Abstract
Abstract. Dynamins are 100-kilodalton guanosine triphosphatases that participate in the formation of nascent vesicles during endocytosis. Here, we have tested if novel dynamin-like proteins are expressed in mammalian cells to support vesicle trafficking processes at cytoplasmic sites distinct from the plasma membrane. Immunological and molecular biological methods were used to isolate a cDNA clone encoding an 80-kilodalton novel dynamin-like protein, DLP1, that shares up to 42% homology with other dynamin-related proteins. DLP1 is expressed in all tissues examined and contains two alternatively spliced regions that are differentially expressed in a tissue-specific manner. DLP1 is enriched in subcellular membrane fractions of cytoplasmic vesicles and endoplasmic reticulum. Morphological studies of DLP1 in cultured cells using either a specific antibody or an expressed green fluorescent protein (GFP)- DLP1 fusion protein revealed that DLP1 associates with punctate cytoplasmic vesicles that do not colocalize with conventional dynamin, clathrin, or endocytic ligands. Remarkably, DLP1-positive structures coalign with microtubules and, most strikingly, with endoplasmic reticulum tubules as verified by double labeling with antibodies to calnexin and Rab1 as well as by immunoelectron microscopy. These observations provide the first evidence that a novel dynamin-like protein is expressed in mammalian cells where it associates with a secretory, rather than endocytic membrane compartment.
Dynamin is a 100-kD large GTPase that participates in the early stages of endocytosis, specifically in the liberation of invaginated nascent vesicles from the plasma membrane (Herskovits et al., 1993a ; Damke et al., 1994, 1995; Hinshaw and Schmid, 1995; Takei et al., 1995; Urrutia et al., 1997). Three distinct dynamin genes have been identified in mammals thus far. A neuron-specific form of dynamin, dynamin I (Dyn1), was originally isolated from mammalian brain (Shpetner and Vallee, 1989; Obar et al., 1990), and additional isoforms of dynamin were subsequently identified from other tissues. Dynamin II is expressed in all tissues (Cook et al., 1994; Sontag et al., 1994), whereas dynamin III is expressed in brain, testis, and lung (Nakata et al., 1993; Cook et al., 1996). The three rat dynamin isoforms share ∼78% amino acid homology overall with the highest homology (88%) found in the NH2-terminal 300 amino acids (aa),1 which include a tripartite GTP-binding region. Homology among dynamin isoforms is reduced significantly in the COOH-terminal proline-rich region that has been found to bind to other macromolecules in vitro such as microtubules, phospholipids, and SH3 domain–containing proteins (Scaife and Margolis, 1990; Maeda et al., 1992; Shpetner and Vallee, 1992; Gout et al., 1993; Herskovits et al., 1993b ; Tuma et al., 1993; Miki et al., 1994; Scaife et al., 1994; Seedorf et al., 1994). Each of the three dynamin genes is known to be expressed in at least four alternatively spliced forms (Robinson et al., 1994; Urrutia et al., 1997). Thus, because neurons express all three of the identified dynamin genes, they are predicted to possess at least 12 different forms of dynamins.
In addition to these mammalian dynamins, two dynamin-related proteins, Vps1p and Dnm1p, have been identified and characterized in yeast. These proteins share >55% homology with the NH2-terminal domains of rat dynamins and have been implicated in vesicle sorting and trafficking at cellular sites other than the plasma membrane. Vps1p is an 80-kD protein that is associated with the Golgi and is required for proper sorting of proteins to the yeast vacuole (Rothman et al., 1990; Vater et al., 1992; Wilsbach and Payne, 1993; Nothwehr et al., 1995). In contrast, the 85-kD Dnm1p is believed to function in the late endocytic pathway because cells possessing a disrupted gene exhibit normal endocytic uptake of ligands, whereas subsequent transfer of ligands from early to late endosomes is inhibited (Gammie et al., 1995). Recently, a 68-kD plant dynamin-like protein, phragmoplastin, has been identified in soybean (Gu and Verma, 1996) and characterized to be involved in vesicle-mediated cell division plate formation in plant cells (Gu and Verma, 1997).
According to studies described above, it becomes clear that different dynamins and dynamin-related proteins play their roles in different membranous organelles. Because vesicle budding events occur at most intracellular membranous organelles, it is likely that a function equivalent to that of dynamin at the plasma membrane is required at other membranous organelles. These functions are likely to be mediated by other dynamins or dynamin-related proteins described above. A recent study has provided insight into distinct sites of dynamin function in mammalian cells. Using biochemical, morphological, and vesicle immunoisolation techniques, it has been shown that a dynamin is associated with the Golgi apparatus (Henley and McNiven, 1996; Maier et al., 1996). Whether this represents an isoform of dynamin II or a yet unidentified dynamin that may act to liberate nascent secretory vesicles from the Golgi cisternae is unknown. Furthermore, mammalian homologues of Vps1p or Dnm1p have not been identified yet. From the observations described above, it is likely that additional unidentified dynamins or dynamin-related proteins are expressed in mammalian cells to perform a variety of vesicle trafficking functions. This prediction is consistent with the observations made for numerous other cytoskeletal and membrane-associated proteins such as the kinesins (Hirokawa, 1996; Moore and Endow, 1996), cytoplasmic myosins (Mooseker and Cheney, 1995), ADP ribosylation factors (ARFs; Goud, 1992; Clark et al., 1993), annexins (Raynal and Pollard, 1994; Smith and Moss, 1994), and adaptors (Robinson, 1997).
In an effort to identify additional dynamins or dynamin-like proteins in mammalian tissues, we have isolated an 80-kD protein immunologically related to dynamin (Yoon, Y., and M. A. McNiven. 1996. Mol. Biol. Cell. 7:82a). This protein, termed DLP1 (dynamin-like protein 1), shares homology with dynamins and other dynamin-related proteins while associating with endoplasmic reticulum and a population of cytoplasmic vesicles. The identification of a novel mammalian dynamin-like protein reported here provides the first evidence that the mammalian dynamin family of proteins is diverse and likely to support vesicle trafficking at multiple cytoplasmic locations.
Materials and Methods
Cell Culture and Tissues
Mouse hepatocytes (normal mouse liver cell line BNL CL.2; American Type Culture Collection [ATCC], Rockville, MD) and primary human foreskin fibroblasts were grown in MEM α medium with l-glutamine, ribonucleosides, and deoxyribonucleosides (GIBCO BRL, Gaithersburg, MD) containing 10% fetal bovine serum. The cultured normal rat liver cell line clone 9 (ATCC CRL-1439) was maintained in Ham's F-12K medium containing 10% fetal bovine serum. The cultured human cholangiocyte cell line (H-69; Grubman et al., 1994) was provided by Dr. Nicholas F. LaRusso (Mayo Clinic, Rochester, MN). Tissues used for this study were harvested from adult male Sprague-Dawley rats (Harlan, Madison, WI).
Antibodies
Antidynamin antibodies MC12 and MC63 were prepared as described previously (Henley and McNiven, 1996). For anti-DLP1 antibodies, three DLP1-specific peptides were selected: aa 266–294 for DLP-N (NNKKSVTDSIRDEYAFLQKKYPSLANRNG), aa 523–545 for DLP-MID (NNIEEQRRNRLARELPSAVSRDK), and aa 716–738 for DLP-C (DDLLTESEDMAQRRKEAADMLKA). These peptides were synthesized and conjugated to keyhole limpet hemocyanin, injected into New Zealand white rabbits, and then the antisera was collected. The crude antisera were affinity purified using corresponding HPLC-purified peptides immobilized on agarose columns. Polyclonal antibody to Rab1 was raised against a specific synthetic peptide, ATAGGAEKSNVKIQSTPVKQSGGGCC, in our laboratory and affinity purified as above. Polyclonal antibodies to Rab5 and TGN38 were prepared in our laboratory using the same methods. Mouse monoclonal anti–α-adaptin antibody (clone 100/2) was purchased from Sigma Chemical Co. (St. Louis, MO), anti–α-tubulin antibody from Amersham Corp. (Arlington Heights, IL), and anticalnexin antibody from Affinity BioReagents, Inc. (Golden, CO). Mouse monoclonal anti-clathrin heavy chain antibody (X22) was prepared from culture supernatant of a hybridoma cell line (ATCC CRL-2228). Mouse monoclonal antidynamin antibody (hudy-1) was provided by Dr. S.L. Schmid (Scripps Research Institute, La Jolla, CA). The following rabbit polyclonal antibodies were provided as gifts: anti–γ-adaptin from Dr. L. Traub (Washington University, St. Louis, MO), anti–β-galactosidase from Dr. N. LaRusso (Mayo Clinic, Rochester, MN), anti-Sec23p from Dr. J.-P. Paccaud (University of Geneva, Geneva, Switzerland), anti-Rab8 from Dr. D.D. Sabatini (New York University, NY), anti-p47 (μ3) from Dr. M.S. Robinson (University of Cambridge, Cambridge, UK). For secondary antibodies: FITC-, TRITC- (Kirkegaard and Perry Laboratories, Gaithersburg, MD), or Texas red-conjugated (Molecular Probes Inc., Eugene, OR) goat anti–rabbit and goat anti–mouse IgG and Cy3-conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) were used for immunocytochemistry. HRP-conjugated goat anti–rabbit and goat anti–mouse IgG (Biosource International, Camarillo, CA) were used for Western blotting.
