Abstract
In vitro transcription/translation of actin cDNA and analysis of the translation products by native-PAGE was used to study the maturation pathway of actin. During the course of actin synthesis, several distinct actin-containing species were observed and the composition of each determined by immunological procedures. After synthesis of the first ∼145 amino acids, the nascent ribosome-associated actin chain binds to the recently identified heteromeric chaperone protein, prefoldin (PFD). PFD remains bound to the relatively unfolded actin polypeptide until its posttranslational delivery to cytosolic chaperonin (CCT). We show that α- and β-tubulin follow a similar maturation pathway, but to date find no evidence for an interaction between PFD and several noncytoskeletal proteins. We conclude that PFD functions by selectively targeting nascent actin and tubulin chains pending their transfer to CCT for final folding and/or assembly.
Keywords: protein folding, molecular chaperones, cytoskeletal proteins, protein synthesis, actin, tubulin
Many proteins undergo spontaneous refolding to their native conformation following denaturation with reagents such as acid, guanidine, or urea. The rate and efficiency of the refolding reaction varies significantly, depending on the particular protein in question. Initially thought to be a spontaneous process dictated only by the primary sequence of the particular polypeptide (Anfinsen, 1973), many protein folding events are now known to require the participation of additional components termed molecular chaperones (recently reviewed in Hartl, 1996; Johnson and Craig, 1997; Netzer and Hartl, 1998). While not conveying any information for the folding reaction itself, molecular chaperones increase the probability of productive folding by stabilizing critical intermediates in the folding pathway.
Molecular chaperones working in tandem have been suggested to participate in the maturation pathway of certain newly synthesized proteins. For example, a pathway of successive chaperone interactions has been suggested for the maturation of some proteins in the eukaryotic cytosol, based on both folding reactions and translation experiments done in vitro. At least three distinct chaperones have been implicated in the folding of luciferase. Nascent luciferase molecules were reported to initially interact with an eukaryotic homologue of the bacterial DnaJ protein, referred to as hsp40 (Frydman et al., 1994). Binding to hsp40 enables the recruitment and binding of hsp70, which stabilizes the unfolded nascent polypeptide. Others also have reported that eukaryotic hsp70 binds cotranslationally to a wide variety of nascent polypeptide chains (Beckmann et al., 1990; Nelson et al., 1992; Frydman et al., 1994; Hansen et al., 1994; Frydman and Hartl, 1996; Eggers et al., 1997; Pfund et al., 1998). In the case of luciferase, after interaction with hsp70, the nascent chain becomes a target for the cytosolic chaperonin, referred to variously as CCT1 (for cytosolic chaperonin containing TCP-1), TriC, or c-cpn, to which it is transferred cotranslationally (Frydman et al., 1994). Subsequent folding to the native state is thought to occur within the central cavity of the CCT particle again after one or more cycles of ATP-dependent binding and release (Frydman and Hartl, 1996; Farr et al., 1997). A similar pathway involving the sequential action of these chaperones has been reported for actin biogenesis (Frydman and Hartl, 1996).
Recently, other components have been identified and shown to participate in the maturation pathway of actin and members of the tubulin family. For example, a heteromeric protein complex has been genetically (Geissler et al., 1999) and biochemically (Vainberg et al., 1998; Siegers et al., 1999) identified and shown to play an important role in promoting the formation of functional actins and tubulins. Termed prefoldin (PFD) in animal cells, this complex has been shown to transfer full-length unfolded target proteins (actin or tubulin) to CCT. Similarly in yeast, the cognate complex, GimC, has been shown to function in concert with CCT to facilitate actin folding in vivo (Siegers et al., 1999).
Because PFD binds unfolded target proteins and transfers them to CCT, it could act at many different points in protein biogenesis, including binding to nascent chains, interaction with nonnative proteins discharged from CCT as part of the ATP-dependent chaperonin cycle, or the rescue of proteins that have become denatured by heat or other stresses. Here, we report a detailed examination of the pathway of actin maturation during in vitro translation. We show that an early and very prominent intermediate contains nascent actin complexed with PFD. Binding of PFD to actin nascent chains occurs after synthesis of the NH2-terminal ∼145 amino acids. Actin molecules bound to PFD are nonnative, are not protected from added proteases as a consequence of association with PFD, and cannot be released from PFD by incubation with ATP. The nascent actin–PFD complex likely persists until the synthesis of actin is complete. The full-length actin bound to PFD is then transferred to CCT where folding to the native state occurs. We show that a similar maturation pathway is operative for both α- and β-tubulin.
Materials and Methods
Materials
Rabbit reticulocyte lysates were prepared and treated with micrococcal nuclease (Calbiochem-Novabiochem Corp.) as described in Jackson and Hunt (1983). Dr. D. Melton (Harvard University, Cambridge, MA) provided the SP6 RNA polymerase expression plasmid pSP64T (Krieg and Melton, 1984). Edeine was the gift of Dr. H. Lodish (Massachusetts Institute of Technology, Cambridge, MA). SP6 RNA polymerase and RNase inhibitor (RNASIN) were from Promega Corp. T7 RNA polymerase was from New England Biolabs. [35S]Methionine (Translabel, 1,000 Ci/mMol) was obtained from New England Nuclear. Apyrase and soybean trypsin inhibitor were from Sigma Chemical Co. Rabbit polyclonal anti-PFD subunit 6 (PFD 6) was described previously (Vainberg et al., 1998). A purified rat mAb, 23C, recognizing TCP-1α, was used for the analysis of CCT (Lewis et al., 1992). Purified rabbit skeletal muscle actin was obtained from Cytoskeleton Inc. AffiGel-10 was from Bio-Rad. DNase I was from Worthington Biochemicals.
