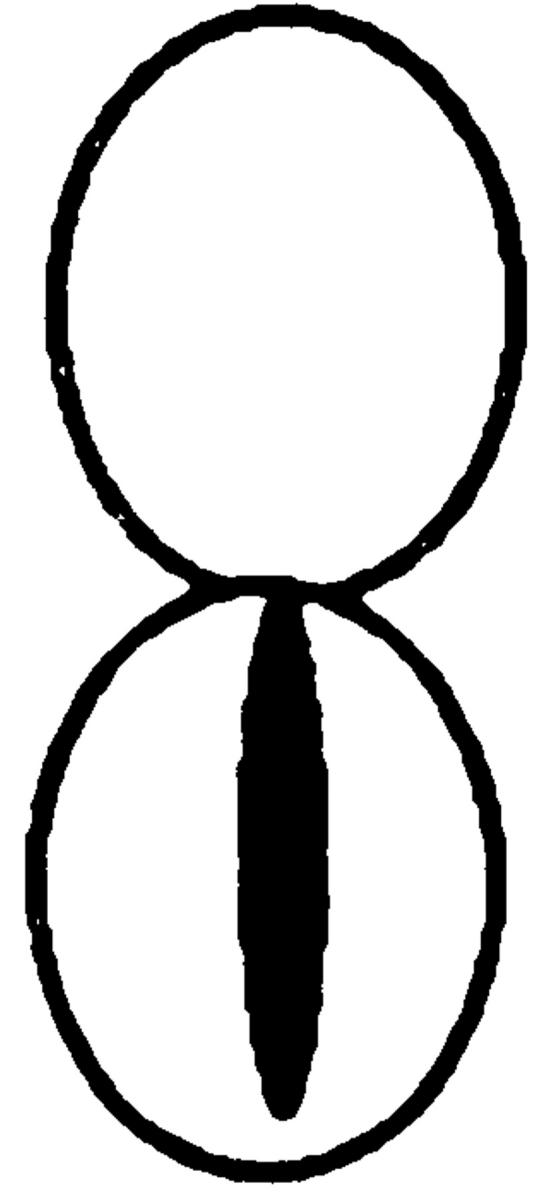Abstract
In the yeast Saccharomyces cerevisiae, positioning of the mitotic spindle requires both the cytoplasmic microtubules and actin. Kar9p is a novel cortical protein that is required for the correct position of the mitotic spindle and the orientation of the cytoplasmic microtubules. Green fluorescent protein (GFP)– Kar9p localizes to a single spot at the tip of the growing bud and the mating projection. However, the cortical localization of Kar9p does not require microtubules (Miller, R.K., and M.D. Rose. 1998. J. Cell Biol. 140: 377), suggesting that Kar9p interacts with other proteins at the cortex. To investigate Kar9p's cortical interactions, we treated cells with the actin-depolymerizing drug, latrunculin-A. In both shmoos and mitotic cells, Kar9p's cortical localization was completely dependent on polymerized actin. Kar9p localization was also altered by mutations in four genes, spa2Δ, pea2Δ, bud6Δ, and bni1Δ, required for normal polarization and actin cytoskeleton functions and, of these, bni1Δ affected Kar9p localization most severely. Like kar9Δ, bni1Δ mutants exhibited nuclear positioning defects during mitosis and in shmoos. Furthermore, like kar9Δ, the bni1Δ mutant exhibited misoriented cytoplasmic microtubules in shmoos. Genetic analysis placed BNI1 in the KAR9 pathway for nuclear migration. However, analysis of kar9Δ bni1Δ double mutants suggested that Kar9p retained some function in bni1Δ mitotic cells. Unlike the polarization mutants, kar9Δ shmoos had a normal morphology and diploids budded in the correct bipolar pattern. Furthermore, Bni1p localized normally in kar9Δ. We conclude that Kar9p's function is specific for cytoplasmic microtubule orientation and that Kar9p's role in nuclear positioning is to coordinate the interactions between the actin and microtubule networks.
Keywords: mitosis, nuclear migration, Spa2, Bni1p, formin
The simple budding yeast, Saccharomyces cerevisiae, is a highly polarized cell. During mitosis, almost all cell surface growth and secretion occur in the nascent bud (Field and Schekman, 1980). Underlying the polarized growth and secretion is a highly polarized actin cytoskeleton (Bretscher et al., 1994; Welch et al., 1994). Cortical patches of actin are found clustered in the bud and actin cables run from the mother cell towards the bud. At each division, the choice of position of the nascent bud is under strict genetic control. Haploids preferentially bud at the end marked by their birth scar, the axial pattern, and diploids tend to alternate between the two ends of the cell, the bipolar pattern (Freifelder, 1960; Chant and Pringle, 1995; Yang et al., 1997). During mating, the established axis of haploid cell division is perturbed such that the new growth is directed towards the tip of the mating projection. In response to a gradient of mating pheromone, the axis of growth re-orients towards the highest concentration of mating pheromone (Segall, 1993). Coincident with the change in the growth axis, the actin cytoskeleton redistributes such that actin cortical patches become clustered in the tip of the projection and cables run to the tip (Hasek et al., 1987; Read et al., 1992).
For efficient cell division and conjugation, the nucleus must also become oriented with respect to the growth axis of the cell. In haploid cells, the telophase of the previous cell cycle leaves the nuclear envelope–embedded microtubule organizing center (called the spindle pole body or SPB)1 oriented away from the next site of bud emergence. Thus, the haploid nucleus must rotate up to 180° each cell cycle. Before anaphase, the nucleus also migrates up to the neck of the nascent bud. The result of these movements is that the nuclear spindle becomes aligned with the growth axis adjacent to the bud, so that anaphase spindle elongation occurs through the neck and into the bud (Kahana et al., 1995; Yeh et al., 1995). Defects in nuclear orientation and migration result in misoriented spindles, anaphase in the mother cell, and binucleate mother cells.
During mating, the nucleus becomes oriented with respect to the mating projection. The nucleus moves up close to the mating projection with the spindle pole body pointing towards the tip of the projection (Byers and Goetsch, 1975; Hasek et al., 1987; Rose and Fink, 1987; Meluh and Rose, 1990). Defects in nuclear orientation or migration during mating lead to zygotes in which the two haploid nuclei fail to congress and fuse (for review see Rose, 1996).
Nuclear orientation and migration are dependent on microtubules that are attached to the nucleus at the SPB (Byers, 1981). During mitosis, mutations in the gene for β-tubulin, TUB2, were used to demonstrate that the cytoplasmic microtubules specifically were required to position and maintain the nucleus near the bud neck (Palmer et al., 1992; Sullivan and Huffaker, 1992). During mating as well, microtubule depolymerizing drugs and mutations have shown the requirement for microtubules in nuclear orientation and migration (for review see Rose, 1996). In each case, cytoplasmic microtubules are seen to enter the bud or mating projection and apparently make contact with cortical sites (Carminati and Stearns, 1997; Miller and Rose, 1998). Thus, it seems likely that interactions between cortical proteins and the cytoplasmic microtubules are a key element in nuclear migration and orientation.
In addition, the actin cytoskeleton has also been implicated in nuclear migration. Using a temperature-sensitive act1-4 allele, Koshland and colleagues found that preoriented spindles become misoriented upon loss of actin structures (Palmer et al., 1992). They suggested that the dependence of spindle orientation upon both microtubules and actin was consistent with cytoplasmic microtubules becoming tethered to specific sites on the cell cortex (Palmer et al., 1992). Similar models have been used to explain related phenomena in other organisms, including the early centrosomal rotations in Caenorhabditis elegans (Hyman and White, 1987) and the asymmetric positioning of the mitotic spindle in Chaetopterus oocytes (Lutz et al., 1988).
Several lines of evidence point to Kar9p as a good candidate to mediate the proposed interaction between actin and microtubules. Originally identified in a screen for bilateral karyogamy mutants (Kurihara et al., 1994), Kar9p plays an important role in nuclear migration in both mating and mitosis (Miller et al., 1998). Mutations in kar9 result in specific defects in cytoplasmic microtubule orientation into the mating projection and the bud. Green fluorescent protein (GFP)–Kar9p localizes as a single dot at the tip of the mating projection and at the tip of the growing bud, coincident with the tip of a bundle of cortically directed cytoplasmic microtubules. However, Kar9p localization at the cortex is independent of microtubules.
In addition to Kar9p, several microtubule motor proteins have been implicated in nuclear migration. The kinesin-related protein, Kip3p is involved in the initial alignment of the preanaphase nucleus at the bud neck (DeZwaan et al., 1997). Because kip3Δ causes very long and misoriented cytoplasmic microtubules (Cottingham and Hoyt, 1997; Miller et al., 1998), Kip3p may act by the destabilization of cytoplasmic microtubules. Cytoplasmic dynein function is required after Kip3p for the translocation of the nucleus through the bud neck (Kahana et al., 1995; Yeh et al., 1995; DeZwaan et al., 1997). Components of the dynein-activating (dynactin) complex including Jnm1p (McMillan and Tatchell, 1994), Act5p (Clark and Meyer, 1994; Muhua et al., 1994), and Nip100p (Kahana et al., 1998) are likely to contribute to the dynein-directed step in nuclear migration. Finally, another kinesin-related motor protein, Kip2p, is required for nuclear migration (Cottingham and Hoyt, 1997; Miller et al., 1998). Since kip2 mutants have very short or absent cytoplasmic microtubules, Kip2p may function to stabilize microtubules in vivo and appears to act in opposition to Kip3p.
Abundant genetic data suggests that there are at least two major, partially independent pathways for nuclear orientation and migration (Miller et al., 1998; Miller and Rose, 1998). Notably, with the exception of actin and tubulin, none of the genes involved in nuclear migration are essential for viability, and single deletion mutations do not result in severe defects. However, crosses between the various mutants suggest that Kar9p and Kip3p act in one pathway, whereas dynein, components of the dynactin complex, and Kip2p act in a second pathway.
In this paper, we examine the determinants for the localization of Kar9p to the cortex. We find that Kar9p localization is strongly dependent upon actin. In addition, Kar9p localization is dependent upon several actin-interacting polarization proteins, Spa2p, Pea2p, Bud6p, and Bni1p. Each of these proteins is found localized at the tip of the shmoo projection and tip of the growing bud (Snyder, 1989; Jansen et al., 1996; Valtz and Herskowitz, 1996; Amberg et al., 1997; Evangelista et al., 1997). Several lines of evidence suggest that these proteins act together in the cell. First, mutations in these genes cause similar defects in the diploid bipolar budding pattern (Snyder, 1989; Valtz and Herskowitz, 1996; Zahner et al., 1996; Amberg et al., 1997). Second, biochemical methods demonstrate that Spa2p, Pea2p, and Bud6p are associated with each other in a complex in vivo (Sheu et al., 1998). Interestingly, Spa2p has also been implicated in the signaling pathways for two mitogen-activated protein kinase cascades (Sheu et al., 1998). Bud6p was found to interact with actin by two-hybrid studies (Amberg et al., 1997), raising the possibility that these three proteins may function together to regulate the actin cytoskeleton (Amberg et al., 1997; Sheu et al., 1998).
Bni1p shows the strongest effects on Kar9p localization. Bni1p is a member of the widely conserved formin family of proteins, which includes mouse limb deformity (Woychik et al., 1990), Drosophila diaphanous (Castrillon and Wasserman, 1994) and cappuccino (Emmons et al., 1995), Aspergillus SepA (Harris et al., 1997) and FigA (Marhoul and Adams, 1995), and Schizosaccharomyces pombe Fus1 (Petersen et al., 1998). Bni1p binds to the actin-binding proteins, Bud6p and profilin (Evangelista et al., 1997; Imamura et al., 1997) and is a target of the rho-related GTPases, Cdc42p and Rho1p (Kohno et al., 1996; Evangelista et al., 1997; Imamura et al., 1997). Thus, Bni1p plays an important role in regulating the actin cytoskeleton.
In this and an accompanying paper (Lee et al., 1999), it is shown that bni1Δ mutants also exhibit a defect in the migration of the nucleus to the bud neck, consistent with their role in Kar9p localization. We also demonstrate the presence of microtubule orientation defects in mating bni1Δ mutants. Furthermore, genetic analysis places BNI1 in the KAR9 pathway for nuclear migration. However, our data suggest that Kar9p is at least partially active in bni1Δ mitotic cells. Furthermore, Kar9p is not required for at least two of Bni1p's functions in the cell. Taken together, these data suggest that Kar9p interacts with Bni1p as part of the link between microtubule-dependent nuclear movement and the cortical actin-dependent positional information.