Isolation of DLP1 and Peptide Microsequencing
To isolate DLP1, a large scale immunoprecipitation was performed using the antidynamin antibody MC12 from rat liver homogenate according to previously described methods (Henley and McNiven, 1996). The immunoprecipitates were run on 5–15% gradient SDS-PAGE and proteins were visualized as described by Rosenfeld et al. (1992). A protein band corresponding to the 80-kD protein was cut out of the gel. Tryptic digestion, peptide elution, and microsequencing were performed by the Mayo Protein Core Facility.
RT-PCR
Total RNA was isolated from rat tissues by guanidium thiocyanate-phenol-chloroform treatment followed by isopropanol precipitation (Chomczynski and Sacchi, 1987). A cDNA was prepared with Moloney murine leukemia virus reverse transcriptase (New England Biolabs Inc., Beverly, MA) and random primers. For DLP1 cloning, PCR was performed with cDNA as a template for 35 cycles at 95°C for 1 min, 52°C for 2 min, and 72°C for 1 min. Degenerate primers were 5′-AGYTCRGTGCTSGARAGYTTRGTNGG-3′ (256-fold) and 5′-GAYTTYGAYGARATHMGNCARGARATH-3′ (4,608-fold) for forward primers and 5′-TTNACCACRTTKGGNGARAANAC-3′ (512-fold) for the reverse primer. For the PCR at the alternative splicing regions, reactions were carried out for 25 cycles at 95°C for 1 min, 56°C for 2 min, and 72°C for 1 min with primers 5′-CTCAGTGCTGGAAAGCCTAGT-3′ and 5′-CATGATGCCATAGTTGAAGTA-3′ for forward primers and 5′-TTTACCCCATTCTTCTGCTTC-3′ and 5′-AATTCCACCACCTGCAGATGC-3′ for reverse primers.
Cloning and Sequencing
Rat brain and liver λ-ZAPII cDNA libraries (Stratagene, La Jolla, CA) were screened using a 32P-labeled DNA probe that was obtained by reverse transcriptase (RT)–PCR. Recombinant plaques were transferred onto nitrocellulose filters (Schleicher & Schuell, Inc., Keene, NH) and cross-linked at 120 mJ/cm2 in a UV cross-linker (Fisher Scientific Co., Pittsburgh, PA). Filters were prehybridized for 4 h at 65°C in 6× SSC containing 0.25% nonfat dried milk, then hybridized overnight in the same solution containing random-primed 32P-labeled DNA probes. Washing was performed with a final stringency of 0.2× SSC/0.02% SDS at 68°C. Positive plaques were isolated and further purified by secondary and tertiary screenings under the same conditions used in the primary screening. Positive clones were rescued and maintained as plasmids (rat DNA in pBluescript SK−) with helper phage (ExAssist; Stratagene), according to the manufacturer's manual. DNA sequencing was performed by an automated DNA sequencer in the Mayo Molecular Biology Core Facility. Sequence comparisons and other sequence analyses were performed using DNASTAR sequence analysis software (DNASTAR Inc., Madison, WI).
Indirect Immunofluorescence Microscopy
Immunofluorescence microscopy was performed as described in previous publications (Marks et al., 1995; Henley and McNiven, 1996). To double stain cells with fluorescently labeled dextran, cells were incubated with medium containing 100 mM lysine-fixable FITC-conjugated dextran (3,000 MW; Molecular Probes, Inc.) for 1.5 h at 37°C, acid washed for 1 min with cold HBSS (Sigma Chemical Co.) adjusted to pH 3.0 with acetic acid, and then rinsed with five changes of cold normal HBSS (pH 7.2). Cells were then fixed and processed for indirect immunofluorescence microscopy. To label endosomes containing transferrin, cells were incubated in serum-free medium (DME plus 0.02% BSA) for 30 min before incubation with 5 μg/ml FITC-conjugated human transferrin (Molecular Probes, Inc.) for 15 min at 37°C. Rinsing and processing for indirect immunofluorescence were the same as in dextran labeling. For immunofluorescence staining with cells expressing GFP-DLP1, cells were prepermeabilized with 0.01% digitonin for 30 s in microtubule stabilizing buffer (100 mM Pipes, pH 6.9, 5 mM MgSO4, 1 mM EGTA, 20% glycerol) and processed for standard indirect immunofluorescence microscopy.
Immunoelectron Microscopy
All steps were carried out essentially as described previously (Dahan et al., 1994). In brief, overnight fasted male or female Sprague-Dawley rats were anesthetized with Somnotol. Livers were perfusion fixed via the portal vein with 4% paraformaldehyde/0.5% glutaraldehyde in 0.1 M PO4 buffer (pH 7.5) for 10 min. Small 1-mm3 pieces of liver were dissected out and left in the fixative at 4°C for 1 h and then washed in 4–5 changes of ice-cold 4% sucrose/0.1 M PO4 buffer. Tissues samples were then cryoprotected in 2.3 M sucrose/0.1 M PO4, mounted on nickel stubs, and quick-frozen in liquid nitrogen. Cryosections were cut and labeled the same day as described previously (Dahan et al., 1994). Antibody dilutions were 1:2 for rabbit anti–rat DLP-N and 1:20 for the goat anti–rabbit secondary gold (10 nm) conjugate. After immunolabeling procedures, sections were stained with uranyl acetate oxalate, washed in two changes of water, and then transferred to drops of methyl cellulose containing 0.4% aqueous uranyl acetate on ice and then air dried. Sections were viewed in an electron microscope at 80 kV (Philips Electron Optics, Mahwah, NJ). Labeling density measurements were evaluated by dividing the number of gold particles over a particular structure by its profile area in μm2 as determined on the measuring tablet of a Zeiss MOP-3 digitizer (Carl Zeiss Inc., Don Mills, Ontario).
Liver Subcellular Fractionation
Subcellular fractionation was performed as described by Fleischer and Kervina (1974) and illustrated as a simplified scheme shown in Diagram 1. In brief, rat livers were harvested and homogenized in 5 vol (vol/wt) of buffer H (0.25 M sucrose in 10 mM Hepes, pH 7.5). The homogenate was spun at 960 g for 10 min. The supernatant (S1) was saved to isolate Golgi fraction, microsomes, and cytosol. For the fractionation of nuclei, mitochondria, and plasma membranes, the pellet (P1) was resuspended to a final sucrose concentration of 1.6 M, overlaid with two-thirds vol of buffer H, and spun at 71,000 g for 70 min in a Beckman SW28 rotor (Beckman Instruments, Inc., Fullerton, CA). While the nuclear fraction was collected as the pellet, membranes enriched at the interface between 0.25 and 1.6 M sucrose were collected and washed twice. The sucrose concentration of resuspended membranes was adjusted to 1.45 M, layered under buffer H, and spun at 68,000 g for 60 min in a Beckman Ti70 rotor. Mitochondrial fractions were recovered as a pellet while the plasma membrane was enriched at the interface. For the fractionation of Golgi apparatus, microsomes, and cytosol, S1 was spun at 34,000 g for 10 min and the pellet was discarded. The supernatant (S2) was spun at 50,000 g for 30 min in the Beckman Ti70, and the resulting supernatant (S3) was spun again at 200,000 g for 60 min. The supernatant (S4) was collected as a cytosolic fraction and the pellet (P4) as the light microsomal fraction. P3 was resuspended gently using a homogenizer in 10 mM Hepes, pH 7.4, containing 52% sucrose, then the sucrose concentration was adjusted to 43.7%. Sucrose concentrations of 38.7, 36, 33, and 29% solutions were sequentially layered on top of the 43.7% sucrose, which contained membrane mixtures, and spun at 120,000 g for 53 min in a SW28 rotor. Golgi fractions were recovered from the 29 and 33% sucrose interface, and heavy microsomes were at the bottom of the gradient. To fractionate rough and smooth microsomes (RM and SM, respectively), equal portions of heavy and light microsomes were combined, adjusted to 0.25 M sucrose, and made to 0.015 M CsCl. The mixture was layered on top of 1.3 M sucrose containing 0.015 M CsCl and spun at 300,000 g for 110 min in a Beckman Ti70 rotor. SM were enriched at the interface and RM were collected as a pink sediment at the bottom.