In Vitro Expression Vectors
The sequence at the initiator methionine codon of chicken βII-tubulin was modified to create a NcoI site by Dr. H. Sternlicht (Case Western Reserve University, Cleveland, OH). The β-tubulin, human β-actin (Ponte, 1984), and Aequorea victoria green fluorescent protein (Chalfie et al., 1994) coding sequences were shuttled into the pSP64T derivative, pSPBP4, replacing the preprolactin gene. pSPBP4 contains the SP6 promoter, the β-globin 5′ untranslated region, an NcoI site encoding an initiation codon with an optimal Kozak consensus sequence (ACCATGGG; Kozak, 1984), the gene for bovine preprolactin, and a polylinker, in that order. The expression plasmids used to produce yeast cytosolic invertase, α-tubulin, and firefly luciferase (pGEM-luc) mRNAs were described previously (Ngsee et al., 1989; Gao et al., 1993; Promega Corp.).
In Vitro Transcription and Translation
DNA templates were transcribed after complete linearization with the appropriate restriction enzymes to create full-length and the desired truncated coding regions. GpppG capped mRNA templates (Krieg and Melton, 1984) generated by in vitro transcription using SP6 RNA polymerase as described previously (Hansen et al., 1986) were used without purification. Transcription with T7 RNA polymerase was performed in 40 mM Tris-HCl, pH 7.9, 2 mM spermidine, 10 mM DTT, 12.4 mM MgCl2, 2 mM each of ATP, CTP, UTP, and GpppG, and 0.4 mM GTP at 37°C for 90 min. To promote full-length transcription, the GTP concentration was then increased to 2 mM and the incubation continued for 15 min. T7 transcripts were precipitated with ethanol and resuspended in diethylpyrocarbonate (DEPC) treated water. Rabbit reticulocyte translation reactions (40% lysate) were performed at 22–24°C for the times indicated in 20 mM Hepes, pH 7.4, 100 mM KCl, 2 mM MgCl2, 2 mM dithiothreitol, 0.2 mM spermidine, and 10 mM S-adenosyl-methionine. In the experiment shown in Fig. 3, edeine (10 mM) and 7-methylguanosine monophosphate (7-MeGMP; 4 mM) were added as follows: after 5 min of translation for −mRNA (no added mRNA) and 99 aa actin; after 6 min for 123 aa, 145 aa, 156 aa, and 257 aa actin; and after 9 min for 336 aa and full-length actin. Aliquots of the reactions were removed for analysis as follows (e, early; l, later): e = 8 min, l = 20 min for −mRNA; e = 8 min, l = 12 min for 99 aa actin; e = 9 min, l = 16 min for 123 aa actin; e = 9 min, l = 20 min for 145 aa and 156 aa actins; e = 12 min, l = 25 min for 257 aa actin; e = 12 min, l = 70 min for 336 aa and full-length actins.
Figure 3.
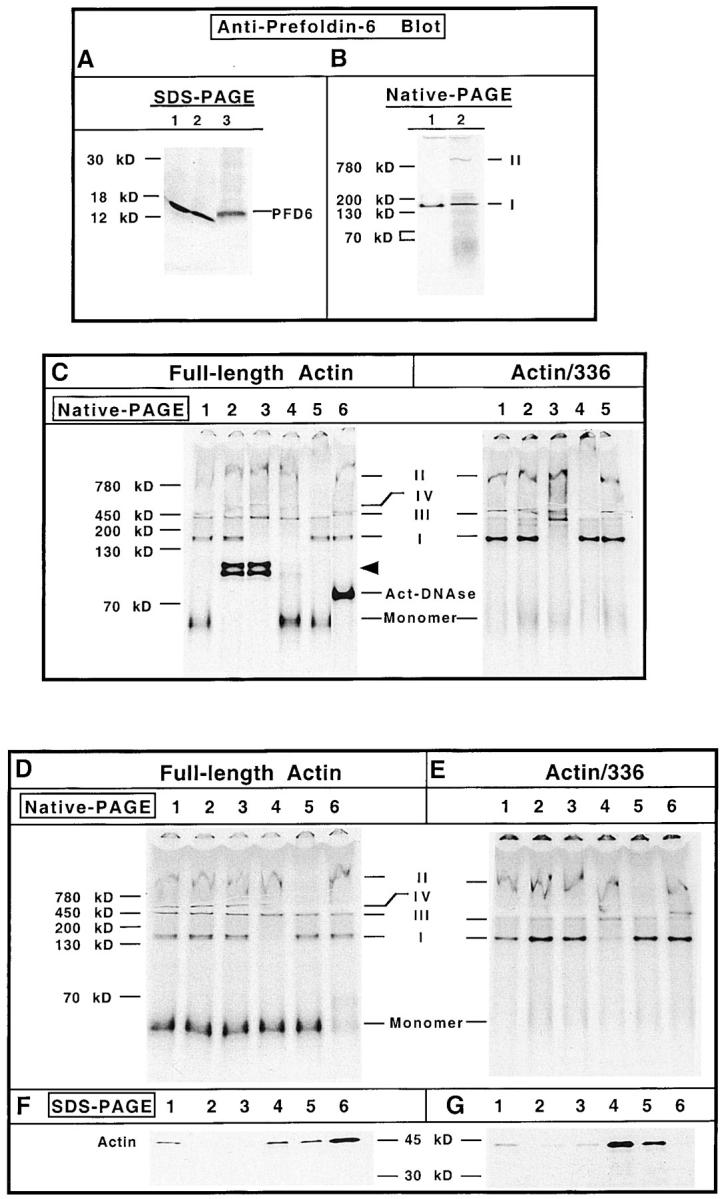
Actin species I contains PFD. (A) HeLa cell lysate or rabbit reticulocyte lysate was examined for content of PFD 6 by SDS-PAGE and Western blot analysis. Lane 1, HeLa cells grown at 37°C; lane 2, HeLa cells 12 h after a 43°C/60 min heat shock treatment; lane 3, rabbit reticulocyte lysate. The position of PFD 6 is shown on the right of the panel. (B) Purified bovine PFD (lane 1) and in vitro translated [35S]methionine-labeled actin (lane 2) were analyzed by native-PAGE, the proteins transferred to nitrocellulose, and the position of purified PFD was determined by immunoblot using the PFD 6 antibody. After extensive washing, the nitrocellulose was placed on film and the position of actin species I and II revealed by autoradiography. The positions of actin species I and II are indicated on the right. (C) Identification of actin-containing complex components by electrophoretic mobility shift assays. Full-length actin mRNA (left) was translated for 15 and 50 min. The reactions were mixed together to create a pool of all actin-containing species. The mRNA encoding 336–amino acid actin was translated for 30 min (right). Before native-PAGE analysis, equal aliquots of the reaction mixtures were incubated with: PBS (control, lane 1); preimmune antiserum (lane 2); anti–PFD 6 serum (lane 3); anti–PFD 6 serum supplemented with purified actin (lane 4); purified anti-CCT mAb (lane 