Materials and Methods
Cell Culture and Fixation Methods
Cells were grown in yeast extract peptone dextrose (YPD) or in synthetic complete (SC) media as previously described (Miller and Rose, 1998). For experiments in which cells were induced with mating pheromone, synthetic α-factor (Department of Molecular Biology, Syn/Seq facility, Princeton University) was added to a final concentration of 10 μg/ml.
For experiments localizing GFP-Kar9p, cultures of each strain containing the plasmid pMR3465 were grown to saturation in SC media without leucine (SC-leucine). The precultures were then diluted 500-fold into SC-leucine media containing 2% raffinose instead of glucose and grown for 15–18 h at 30°C to early exponential phase. Expression of GFP-Kar9p was induced for 2–2.5 h at 30°C by the addition of galactose to 2%. To express GFP-Kar9p in shmoos, α-factor was added at the time of galactose induction. Cells were fixed by the addition of formaldehyde to 3.7% for 10 min and washed twice with PBS.
For localization of GFP-Bni1p, cells were treated as described above, with the exception that they were grown in SC media without uracil and the expression of GFP-Bni1p was induced by the addition of galactose for 4 h.
For determination of the cell-cycle distribution and nuclear position in different mutants, saturated overnight cultures were subcultured into YPD preincubated at either 30° or 14°C. Cells were grown to mid-exponential phase and fixed in methanol:acetic acid (3:1), washed twice in PBS, and stained with 4′,6-diamidino-2-phenylindole (DAPI; Accurate Chemical and Scientific Corp.).
For the analysis of the budding pattern, a/α diploid cells homozygous for the indicated mutations were grown to late exponential phase in YPD at 30°C. Cells were then stained with Calcofluor white (Sigma Chemical Co.) to visualize bud scars as described (Pringle, 1991). Only those cells with three or more bud scars were scored. If the bud scars were located at both poles, the cells were scored as bipolar. All other patterns were scored as random.
Latrunculin-A Treatment
The wild-type strain, MS1556, expressing the GFP-Kar9p plasmid was grown to early exponential phase as described above. Mitotic cells or pheromone-treated cells (shmoos) were concentrated 10-fold, and latrunculin-A (LAT-A) was added to 200 μm (Molecular Probes, Inc.) in DMSO for 10 min. Control cells were treated with an equivalent volume of DMSO. Cells were then collected by centrifugation and fixed for observation of the GFP fluorescence as described above or for staining of actin with rhodamine-phalloidin. For rhodamine-phalloidin staining, cells were collected by centrifugation and resuspended in 0.1% Triton X-100, 3.7% formaldehyde in PBS at room temperature for 30 min. Cells were washed once in PBS and resuspended in 3.7% formaldehyde/PBS at 30°C for 2 h. Cells were washed twice in PBS and resuspended in 90 μl PBS. Cells were stained by the addition of 30 μl rhodamine-phalloidin (0.2 U/μl; Molecular Probes, Inc.) in methanol at room temperature in the dark for 90–120 min. Cells were then washed once in PBS and scored by fluorescence microscopy.
Strain Construction
Yeast strains used in the course of this study are listed in Table I. Plasmids used in strain construction are listed in Table II. Isogenic deletion mutants were constructed in the wild-type strain, MS1556, derived from S288C. All oligonucleotides used for deletion construction were made by the Department of Molecular Biology Syn/Seq Facility, Princeton University. The bni1Δ was made by cutting p321 (Evangelista et al., 1997) with HindIII and XhoI and transforming into MS1556. This resulted in the bni1Δ strains, MS5212 and MS5340.
Table I.
Yeast Strains
| Strain | Genotype | Source | ||
|---|---|---|---|---|
| MS52 | MATα ura3-52 leu2-3 leu2-113 trp1Δ1 | |||
| MS1556 | MAT a ura3-52 leu2-3 leu2-112 ade2-101 his3Δ-200 | |||
| MS4062 | MATα kar9Δ::LEU2 ura3-52 leu2-3 leu2-113 trp1Δ1 | |||
| MS4306 | MAT a kar9Δ::HIS3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS4316 | MATα kar9Δ::LEU2 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS4903 | MATa dhc1Δ::LEU2 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5208 | MAT a spa2Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5209 | MAT a spa2Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5215 | MAT a bni1Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5340 | MAT a bni1Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 (pMR3465) | |||
| MS5229 | MAT a pea2Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5230 | MAT a pea2Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5429 | MATα dhc1Δ::LEU2 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5665 | MATα kip3Δ::HIS3 kar9Δ::LEU2 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5794 | MAT a bnr1Δ::HIS3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5849 | MAT a bud6Δ::HIS3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS5850 | MAT a bud6Δ::HIS3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6127 | MAT a/α kar9Δ::LEU2/+ bni1Δ::URA3/+ kip3Δ::HIS3/+ ura3-52/ura3-52 leu2-3/leu2-3 | |||
| leu2-113/leu2-113 his3Δ-200/his3Δ-200 ade2-101/ade2-101 trp1Δ1/+ | ||||
| MS6148 | MAT a ade2-101 ura3-52 trp1Δ1 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6149 | MATa kar9Δ::LEU2 bni1Δ::URA3 trp1Δ1 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6151 | MATa kar9Δ::LEU2 bni1Δ::URA3 trp1Δ1 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6154 | MATα kar9Δ::LEU2 bni1Δ::URA3 trp1Δ1 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6161 | MATα kar9Δ::LEU2 bni1Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6162 | MATa kar9Δ::LEU2 bni1Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6163 | MATα kar9Δ::LEU2 bni1Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6164 | MATα bni1Δ::URA3 ura3-52 trp1Δ1 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6165 | MATα kar9Δ::LEU2 ura3-52 leu2-3 leu2-113 his3Δ-200 ade2-101 | |||
| MS6166 | MATa kar9Δ::LEU2 bni1Δ::URA3 ura3-52 leu2-3 leu2-113 his3Δ200 ade2-101 | |||
| JY427 | MAT a fus1Δ1 leu2-3 leu2-113 ura3-52 | Fink | ||
| JY424 | MAT a fus2Δ3 his4-34 leu2-3 leu2-113 ura3-52 | Fink | ||
| MY3722 | MAT a fus7-1811 ura3-52 leu2 | |||
| MY3371 | MAT a GAL2 + ura3-52 leu2-Δ1 hap2 mal | Winston | ||
| Y306 | MAT a rsr1::URA3 ura3 leu2 his3 | Bender | ||
| Y373 | MAT a msb1::URA3 ura3 leu2 | Bender | ||
| Y897 | MAT a boi2::URA3 ura3 leu2 | Bender | ||
| YEF1002 | MAT a rga1::HIS3 ura3 leu2 his3 trp1 lys2 | Bi/Pringle | ||
| DDY76 | MAT a ABP1-858::URA3 lys2-801 leu2-3 leu2-113 ura3-52 | Drubin | ||
| DDY495 | MAT a sla1-Δ1::URA3 ura3-53 leu2-3 leu2-113 | Drubin | ||
| DBY7055 | MAT a BUD6/AIP3-Δ2::HIS3 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 | Amberg |
All yeast strains were constructed for this study, unless otherwise indicated.
Table II.
Plasmids and Bacterial Strains
| Strain | Genotype/description | Source | ||
|---|---|---|---|---|
| pMR3465 | PGAL1-GFP-KAR9 LEU2 CEN4 ARS1 AMPR | Rose | ||
| p321 | BNI1 disruption construct | Boone | ||
| p210 | SPA2-3::URA3 disruption construct | Snyder | ||
| pNV44 | PEA2 disruption construct | Herskowitz | ||
| pRS403 | HIS3 | Hieter | ||
| pMR4348 | PGAL1-HA3-BNI1-GFP URA3 CEN AMPR | Boone | ||
| /p1955 |
To construct bnr1Δ::HIS3, all but the first codon of BNR1 was replaced with HIS3 using the one-step gene replacement method (Rothstein, 1983). The disruption fragment was generated by PCR using the following two oligonucleotides (BNR1 sequence is shown in normal font and HIS3 sequence is shown in italics): 5′ TTTTGAAGATTACATAGTGATGAT GATCGTGACACAAAAGCAGATAAAAAAATAGCACAATCATCAGCGATGCCGTTTTAAGAGCTTGGTG 3′ and 5′ AGCGATTGCGAATATTGTCCATTTCTTTATATAAGCTCCACAACTACATAAATACTAAGTC T T CACTA CGAG TTCAAGAGAAAAAAAAAG 3′ . Plasmid pRS403 (Sikorski and Hieter, 1989) was used as the template for the HIS3 portion of the construct. MS1556 was transformed to HIS3 prototrophy to create MS5794. Replacement of BNR1 was confirmed by PCR analysis.
To construct bud6Δ::HIS3, all but the first codon of BUD6 wasreplaced by the HIS3 gene using the one step gene replacement method (Rothstein, 1983). The disruption fragment was generated by PCR using the following forward and reverse primers (BUD6 sequence is in normal font and HIS3 sequence is shown in italics): 5′ ACCCAAGAAAAG- GGAAAAAGAGTAGAAGCTGGCTGCCAAATTGGTGTAGTAATCCT CGTATTATTTTAAAATTAGATG CCG TTTT AAGAGC TT G-GTG 3′ and 5′ TATTGCCGCAAACTTTGTAATAAGCCAAAAG-CACTAA T C T C TTTTCCGTTAGC TTTCATAAAATTAGTGTATTTACGAGTTCAAGAGAAAAAAAAAG 3′. Plasmid, pRS403 (Sikorski and Hieter, 1989), was used as the template for the HIS3 portion of the disruption construct. MS1556 was transformed to yield the BUD6 deletion strain, MS5849 and MS5850. The deletion was confirmed at both the 5′ and 3′ ends by PCR analysis.
To construct a pea2Δ mutation, pNV44 (Valtz and Herskowitz, 1996) was digested with KpnI and BamH1. The resulting 2.0-kb band was gel purified and transformed into the wild-type strain, MS1556, resulting in disruption of the endogenous PEA2 locus by the one-step gene replacement (Rothstein, 1983). Two independent colonies, MS5229 and MS5230, were isolated.
A spa2Δ mutation was made by the one-step gene replacement method. p210 (in which the SacI/SphI fragment of SPA2 was replaced by URA3), a kind gift from Michael Snyder (Yale University, New Haven, CT), was digested with HindIII and SalI. A 4.0-kb band was gel purified and transformed into the wild-type strain, MS1556, creating the SPA2 disruptants, MS5208 and MS5209.
Microscopy
Microscopy was carried out on an Axiophot microscope equipped with a 1.4 NA 100× Neofluor lens (Carl Zeiss, Inc.) or a 1.3 NA 100× UplanFl iris lens (Olympus Corp.). Images were recorded using a SIT Video Camera 3200 with a camera controller C2400 (Hamamatsu Corp.). Images were initially processed using Omnex Image processing unit (Imagen) and captured to computer disk using a Scion image capture board. Adobe Photoshop 4.0 was used to further optimize contrast.
Results
Kar9p Localization Is Actin Dependent
Kar9p is a cortical protein that plays an important role in orienting cytoplasmic microtubules. To understand the mechanism of Kar9p microtubule orientation, we sought to identify the cellular components involved in localizing Kar9p to its cortical site. Previous work demonstrated that Kar9p's cortical localization is independent of microtubules (Miller and Rose, 1998). Therefore, Kar9p's cortical localization must be dependent on other peripheral determinants.
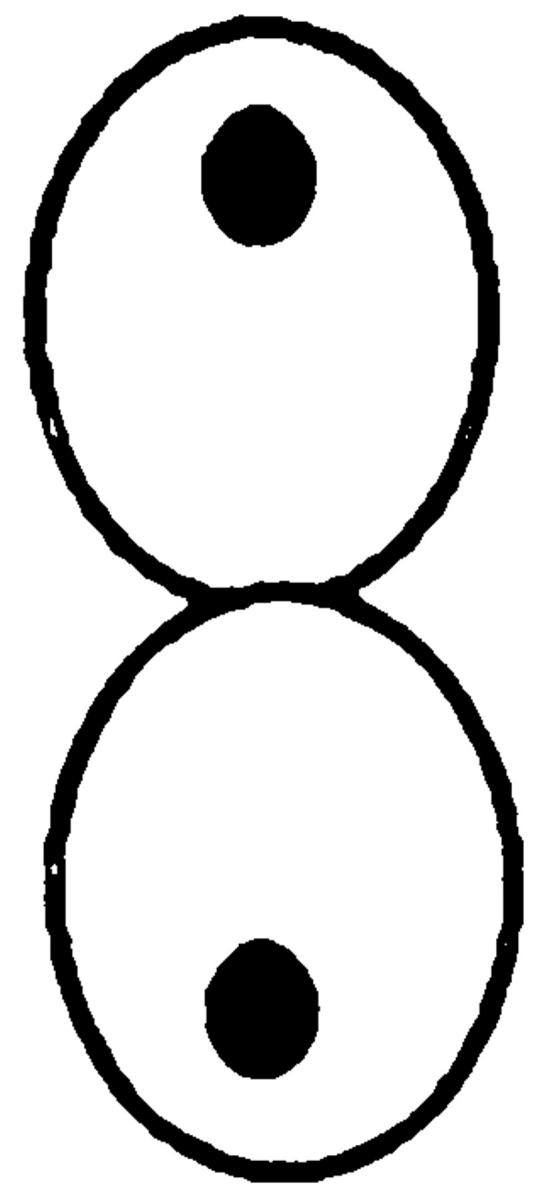
As a first step, we used rhodamine-phalloidin to determine whether GFP-Kar9p colocalizes with cortical actin in pheromone-treated cells. In every case (n = 20), we found GFP-Kar9p coincident with actin at the tip of the shmoo (data not shown). Although in several cases we observed a well-resolved actin dot that coincided with the GFP-Kar9p, most often the cortical actin dots were too concentrated to resolve individual structures. These results suggested that Kar9p might be associated with cortical actin structures.