Liver Microsome Fractionation
Rat liver microsomes were fractionated by methods described previously (Howell et al., 1978; Howell and Palade, 1982) except for the buffer composition. In this experiment, 50 mM imidazole, pH 7.4 and 250 mM sucrose were used for the initial homogenization and total microsome isolation. In brief, rat liver was homogenized and centrifuged at 10,000 g for 10 min to remove cell debris, nuclei, and mitochondria. Total microsomes were obtained by centrifuging the postmitochondrial supernatant at 100,000 g for 90 min. The total microsomes were resuspended, made to 1.22 M sucrose, and loaded under a sucrose step gradient of 1.15, 0.86, and 0.25 M sucrose. The gradient was centrifuged for 3 h at 82,500 g. Light Golgi fraction (GL) was obtained as a material floated to 0.25/0.86 M sucrose interface. Heavy Golgi fraction (GH) was a material at the 0.86/1.15 M sucrose interface, and ER fraction was recovered as a pellet of the centrifugation.
SDS-PAGE and Western and Northern Blot Analyses
SDS-PAGE was conducted using a buffer system based on the method of Laemmli (1970). Western blotting procedure is performed as described previously (Henley and McNiven, 1996) except using 10% nonfat dry milk as a blocking reagent. Northern blot analysis was essentially carried out using the methods described in a previous publication (Török et al., 1996).
Results
Identification of a Novel Dynamin-like Protein
An antibody raised against a highly conserved GTP-binding region of dynamin consistently recognized a protein of 80 kD in addition to dynamin, as assessed by immunoblot analyses of a rat liver extract (Fig. 1). This 80-kD protein was isolated from liver homogenates by immunoprecipitation, and amino acid sequences of several tryptic peptides were determined. These sequences showed striking similarity to dynamin but indicated that the 80-kD protein was a novel protein distinct from dynamin (Fig. 1). Degenerate primers were designed based on the amino acid sequences of the peptides, and a DNA probe was generated by RT-PCR. Screening of a rat cDNA library identified a full-length cDNA coding for a novel dynamin-like protein, DLP1. DLP1 was encoded by an open reading frame of 755 aa with a calculated molecular mass of 83.9 kD (Fig. 2 a; Yoon, Y., and M.A. McNiven. 1996. Mol. Biol. Cell. 7:82a). While we were performing this study, we found that a partial sequence of the same protein from human had been deposited in the EST database (Adams et al., 1995). Amino acid sequence comparison of rat DLP1 with rat Dyn1 and two yeast dynamin-related proteins, Dnm1p and Vps1p, is shown in Fig. 2 a. As for all dynamin family members, there is a high degree of homology between DLP1 and the conventional dynamins within the NH2-terminal domain. The tripartite GTP-binding motif of dynamin is highly conserved in DLP1, strongly indicating that DLP1 is a GTP-binding protein. Like Dnm1p and Vps1p, DLP1 does not have the proline-rich tail of dynamin. All four sequences are completely divergent at the region aligned with the pleckstrin homology (PH) domain of dynamin (aa 510–633 of Dyn1) in this sequence comparison. Homology is partially restored at the extreme COOH-terminal end of DLP1. Portions of this region were selected as having high probability of coiled coil formation by the sequence analysis program: aa 711–752 of DLP1, aa 654– 681 and 710–741 of Dyn1, and aa 666–699 of Vps1p.
Figure 1.
Amino acid sequencing of an 80-kD protein isolated with a pan dynamin antibody revealed strong sequence similarity to the dynamin family. An 80-kD protein recognized by the pan dynamin antibody MC12 was isolated by large scale immunoprecipitation from rat liver homogenate. The immunoprecipitate was run on a 5–15% polyacrylamide gradient gel, and the 80-kD protein band was excised and eluted from the gel. Amino acid sequencing of six tryptic peptides was conducted and compared with rat dynamins I, II, and III as well as Vps1p and Dnm1p. Peptides A to E aligned with the NH2-terminal region of the dynamins, Vps1p and Dnm1p, and peptide F aligned with the COOH terminus of these proteins. Sequence information obtained from these peptides indicated that this protein was strongly related to the dynamins, yet different enough to represent a novel and distinct gene product. Boxes represent conserved residues.
Figure 2.

A full-length clone of a novel dynamin-like protein (DLP1) shares homology with dynamin and other dynamin-related proteins. (A) Amino acid sequence comparison of rat DLP1 with rat dynamin I, yeast Dnm1p, and Vps1p was performed by the Clustal method using the DNASTAR sequence analysis program. Conserved amino acid residues are boxed in black. The three regions with asterisks are GTP-binding elements. There is a high degree of homology within the NH2-terminal domains, especially in the tripartite GTP-binding motif (asterisks), but all four sequences are completely divergent at the region aligned with the PH domain of dynamin I (aa 510–633 of Dyn1). Homology is partially restored at the extreme COOH-terminal end of DLP1. Proline-rich tails are not present in DLP1, Dnm1p, or Vps1p. (B) Table showing percent homology among dynamin-related proteins. Rat DLP1 shares up to 42% homology with other dynamin-related proteins. (C) Phylogenetic tree of the large GTPase superfamily showing that DLP1 is subgrouped with the yeast dynamin-related proteins.
Interestingly, DLP1 shares greater homology with the dynamin-related proteins from Saccharomyces cerevisiae, such as Dnm1p (42%) and Vps1p (40%), than with the conventional mammalian dynamins (36%; Fig. 2 b). Whether DLP1 can be considered a true mammalian homologue of Vps1p or Dnm1p is unclear at this time. According to the phylogenetic tree shown in Fig. 2 c, the dynamin family has been expanded and is subdivided into three groups. In addition to the original group of dynamins, including the Drosophila shibire gene product, a second group includes rat DLP1 described here and three yeast proteins (Dnm1p, Vps1p, and Schizosaccharomyces pombe dynamin-like protein). The third group consists of plant dynamin-like proteins (Dombrowski and Raikhel, 1995; Gu and Verma, 1996).
DLP1 Is Expressed in All Tissues as Multiple Alternatively Spliced Variants
Northern and Western blot analyses of various rat tissues showed that DLP1 is expressed ubiquitously. By Northern analysis (Fig. 3 a), all tissues examined expressed a 4.6-kb transcript. In testis, however, an additional 3-kb message was also detected. This smaller transcript would be large enough to encode a full-length DLP1 if a tissue-specific splicing event occurred outside the coding region. Alternatively, a truncated form of DLP1 may be encoded by this shorter transcript. Three different clones encoding shorter open reading frames were isolated from the library screening. All three were truncated by ∼200 aa at the COOH termini (results not shown). In preliminary studies using RT-PCR with primers specific for the truncated forms, only one of them was found to be expressed. However, this truncated form was expressed ubiquitously and at a much lower level than that of the full-length DLP1 (results not shown) suggesting that it is not encoded by the 3-kb message detected in testis.
Figure 3.
Rat DLP1 is expressed in all tissues examined. (A) Northern blot analysis showing ubiquitous expression of rat DLP1. Total RNA (15 μg/lane) from various rat tissues was hybridized with a DLP1-specific probe. An unknown lower band was detected in testis. The same blot was also hybridized with a GAPDH probe for internal control. (B and C) Western blot analyses showing that DLP1 is present ubiquitously in rat tissues (B) and cultured mouse and human cells (C). Total protein from various rat tissues and tissue cultured cells was run on SDS–polyacrylamide gels, and DLP1 was detected by an affinity-purified anti-DLP1 antibody (DLP-MID). Each lane was loaded with 20 μg total protein, except the brain lane (10 μg). Note that DLP1 in brain, mouse hepatocytes, and human fibroblasts runs differently in SDS-PAGE from DLP1 in other tissues. B, brain; H, heart; Lg, lung; K, kidney; Sp, spleen; T, testis; Lv, liver; P, pancreas; MH, mouse hepatocyte cell line; HF, cultured human fibroblasts.