5); and DNase I (lane 6). DNase shift (lane 6) was omitted for the 336–amino acid actin translation products. The different protein complexes were then analyzed by native-PAGE. A fluorogram of the gel is shown. Molecular mass markers are shown at the left, and the positions of the different actin complexes are indicated in the center. The arrowhead indicates the shift in full-length actin migration due to the presence of actin-binding proteins present in the crude rabbit antisera. (D and F) Identification of actin-containing complex components by immunodepletion with immobilized antichaperone specific antibodies and DNase I. The full-length [35S]methionine-labeled actin translation reaction products (C) were incubated with antichaperone antibodies first bound to protein A–Sepharose, or with DNase I coupled to Affigel-10. After incubation, the samples were clarified and the corresponding supernatants analyzed for the presence of the different actin species by native-PAGE as shown in D. In parallel, the corresponding pellets containing the immobilized antibodies or DNase I were resuspended in Laemmli sample buffer and analyzed for their relative content of radiolabeled actin by SDS-PAGE as shown in F. An aliquot of the in vitro translation products (i.e., starting material) is shown in lane 1; protein A–Sepharose, lane 2; immobilized preimmune antibodies, lane 3; PFD 6 antibody, lane 4; anti-CCT antibody, lane 5; immobilized DNase, lane 6. The positions of the different actin complexes are indicated in the center. (E and G) The [35S]methionine-labeled 336–amino acid actin translation reaction products (C) were incubated with the immobilized antichaperone antibodies or immobilized DNase. Subsequently, the samples were clarified and the supernatants and pellets analyzed by native-PAGE and SDS-PAGE, respectively. Lane designations are the same as in D and F.
Isolation and Analysis of Ribosome-associated Nascent Chain–Chaperone Complexes
The reticulocyte lysate was programmed with mRNA encoding the 336– (or 257–) amino acid NH2-terminal fragment of actin and incubated at 24°C for 8 min. Edeine (10 mM) and 7-MeGMP (4 mM) were added to inhibit new chain initiation. The reaction was further incubated for 9 min (7 min for the 257 amino acid actin) to allow for complete translation of the mRNA template. The nascent chain–ribosome complexes were stabilized by addition of 0.5 mM cycloheximide, ATP levels were depleted by incubation for 10 min at 24°C with apyrase (50 U/ml) and 25 mg/ml soybean trypsin inhibitor. The mixture was applied to a gradient of sucrose consisting of 0.2 ml of 68% sucrose overlayered with 0.5 ml 20% sucrose, both containing 20 mM Hepes, pH 7.4, 2.1 mM MgCl2, 100 mM KCl, 2 mM reduced glutathione, 0.1 mM EDTA, and 0.2 mM cycloheximide. The gradient was centrifuged in a Beckman TLA100 SW55 ultracentrifuge rotor at 50,000 rpm for 90 min at 4°C. After centrifugation, the gradient was fractionated from the top. An aliquot of each gradient fraction was incubated for 10 min with 0.5 mM puromycin and 20 μg/ml RNase A, and then analyzed by Tricine SDS-PAGE. RNA was isolated from each of the gradient fractions by extraction with phenol/chloroform and precipitation with ethanol. The RNA samples were suspended in DEPC-treated water and denatured by incubation at 60°C in 7 M urea RNA sample buffer. The RNA was fractionated by 5% PAGE containing 7 M urea, the gel stained with ethidium bromide, and the UV transilluminated image photographed. The tRNA and high molecular weight ribosomal RNAs recovered from each fraction were quantified using NIH Image software. For native-PAGE analysis, aliquots of the starting materials applied to the gradient and of each gradient fraction were incubated with puromycin (0.5 mM), apyrase (50 U/ml), and soybean trypsin inhibitor (25 mg/ml) for 10 min at room temperature, followed by digestion with RNase A (20 μg/ml) in native-PAGE sample buffer for 10 min before analysis by native-PAGE as described below.