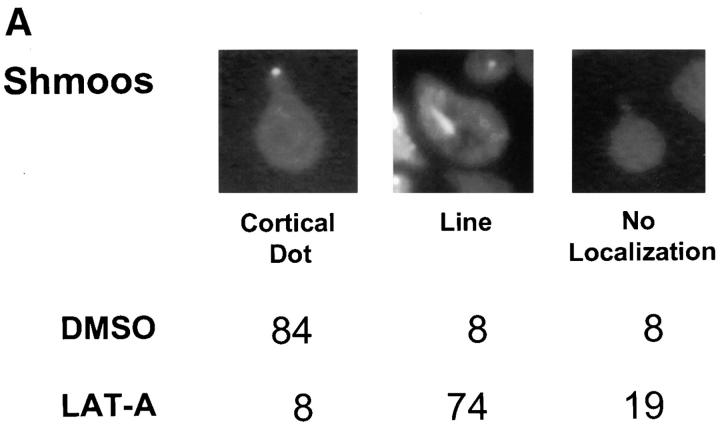
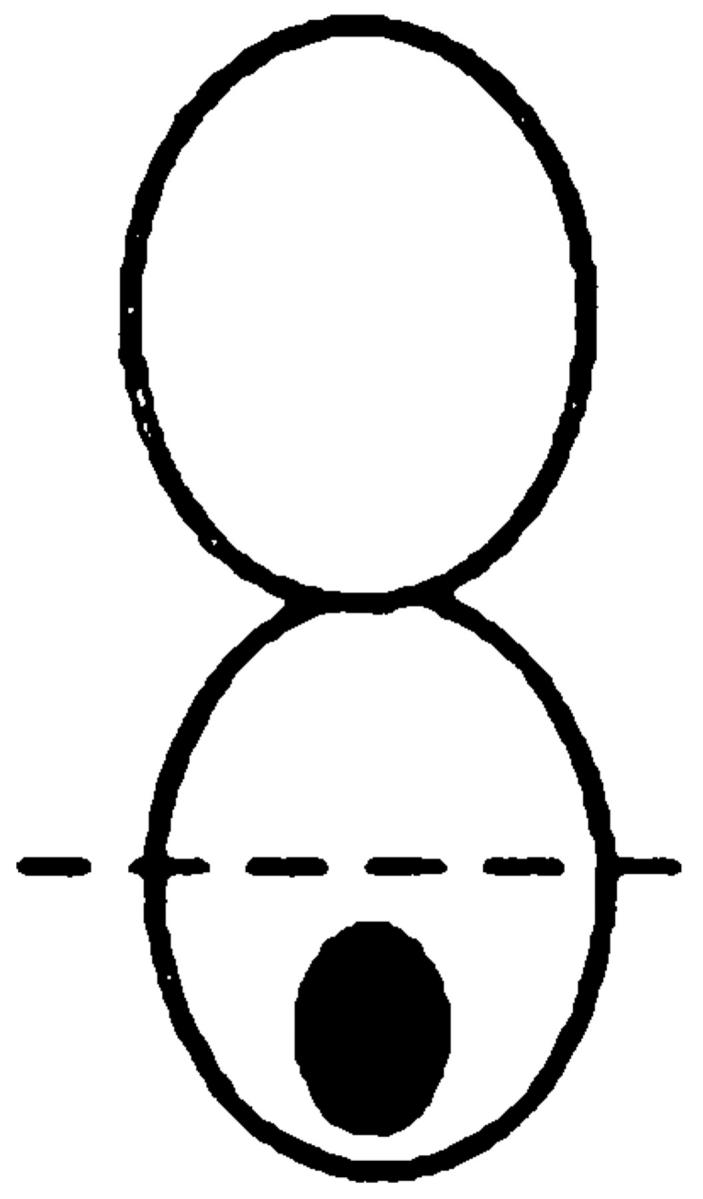
To determine whether the actin cytoskeleton is required for Kar9p localization, we used the actin-depolymerizing drug LAT-A. Confirming earlier work (Ayscough et al., 1997), we found that the actin network was 100% depolymerized within 10 min after treatment with LAT-A, as shown by staining with rhodamine-phalloidin (data not shown). We first examined the localization of a functional GFP-Kar9p construct in pheromone-treated cells. In the control experiments, 84% of shmoos exhibited a single dot of GFP-Kar9p fluorescence at the tip of the mating projection (Fig. 1 A). In contrast, when shmoos were treated with LAT-A, only 8% of the shmoos exhibited the cortical dot at the tip of the projection. Instead, in 74% of the LAT-A–treated shmoos, GFP-Kar9p fluorescence was found in a line extending from the nucleus. Based on previous experiments, we expect that this corresponds to cytoplasmic microtubules (Miller and Rose, 1998). In addition, 19% of the LAT-A–treated shmoos exhibited no localization of the GFP-Kar9p fluorescence. Thus, depolymerization of the actin cytoskeleton results in rapid loss of the cortical localization of Kar9p and redistribution onto the cytoplasmic microtubules.
Figure 1.
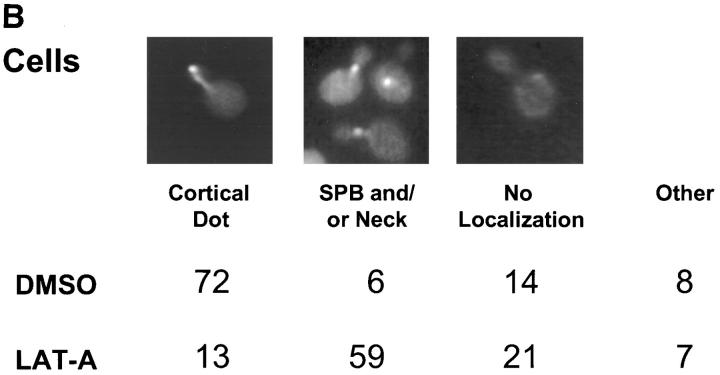
In both shmoos and cells, Kar9p localization is actin dependent. Wild-type shmoos and mitotic cells (MS1556) expressing GFP-Kar9p (pMR3465) (2–2.5 h) were treated with LAT-A for 10 min, as described in Materials and Methods. The percentage of mitotic cells and shmoos exhibiting each pattern of GFP-Kar9p localization was determined after microscopic examination. Only mitotic cells with medium to large buds were scored. n > 100 cells for each experimental condition. The panel depicting the example of the cortical dot also shows a line of GFP-Kar9p localization extending from it. Approximately 10% of cells with cortical dots also show the line pattern. Previous work (Miller and Rose, 1998) has shown that the line represents microtubule association.
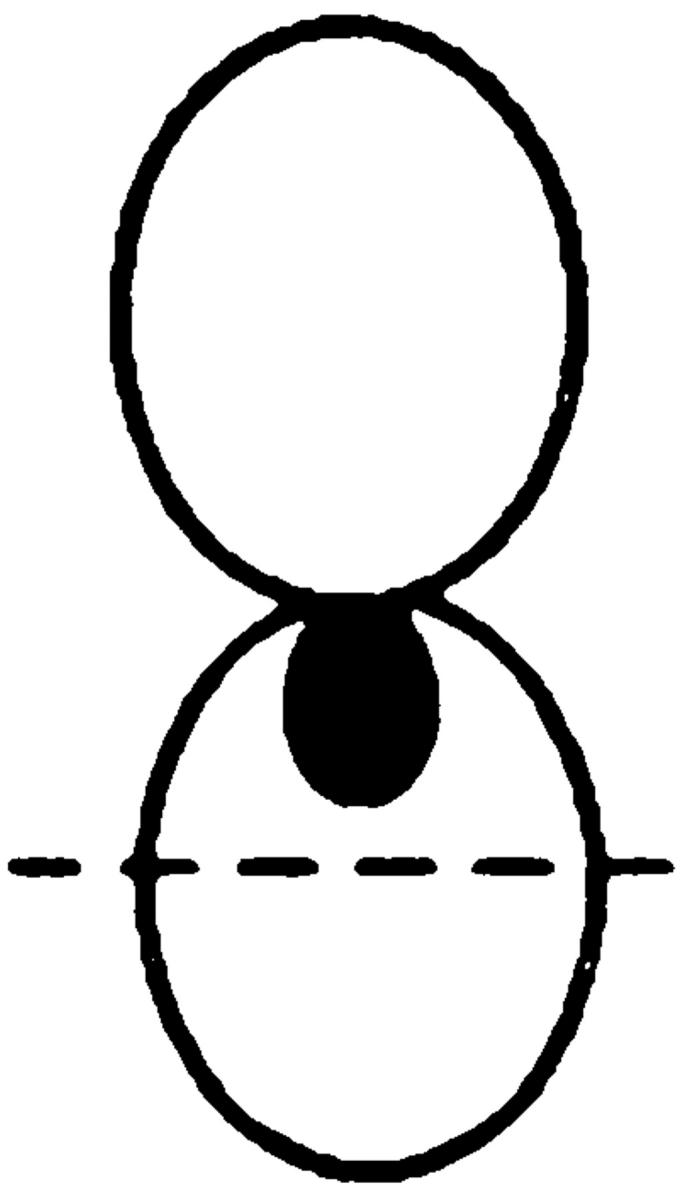
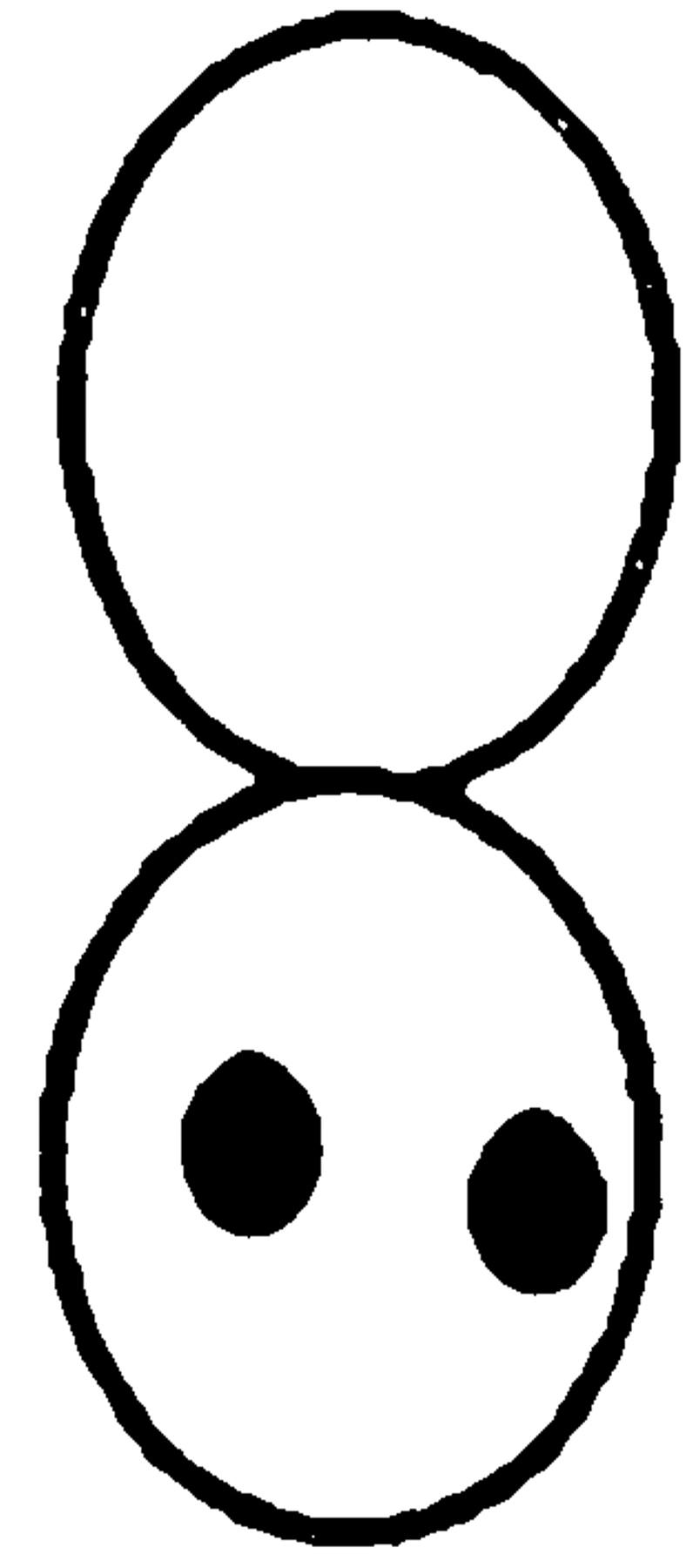
We next examined the requirement for polymerized actin for Kar9p localization in mitotically dividing cells. Unbudded and most post-anaphase large budded cells do not exhibit localized GFP-Kar9p (Miller and Rose, 1998), so we restricted our observations to cells with medium to large buds. In the control cells, 72% displayed a single dot of GFP-Kar9p fluorescence at the tip of the bud (Fig. 1 B). When mitotic cells were treated with LAT-A, only 13% exhibited the dot at the tip of the bid. Instead, 59% of the cells exhibited a dot of GFP-Kar9p fluorescence at two alternate locations, coincident with the DAPI staining and/ or close to the mother-bud neck (Fig. 1 B). The GFP fluorescence coincident with the nuclear DAPI stain is likely to correspond to SPB localization. Another 21% of the LAT-A–treated mitotic cells exhibited no localized GFP-Kar9p fluorescence. Thus, in >80% of LAT-A–treated cells, GFP-Kar9p localization was not found at the tip of the bud. From these results, we conclude that GFP-Kar9p localization in both shmoos and mitotically dividing cells is dependent on polymerized actin.