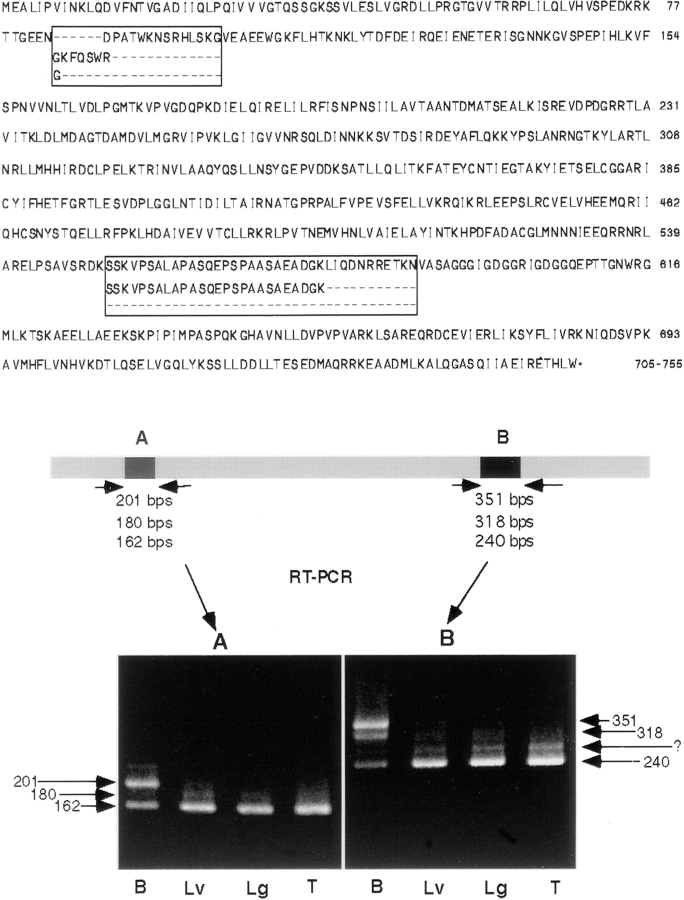
To define the distribution of DLP1 protein in tissues and cells, peptides to sequences representing the middle, NH2-terminal, and COOH-terminal regions of DLP1 were synthesized and specific antibodies were raised (see Materials and Methods). Immunoblot analyses of cell and tissue homogenates were conducted, and they confirmed the observations made from the Northern analyses that DLP1 is expressed in a wide variety of cell types and tissues (Fig. 3, b and c). Interestingly, brain DLP1 ran significantly higher on SDS-PAGE than DLP1 from other tissues (Fig. 3 b). It was also noted that the most prominent DLP1 band from both cultured mouse hepatocytes and human fibroblasts ran slightly lower than DLP1 from the rat nonneuronal tissues (Fig. 3 c). To test for the existence of alternatively spliced forms of DLP1 that could explain the multiple molecular mass seen between brain and other tissues, additional DNA sequencing of positive clones was performed and revealed two alternatively spliced regions (Fig. 4). In each region, at least three differentially spliced forms were found. In addition, RT-PCR was performed with RNA from four different tissues using primers specific for the flanking sequences of two alternative splicing regions. The predicted sizes of the different splicing variants were 201, 180, and 162 bp at the NH2-terminal splicing region and 351, 318, and 240 bp at the COOH-terminal region (Fig. 4). We found that brain preferentially expressed the longer forms at both regions, whereas other tissues expressed shorter forms almost exclusively (Fig. 4). RT-PCR with brain mRNA mainly amplified a 201-bp fragment at the NH2-terminal splicing region and a 351-bp fragment at the COOH-terminal splicing region with some amplification of the shorter forms. RT-PCR with RNA from liver, lung, and testis predominantly amplified 162- and 240-bp fragments at NH2-terminal and COOH-terminal splicing regions, respectively. Additional faint bands were detected by the RT-PCR, suggesting that there may be more splicing variants in these regions. The calculated molecular mass of the brain DLP1 form is 83.9 kD and that of other tissues is 78.6 kD.
Figure 4.
DLP1 has two short alternatively spliced regions that are differentially expressed in tissues. (Top) Amino acid sequence of rat DLP1 showing two alternatively spliced regions. Each region has at least three differentially spliced forms. (Bottom) Brain expresses longer forms of DLP1 in both alternative splicing regions, and other tissues predominantly express the shorter forms. Two sets of specific PCR primers flanking two splicing regions, A and B, were used for RT-PCR with RNA from four different rat tissues (brain, liver, lung, and testis). Details in text.
Subcellular Localization of DLP1
To identify the cytoplasmic compartment with which DLP1 associates, indirect immunofluorescence microscopy was performed on a cultured rat hepatocyte cell line (clone 9) and human fibroblasts using the affinity-purified, polyclonal peptide antibodies described above. As shown in Fig. 5, antibodies to DLP1 stain punctate vesicular structures that are concentrated at the perinuclear region and extend out into the peripheral cytoplasm in linear arrays as if associated with cytoskeletal elements. Double fluorescence labeling of cells for DLP1 and endocytosed fluorescent dextran or transferrin did not reveal a striking colocalization (Fig. 6 a), suggesting that DLP1 is not associated with endocytic or lysosomal compartments.
Figure 5.
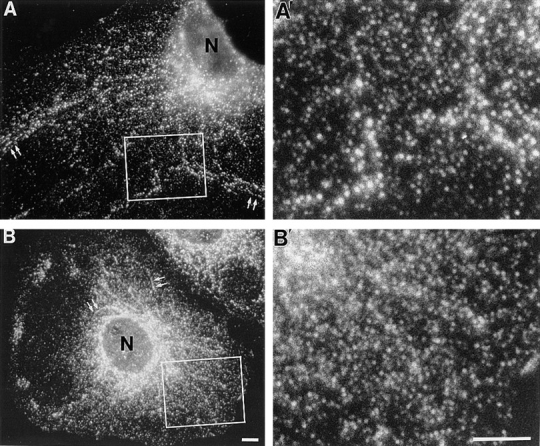
DLP1 is associated with numerous punctate vesicular structures in cultured mammalian cells. Immunofluorescence microscopy of two distinct cultured cell lines, human fibroblasts (A) and a rat hepatocyte cell line, clone 9 (B), stained with affinity-purified antibodies to DLP1 (DLP-N). DLP1 is associated with numerous vesicular structures that are distributed throughout the cytoplasm and appear aligned along cytoskeletal filaments that extend outward from perinuclear foci (arrows). Higher magnification images of boxed regions in A and B are shown in A′ and B′, respectively, and strongly suggest a vesicular association. N, nucleus. Bar, 10 μm.
Figure 6.

DLP1-associated organelles do not colocalize with multiple endocytic organelle markers but are fractionated into light membrane and ER fractions. (A) Cultured mouse hepatocytes (top two panels of A) or human cholangiocytes (bottom two panels of A) were double stained with antibodies to DLP1 (DLP1-N) and FITC-conjugated dextran (Dx) to label endocytic compartments and lysosomes, and FITC-conjugated transferrin (Tf) to label the recycling endosomal compartment. Little, if any, colocalization between DLP1 and the labeled endocytic compartments is observed. N, nucleus. (B) Immunoblots of rat liver subcellular fractions with DLP1 and various endocytic marker antibodies. Rat liver membranes were fractionated by differential centrifugation and sucrose gradient centrifugation. Each fraction was separated by SDS-PAGE and immunoblotted with anti-DLP1 antibody (DLP-MID) and endocytic marker antibodies including: MC63 for conventional dynamin, anti–α-adaptin, anti-Rab5, and anti–β-galactosidase. While DLP1 is abundant in cytosol and a light vesicle fraction (LM *), conventional dynamin is enriched in the plasma membrane (PM) with little seen in the LM fraction. α-adaptin and Rab5 are enriched in the plasma membrane fraction, and β-galactosidase is in multiple fractions. (C) Same fractions (as prepared in B) were immunoblotted with anti-DLP1 antibody (DLP-MID) and marker antibodies for secretory vesicles including anti–γ-adaptin for Golgi-associated clathrin-coated vesicles, anti-Sec23p for COPII-coated vesicles, anti-Rab8 for post-Golgi secretory vesicles, and anti-μ3 for vesicles associated with the newly identified nonendocytic adaptor complex AP-3. There is substantial coenrichment of DLP1 with the secretory vesicle marker proteins in the LM fraction. Cyt, cytosol; PM, plasma membranes; LM, light microsomes; G, Golgi fraction. (D) Rat liver total microsomes were fractionated by sucrose step gradient centrifugation and fractions were immunoblotted with anti-DLP1 (DLP-MID). DLP1 was enriched in the ER and 1.22 M sucrose fraction, in addition to GL. LS, low speed spin supernatant; LP, low speed spin pellet, TM, total microsomes; GL, light Golgi fraction; GH, heavy Golgi fraction; 1.15, 1.15 M sucrose fraction; 1.22, 1.22 M sucrose fraction; ER, endoplasmic reticulum fraction. (E) Extended fractionation for the liver microsomes (as prepared in B and C). Fractions were immunoblotted with anti-DLP1 and anti-Rab1, showing DLP1 enrichment in ER. HM, heavy microsomes; SM, smooth microsomes; RM, rough microsomes. 40 μg protein from each fraction was loaded onto the gels in B–E. Bars, 20 μm.
Immunoblot analyses of rat liver subcellular membrane fractions supported the immunofluorescence observations and suggested that DLP1 is associated with small nonendocytic organelles. Although a large portion of DLP1 was found to be cytosolic, a substantial amount was associated with a light microsomal fraction containing light membranous cytoplasmic vesicles (Fig. 6, b and c). To characterize this fraction, we immunoblotted subcellular fractions with antibodies to endocytic or lysosomal compartments. The endocytic markers dynamin and α-adaptin were enriched in the plasma membrane fraction as predicted, whereas Rab5 and the lysosomal marker β-galactosidase was found in multiple fractions (Fig. 6 b). Importantly, conventional dynamin was enriched in both plasma membrane and Golgi membrane fractions with little found in the DLP1-containing light microsomal fraction.