SDS-PAGE
SDS-PAGE analysis was performed using the gel formulations of Blattler et al. (1972) or when indicated, a Tris/Tricine system (Schagger and von Jagow, 1987). When peptidyl-tRNA species were to be examined, the translation products were heated in Laemmli sample buffer, pH 6.8 (Laemmli, 1970). tRNA free polypeptides were analyzed after the peptidyl-tRNA species were dissociated by incubation with 0.5 mM puromycin at room temperature for 10 min, then heated in SDS-PAGE loading buffer, pH 11.3.
Native-PAGE
The discontinuous gradient native gels used (Schagger and von Jagow, 1991) were composed of two (5.5% on top of 12.5% for Figs. 1, 2 A, and 7) or three layers (5.5% on top of 12.5% on top of 15% for Figs. 2 C, 3, 4, and 5). The upper chamber (cathode) buffer consisted of 50 mM Tricine and 15 mM BisTris, pH 7.0. The lower chamber (anode) buffer chamber contained 50 mM BisTris, pH 7.0. Reaction mixtures were adjusted with native gel loading buffer to final concentrations of 50 mM BisTris, pH 7.0, 5% glycerol, 2 mM reduced glutathione, 1 mM each methionine and cysteine, and a small amount of tracking dye, Ponceau S. The gels were run initially at 50 V for 20 min (∼5 mA), and then with a maximum current of 13 mA at 4°C until the tracking dye migrated to the bottom of the gel. The position of migration of native molecular mass standards is indicated in each figure: soybean trypsin inhibitor, 21 kD; BSA monomer, 70 kD; BSA dimer, 130 kD; BSA trimer, 200 kD; ferritin, 450 kD; and α2-macroglobulin, 780 kD. Proteins were visualized either by Coomassie blue staining or by fluorography. Radiolabeled proteins (where indicated) were quantified by scanning the x-ray film image and processing with NIH Image software.
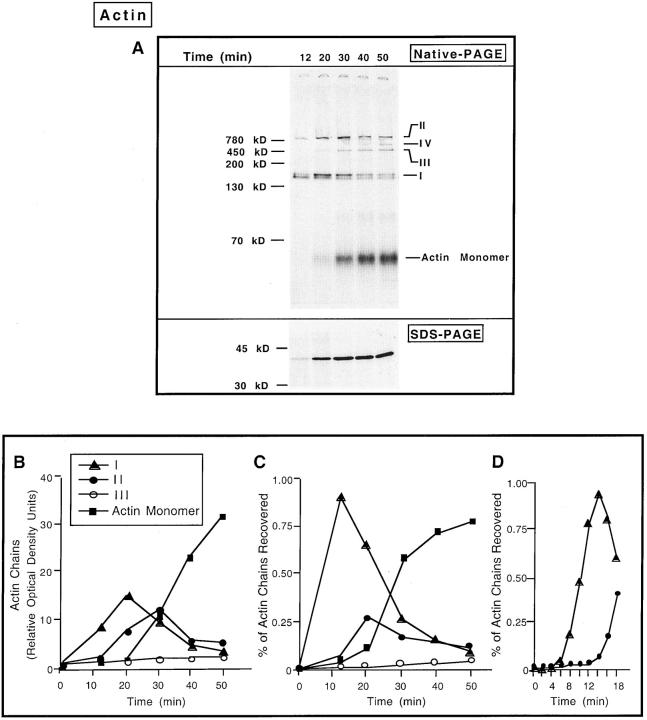
Figure 1.
Intermediates in the biogenesis of full-length β-actin. (A) The reticulocyte lysate was programmed with full-length mRNA encoding β-actin in the presence of [35S]methionine. Edeine (10 mM) and 7-MeGMP (4 mM) were added after 12 min to block new initiation of translation. Aliquots were removed at various times (indicated at the top of the panel), the samples incubated with apyrase (to deplete ATP) and puromycin (to release nascent chains), and analyzed by native-PAGE (top). The migration of native molecular mass markers is shown on the left. The positions of migration of actin monomer along with four other actin-containing species (I, II, III, and IV) are shown on the right. The same samples, analyzed by SDS-PAGE, are shown in the lower panel. The amount of total material applied to the native gel is twice that loaded on the SDS-PAGE. Fluorograms of the gels are shown after 2.5 h of exposure. (B) Kinetics of appearance of the species detected after native-PAGE (species I, II, III, and monomeric actin). The optical density of each band from the x-ray film was calculated with NIH Image software. The optical density units derived for each species are plotted as a function of time. (C) Kinetics of appearance of the species detected after native-PAGE, plotted as follows: optical density data from B for each species were divided by the sum of the optical density units of all the radiolabeled species recovered in the gel for a given time point. The percentage of each actin species (I, II, III, and monomeric actin) is plotted as a function of time. (D) To examine the earliest forms of nascent actin, actin mRNA was translated for 2 min, and then edeine and 7-MeGMP were added. Aliquots were removed every 2 min and the actin-containing complexes fractionated by native-PAGE. The percentages of actin chains in species I or II were determined as described in C.