Kar9p Localization Is Affected by the Polarization Mutants, spa2Δ, pea2Δ, bud6Δ, and bni1Δ
Although Kar9p's localization was actin dependent, it was found only as a single dot and therefore did not colocalize with other cortical actin patches at the shmoo tip and bud cortex. Therefore, we reasoned that additional cortical actin-associated proteins might restrict Kar9p localization to one site. In this case, mutations in a subset of specific actin-associated proteins would eliminate or alter Kar9p localization. To test this idea, we used pheromone-treated cells to assay GFP-Kar9p localization in mutants that affect actin, polarization, and/or mating. Most mutations had no effect on GFP-Kar9p localization to the tip of the shmoo projection. These included genes affecting actin function, ABP1 (Drubin et al., 1988, 1990) and SLA1 (Holtzman et al., 1993); genes involved in GTPase signaling, BOI2 (Bender et al., 1996), RSR1/BUD1 (Bender and Pringle, 1989), RGA1 (Stevenson et al., 1995), and MSB1 (Bender and Pringle, 1989); and genes affecting cell fusion, RVS161 (Brizzio et al., 1998), FUS1 (Trueheart and Fink, 1989), and FUS2 (Trueheart et al., 1987). Furthermore, GFP-Kar9p localization in mitotic cells was not affected by mutations in the genes encoding two microtubule-dependent motor proteins, KIP3 and DHC1 (Miller et al., 1998).
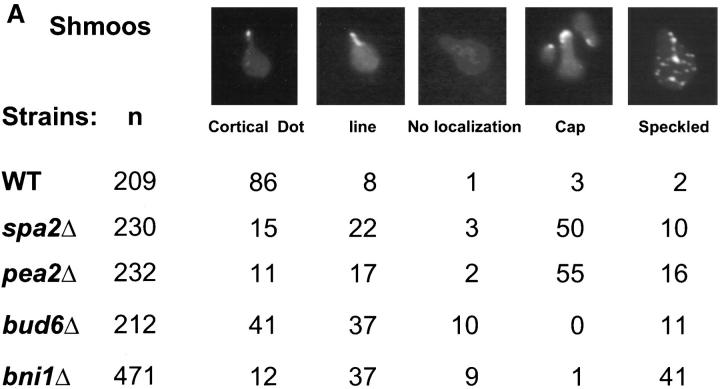
However, GFP-Kar9p localization in shmoos was affected by four mutations implicated in actin organization and polarization, spa2, pea2, bud6, and bni1. SPA2 and PEA2 are required for polarization in mating cells and mutations result in broader peanut-shaped shmoo projections (Valtz and Herskowitz, 1996). In mitotic cells, SPA2 and PEA2 are required for the diploid-specific bipolar budding pattern. Spa2p and Pea2p physically interact with each other and are interdependent for their own localization to the tips of shmoo projections (Valtz and Herskowitz, 1996). In the wild-type strain, 86% of the cells displayed GFP-Kar9p localization as a single dot at the tip of the shmoo projection. In contrast, only 15% of spa2Δ and 11% of pea2Δ shmoos exhibited GFP-Kar9p fluorescence as a single dot. Instead, 50% of spa2Δ and 55% of pea2Δ shmoos showed a new pattern in which GFP-Kar9p was found in a broad region over the shmoo tip. In addition, both mutants showed a significant number of cells with GFP-Kar9p in a line or in multiple “speckles” (Fig. 2 A).
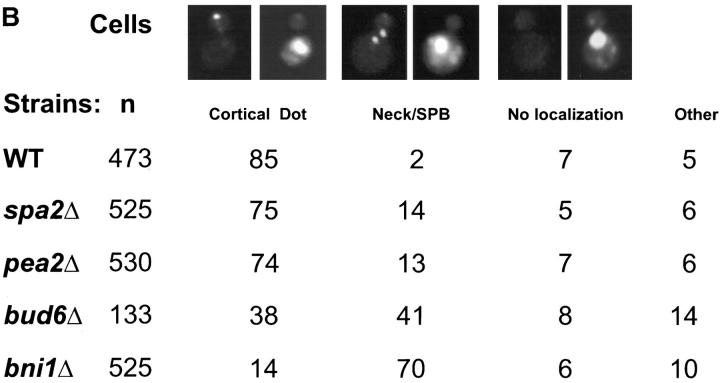
Figure 2.
GFP-Kar9p shows aberrant localization in polarization mutants. GFP-Kar9p expression was induced in shmoos by the simultaneous addition of alpha factor and galactose to early exponentially growing cultures. Both shmoos and mitotically dividing cells were induced for GFP-Kar9p expression for 2–2.5 h by the addition of galactose (2%). The following strains containing PGAL1-GFP-KAR9 LEU2 CEN4 ARS1 AMPR (pMR3465) were used: wild type (MS1556), spa2Δ (MS5209 and MS5208), pea2Δ (MS5229 and MS5230), bni1Δ (MS5215 and MS5340), and bud6Δ (MS5850). Cells were fixed with formaldehyde and scored for GFP-Kar9p localization patterns by microscopy. In the shmoo assay, only cells with a defined shmoo projection were scored.
BUD6 is also required for the diploid-specific bipolar budding pattern and Bud6p (Aip3p) has been shown to interact with actin (Amberg et al., 1997). In the bud6Δ strain, only 41% of shmoos exhibited Kar9p fluorescence as a single dot (Fig. 2 A). Instead, a large percentage of the bud6Δ shmoos exhibited GFP-Kar9p fluorescence as a line (37%, compared with 8% in wild type, Fig. 2 A).
BNI1 encodes a formin homologue that has been implicated in the regulation of the actin cytoskeleton and cell polarization (Kohno et al., 1996; Evangelista et al., 1997), as well as bipolar budding (Zahner et al., 1996). Bni1-GFP also localizes to the shmoo tip (Evangelista et al., 1997). When GFP-Kar9p localization was examined in bni1Δ shmoos, only 12% exhibited Kar9p fluorescence as a cortical dot. Instead, 37% of the bni1Δ shmoos exhibited GFP-Kar9p in a line pattern and 41% in a speckled pattern throughout the shmoo (Fig 2 A).
Because Kar9p also functions to orient cytoplasmic microtubules during vegetative growth (Miller and Rose, 1998), we sought to determine whether these four mutants also affected Kar9p localization during mitosis. In 85% of wild-type medium-to-large budded cells, GFP-Kar9p localized to a single dot at the tip of the bud. In the spa2Δ and pea2Δ cells, there was a modest reduction in the frequency of GFP-Kar9p localization to the bud tip, down to 75 and 74%, respectively (Fig. 2 B). Although modest, the reduction was observed in three independent experiments. Only 38% of the bud6Δ cells had a dot of GFP-Kar9p fluorescence at the tip of the bud. In 41% of the bud6Δ cells, GFP-Kar9p was mislocalized (Fig. 2 B) to the mother-bud neck and/or a region coincident with DAPI staining. Finally, in the bni1Δ cells, only 14% of cells had a dot of GFP-Kar9p at the tip of the bud. Instead, 70% of these cells had Kar9p fluorescence at the mother-bud neck and/ or a region coincident with DAPI staining (Fig. 2 B). Therefore, all four mutations affected Kar9p localization, although only in the bni1Δ strain was the magnitude similar to that of actin depolymerization.
Because the deletion of BNI1 had such a strong effect on Kar9p localization, we next determined whether the two proteins are interdependent for localization. Accordingly, we examined the localization of Bni1p using a fully functional GFP-Bni1p fusion (a generous gift from Charlie Boone, Queens University, Kingston, Canada) in kar9Δ mutant shmoos and mitotic cells. In comparing kar9Δ to wild type, there were no obvious differences in the localization pattern of GFP-Bni1p to the presumptive bud site or the tip of the growing bud (data not shown). Therefore, Bni1p's association with the cortex is not dependent on Kar9p.
BNI1-related (BNR1) is another yeast gene encoding a formin homology domain protein. Bnr1p shares some functions with Bni1p (Imamura et al., 1997). We therefore determined whether BNR1 was also required for GFP-Kar9p localization. In both shmoos and mitotic cells, GFP-Kar9p localization in an isogenic bnr1Δ strain was indistinguishable from wild type (data not shown). Thus, localization of Kar9p to the cortex is a specific function of Bni1p.