Because the distribution of DLP1 did not coincide significantly with endocytic compartments, we next probed liver subcellular fractions with markers for the secretory pathway. The markers used include antibodies to γ-adaptin, a subunit of the AP-1 adaptor complex that participates in TGN-derived, clathrin-coated vesicle formation (Ahle et al., 1988; Robinson, 1992); COPII (Sec23p), a marker for ER-to-Golgi transport vesicles (Barlowe et al., 1994); Rab8, a marker for a subpopulation of post-Golgi secretory vesicles (Huber et al., 1993); and μ3, a subunit of the recently identified adaptor complex AP-3 that may be involved in nonclathrin-coated vesicle formation from the TGN (Simpson et al., 1996; Dell'Angelica et al., 1997; Robinson, 1997). As shown in Fig. 6 c, multiple secretory membrane proteins were significantly enriched in the DLP1 light microsomal fraction. The coenrichment of these secretory vesicle proteins with DLP1 in the light microsomal fraction suggests that DLP1 may associate with a secretory compartment.
It is possible, although unlikely, based on the immunofluorescence images, that DLP1 forms large protein aggregates and pellets with membranes as opposed to being vesicle-associated in the differential centrifugation. To address this concern, we performed a second, well-characterized density gradient centrifugation method in which vesicles float upward in the sucrose gradient as opposed to pelleting (Ehrenreich et al., 1973; Howell et al., 1978; Howell and Palade, 1982). With this method, we found that, in addition to being cytosolic, DLP1 was enriched in light Golgi and ER fractions (Fig. 6 d). The light Golgi fraction, which contains multiple small vesicles, was obtained by floatation of total liver microsomes from the 1.22 M sucrose load (see Materials and Methods), indicating that DLP1 is associated with vesicle membranes. To further verify the cofractionation of DLP1 with the ER, an extended fractionation of microsomal membranes was conducted by the method used in Fig. 6, b and c. Immunoblotting of these fractions again revealed DLP1 enrichment in the rough microsomal fraction (RM) where ER was enriched (Fig. 6 e). In this fractionation, Rab1, a marker for ER-to-Golgi transport, was enriched in Golgi, smooth ER (SM), and rough ER (RM), and ribophorin II, a resident ER protein in SM and RM (results not shown) as previously shown (Plutner et al., 1991).
In the experiments described above, we have applied two different subcellular fractionation methods and found that DLP1 is enriched in ER fraction and a light vesicle membrane compartment as compared with dynamin that is enriched in plasma membrane and Golgi fractions. To confirm that dynamin and DLP1 are on different membrane compartments, we performed double label immunofluorescence microscopy of cultured hepatocytes with antibodies to DLP1, dynamin, and clathrin. As shown in Fig. 7 a, there is no significant colocalization between DLP1 and dynamin. Whereas DLP1 antibodies stained smaller vesicular structures concentrated in the perinuclear region, dynamin antibodies (hudy-1) recognized larger punctate structures, presumably clathrin-coated pits on the plasma membrane (Damke et al., 1994). Additional double label immunofluorescence microscopy using DLP1 and clathrin heavy chain antibodies showed that DLP1 is not localized to the clathrin-containing organelles that are present at the TGN and the plasma membrane (Fig. 7 b). These results, in combination with the immunoblot analyses of subcellular fractions, suggest that DLP1 is not associated with an endocytic compartment and has a localization distinct from that of conventional dynamin.
Figure 7.
DLP1 does not localize with dynamin- or clathrin-associated organelles. Clone 9 cells were labeled with anti-DLP1 (DLP-N) and anti-dynamin (hudy-1) antibodies (A), or with anti-DLP1 (DLP-N) and anti-clathrin heavy chain antibodies (B). In both images, the DLP1 staining (red fluorescence) does not colocalize with dynamin or clathrin (green fluorescence). Insets show higher magnification images of boxed regions N, nucleus. Bars, 10 μm.
DLP1-positive Structures Associate with Microtubules and Tubules of the Endoplasmic Reticulum
The immunofluorescence micrographs of cellular DLP1 shown in Fig. 5 suggested that vesicles that associate with DLP1 aligned along linear structures reminiscent of microtubule arrays emanating from a perinuclear centrosome. To further define this linear organization, double immunofluorescence microscopy was performed using antibodies to DLP1 and tubulin. As shown in Fig. 8, a large portion of DLP1-positive vesicles coincided with microtubules. This association is most striking in the color double-stained images (Fig. 8, c and inset) that reveal discrete DLP1-positive vesicles bound along single microtubules.
Figure 8.
Distribution of DLP1-vesicles are coincident with microtubule arrays in cultured cells. Double immunofluorescence microscopy of cultured human fibroblasts (A and B) and clone 9 cells (C) with antibodies to DLP1 and α-tubulin. There is high degree of overlap (arrows) between the distribution of DLP1-stained vesicles (A) and the microtubule array (B). (C) A clone 9 cell stained for DLP1 (red) and microtubules (green) showing DLP1-vesicles aligned along the microtubule network. Inset is a higher magnification image of the boxed region revealing the intimate association between the DLP1 vesicles and microtubules. N, nucleus. Bars: (B) 20 μm; (C and inset) 10 μm.
Although we observed significant overlap between the DLP1 and microtubules, it was unclear as to why long prominent chains of DLP1-positive structures appeared to align along some microtubule strands but not others. This suggested that the DLP1 vesicles might selectively associate with a membrane organelle that extends along the microtubule proper. Based on the fact that DLP1 is enriched in ER containing subcellular fractions as opposed to endocytic compartments (Fig. 6), and that ER tubules are known to align along microtubule networks (Terasaki et al., 1986; Lee et al., 1989), we conducted double immunofluorescence staining for DLP1 and two ER markers, calnexin and Rab 1. Calnexin is a resident chaperone protein of the ER (Rajagopalan et al., 1994) while Rab1 has been implicated in regulation of vesicle transport between the ER and the Golgi apparatus (Plutner et al., 1991; Tisdale et al., 1992; Davidson and Balch, 1993; Nuoffer et al., 1994; Pind et al., 1994). As shown in Fig. 9, we observed a remarkable overlap between DLP1 and calnexin-positive ER tubules. Although DLP1 vesicles were observed throughout the cytoplasm, the coalignment between the ER and the long extended arrays of DLP1-positive structures was nearly absolute and very striking. In high magnification images (Fig. 9, inset), one can observe many DLP1 punctate spots situated along a single ER cisterna. Next, we compared the DLP1 distribution to that of Rab1. Because our antibodies to both Rab1 and DLP1 were made in rabbits making double staining difficult, we used clone 9 cells stably transfected to express DLP1 fused to the green fluorescent protein (GFP-DLP1; K.R. Pitts, Y. Yoon, and M.A. McNiven, manuscript in preparation). Because GFP-DLP1 in these cells was overexpressed and filled the cytoplasm, the cells were permeabilized before fixation to allow cytosol and many of the individual vesicles to leak out of the cell, resulting in a reduced cytoplasmic fluorescence. After permeabilization, fixation, and immunostaining for Rab1 of the transfected cells, we observed prominent linear distributions of GFP-DLP1 spots that colocalized with the Rab1-positive compartment (Fig. 10). Since the Rab1 has been shown to distribute at ER as well as Golgi regions (Plutner et al., 1991) and GFP-DLP1 did not localize with Golgi markers by immunofluorescence (results not shown), the Rab1-positive GFP-DLP1 tubules are ER cisternae, confirming DLP1 localization at ER tubules. Interestingly, the two proteins appear to coalign with each other but do not overlap (Fig. 10, c and d). Thus, one can observe a consistent alternating pattern of Rab1- and DLP1-positive structures.
Figure 9.

DLP1 localizes to tubules of the endoplasmic reticulum. Double immunofluorescence microscopy of cultured human fibroblasts with antibodies to DLP1 (A and D) and calnexin (B and E). DLP1 spots are organized into long tracks that extend out into the peripheral cytoplasm and coalign perfectly with ER tubules (arrows). Overlap images are seen in C and F. Note that the linear extension of DLP1 is only observed when associated with the ER tubule. N, nucleus. Bars, 10 μm.
Figure 10.


DLP1 is associated with Rab1-positive ER tubules. Clone 9 cells expressing GFP-DLP1 were prepermeabilized with digitonin before fixation and immunostaining with anti-Rab1 antibodies. GFP-DLP1 distributes along the linear structures (A). These linear structures are ER tubules to which Rab1 is associated (B). (C) Overlaying image of GFP-DLP1 and Rab1 staining showing DLP1 and Rab1 are localized on the same ER tubules. (D) Enlarged image of boxed region in C. Note that most GFP-DLP1 spots alternate with Rab1 along the ER tubules. N, nucleus. Bars, 10 μm.
