 for Mac OSX (The Institute for Genomic Research MultiExperiment Viewer) (25). A gradient was created from Z of 0 (white) to Z ≥ 10 (blue).
Peptide Array Blots.
Peptide library arrays (SPOT arrays) were from Massachusetts Institute of Technology Biopolymers Laboratory. The top half of the array contained peptides of the sequence XXLS*QXXX, whereas the bottom half contained peptides of the sequence XXLT*QXXX, with X being an amino acid varied at each position across the blot. The first column (X) contains the minimum motif, whereas subsequent columns test addition of a fixed amino acid at residues relative to the phosphorylation site. Arrays were blotted by using standard Western blotting protocols. The SPOT array blots were digitized by using an Epson 1240 scanner, and intensities were quantified by using Scion Image Quant 1.63 for Mac. Total intensity was found for each row (each position relative to the phosphorylation site), and intensity of each individual spot was expressed as a percentage of the total intensity.
Semiquantitative Data Collection.
Immunoaffinity purification/LC-MS/MS analysis was performed as described. Precursor peptide ion chromatographic peak apex measurements were generated and compared as described in SI Text.
SILAC Data Collection.
SILAC analysis was performed by using standard protocols outlined in SI Text.
Immunoprecipitation with Covalently Coupled Antibody–Protein A Complexes.
ATM/R substrate motif antibodies were coupled to beads and used in immunoprecipitation using standard methods as detailed in SI Text.
Supplementary Material
Acknowledgments
We thank Ting Lei Gu for advice in directing this research and help in preparation of this manuscript and Jon Kornhauser for help in correcting protein class assignments.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707579104/DC1.
References
- 1.Hartwell LH, Weinert TA. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 2.Zhou BB, Elledge SJ. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 3.Abraham RT. DNA Rep. 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kastan MB, Lim DS. Nat Rev Mol Cell Biol. 2000;1:179–186. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 7.Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 8.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Proc Natl Acad Sci USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 10.Zhao H, Piwnica-Worms H. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traven A, Heierhorst J. BioEssays. 2005;27:397–407. doi: 10.1002/bies.20204. [DOI] [PubMed] [Google Scholar]
- 12.Kim ST, Lim DS, Canman CE, Kastan MB. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 13.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 14.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 15.Manning BD, Cantley LC. Sci STKE. 2002:PE49. doi: 10.1126/stke.2002.162.pe49. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 17.Livingstone M, Ruan H, Weiner J, Clauser KR, Strack P, Jin S, Williams A, Greulich H, Gardner J, Venere M, et al. Cancer Res. 2005;65:7533–7540. doi: 10.1158/0008-5472.CAN-04-3729. [DOI] [PubMed] [Google Scholar]
- 18.Eng JK, McCormack AL, Yates JR., III J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 19.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 20.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Mol Cell Proteom. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 21.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 22.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 23.Walters DK, Mercher T, Gu TL, O'Hare T, Tyner JW, Loriaux M, Goss VL, Lee KA, Eide CA, Wong MJ, et al. Cancer Cell. 2006;10:65–75. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Obenauer JC, Cantley LC, Yaffe MB. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.