bni1Δ Mutants Exhibit Nuclear Migration Defects
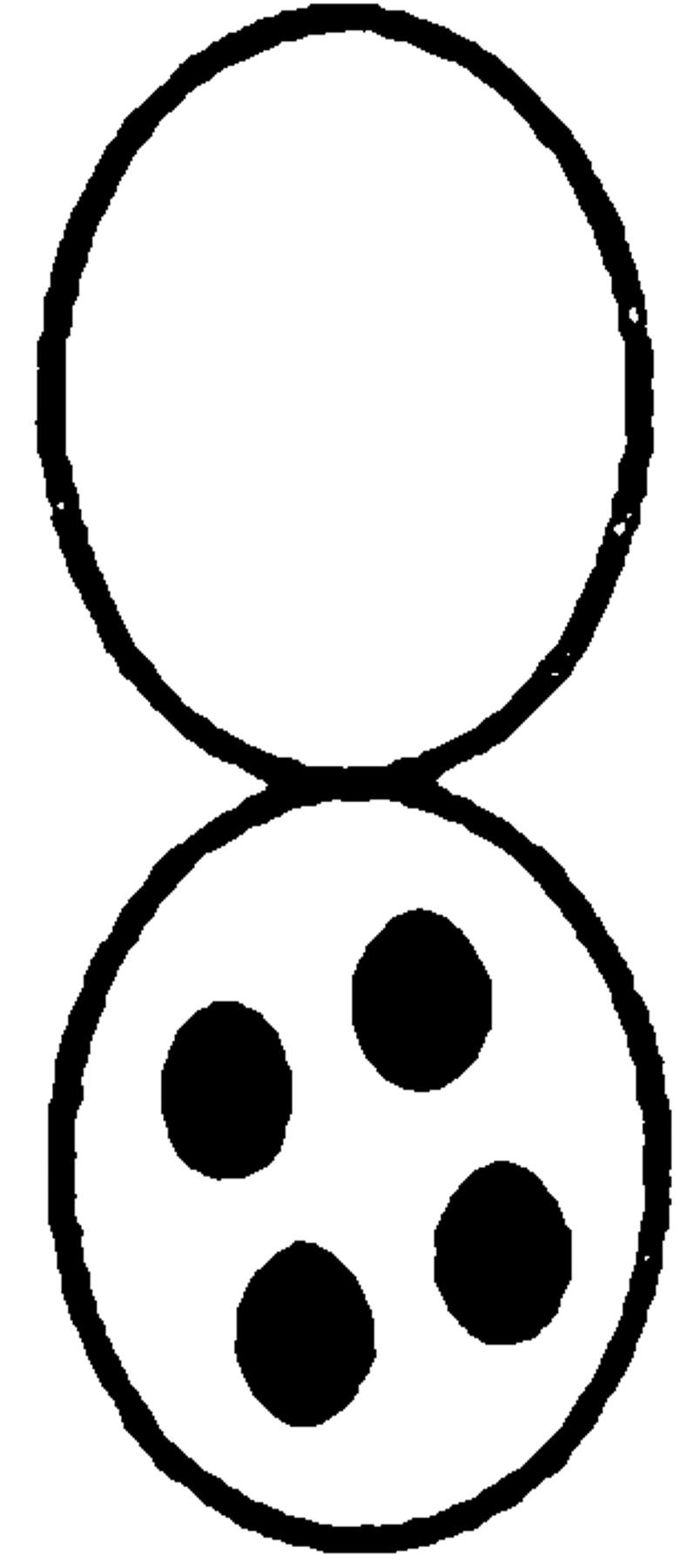
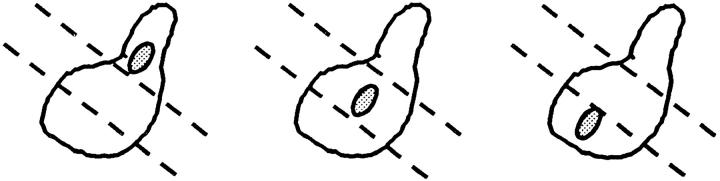
Since spa2Δ, pea2Δ, bud6Δ, and bni1Δ mutants resulted in an altered localization for GFP-Kar9p, we wanted to determine whether these mutants also exhibited defects in nuclear migration like kar9Δ. Nuclear position was scored in asynchronous cultures using the DNA-specific fluorescent dye, DAPI (Table III). In the wild-type control, 20% of the cultures were large budded and, of these, 96% showed normal nuclear positions (i.e., located at the bud neck, or in anaphase across the bud neck, or in telophase with one nucleus each in the mother and bud). In contrast, 40% of kar9Δ mutants were large-budded (Table III), suggesting that kar9Δ has a cell cycle delay in mitosis. Consistent with our previous report (Miller and Rose, 1998), nuclear positioning was aberrant in up to 50% of the kar9Δ large-budded cells (i.e., nuclei not located at the bud neck, or anaphase or telophase occurring in the mother cell). The spa2Δ, pea2Δ, and bud6Δ mutants exhibited no obvious defects in nuclear position within the large-budded cells. However, like kar9Δ, the bni1Δ strain exhibited a significant defect in nuclear position; in 23% of the large-budded cells, anaphase was occurring within the mother cell. In contrast, only 2% of wild-type large-budded cells exhibited this phenotype (Table III). Unlike kar9Δ, only 5% of bni1Δ large-budded cells contained two nuclei within the mother cell (compared with 18% in kar9Δ). Similar results were obtained when the spa2Δ, pea2Δ, bud6Δ, and bni1Δ cultures were grown at 14°C (data not shown). Therefore, we conclude that bni1Δ is a nuclear migration mutant. However, the nuclear migration defect of bni1Δ is not as severe as that of kar9Δ.
Table III.
Nuclear Migration Defects of Mutants
The percentage of nuclear positioning defects in mutant cultures. Cultures of wild type (MS1556), kar9Δ (MS4306), bni1Δ (MS5215), bud6Δ (MS5849), spa2Δ (MS5208), and pea2Δ (MS5229) were grown to early exponential phase at 30°C. The position of nuclear DNA was visualized by DAPI to visualize nuclear material. Each strain was scored for bud size as unbudded, small, medium, or large budded. Large-budded cells were also examined for their nuclear phenotype and scored as either normal or abnormal (nuclear morphology corresponding to the phenotypes schematically depicted). The percentages of bud size are from the total cells counted (n > 300). The percentages for the nuclear phenotypes are derived from the percent of large-budded cells. The percentage of defects in large-budded cells is shown in the Σ % LB abnormal column. A representative experiment is shown.
We next examined nuclear position in cells treated with mating pheromone. In wild type, the nuclei migrated to the base of the shmoo projection in 66% of the cells. As previously reported (Miller and Rose, 1998), the kar9Δ mutants exhibited a severe defect in nuclear migration to the shmoo projection, with 71% of the nuclei found in the center of the shmoo body (Table IV). The spa2Δ and pea2Δ mutants did not exhibit a nuclear migration defect, and may have localized better than the wild type, consistent with the continued localization of Kar9p at the shmoo tip. However, in 58% of the bud6Δ shmoos and 72% of the bni1Δ shmoos, the nuclei failed to migrate into the shmoo projection and were instead positioned in the center of the shmoo body (Table IV). Therefore, during mating, bud6Δ mutants exhibited a moderate defect and bni1Δ mutants exhibited a major defect in nuclear positioning.
Table IV.
Nuclear Position in Mutant Shmoos
|
||||||
|---|---|---|---|---|---|---|
| Nucleus | In/at neck | In center | At bottom | |||
| Wild type | 66 | 34 | 0 | |||
| kar9Δ | 1 | 71 | 28 | |||
| spa2Δ | 85 | 15 | 0 | |||
| pea2Δ | 83 | 15 | 2 | |||
| bud6Δ | 42 | 58 | 0 | |||
| bni1Δ | 15 | 72 | 13 | |||
The percentage of nuclear positioning defects in mutant shmoos. Wild type (MS1556) and mutant strains kar9Δ (MS4306), spa2Δ (MS5208), pea2Δ (MS5229, bud6Δ (MS5849), and bni1Δ (MS5215) were induced to form shmoos with alpha factor for 2 h. Cells were fixed, stained with DAPI, and then scored for nuclear position. n = 100 cells for each strain.
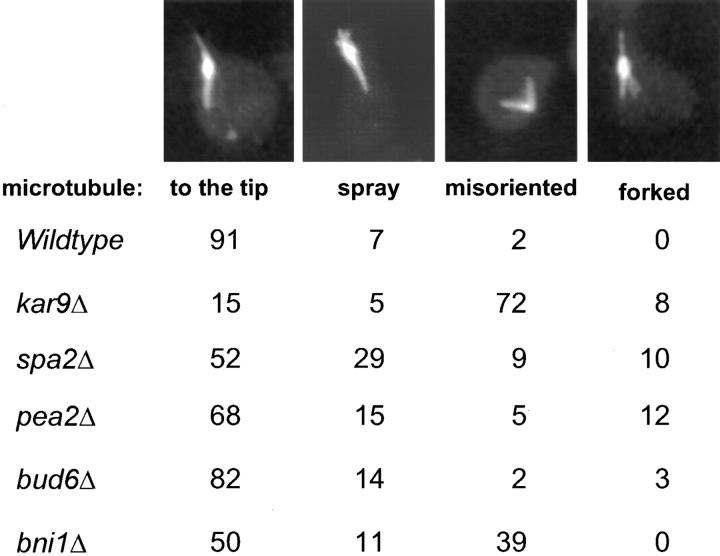
Microtubule Morphologies in Mutants
We next examined the microtubule morphologies of the mutants. In >90% of wild-type shmoos, the cytoplasmic microtubules were present as a single bundle that extended from the nucleus to the tip of the shmoo projection (Fig. 3). As previously reported, only 15% of the kar9Δ shmoos contained cytoplasmic microtubules that extended to the shmoo tip. In most kar9Δ shmoos, the cytoplasmic microtubule bundle was misoriented and extended in random directions. In the spa2Δ mutant shmoos, only 52% showed a single bundle of microtubules oriented towards the shmoo tip. Instead, 29% of the spa2Δ mutant shmoos exhibited a “spray” of microtubules extending to the shmoo tip, and 10% showed supernumerary microtubules oriented away from the shmoo tip (“forked pattern,” Fig. 3). The pea2Δ mutant showed a similar pattern, albeit significantly less severe than that of spa2Δ. The presence of additional cytoplasmic microtubules making contact with the cortex is consistent with Kar9p localizing over a broader region of the shmoo tip.
Figure 3.
Microtubule orientation in shmoos. Wild-type (MS1556) and mutant strains kar9Δ (MS4306), spa2Δ (MS5208), pea2Δ (MS5229), bud6Δ (MS5849), and bni1Δ (MS5215) were induced to form shmoos with alpha factor for 2 h. Cells were then prepared for indirect immunofluorescence as described in Materials and Methods.
The bni1Δ strain also showed a significant defect in microtubule orientation in the shmoo. Like spa2Δ, only 50% of bni1Δ shmoos exhibited the normal microtubule pattern. However, unlike spa2Δ, 39% of the bni1Δ mutant shmoos exhibited misoriented microtubules similar to the pattern seen with the kar9Δ (Fig. 3). Thus, in mating cells, the defect in microtubule orientation in the bni1Δ mutants is consistent with the aberrant localization of Kar9p.
We next examined the microtubule morphology in bni1Δ mitotic cells. For this purpose, we focused on those cells exhibiting a nuclear positioning defect such that anaphase was occurring within the mother cell. As reported previously (Miller and Rose, 1998), when the rare wild-type cells with anaphase in the mother were examined, the cytoplasmic microtubules nearly always extended into the bud (90%, Table V). In contrast, in the kar9Δ mutant, the cytoplasmic microtubule bundles infrequently extended into the bud (31%) and were more often found in the mother cell (48%). However, in 79% of the bni1Δ mutant, the cytoplasmic microtubules were correctly extended into the bud.
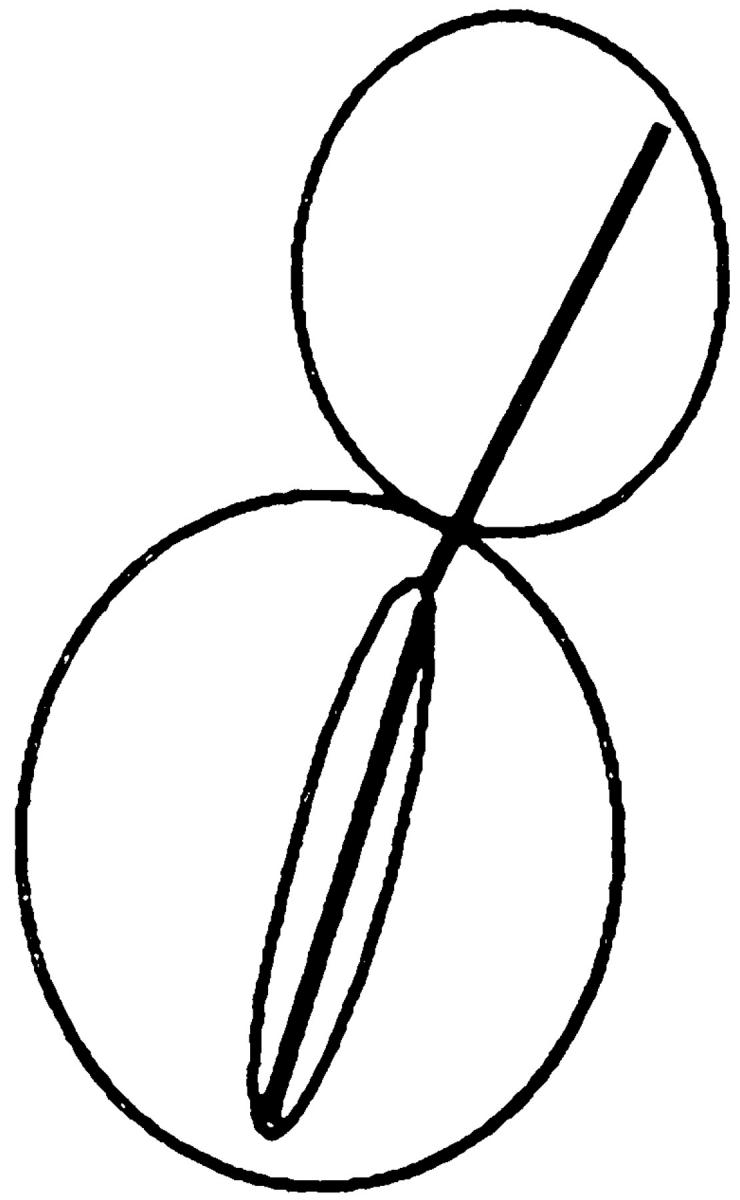
Table V.