As a final confirmation of DLP1 localization to ER tubules, immunogold labeling of hepatocytes was performed on ultrathin frozen sections of rat liver. Fig. 11 is a collection of representative images showing a prominent labeling of ER cisternae and tubular networks by DLP1 antibodies as compared with other organelles. There was little or no labeling of Golgi stacks proper but some gold particles were found on closely associated tubular networks. Table I provides a quantitation of gold particles on different cytoplasmic organelles, revealing that DLP1 is predominantly found along tubular reticular membranes of ER. Golgi stacks are depleted in DLP1 immunoreactivity as revealed by a labeling density similar to background. Thus, taken together, the biochemical and morphological observations reported here are consistent with the premise that DLP1 is associated with both ER tubules and cytoplasmic vesicles.
Figure 11.

Immunogold labeling of DLP1 showing DLP1 on ER cisternae. Ultrathin cryosections of rat liver were immunolabeled with rabbit anti-DLP1 (DLP-N) followed by goat anti–rabbit IgG 10 nm gold. DLP1 labeling is observed over parallel ER cisternae (rER and arrowheads), and tubular reticular network profiles (small arrows), some of which surround the Golgi apparatus (G). Golgi stacks are largely devoid of DLP1 immunoreactivity as are peroxisomes (P), mitochondria (M), and lysosomes (L). LAT, lateral plasma membrane; bc, bile canalicular plasma membrane. Bars, 400 nm.
Table I.
Intracellular Distribution of DLP1 in Rat Liver Hepatocytes In Situ*
| Compartments examined (n) | Labeling density‡ | |||
|---|---|---|---|---|
| No. gold/profile area | No. gold/profile perimeter | |||
| Golgi stacks§ (87) | 3.32 | — | ||
| ER tubular networks‖ (381) | 8.31 | — | ||
| Mitochondria/Peroxisomes (159) | 1.89 | — | ||
| Golgi cisternae ¶ (27) | — | 0.19 | ||
| ER cisternae ¶ (29) | — | 0.51 | ||
| Bile Canalicular/Lateral | — | 0.20 | ||
| Plasma Membranes ¶ (45) | ||||
| Mitochondrial/Perixisomal | — | 0.06 | ||
| Membranes ¶ (14) | ||||
n, no. of micrograph.
Rat liver cryosections were imunolabeled with rabbit anti-DLP1 (DLP-N) followed by goat anti–rabbit IgG–gold (10 nm). 35 micrographs of hepatocyte sections (final magnification, 36000×) were evaluated for gold particle labeling density in the indicated compartments.
Labeling density was evaluated by dividing the total number of gold particles over a particular compartment (e.g., Golgi stacks) by the total profile area of that compartment (in μm2) or total profile membrane perimeter length (μm). Only gold particles within 20 nm of membranous profiles (for Golgi and ER profiles) were scored.
This compartment consists of stacks of Golgi apparatus.
Consists of long continuous cisternae, 30–50 nm in thickness (rough ER) and membranous tubular reticular profiles in the cytoplasm (smooth ER) and at the periphery of Golgi stacks.
In a second quantitative analysis, gold particle labeling was determined per membrane length of compartments whose delimiting membrane is clearly visualized on cryosections (i.e., individual Golgi and ER cisternae, the bile canalicalicular and lateral plasma membranes, mitochondrial outer membrane, and peroxisomal membrane). DLP1 is predominantly found along rough ER cisternae and vesicular tubular membranes of smooth ER around the Golgi apparatus.
Discussion
A Novel Dynamin-like Protein in Mammalian Cells
In this study, we have identified a novel dynamin-like protein, DLP1, that shares 34–37% and 40–42% overall homology to conventional dynamins and yeast dynamin- related proteins, respectively (Yoon, Y., and M.A. McNiven. 1996. Mol. Biol. Cell. 7:82a). According to a sequence comparison of DLP1 with dynamin, Vps1p, and Dnm1p, the NH2-terminal enzymatic domains of these proteins are highly conserved except for a stretch of 25–45 aa between the first and second GTP-binding elements of DLP1, Dnm1p, and Vps1p. This amino acid cassette is not present in dynamin and contains the first of two alternatively spliced sites of DLP1 that will be discussed later. Sequence homology among these proteins is very low at the region aligned with the dynamin PH domain (aa 510–633 of Dyn1) in the comparison shown in Fig. 2 a. The PH domain has been suggested to mediate membrane binding of dynamin and other PH domain–containing proteins (Lemmon et al., 1996; Salim et al., 1996). It is unclear whether this region of DLP1 forms a PH domain because it cannot be predicted by primary amino acid sequence comparison (Downing et al., 1994; Ferguson et al., 1994; Lemmon et al., 1996). However, secondary structure predictions of both DLP1 and Dnm1p show similar features in this region to that of known PH domains that contain alternating structures of several β-sheets and variable loops (Ferguson et al., 1994). The divergent sequence in this region suggests that this domain may render different organelle specificity to the dynamins and dynamin-related proteins. Finally, the extreme COOH-terminal region of DLP1 shows a small but significant restoration of homology to both dynamin and the two yeast proteins. This tail region of DLP1 is predicted to form a coiled coil, suggesting that it may participate in the formation of polymeric structures with other proteins.
A Ubiquitous Dynamin-like Protein That Is Alternatively Spliced in a Tissue-specific Manner
Northern and Western blot analyses indicate that DLP1 is expressed ubiquitously, but a tissue-specific expression of different alternatively spliced variants exists. Two alternative splicing regions were found in DLP1. As mentioned, the first is in the amino acid cassette that is not present in the dynamins. The second is in the region aligned with the PH domain of dynamin. Both of these regions are highly divergent from the dynamins and the other dynamin- related proteins. Further sequence analysis indicates that these alternatively spliced regions have a high surface probability. Thus, these regions are likely to be exposed and may bind to membrane lipids or other proteins. Therefore, different splicing variants produced in these regions may have different binding properties. Such alterations in binding properties by alternative splicing have been reported for many proteins, such as agrin, in which a 4-aa insertion creates a specific heparin-binding site (Gesemann et al., 1996). Interestingly, DLP1 exhibits tissue-specific alternative splicing, with brain expressing a larger form that includes an additional 50 aa at two splicing regions (Figs. 3 b and 4). Whether these insertions in the brain form of DLP1 modify its distribution or function for rapid secretory or endocytic processes in neurons remains to be tested.
A Vesicle-associated Dynamin-like Protein with a Distribution Distinct from Conventional Dynamin and Endocytic Markers
We have analyzed the cytoplasmic distribution of DLP1 using multiple methods including immunoblotting of subcellular fractions (Fig. 6), immunofluorescence microscopy (Figs. 5, 7–9), the expression of GFP-tagged DLP1 in cultured cells (Fig. 10), and immunogold labeling (Fig. 11). By morphological criteria, antibodies to DLP1 clearly label a vesicular compartment. Further, GFP-DLP1 expressed in cultured cells shows a vesicular distribution that is near identical to that observed by antibody staining. In addition to this striking vesicular morphology, Western blot analysis of rat liver membrane fractions revealed that DLP1 is soluble and a significant portion is associated with a light membrane vesicle fraction (Fig. 6, b and c). To insure that this fractionation represents a true association with vesicles as opposed to protein aggregates, we used another sucrose gradient centrifugation method (Fig. 6 d). Regardless of the fractionation method implemented, we consistently observed DLP1 associated with lighter membranes or vesicles. In addition, DLP1-positive vesicles did not colocalize with endocytic vesicles containing fluorescently tagged dextran and transferrin in cultured cells (Fig. 6 a). Finally, immunoblot analyses of rat liver membrane fractions for DLP1 and endocytic marker proteins such as α-adaptin, Rab 5, and β-galactosidase (Fig. 6 b), as well as transferrin, Rab4, Rab7, and Rab9 (data not shown), suggest that DLP1 is not enriched in fractions containing these endocytic markers. Dynamin was found predominantly in plasma membrane and Golgi fractions, which is consistent with its proposed participation in the formation of vesicles from the plasma membrane (Kosaka and Ikeda, 1983a ,b; van der Bliek et al., 1993; Damke et al., 1994; Takei et al., 1995) and the Golgi apparatus (Henley and McNiven, 1996; Maier et al., 1996). In support of the immunoblotting data, double immunofluorescence staining of cultured cells with antibodies to DLP1 and clathrin or dynamin was performed (Fig. 7). From these studies we conclude that DLP1 clearly associates with a population of vesicles that is both smaller in size and distinct from those associated with clathrin and dynamin at the plasma membrane.