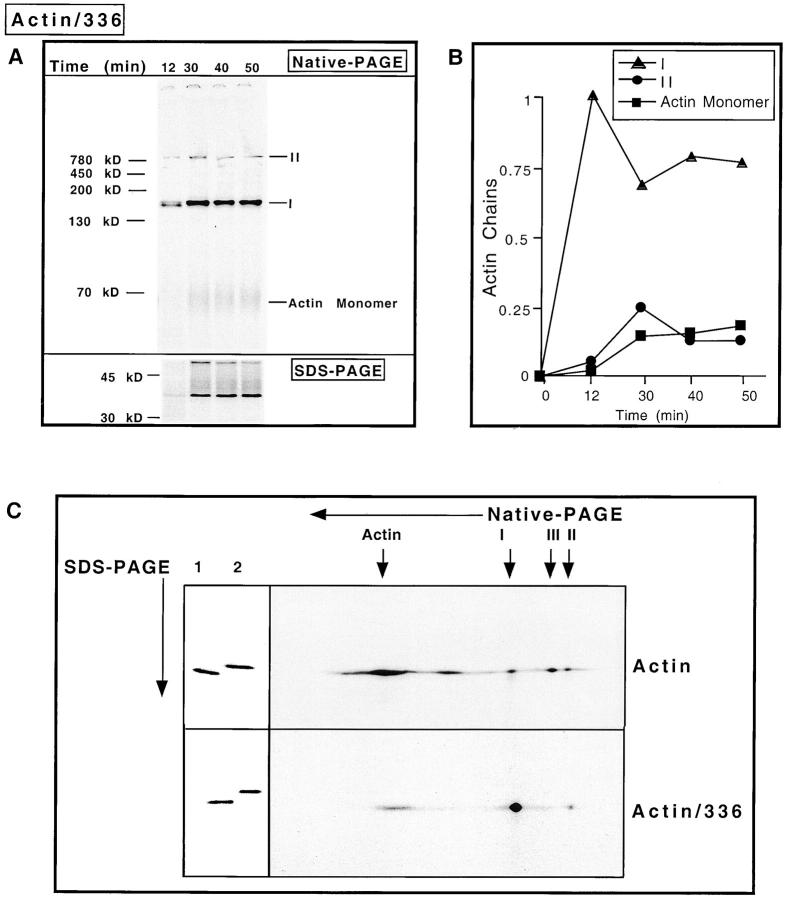
Figure 2.
COOH-terminal truncated actin accumulates as the species I complex. (A) The reticulocyte lysate was programmed with mRNA encoding a 336–amino acid NH2-terminal fragment of actin (full-length actin is 374 amino acids) and the reaction products analyzed as in Fig. 1 A. The amount of total material applied to the native-PAGE (top) is twice that of the amount loaded on the SDS-PAGE (bottom). Fluorograms of the gels are shown after 2.5 h of exposure. The migration of native molecular mass markers is shown on the left and the actin species shown at the right. (B) Kinetics of appearance of the species detected after native-PAGE was plotted as follows: optical density value derived for each species was divided by the sum of the optical density units of all the radiolabeled species recovered in the gel for a given time point. The percentage of each actin species (I, II, and monomeric actin) is plotted as a function of time. (C) In vitro translation products of either full-length actin mRNA (Actin), or the 336–amino acid actin truncation mRNA (Actin/336) were analyzed in the first dimension by native-PAGE. The lane containing the reaction material was excised and then analyzed by SDS-PAGE as described in Materials and Methods. The starting materials, actin/336 or full-length actin, were loaded in lanes 1 and 2, respectively (left). Fluorograms of the gels are shown.
Assay for Altered Mobility Induced by Antibody or Ligand and Immunodepletion
Antibody Binding Induced Shift.
Positive identification of components present within nascent chain–chaperone complexes was performed by an electrophoretic mobility shift assay with the nascent chain serving as the radiolabeled probe. Antibodies to the chaperone protein of interest or control antibodies (e.g., preimmune serum) were added to the puromycin-released and ATP-depleted reticulocyte lysate translation products. After incubation for 15–20 min at room temperature, native gel sample buffer was added and the samples analyzed by native-PAGE.
Immunodepletion and DNase I–Sepharose Binding.
The puromycin-released and ATP-depleted reticulocyte lysate translation products were mixed with antibodies (preimmune, anti–PFD 6, or anti–TCP-1α) previously bound to protein A–Sepharose or immobilized DNase I in native gel sample buffer containing 100 μg/ml BSA. After 45 min at 4°C with occasional mixing, the Sepharose beads were removed by centrifugation and the supernatant solution was analyzed by native-PAGE. Sepharose beads were washed four times in PBS, the bound proteins eluted by heating in Laemmli sample buffer, and analyzed by SDS-PAGE.
DNase I Binding Induced Shift.
In vitro translated actin was incubated in the reticulocyte lysate supplemented with 400 μg/ml of DNase I at room temperature for 15 min. Native-PAGE loading buffer was added and the mixture analyzed by native-PAGE.