Microtubule Orientation in kar9Δ bni1Δ Double Mutants
|
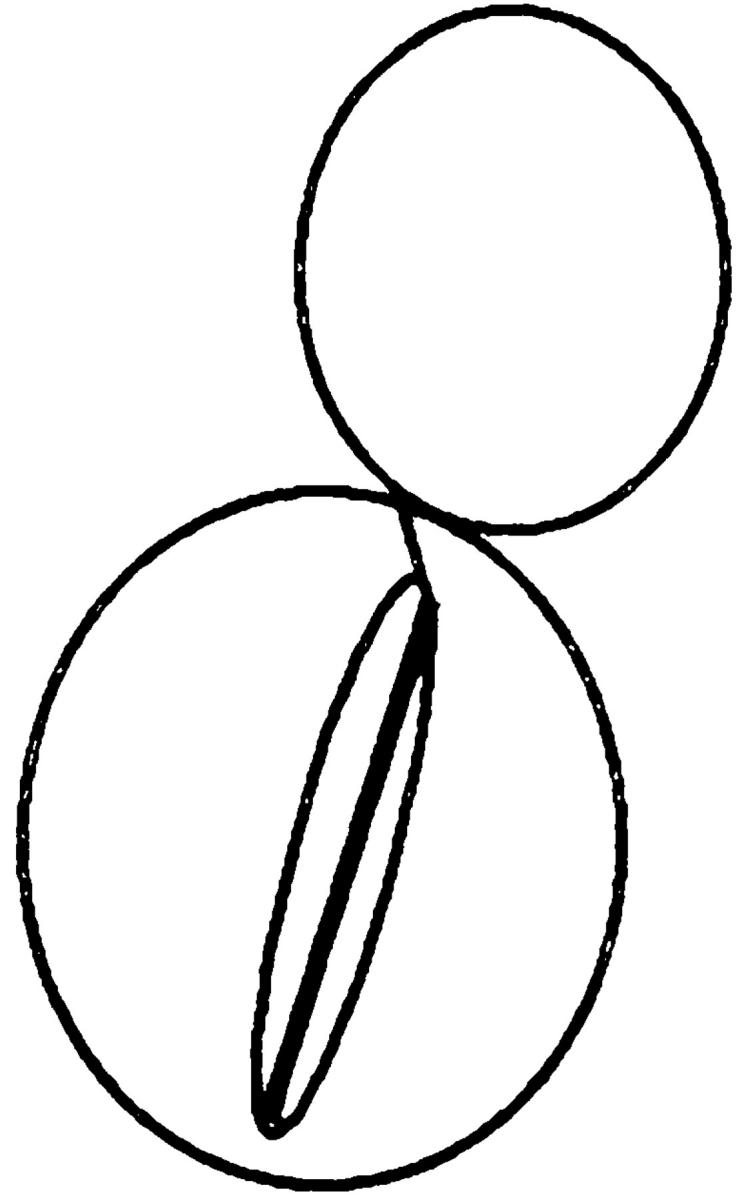
|
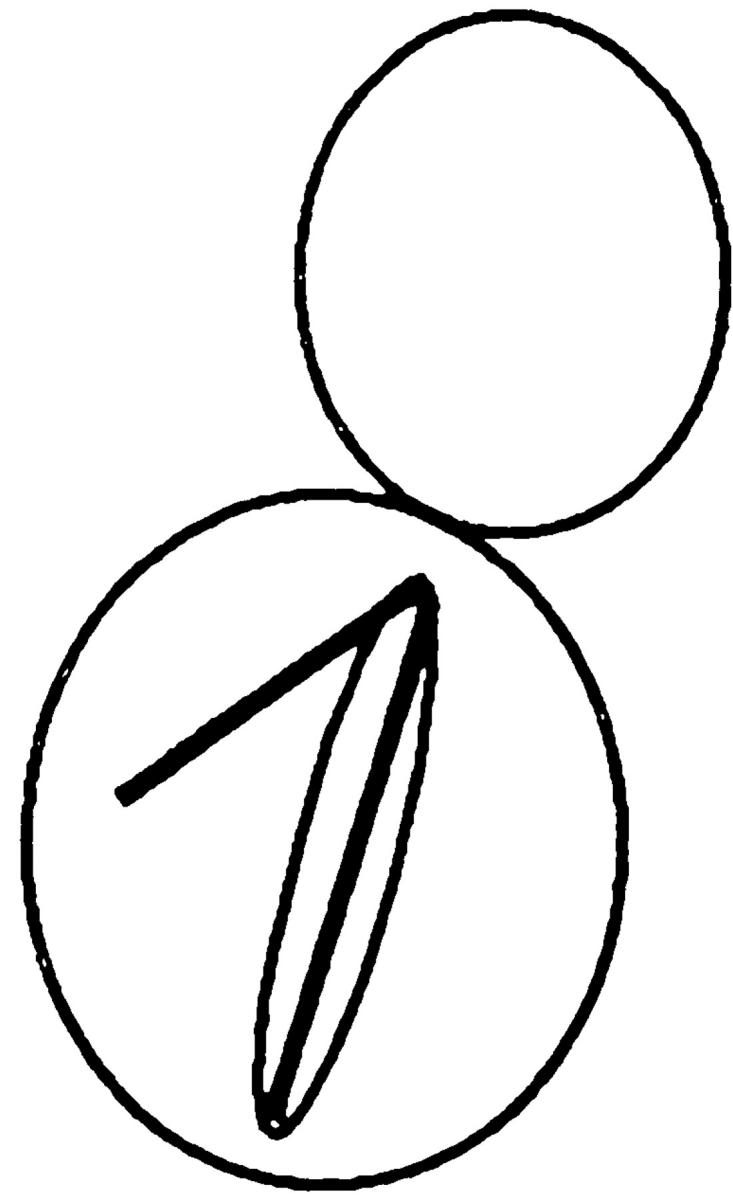
|
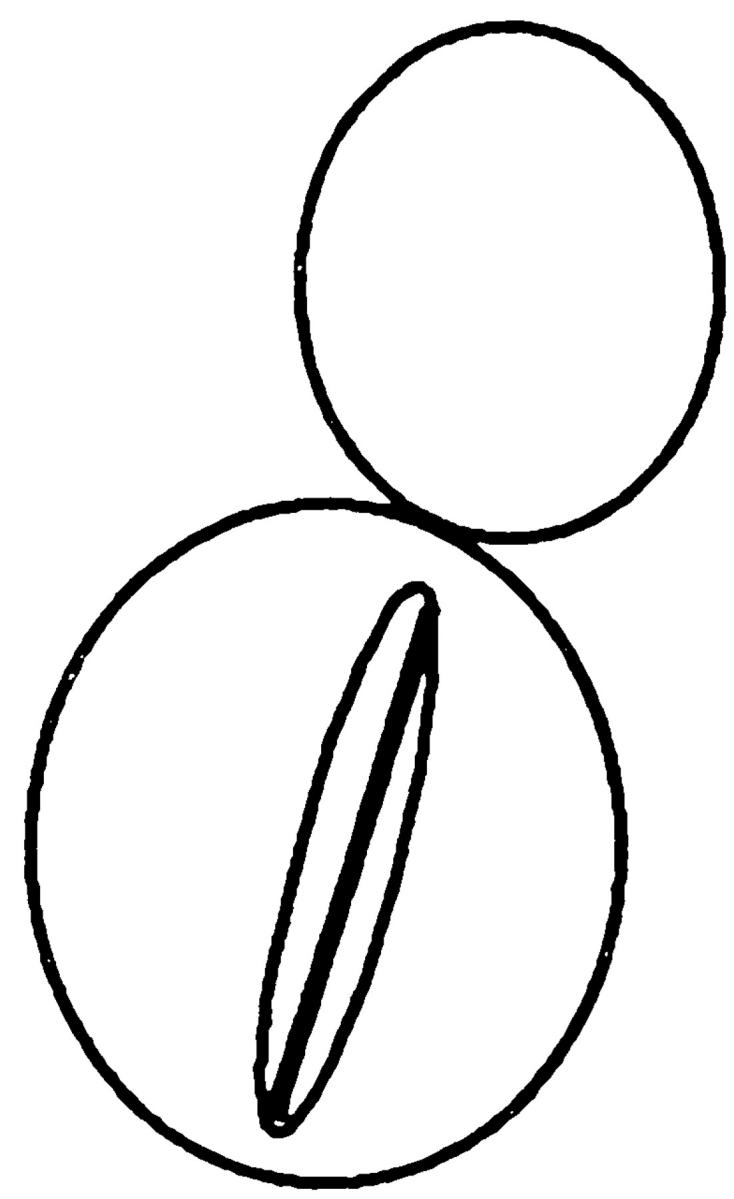
|
|||||
|---|---|---|---|---|---|---|---|---|
| Nucleus | cMTs extending into bud | cMTs to neck | cMTs not extending into bud | No cMTs | ||||
| Wild type | 90 | 0 | 3 | 7 | ||||
| kar9Δ | 31 | 17 | 48 | 4 | ||||
| bni1Δ | 79 | 8 | 7 | 6 | ||||
| kar9Δ bni1Δ | 41 | 10 | 49 | 1 |
Kar9p is functioning in a bni1Δ mutant. In cells stained with DAPI and antitubulin, the percentage of cells with microtubule orientation defects was scored in cells with anaphase within the mother cell. The following strains derived from MS6127 were used: wild-type strain (MS6148), kar9Δ (MS6165), bni1Δ (MS6164), and kar9Δ bni1Δ strains (MS6151, MS6144, MS6162, MS6163). Cells were grown to early exponential phase at 30°C, and then fixed for indirect immunofluorescence using the tubulin antibody, YOL1/34. n = 100 cells each for wild-type, bni1Δ, and kar9Δ strains. Values for kar9Δ bni1Δ are the average obtained from analysis of four independent strains (n = 402).
This result suggests that either the bni1Δ mutation also suppresses the microtubule orientation defect created by mislocalized Kar9p or that enough Kar9p was correctly localized in the bni1Δ mutant to facilitate microtubule orientation. To differentiate between these two models, we created kar9Δ bni1Δ double mutants and examined them for defects in nuclear positioning and cytoplasmic microtubule orientation. In 49% of the kar9Δ bni1Δ double mutant cells, cytoplasmic microtubules were misoriented (Table V) like the kar9Δ mutant. Therefore, Kar9p was still responsible for the normal microtubule orientation found in the bni1Δ single mutant cells, in spite of the defect in Kar9p localization. These results suggest that Kar9p localization must be partially independent of Bni1p. This result would also be consistent with the weaker defect in nuclear migration seen for bni1Δ compared with kar9Δ.
We next examined the double mutants for defects in nuclear positioning. Compared with the kar9Δ single mutant, the kar9Δ bni1Δ double mutant large-budded cells were slightly more severe for defects in nuclear positioning (47 vs. 33% aberrant, Table VI). In unbudded cells, however, a significantly more severe defect was observed. In unbudded cells, very low percentages of abnormal cells were found in wild-type, kar9Δ, and bni1Δ single mutants (0, 4, and 0.5%, respectively). However, in the kar9Δ bni1Δ double mutant, 22% of the cells were either multinucleate or anucleate. Therefore, the double mutant cells often appeared to progress through cytokinesis, which almost never occurred with the single mutants.
Table VI.
Nuclear Migration Defects of Mutants
The bni1Δ kar9Δ double mutant exhibits a slightly worse phenotype than the single mutants alone. The wild-type strain (MS6148), bni1Δ (MS6164), kar9Δ (MS6165), and five bni1Δ kar9Δ strains (MS6166, MS6161, MS6154, MS6149, and MS6151) were grown to early exponential phase, zymolyased, stained with DAPI, and prepared as described in Table V. No obvious nuclear positioning defect was observed in small- and medium-budded cells of wild-type, bni1Δ, or kar9Δ strains. In contrast, 4% of medium-budded bni1Δ kar9Δ cells contained two nuclei (n = 195, data not shown). Σ, the sum of abnormal phenotypes.
Genetic Analysis Places Bni1p in the Kar9p Pathway for Nuclear Migration
Because of the synthetic lethality between kar9Δ and dhc1Δ, Kar9p and dynein are thought to function in separate, partially redundant pathways for nuclear migration (Miller and Rose, 1998). Because spa2Δ, pea2Δ, bud6Δ, and bni1Δ mutants each altered GFP-Kar9p localization, we wanted to know whether any of these mutations would show genetic interactions similar to those observed with kar9Δ. Isogenic marked deletions of each gene were created and crossed to marked kar9 and dhc1 deletions. The viability of the double mutants was assayed for growth 2–3 d after spore germination. The results of the crosses are summarized in Table VII. When spa2Δ, pea2Δ, bud6Δ, or bni1Δ were crossed with kar9Δ, all four double-mutant combinations were viable (Table VII, 1–4). Similarly, the spa2Δ dhc1Δ, and pea2Δ dhc1Δ double mutants were viable (Table VII, 6–8). The bud6Δ dhc1Δ double mutants were viable, but exibited a mild growth defect (Table VII, 8). However, when bni1Δ was crossed to dhc1Δ, all the predicted double mutants were barely viable and formed microcolonies (Table VII, 9). Similar microcolonies were also observed in synthetic lethal genetic interactions seen with kar9Δ, including dhc1Δ, bik1Δ, and kip2Δ (Miller et al., 1998; Miller and Rose, 1998). Therefore, we consider this to be a synthetic lethal interaction and conclude that Bni1p acts in the Kar9p pathway for nuclear migration.