An ER-associated Dynamin-like Protein
Although DLP1 does not appear to associate with an endocytic compartment, additional data using a variety of different experimental approaches strongly suggest that this dynamin-like protein associates with the ER and small discrete vesicles that associate with microtubules. This conclusion is based on the immunoblotting of subcellular fractions (Fig. 6), as well as the fact that the long strings of DLP1 spots are always associated with the ER tubules (Figs. 9 and 10), whereas individual vesicles are on single microtubules (Fig. 8). It is important to emphasize that the localization of DLP1 at the ER tubules is not coincidental overlap, as supported by the immunoelectron microscopy data described in Fig. 11.
Presently, the identity and composition of the DLP1-positive cytoplasmic vesicles is not clear although biochemical characterization is currently underway. It is interesting that DLP1 and Rab1 are on the same ER cisterna but do not overlap with each other (Fig. 10). In fact, both antigens appear as distinct patches or vesicles that align along the ER in a striking alternating pattern. Whether Rab1 and DLP1 function sequentially in the same process or totally distinct processes is unclear. It is most attractive to speculate that DLP1 and Rab1 act together to mediate the formation and liberation of vesicles from the ER where they could become free to attach to microtubules for subsequent transport to the Golgi complex via pre-Golgi compartments (Presley et al., 1997). Indeed, studies using time-lapse fluorescence microscopy of GFP-DLP1 in living cells demonstrate that these vesicles are motile and move unidirectionally from the cell periphery toward the perinuclear region (results not shown). This observation is consistent with the prediction that DLP1 vesicles are transported from peripheral ER elements to the Golgi compartment via microtubule tracks. Future studies using inhibitory DLP1 antibodies and mutant DLP1 expression constructs will test these predictions and define if this protein participates in anterograde and/or retrograde vesicle flux through the early secretory pathway. Taken together, the biochemical and morphological data presented here raise the exciting possibility that DLP1 is an ER equivalent of the conventional plasma membrane dynamin and participates in the formation of nascent secretory vesicles from the ER cisternae. Functional studies are underway to test this prediction.
The identification and initial characterization of the dynamin-like protein reported here supports the central premise that the dynamin family of proteins may be extensive in number as proposed by McNiven and coworkers (Henley and McNiven, 1996; Urrutia et al., 1997). Based on these findings and the recent proliferation of many different protein families, such as the cytoplasmic myosins, kinesins, adaptins, ARFs, Rabs, syntaxins, and others that are involved in protein trafficking, the number of dynamin family members will likely continue to expand in the near future. Further investigations will define whether DLP1 participates in vesicle scission or other vesicle trafficking processes such as targeting or transport.
Figure.
Diagram 1.
Footnotes
1. Abbreviations used in this paper: aa, amino acids; GH and GL, heavy and light Golgi fraction, respectively; PH, pleckstrin homology; RM and SM, rough and smooth microsomes, respectively; RT-PCR, reverse transcriptase–PCR.
We are especially grateful to Mrs. B. Oswald for the initial identification and isolation of DLP1, antibody purification, and other technical assistance. We thank Dr. S.L. Schmid for providing the antidynamin antibody, hudy 1; Dr. L. Traub for anti–γ-adaptin antibody; Dr. J.-P. Paccaud for anti-Sec23p; Dr. D.D. Sabatini for anti-Rab8; Dr. M.S. Robinson for anti-μ3; and Dr. N.F. LaRusso for anti–β-galactosidase antibody and the human cholangiocyte cell line. Cryosectioning of liver tissue by Mrs. J. Mui in Dr. J. Bergeron's laboratory at McGill University is gratefully acknowledged. We are also grateful to Mr. E. Krueger for helping with photographic techniques, and to Dr. J.R. Henley and Ms. R.R. Torgerson for helpful comments and reading the manuscript.
This study was supported by National Institutes of Health (NIH) training grant (DK07198) and National Research Service Award postdoctoral fellowship from National Institute of Diabetes and Digestive and Kidney Diseases (DK09574) to Y. Yoon and NIH grant (DK44650) to M.A. McNiven.
Address correspondence to Mark A. McNiven, Center for Basic Research in Digestive Diseases and Department of Biochemistry and Molecular Biology, Mayo Clinic and Foundation, 200 First St. SW, Rochester, MN 55905. Tel.: (507) 284-0683. Fax: (507) 284-0762. E-mail: mcniven.mark@mayo.edu
Dr. Dahan's current address is Center for Basic Research in Digestive Diseases, Mayo Clinic, Rochester, Minnesota 55905.
References
- Adams, M., A. Kerlavage, R. Fleischmann, R. Fuldner, C. Bult, N. Lee, E. Kirkness, K. Weinstock, J. Gocayne, O. White, et al. 1995. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 377(Suppl.):3–174. [PubMed]
- Ahle S, Mann A, Eichelsbacher U, Ungewickell E. Structural relationships between clathrin assembly proteins from the Golgi and the plasma membrane. EMBO (Eur Mol Biol Organ) J. 1988;7:919–929. doi: 10.1002/j.1460-2075.1988.tb02897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Clark J, Moore L, Krasinskas A, Way J, Battey J, Tamkun J, Kahn RA. Selective amplification of additional members of the ADP-ribosylation factor (ARF) family: cloning of additional human and DrosophilaARF-like genes. Proc Natl Acad Sci USA. 1993;90:8952–8956. doi: 10.1073/pnas.90.19.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook TA, Urrutia R, McNiven MA. Identification of dynamin 2, an isoform ubiquitously expressed in rat tissues. Proc Natl Acad Sci USA. 1994;91:644–648. doi: 10.1073/pnas.91.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook TA, Mesa K, Urrutia R. Three dynamin-encoding genes are differentially expressed in developing rat brain. J Neurochem. 1996;67:927–931. doi: 10.1046/j.1471-4159.1996.67030927.x. [DOI] [PubMed] [Google Scholar]
- Dahan S, Ahluwalia J, Wang L, Posner B, Bergeron J. Concentration of intracellular hepatic apolipoprotein E in Golgi apparatus saccular distensions and endosomes. J Cell Biol. 1994;127:1859–1869. doi: 10.1083/jcb.127.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, van der Bliek AM, Schmid SL. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson H, Balch W. Differential inhibition of multiple vesicular transport steps between the endoplasmic reticulum and trans Golgi network. J Biol Chem. 1993;268:4216–4226. [PubMed] [Google Scholar]
- Dell'Angelica E, Ooi C, Bonifacino J. β3A-adaptin, a subunit of the adaptor-like complex AP-3. J Biol Chem. 1997;272:15078–15084. doi: 10.1074/jbc.272.24.15078. [DOI] [PubMed] [Google Scholar]
- Dombrowski JE, Raikhel NV. Isolation of a cDNA encoding a novel GTP-binding protein of Arabidopsis thaliana. . Plant Mol Biol. 1995;28:1121–1126. doi: 10.1007/BF00032672. [DOI] [PubMed] [Google Scholar]
- Downing AK, Driscoll PC, Gout I, Salim K, Zvelebil MJ, Waterfield MD. Three-dimensional solution structure of the pleckstrin homology domain from dynamin. Curr Biol. 1994;4:884–891. doi: 10.1016/s0960-9822(00)00197-4. [DOI] [PubMed] [Google Scholar]
- Ehrenreich J, Bergeron J, Siekevitz P, Palade G. Golgi fractions prepared from rat liver homogenates: I. Isolation procedure and morphological characterization. J Cell Biol. 1973;59:45–72. doi: 10.1083/jcb.59.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Crystal structure at 2.2 Å resolution of the pleckstrin homology domain from human dynamin. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- Fleischer S, Kervina M. Subcellular fractionation of rat liver. Methods Enzymol. 1974;31:6–41. doi: 10.1016/0076-6879(74)31005-1. [DOI] [PubMed] [Google Scholar]
- Gammie AE, Kurihara LJ, Vallee RB, Rose MD. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesemann M, Cavalli V, Denzer AJ, Brancaccio A, Schumacher B, Ruegg MA. Alternative splicing of agrin alters its binding to heparin, dystroglycan, and the putative agrin receptor. Neuron. 1996;16:755–767. doi: 10.1016/s0896-6273(00)80096-3. [DOI] [PubMed] [Google Scholar]
- Goud B. Small GTP-binding proteins as compartmental markers. Semin Cell Biol. 1992;3:301–307. doi: 10.1016/1043-4682(92)90017-p. [DOI] [PubMed] [Google Scholar]
- Gout I, Dhand R, Hiles ID, Fry MJ, Panayotou G, Das P, Truong O, Totty NF, Hsuan J, Booker GW, et al. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- Grubman SA, Perrone RD, Lee DW, Murray SL, Rogers LC, Wolkoff LI, Mulberg AE, Cherington V, Jefferson DM. Regulation of intracellular pH by immortalized human intrahepatic biliary epithelial cell lines. Am J Physiol. 1994;266:G1060–G1070. doi: 10.1152/ajpgi.1994.266.6.G1060. [DOI] [PubMed] [Google Scholar]
- Gu X, Verma DPS. Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO (Eur Mol Biol Organ) J. 1996;15:695–704. [PMC free article] [PubMed] [Google Scholar]