Two-Dimensional Native/Denaturing-PAGE
Translation reaction products were fractionated by native-PAGE as described above. The lanes were excised from the gel and incubated for 10 min in two times concentrated Laemmli sample buffer at 60°C. The entire gel lane then was applied to and electrophoresed through a second dimension of 12.5% SDS-PAGE.
Results
Order of Events in the Actin Folding Pathway
The pathway of actin maturation was investigated using translation in vitro. Full-length actin mRNA was translated in the presence of [35S]methionine and the reaction products analyzed by both SDS-PAGE and native-PAGE. In the first experiment, full-length actin mRNA was translated for 12 min. Inhibitors of translational initiation (edeine and 7-MeGMP) were added and the translation reactions continued to allow for polypeptide chain elongation and completion. At various times, aliquots were removed and incubated with apyrase (to deplete ATP levels and thereby stabilize chaperone–nascent chain complexes) and puromycin (to release unfinished nascent chains from the ribosome). Analysis of the reaction products by SDS-PAGE revealed a small amount of full-length actin after 12 min of translation (Fig. 1 A, SDS-PAGE). Maximal amounts of full-length actin had accumulated between 20 and 30 min. Native-PAGE analysis showed that these reaction products consisted of full-length monomeric actin together with at least four actin-containing complexes with apparent masses between 150 and 800 kD (indicated as I, II, III, and IV in Fig. 1 A, native-PAGE). The monomeric actin species had acquired its native folded state as evidenced by its ability to interact with DNase I, an assay used to monitor the folding status of actin (see Fig. 3, C and D; Lazarides and Lindberg, 1974).
Quantitation of the different actin complexes accumulating over time (i.e., native-PAGE in Fig. 1 A) was performed and is presented in two different ways in Fig. 1, B and C. Such analyses showed that the ∼150-kD complex (species I) was the first complex formed. Subsequently, the ∼800-kD complex (species II) was observed to increase. Finally, species III and IV were found to form later in time. Based on its apparent mass of ∼800 kD, species II represents actin bound to the CCT, which has been described by others. The nature of the other complexes, in particular species I, is not known and is the major focus of the present study. Note that by 40 min the levels of both species I and II had markedly diminished, and that full-length monomeric actin was now the predominant species. Curiously, the levels of the ∼500-kD complex (species III in Fig. 1 A) did not change significantly over the course of the last 20–30 min of translation. Given their low levels and relatively late appearance (i.e., they rise only after the native full-length actin monomer is present), it is unlikely that species III and IV represent intermediates in the pathway of actin monomer folding. Hence, we will restrict our focus here to the characterization of species I and II.
In a second experiment of this type, full-length actin mRNA was translated for only 2 min before addition of the inhibitors of translation. Analysis of the reaction products over the course of the next 16 min via native-PAGE revealed that the formation of actin species I preceded that of species II, the actin–CCT complex (Fig. 1 C).
If the actin chains in species I were engaged with a molecular chaperone component, then an actin molecule unable to attain the native conformation might be expected to display a longer residence time with its particular chaperone. Previously, it has been shown that removal of the last 20 amino acids of yeast actin, which are essential for holding together the three noncontiguous strands of subdomain I, resulted in the loss of actin's structural integrity (Wertman et al., 1992; Xia and Peng, 1995). To determine whether actin would accumulate in the species I complex if the acquisition of the native state was prevented, an mRNA lacking a stop codon and encoding an actin chain lacking the COOH-terminal 38 amino acids was generated and translated in vitro. In this experiment, synthesis of the NH2-terminal 336 amino acids will proceed (full-length actin is 374 amino acids), but in the absence of a translational stop signal the nascent chain remains bound to the ribosome. Addition of puromycin should then result in the release of nascent chains and allow for the analysis of the mass of the nascent actin chain. Translation of the truncated actin mRNA was allowed to proceed for 12 min in the presence of [35S]methionine before the addition of translational initiation inhibitors. At various times thereafter, aliquots were removed, puromycin was added, ATP levels depleted via addition of apyrase, and the material analyzed by native-PAGE (Fig. 2 A). Actin species I was the first and most predominant species observed. Only small amounts of the actin–CCT complex (species II) were found. Subsequent to the appearance of species I and II, low levels of the truncated actin molecule were observed to migrate near the bottom of the gel, presumably having folded into some type of monomeric-like species. Quantitation of the different actin-containing species present in Fig. 2 A revealed that the majority of the truncated actin molecule accumulated as the species I complex (Fig. 2 B).
Two-dimensional gel analysis (native-PAGE in the first dimension followed by SDS-PAGE in the second dimension) revealed that full-length actin could be found within all of the species (Fig. 2 C). However, in the case of the 336–amino acid actin translation product, the vast majority of the truncated actin chains was present within species I. Here, very little of the truncated actin chains were found present within the CCT containing complex, species II.
The Actin Species I Complex Contains PFD
A heteromeric chaperone termed PFD has been described that can form a complex with chaotrope denatured full-length actin, and then deliver this target to CCT where folding to the native state commences (Vainberg et al., 1998). To see whether species I might represent actin bound to PFD, an antibody to the bacterially expressed human PFD 6 was prepared and characterized. When an immunoblot of HeLa cell and rabbit reticulocyte lysate proteins was probed with the anti–PFD 6 antibody, a single polypeptide of the appropriate size for the PFD 6 species (∼14 kD, as described by Vainberg et al., 1998) was detected (Fig. 3 A).