Table VII.
Viability of Mutants in Combination with kar9Δ or dhc1Δ
| Mutant combination | Tetrads analyzed | Number of predicted mutants | Mutants dead or forming microcolonies | Viability of double or triple mutant | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | spa2Δ kar9Δ | 8 | 7 | 0% | Viable | |||||
| 2. | pea2Δ kar9Δ | 13 | 16 | 0% | Viable | |||||
| 3. | bud6Δ kar9Δ | 27 | 28 | 0% | Viable | |||||
| 4. | bni1Δ kar9Δ | 11 | 8 | 0% | Viable | |||||
| 5. | bnr1Δ kar9Δ | 23 | 28 | 11% | Viable | |||||
| 6. | spa2Δ dhc1Δ | 23 | 18 | 11% | Viable | |||||
| 7. | pea2Δ dhc1Δ | 18 | 18 | 6% | Viable | |||||
| 8. | bud6Δ dhc1Δ | 24 | 26 | 8% | Viable* | |||||
| 9. | bni1Δ dhc1Δ | 19 | 20 | 100% | SL | |||||
| 10. | bnr1Δ dhc1Δ | 23 | 20 | 5% | Viable | |||||
| 11. | bni1Δ kip3Δ | 38 | 14 | 0% | Viable | |||||
| 12. | bni1Δ kar9Δ kip3Δ | 38 | 19 | 5% | Viable | |||||
Standard meiotic crosses were used to construct the indicated double mutants. The size of the colonies was scored 2–3 d after germination at 30°C. SL indicates that synthetic lethality was observed.
The bud6Δ dhc1Δ double mutant exhibited a decreased growth rate. The following crosses were carried out to create the indicated double mutants: 1. spa2Δ kar9Δ, MS5208 × MS4062; 2. pea2Δ kar9Δ, MS5229 × MS4062; 3. bud6Δ kar9Δ, MS5849 × MS4316; 4. bni1Δ kar9Δ, MS5215 × MS4062; 5. bnr1Δ kar9Δ, MS5794 × MS4316; 6. spa2Δ dhc1Δ, MS5208 × MS5429; 7. pea2Δ dhc1Δ, MS5229 × MS5429; 8. bud6Δ dhc1Δ, MS5849 × MS5429; 9. bni1Δ dhc1Δ, MS5215 × MS5429; 10. bnr1Δ dhc1Δ, MS5794 × MS5429; 11. bni1Δ kip3Δ, MS5665 × MS5215; 12. bni1Δ kar9Δ kip3Δ, MS5665 × MS5215.
As a control, crosses were performed using a deletion of the related formin homology domain protein, Bnr1p. The bnr1Δ mutation was not synthetically lethal in combination with kar9Δ or dhc1Δ (Table VII, 5 and 10). Therefore, the interaction seen between dhc1Δ and bni1Δ is specific to Bni1p and not a general feature of formin homology proteins.
In previous work, Kip3p was shown to function in the KAR9 pathway for nuclear migration (Miller et al., 1998). Because Bni1p also appears to function in the KAR9 pathway, we crossed the bni1Δ to the kip3Δ to determine whether Bni1p and Kip3p might also function in a common pathway. We found that the bni1Δ kip3Δ double mutant is viable, as was also reported by Lee et al. (1999). This suggests that Kar9p, Kip3p, and Bni1p function in a common pathway. Accordingly, the kar9Δ bni1Δ kip3Δ triple mutant would also be predicted to be viable. We found that the kar9Δ bni1Δ kip3Δ triple mutant was indeed viable (Table VII, 12), supporting the hypothesis that Kar9p, Kip3p, and Bni1p function together in a common pathway.
kar9Δ Does Not Exhibit Bipolar Budding Defects
The four mutations that affect Kar9p localization, spa2Δ, pea2Δ, bud6Δ, and bni1Δ, share an additional common phenotype of being defective in the diploid-specific bipolar budding pattern (Snyder, 1989; Valtz and Herskowitz, 1996; Zahner et al., 1996; Amberg et al., 1997). This provided an additional way of addressing whether Kar9p is simply a Bni1p-associated protein or functions specifically for microtubule orientation. Therefore, we examined whether kar9Δ mutants are also defective for bipolar budding. Diploid cells with three or more bud scars were scored to determine whether they showed the bipolar or random budding pattern (see Materials and Methods). In the wild-type control strain, 95% of the cells exhibited a bipolar budding pattern and 5% showed a random pattern (n = 263). In the pea2Δ strain, only 28% of the cells exhibited the bipolar budding pattern, while 72% budded randomly (n = 201). Unlike the pea2Δ mutant, the kar9Δ mutant exhibited the wild-type budding pattern (90% bipolar, n = 300). The kar9Δ mutants also did not display a defect in the haploid-specific axial budding pattern (data not shown). Taken together with the observation that kar9Δ mutant shmoos do not exhibit polarization defects, and that Bni1p localizes normally in the kar9Δ mutants, these results argue strongly that Kar9p's function with Bni1p is specific to the nuclear migration pathway.
Discussion
In yeast, both actin and the cytoplasmic microtubules have been implicated in normal spindle orientation and movement, suggesting a possible interaction between these two cytoskeletal networks (Palmer et al., 1992; Read et al., 1992; Sullivan and Huffaker, 1992). However, the mechanism by which the actin cytoskeleton might contribute to spindle orientation has not been clear. Studies on centrosomal rotation in C. elegans (Hyman, 1989) and nuclear movement in yeast (Carminati and Stearns, 1997) have also shown the involvement of cortical factors in microtubule positioning and orientation. However, to date, only Kar9p has been identified as a likely candidate for one of the cortical proteins required for cytoplasmic microtubule orientation. In this study, we demonstrated that Kar9p localization is dependent upon the actin cytoskeleton. We have also shown that Kar9p localization is strongly influenced by Bni1p, a formin involved in actin cytoskeletal functions, polarization, budding, and cell fusion processes. Like Kar9p, Bni1p was found to participate in nuclear migration processes during both shmooing and mitosis. Genetic analysis suggests that Bni1p functions in the Kar9p pathway rather than the dynein pathway for nuclear migration. Taken together, these data suggest that Kar9p and Bni1p functionally link the actin and microtubule networks to promote proper spindle orientation and nuclear migration.
Three other proteins were found to influence Kar9p localization in mating cells, Bud6p, Spa2p, and Pea2p. Spa2p and Pea2p are required for polarization in mating cells and in their absence the shmoo tip is broader. In these two mutants Kar9p localization was spread over a large area of the shmoo tip. Thus, it seems likely that Pea2p and Spa2p and other proteins help to cluster Kar9p into a single focus at the shmoo tip.
Taken together, these observations suggest a model for nuclear orientation in mitotic and mating cells. As the nascent bud emerges, actin patches become associated with the bud cortex. Similarly, actin patches associate with the cortex at the tip of the mating projection. One or a subset of actin patches becomes integrated into a specialized site that we will refer to as the “polarization complex,” normally at or near the tip of the bud and the mating projection. Additional actin-associated proteins playing roles in polarized growth would be recruited to the specialized polarization complex. Among these would be Bni1p, Spa2p, and Pea2p. Each of these proteins would interact with a subset of other components in the complex and each may have cortical associations that are independent of actin. Some of the proteins, perhaps including Bni1p, may play key roles in cross-linking multiple proteins and thereby play a major “scaffolding” role. Loss of such proteins would affect the localization of several proteins to the complex. The specific proteins required might also differ between mating and mitosis. Finally, other proteins, such as Kar9p, would be recruited to more peripheral locations on the polarization complex. Such peripheral proteins may serve as “docking sites” or “adapters” for extrinsic elements such as the microtubules that need to identify and interact with the cortical polarization complex. Thus, the assembly and maintenance of the actin-associated polarization complex provides a unique molecular “address” within the cell that can be “read” by different cellular systems through specific adapter molecules.
BNI1 Is Required for Nuclear Migration
We have identified Bni1p as being required for nuclear migration in mating and mitosis because of its role in Kar9p localization. In pheromone-treated cells, the bni1Δ mutant exhibits a strong defect in microtubule orientation, consistent with the defect in Kar9p localization.
The role of Bni1p in mitotic cells is more complex. Although Kar9p was mislocalized away from the bud tip in mitotic bni1Δ mutant cells, the overall effect was less dramatic. In the mitotic cells, Kar9p localization was shifted to secondary sites at the SPB and/or bud site and the cytoplasmic microtubules were not grossly misoriented by the onset of anaphase. Several explanations for this are possible. First, some 14% of the bni1Δ cells showed detectable Kar9p at the tip of the bud. It is possible that different levels of Kar9p at the bud tip are required for microtubule orientation and subsequent nuclear migration. Perhaps there is enough Kar9p at the bud tip to allow correct microtubule alignment, but not enough Kar9p to activate some other component necessary for nuclear migration, such as Kip3p. Second, although Kar9p was not localized to the bud tip in the mutant, it was localized near the bud neck. Microtubules contacting Kar9p at the bud neck would be properly oriented to eventually contact the bud cortex after continued assembly. This would be consistent with the analysis of the double mutant that indicated that in the bni1Δ mutant the orientation of the cytoplasmic microtubules was still dependent on Kar9p.
The residual localization of GFP-Kar9p in the bni1Δ, the dependence of microtubule orientation on Kar9p in the bni1Δ, and the fact that the kar9Δ mutant exhibits a more severe nuclear migration defect than the bni1Δ mutant suggests that Kar9p is at least partially active in the absence of Bni1p. Taken together, the simplest explanation for these observations is that Kar9p has substantial residual localization to the cortex, independent of Bni1p. In the absence of Bni1p, the level of GFP-Kar9p at the cortex may simply fall below the limit of detection in most cells. Nevertheless, the reduction in Kar9p localization would be sufficient to compromise the Kar9p pathway for nuclear migration. One prediction of these results is that Kar9p localization would be partially dependent on additional cortical proteins.
Independently, Lee et al. (1999) have also identified Bni1p as being required for nuclear migration. In their work, dynamic studies of nuclear migration and microtubule dynamics clearly show a defect in cytoplasmic microtubule function. In addition, they also find that Kip3 and Bni1p are likely to be part of the same nuclear migration pathway.
In contrast to bni1Δ, there was no obvious defect in nuclear migration in the spa2Δ, pea2Δ, and bud6Δ cells. This is not surprising given their milder defects in Kar9p localization. Our results showing that bud6Δ does not exhibit a significant nuclear migration defect are in contrast to results previously reported (Amberg et al., 1997). This may be due to differences in strain background or partially toxic effects of the disruption mutant used in that study. In support of this, we find that the bud6Δ::URA3 strain (DBY7055) did exhibit a synthetic phenotype and temperature sensitivity in combination with dhc1Δ.
Bni1p Has Functions Distinct from Kar9p
Although kar9 and bni1 mutants exhibit several similarities and genetically lie in the same pathway for nuclear migration, several lines of evidence suggest that they also have distinct functions. First, unlike bni1Δ mutants (Evangelista et al., 1997), kar9Δ mutants do not exhibit defects in actin localization (Miller and Rose, 1998). Second, kar9Δ mutants do not exhibit defects in the bipolar budding pattern. Third, unlike bni1Δ mutants (Evangelista et al., 1997), kar9Δ did not form round-shaped shmoos or show other obvious defects in polarization. Fourth, unlike bni1Δ mutants, kar9Δ mutants do not exhibit a severe mating defect (Kurihara et al., 1994). Fifth, although Bni1p is required for efficient Kar9p localization, Kar9p is not required for GFP-Bni1p localization in mitotically dividing cells or shmoos. Thus, Bni1p mediates a series of functions in mating and mitosis that are not part of Kar9p's repertoire of functions.
If Bni1p's only function in nuclear migration were to localize Kar9p, then the bni1Δ kar9ΔΔ double mutant should not be more severe than the kar9Δ single mutant. However, the double mutant has a more severe phenotype than either single mutant alone, as evidenced both by a slight increase in the number of aberrant large-budded cells and by an increase in the percentage of multinucleate and anucleate unbudded cells (Table VI). Thus, Bni1p may have a secondary role in nuclear migration in addition to the localization of Kar9p. However, if Bni1p does have a secondary role independent of Kar9p, its contribution to nuclear migration must be relatively minor. Otherwise, the additive effects of the two independent defects should lead to a severe growth defect for the double mutant. Instead, we find that the double mutant grows no worse than the single mutants, identical to the wild type.