- Gu X, Verma D. Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell. 1997;9:157–169. doi: 10.1105/tpc.9.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley JR, McNiven MA. Association of a dynamin-like protein with the Golgi apparatus in mammalian cells. J Cell Biol. 1996;133:761–775. doi: 10.1083/jcb.133.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits JS, Burgess CC, Obar RA, Vallee RB. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993a;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits JS, Shpetner HS, Burgess CC, Vallee RB. Microtubules and Src homology 3 domains stimulate the dynamin GTPase via its C-terminal domain. Proc Natl Acad Sci USA. 1993b;90:11468–11472. doi: 10.1073/pnas.90.24.11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Organelle transport along microtubules-the role of KIFs. Trends Cell Biol. 1996;6:135–141. doi: 10.1016/0962-8924(96)10003-9. [DOI] [PubMed] [Google Scholar]
- Howell K, Palade G. Hepatic Golgi fractions resolved into membrane and content subfractions. J Cell Biol. 1982;92:822–832. doi: 10.1083/jcb.92.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell K, Ito A, Palade G. Endoplasmic reticulum marker enzymes in Golgi fractions-what does this mean? . J Cell Biol. 1978;79:581–589. doi: 10.1083/jcb.79.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber LA, Pimplikar S, Parton RG, Virta H, Zerial M, Simons K. Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J Cell Biol. 1993;123:35–45. doi: 10.1083/jcb.123.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Ikeda K. Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single gene mutation in Drosophila. . J Neurobiol. 1983a;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- Kosaka, T., and K. Ikeda. 1983b Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J. Cell Biol. 97:499–507. [DOI] [PMC free article] [PubMed]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee C, Ferguson M, Chen L. Construction of the endoplasmic reticulum. J Cell Biol. 1989;109:2045–2055. doi: 10.1083/jcb.109.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Schlessinger J. PH domain: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- Maeda K, Nakata T, Noda Y, Sato-Yoshitake R, Hirokawa N. Interaction of dynamin with microtubules: its structure and GTPase activity investigated by using highly purified dynamin. Mol Biol Cell. 1992;3:1181–1194. doi: 10.1091/mbc.3.10.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier O, Knoblich M, Westermann P. Dynamin II binds to the trans-Golgi network. Biochem Biophys Res Commun. 1996;223:229–233. doi: 10.1006/bbrc.1996.0876. [DOI] [PubMed] [Google Scholar]
- Marks D, LaRusso N, McNiven M. Isolation of the microtubule motor kinesin from rat liver: selective inhibition by cholestatic bile acids. Gastroenterology. 1995;108:824–833. doi: 10.1016/0016-5085(95)90457-3. [DOI] [PubMed] [Google Scholar]
- Miki H, Miura K, Matuoka K, Nakata T, Hirokawa N, Orita S, Kaibuchi K, Takai Y, Takenawa T. Association of Ash/Grb-2 with dynamin through the Src homology 3 domain. J Biol Chem. 1994;269:5489–5492. [PubMed] [Google Scholar]
- Moore JD, Endow SA. Kinesin proteins: a phylum of motors for microtubule-based motility. BioEssays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Mooseker MS, Cheney RE. Unconventional myosins. Ann Rev Cell Develop Biol. 1995;11:633–675. doi: 10.1146/annurev.cb.11.110195.003221. [DOI] [PubMed] [Google Scholar]
- Nakata T, Takemura R, Hirokawa N. A novel member of the dynamin family of GTP-binding proteins is expressed specifically in the testis. J Cell Sci. 1993;105:1–5. doi: 10.1242/jcs.105.1.1. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C, Davidson H, Matteson J, Meinkoth J, Balch W. A GDP-bound form of Rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J Cell Biol. 1994;125:225–237. doi: 10.1083/jcb.125.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar RA, Collins CA, Hammarback JA, Shpetner HS, Vallee RB. Molecular cloning of the microtubule-associated mechanochemical enzyme dynamin reveals homology with a new family of GTP-binding proteins. Nature. 1990;347:256–261. doi: 10.1038/347256a0. [DOI] [PubMed] [Google Scholar]
- Pind S, Nuoffer C, McCaffery J, Plutner H, Davidson H, Farquhar M, Balch W. Rab1 and Ca2+are required for the fusion of carrier vesicles mediating endoplasmic reticulum to Golgi transport. J Cell Biol. 1994;125:239–252. doi: 10.1083/jcb.125.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutner H, Cox A, Pind S, Khosravi-Far R, Bourne J, Schwaninger R, Der C, Balch W. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J Cell Biol. 1991;115:31–43. doi: 10.1083/jcb.115.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J, Cole N, Schroer T, Hirschberg K, Zaal K, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Xu Y, Brenner M. Retention of unassembled components of integral membrane proteins by calnexin. Science. 1994;263:387–390. doi: 10.1126/science.8278814. [DOI] [PubMed] [Google Scholar]
- Raynal P, Pollard HB. Annexins: the problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim Biophys Acta. 1994;1197:63–93. doi: 10.1016/0304-4157(94)90019-1. [DOI] [PubMed] [Google Scholar]
- Robinson MS. Adaptins. Trends Cell Biol. 1992;2:293–297. doi: 10.1016/0962-8924(92)90118-7. [DOI] [PubMed] [Google Scholar]
- Robinson MS. Coats and budding. Trends Cell Biol. 1997;7:99–102. doi: 10.1016/S0962-8924(96)10048-9. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Liu J-P, Powell KA, Fykse EM, Südhof TC. Phosphorylation of dynamin I and synaptic-vesicle recycling. Trends Neurosci. 1994;17:348–353. doi: 10.1016/0166-2236(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Rothman JH, Raymond CK, Gilbert T, O'Hara PJ, Stevens TH. A putative GTP-binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell. 1990;61:1063–1074. doi: 10.1016/0092-8674(90)90070-u. [DOI] [PubMed] [Google Scholar]
- Salim K, Bottomley MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, Margolis RL, Gigg R, Smith CI, Driscoll PC, et al. Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO (Eur Mol Biol Organ) J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- Scaife R, Margolis RL. Biochemical and immunochemical analysis of rat brain dynamin interaction with microtubules and organelles in vivo and in vitro. J Cell Biol. 1990;111:3023–3033. doi: 10.1083/jcb.111.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaife R, Gout I, Waterfield MD, Margolis RL. Growth factor- induced binding of dynamin to signal transduction proteins involves sorting to distinct and separate proline-rich dynamin sequences. EMBO (Eur Mol Biol Organ) J. 1994;13:2574–2582. doi: 10.1002/j.1460-2075.1994.tb06547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf K, Kostka G, Lammers R, Bashkin P, Daly R, Burgess WH, van der Bliek AM, Schlessinger J, Ullrich A. Dynamin binds to SH3 domains of phospholipase Cγ and GRB-2. J Biol Chem. 1994;269:16009–16014. [PubMed] [Google Scholar]
- Shpetner HS, Vallee RB. Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell. 1989;59:421–432. doi: 10.1016/0092-8674(89)90027-5. [DOI] [PubMed] [Google Scholar]
- Shpetner HS, Vallee RB. Dynamin is a GTPase stimulated to high levels of activity by microtubules. Nature. 1992;355:733–735. doi: 10.1038/355733a0. [DOI] [PubMed] [Google Scholar]
- Simpson F, Bright NA, West MA, Newman LS, Darnell RB, Robinson MS. A novel adaptor-related protein complex. J Cell Biol. 1996;133:749–760. doi: 10.1083/jcb.133.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Moss SE. Structural evolution of the annexin supergene family. Trends Genet. 1994;10:241–246. doi: 10.1016/0168-9525(94)90171-6. [DOI] [PubMed] [Google Scholar]
- Sontag J-M, Fykse EM, Ushkaryov Y, Liu J-P, Robinson PJ, Südhof TC. Differential expression and regulation of multiple dynamins. J Biol Chem. 1994;269:4547–4554. [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-γS in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Terasaki M, Chen L, Fujiwara K. Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol. 1986;103:1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale E, Bourne J, Khosravi-Far R, Der C, Balch W. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török N, Urrutia R, Nakamura T, McNiven M. Upregulation of molecular motor-encoding genes during HGF and EGF-induced cell motility. Am J Cell Phys. 1996;167:422–433. doi: 10.1002/(SICI)1097-4652(199606)167:3<422::AID-JCP6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Tuma PL, Stachniak MC, Collins CA. Activation of dynamin GTPase by acidic phospholipids and endogenous rat brain vesicles. J Biol Chem. 1993;268:17240–17246. [PubMed] [Google Scholar]
- Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: redundant or distinct functions for an expanding family or related GTPases? . Proc Natl Acad Sci. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbach K, Payne GS. Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1993;12:3049–3059. doi: 10.1002/j.1460-2075.1993.tb05974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]