We compared the native-PAGE migration of purified PFD with that of actin species I and II produced during the in vitro translation reaction. Following native-PAGE, the proteins were transferred to nitrocellulose and probed with the anti–PFD 6 antibody. After the enhanced chemiluminescence (ECL) image was developed, the nitrocellulose was extensively washed (to remove the ECL reagents), dried, and then reexposed to film to detect the [35S]methionine-labeled actin species. As shown in Fig. 3 B, the PFD complex recognized by the antibody (lane 1) migrated in a manner similar to that observed for radiolabeled actin species I (lane 2). These observations are consistent with the idea, but do not prove, that species I consists, at least in part, of actin bound to PFD.
Gel mobility shifts and immunodepletion experiments were used to further confirm the identity of species I and II. The migration of the different actin-containing species was unaffected when the material was incubated with PBS (Fig. 3 C, lane 1). Incubation of this material with preimmune rabbit serum affected only the migration of native monomeric actin (Fig. 3 C, lane 2), likely due to the presence of the actin-binding proteins gelsolin and vitamin D–binding protein within the crude rabbit polyclonal serum (Lee and Galbraith, 1992). Presumably the interaction of full-length actin with these two different serum proteins accounts for the doublet observed. When the anti–PFD 6 rabbit antiserum was added to the translation reaction the full-length monomeric actin species was gel shifted (due to the presence of the actin-binding proteins gelsolin and vitamin D–binding protein). Note, however, that the species I band now also disappeared upon addition of anti-PFD serum (Fig. 3 C, lane 3). Thus, species I contains actin and PFD. If the PFD 6 antiserum was first preincubated with purified native actin, the mobility of the radiolabeled native actin monomer was no longer altered, yet species I still disappeared (Fig. 3 C, lane 4). Thus, addition of native actin was sufficient to titrate all of the native actin-binding proteins present within the crude rabbit sera. When a purified mouse mAb to the CCT α subunit (antibody 23C) was added, the large (∼800-kD) species II complex disappeared (Fig. 3 C, lane 5). Finally, the radiolabeled actin monomer produced in these reactions was observed to gel shift upon addition of DNase I, demonstrating that this actin species had indeed reached the native state (Fig. 3 C, lane 6).
Analysis of the 336–amino acid actin translation products again revealed that the actin species I shifted its gel mobility only upon addition of the PFD 6 rabbit serum (Fig. 3 C, lane 3). In addition, the antibody to CCT resulted in the disappearance of the larger actin species II (Fig. 3 C, lane 4). Note that the addition of either preimmune serum (Fig. 3 C, lane 2) or DNase I (Fig. 3 C, lane 5) now had no effect on the position of any of the species in the gel, consistent with there being no properly folded full-length actin present.
We confirmed the results of the mobility shift experiments using the different antibodies or DNase first immobilized on Sepharose beads. For example, incubation of the translation reactions with immobilized anti-PFD serum again resulted in the disappearance of the actin I species (Fig. 3 D, lane 4). Analysis of the material bound to the immobilized PFD antiserum by SDS-PAGE confirmed the presence of radiolabeled actin (Fig. 3 F, lane 4). Immobilized antibodies to CCT effectively resulted in the disappearance of actin species II (Fig. 3 D, lane 5) while the immobilized DNase effectively removed all monomeric actin (Fig. 3 D, lane 6). In each case the immobilized CCT antibody or DNase beads now contained significant amounts of the radiolabeled actin species (Fig. 3 F, lanes 5 and 6).
Very similar results were obtained when the immunodepletion analyses were performed on the 336 actin translation product. Again, immobilized antibodies to PFD and CCT resulted in the specific depletion of species I and II, respectively (Fig. 3 E, lanes 4 and 5). Note that none of the 336–amino actin translation product was captured by the immobilized DNase, indicative that this actin truncation product is unable to adopt its native conformation (Fig. 3 G, lane 6).
Interestingly, the ∼500-kD complex we termed species IV, which only formed when full-length actin mRNA was translated (see Fig. 1 A), was also somewhat depleted upon incubation with the anti-CCT antibody (most noticeable in Fig. 3 D, lane 5). Actin bound to this smaller complex containing at least one of the subunits of CCT appears to be native or native-like, since the immobilized DNase I also resulted in the disappearance of actin species IV (Fig. 3 D, lane 6). Further work will be necessary to characterize this species, as well as the species we have labeled species III. Taken together, these observations demonstrate that species I consists of actin bound to PFD, while actin species II represents actin bound to the CCT.