That anucleate cells are observed in the bni1Δ kar9Δ double mutant combination raises the additional possibility that Kar9p and/or Bni1p may play an inhibitory role in cytokinesis. This might be true if one or the other is part of the checkpoint mechanism that coordinates nuclear migration and cytokinesis. In addition to bud tip localization, both Bni1p and Kar9p are also localized at the mother-bud neck in a minor fraction of large budded cells (Evangelista and Boone, personal communication; Fujiwara et al., 1998; Miller and Rose, 1998). More likely, the increased severity of the double mutant may cause some cells to eventually complete cytokinesis without proper nuclear migration.
Kar9p Localization by Bni1p Is Highly Specific
Consistent with the observation that they all contribute to polarization, Spa2p, Pea2p, and Bud6p have been isolated as a stable complex (Sheu et al., 1998). Furthermore, Spa2p interacts with Bni1p by two-hybrid analysis (Fujiwara et al., 1998). Yet mutations in these proteins affected Kar9p localization differently, with only mild mitotic defects associated with spa2Δ and pea2Δ, and none apparent for bud6Δ. In shmoos and mitotic cells, the most severe effect on Kar9p localization occurred in the bni1Δ mutants. Nevertheless, all four of these mutants affect polarization in a similar manner and to a similar degree. Thus, the effects of bni1Δ on Kar9p localization are not likely to be due to general defects in polarization.
All four mutants, spa2Δ, pea2Δ, bud6Δ, and bni1Δ, also affect cell fusion and bipolar budding. However, another bipolar budding mutant, rvs161Δ, exhibited no defects in Kar9p localization. In addition, Kar9p localization was not altered in the cell fusion mutants fus1Δ and fus2Δ. Thus, alterations in Kar9p localization are not due to general disruptions in either bipolar budding or the cell fusion process.
Several other observations speak to the specificity of the effects of these mutations on Kar9p localization. Bnr1p is another formin family protein that shares overlapping functions with Bni1p (Imamura et al., 1997). Kar9p localized normally in bnr1Δ mutants (data not shown) and also did not show any genetic interactions with kar9Δ or dhc1Δ, indicating that the effects on Kar9p localization are not general to all formin proteins, but specific to Bni1p.
Other mutants such as sla1Δ that affect the assembly of the actin cytoskeleton (Holtzman et al., 1993) also did not alter Kar9p localization in shmoos. This argues that the specialized interaction of Spa2p, Pea2p, Bud6p, and Bni1p with the actin cytoskeleton is the important feature for Kar9p localization.
Differences between Mitotic and Mating Cells
The localization patterns for Spa2p and Pea2p are interdependent, suggesting that their functions are closely interrelated (Valtz and Herskowitz, 1996). Consistent with this, we find that GFP-Kar9p mislocalization was very similar in the spa2Δ and pea2Δ mutants. Thus, with respect to Kar9p localization, these two mutants can be referred to as the “spa2 class.”
Interestingly, the spa2 class showed a dramatic difference in the severity of Kar9p mislocalization comparing shmoos and mitotically dividing cells. In mitotic cells, mislocalization was apparent in only ∼10% of cells. In contrast, in most shmoos, Kar9p localization was altered from a single dot to a cap-like pattern. Thus, Spa2p and Pea2p likely play very different roles in polarization and microtubule orientation in mating cells.
The finding that Kar9p localization is often spread out over a broader area in spa2Δ or pea2Δ shmoos is consistent with the proposed role for Spa2p and Pea2p in restricting the zone of polarization in shmoos (Valtz and Herskowitz, 1996). One prediction from such a model would be that a narrowed site of polarization would restrict the area of microtubule interaction with the cortex by restricting Kar9p localization to a dot. Consistent with this, we found a “splayed-out” microtubule pattern in a subset of spa2Δ mutants.
Two models might explain the altered patterns of Kar9p localization. In one model, Spa2p and Pea2p may play a direct role in limiting Kar9p localization to a tight focus by helping aggregate the Bni1p/Kar9p complex. Alternatively, Spa2p and Pea2p might play an indirect role in Kar9p localization via their effects on shmoo morphology. In either case, correct polarization might be required to define a specialized region of the mating projection that is primed for cell and nuclear fusion. Such a primed region would facilitate the alignment of cytoplasmic microtubules, allowing nuclear congression to follow rapidly after cell fusion. In spa2Δ and pea2Δ mutant shmoos, the primed region would be more diffuse over the blunter mutant shmoo projection. In support of the indirect model, in spa2Δ prezygotes the clustering of vesicles along the zone of cell fusion is disrupted (Gammie et al., 1998). Regardless of the specific mechanism, these results highlight the differences in cell polarization between mating and mitotic cells.
In contrast, both the bni1Δ mutation and actin depolymerization resulted in strong defects in Kar9p localization in both shmoos and mitotic cells. Although the bni1Δ mutant was defective for nuclear migration in both shmoos and mitotic cells, there were differences in the effects on microtubule orientation. In bni1Δ shmoos, the microtubules were clearly misoriented, whereas mitotic cells did not exhibit an obvious defect. One interpretation might be that correct Kar9p localization is more important for microtubule orientation in mating cells than in mitotic cells. Alternatively, the difference between the two cells may arise from the different patterns of mislocalized Kar9p. In the bni1Δ shmoos, GFP-Kar9p was present in a diffuse speckled pattern. In the bni1Δ mutant, GFP-Kar9p mislocalized to secondary sites at the SPB and bud neck, where it might still be able to provide some orientation to the microtubules.
Coordinating Cytoskeletal Systems
Abundant evidence has shown that Bni1p, specifically, and formins, generally, are involved in organizing the actin cytoskeleton. In addition to Spa2p and Bud6p, Bni1p interacts with profilin and the small G-protein Rho1p (Kohno et al., 1996; Evangelista et al., 1997). It seems likely that formins may play a general role in linking the actin and microtubule cytoskeletons. For example, mutations in the Drosophila formin, cappuccino result in mislocalized molecular determinants and abnormal microtubule patterns (Theurkauf, 1994; Emmons et al., 1995).
In conclusion, we report here that Kar9p localization is dependent upon actin and the polarization protein, Bni1p. This provides the first evidence in yeast for a functional link between the actin and microtubule cytoskeletons. These data suggest a mechanism by which the polarized information contained within Kar9p is transferred into the vectorial movement of spindle orientation and nuclear movement into the bud. This work provides an avenue into future investigations as to how nuclear positioning is coordinated with the growth of the bud and cytokinesis.
Acknowledgments
We thank David Amberg, Alan Bender, Charlie Boone, Ira Herskowitz, and Mike Snyder for providing strains and disruption plasmids. We also thank Joe Nickels for helpful technical discussions on methods for rhodamine-phalloidin staining. We also thank Doug Koshland for helpful and interesting conversations. We also thank L. Lee and D. Pellman for communicating results before publication.
This work was supported by National Institutes of Health grant GM-37739 (M.D. Rose).
Abbreviations used in this paper
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- LAT-A
latrunculin-A
- SC
synthetic complete
- SPB
spindle pole body
Footnotes
Address correspondence to Mark D. Rose, Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ 08544. Tel.: (609) 258-2804. Fax: (609) 258-6175. E-mail: mrose@molbio.princeton.edu
References
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Pringle JR. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. . Proc Natl Acad Sci USA. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender L, Lo HS, Lee H, Kokojan V, Peterson V, Bender A. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in Yeast. J Cell Biol. 1996;133:879–894. doi: 10.1083/jcb.133.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Drees B, Harsay E, Schott D, Wang T. What are the basic functions of microfilaments? Insights from studies in budding yeast. J Cell Biol. 1994;126:821–825. doi: 10.1083/jcb.126.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio V, Gammie AE, Rose MD. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. . J Cell Biol. 1998;141:567–584. doi: 10.1083/jcb.141.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, W.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 59–96.
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation in Saccharomyces cerevisiae. . J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformitygene. Development (Camb) 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. . J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SW, Meyer DI. ACT3: a putative centractin homologue in S. cerevisiaeis required for proper orientation of the mitotic spindle. J Cell Biol. 1994;127:129–138. doi: 10.1083/jcb.127.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiaeis accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiaeis required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Miller KG, Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Mulholland J, Zhu ZM, Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I. Nature. 1990;343:288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- Emmons S, Phan H, Calley J, Chen W, James B, Manseau L. Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformitylocus. Genes Dev. 1995;9:2482–2494. doi: 10.1101/gad.9.20.2482. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Field C, Schekman R. Localized secretion of acid phosphatase reflects the pattern of cell surface growth in Saccharomyces cerevisiae. . J Cell Biol. 1980;86:123–128. doi: 10.1083/jcb.86.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder D. Bud position in Saccharomyces cerevisiae. . J Bacteriol. 1960;124:511–523. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p–Bni1p–Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. . Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie AE, Brizzio V, Rose MD. Distinct morphological phenotypes of cell fusion mutants. Mol Biol Cell. 1998;9:1395–1410. doi: 10.1091/mbc.9.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SD, Hamer L, Sharpless KE, Hamer JE. The Aspergillus nidulans sepAgene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO (Eur Mol Biol Organ) J. 1997;16:3474–3483. doi: 10.1093/emboj/16.12.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasek J, Rupes I, Svobodova J, Streiblova E. Tubulin and actin topology during zygote formation of Saccharomyces cerevisiae. . J Gen Microbiol. 1987;133:3355–3363. doi: 10.1099/00221287-133-12-3355. [DOI] [PubMed] [Google Scholar]
- Holtzman DA, Yang S, Drubin DG. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. . J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. . J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell–specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Kahana JA, Schlenstedt G, Evanchuk DM, Geiser JR, Hoyt MA, Silver PA. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol Biol Cell. 1998;9:1741–1756. doi: 10.1091/mbc.9.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana JA, Schnapp BJ, Silver PA. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, et al. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Klee SK, Evangelista M, Boone C, Pellman D. Control of mitotic spindle position by the Saccharomyces cerevisiaeformin Bni1p. J Cell Biol. 1999;144:947–961. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz DA, Hamaguchi Y, Inoue S. Micromanipulation studies of the asymmetric positioning of the maturation spindle in Chaetopterus sp.oocytes: I. Anchorage of the spindle to the cortex and migration of a displaced spindle. Cell Motil Cytoskelet. 1988;11:83–96. doi: 10.1002/cm.970110202. [DOI] [PubMed] [Google Scholar]
- Marhoul JF, Adams TH. Identification of developmental regulatory genes in Aspergillus nidulansby overexpression. Genetics. 1995;139:537–547. doi: 10.1093/genetics/139.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiaeis required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Miller RK, Heller KK, Frisen L, Wallack DL, Loayza D, Gammie AE, Rose MD. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol Biol Cell. 1998;9:2051–2068. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Nielsen O, Egel R, Hagan IM. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998;141:1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- Read EB, Okamura HH, Drubin DG. Actin- and tubulin-dependent functions during Saccharomyces cerevisiaemating projection formation. Mol Biol Cell. 1992;3:429–444. doi: 10.1091/mbc.3.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD. Nuclear fusion in the yeast Saccharomyces cerevisiae. . Annu Rev Cell Dev Biol. 1996;12:663–695. doi: 10.1146/annurev.cellbio.12.1.663. [DOI] [PubMed] [Google Scholar]
- Rose MD, Fink GR. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Segall JE. Polarization of yeast cells in spatial gradients of alpha mating factor. Proc Natl Acad Sci USA. 1993;90:8332–8336. doi: 10.1073/pnas.90.18.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieler P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson BJ, Ferguson B, De Virgilio C, Bi E, Pringle JR, Ammerer G, Sprague GF., Jr Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. . Genes Dev. 1995;9:2949–2963. doi: 10.1101/gad.9.23.2949. [DOI] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE. Premature microtubule-dependent cytoplasmic streaming in cappuccino and spiremutant oocytes. Science. 1994;265:2093–2096. doi: 10.1126/science.8091233. [DOI] [PubMed] [Google Scholar]
- Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J, Fink GR. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc Natl Acad Sci USA. 1989;86:9916–9920. doi: 10.1073/pnas.86.24.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtz N, Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J Cell Biol. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Holtzman DA, Drubin DG. The yeast actin cytoskeleton. Curr Opin Cell Biol. 1994;6:110–119. doi: 10.1016/0955-0674(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Woychik RP, Maas RL, Zeller R, Vogt TF, Leder P. ‘Formins': proteins deduced from the alternative transcripts of the limb deformitygene. Nature. 1990;346:850–853. doi: 10.1038/346850a0. [DOI] [PubMed] [Google Scholar]
- Yang S, Ayscough KR, Drubin DG. A role for the actin cytoskeleton of Saccharomyces cerevisiaein bipolar bud-site selection. J Cell Biol. 1997;136:111–123. doi: 10.1083/jcb.136.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. . Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]