PFD Binds Actin Chains after Synthesis of the NH2-terminal 130–140 Amino Acids
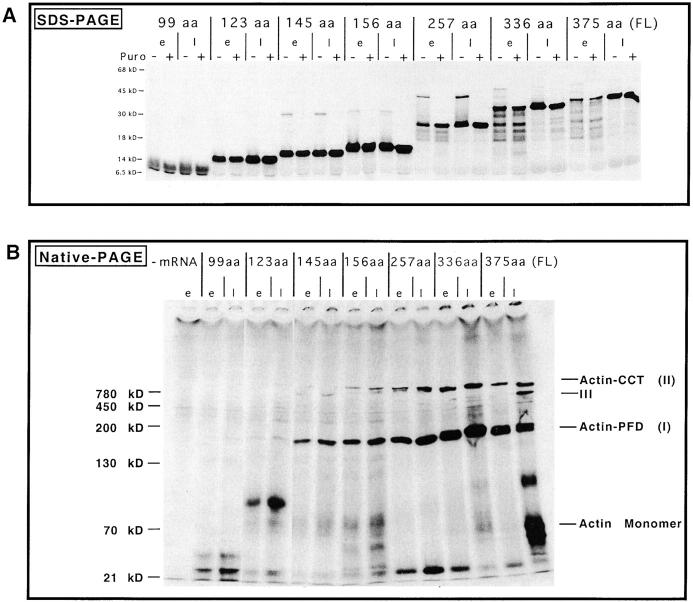
Experiments were designed to determine whether there was a minimum length of the nascent actin chain required to interact with PFD. A series of shorter actin mRNA truncations was translated in vitro, and the products analyzed by native- and denaturing-PAGE. Time points were taken early after the addition of inhibitors of translational initiation, or at a later time when translational elongation was complete (Fig. 4, e, early; or l, late). In each case, the reactions were divided: to one half puromycin was added to release nascent chains from the ribosome, while to the other half SDS-containing buffer was added to strip the nascent chains off the ribosome (Fig. 4, + or − puromycin, respectively). Analysis of the reaction products by SDS-PAGE revealed relatively homogeneous populations of those actin nascent chains containing ≤156 amino acids (Fig. 4 A). In contrast, those reactions programmed with mRNAs encoding longer actin molecules produced significant heterogeneity in the nascent chains early during translation, suggestive of ribosomal pausing during reading of the mRNA (e.g., 336–amino acid actin). Note that in those reactions where puromycin was not added, a portion of the truncated actin molecules migrated very slowly in the SDS gel (e.g., bands migrating at ∼45 kD in translation reactions programmed with the 257 amino acid actin mRNA in Fig. 4 A). These species probably represent actin chains (∼20–25 kD) still bound covalently to tRNA (∼25 kD).
Figure 4.
Native-PAGE analysis of different COOH-terminal truncated actin polypeptides. Reticulocyte lysate was programmed with either full-length actin mRNA or a number of progressively shorter mRNA species encoding COOH-terminal deletions. The predicted number of amino acids translated is shown. Translations were initiated in the presence of [35S]methionine, and then edeine and 7-MeGMP were added to block the initiation of new chains at different times (see Materials and Methods). Aliquots were taken either early (e) or later (l) after edeine and 7-MeGMP addition, to visualize the different species of nascent actin polypeptides. Note that for the SDS-PAGE analysis (A) each reaction was divided: one half was untreated (−), while puromycin and RNase were added to the other half to release nascent chains (+). Products were analyzed by SDS-PAGE. Analysis of reactions via native-PAGE (B), puromycin was added to each of the samples and the material applied to the gel. Shown are fluorographs of the gels, with molecular mass markers indicated on the left. The positions of the different actin species, as well as monomeric actin, are shown at the right in B.
Native-PAGE analysis of the reaction products obtained from the different truncated actin mRNAs revealed when the actin–PFD complex began to form (Fig. 4 B). Translation of the first 99 or 123 amino acids of actin followed by release of nascent chains with puromycin yielded no detectable actin–PFD complex. Interestingly, puromycin release of the 99 amino acid truncation product resulted in the fragment migrating in the gel with an apparent mass of ∼20–25 kD, while the 123 amino acid actin translation product migrated with an apparent mass of ∼100 kD. It is unclear whether these two actin fragments are interacting with one or more unknown proteins, but gel shift experiments using antibodies to CCT or PFD had no effect on the native-PAGE migration of these species (data not shown). The actin–PFD complex was detected only after translation of the first 145 amino acids, thus defining a minimal requirement for binding of the nascent actin chain to the chaperone. All longer actin truncation products were also found preferentially bound to PFD. A fraction of the 257–amino acid truncated actin molecule apparently had released from PFD (note those species of ∼25 kD at the bottom of the native-gel). Presumably, a portion of these chains forms some native-like structure that is not recognized by either CCT or PFD present in the lysate. Relatively low amounts of the different truncated actin products were found in the ∼800-kD CCT complex (actin–CCT).
The Actin–PFD Complex Forms Cotranslationally
The data presented to this point are consistent with the idea that PFD interacts with nascent actin after synthesis of the first 130–145 amino acids. It remained possible, however, that complex formation between actin and PFD may occur in the reticulocyte lysate only after release of the nascent actin chains from the ribosome. To determine whether PFD can bind actin cotranslationally, the 336– and 257– amino acid actin truncations were translated, cycloheximide was added to stabilize the nascent chains, ATP was depleted, and polysomes were isolated by sedimentation through sucrose. The fractions from the polysome gradient were analyzed by SDS-PAGE and native-PAGE. In addition, RNA analysis was done to ensure that we had isolated an enriched polysome population. As shown in Fig. 5 A, the majority of the polysome-bound 336 amino acid actin translation product (marked by the bracket) was found near the bottom of the gradient (i.e., in fractions 6 and 7, these being enriched in high molecular weight RNA). The absence of endogenous globin chains (asterisk) and tRNAs in these fractions indicated that contamination of the polysome fraction by cytosol was low. When released from the purified polysomes by puromycin, the 336 actin translation product again migrated in the native gel with a native molecular mass of ∼150 kD (i.e., similar to that observed when the chains were released in the context of whole reticulocyte lysate; Fig. 5 B).
Figure 5.
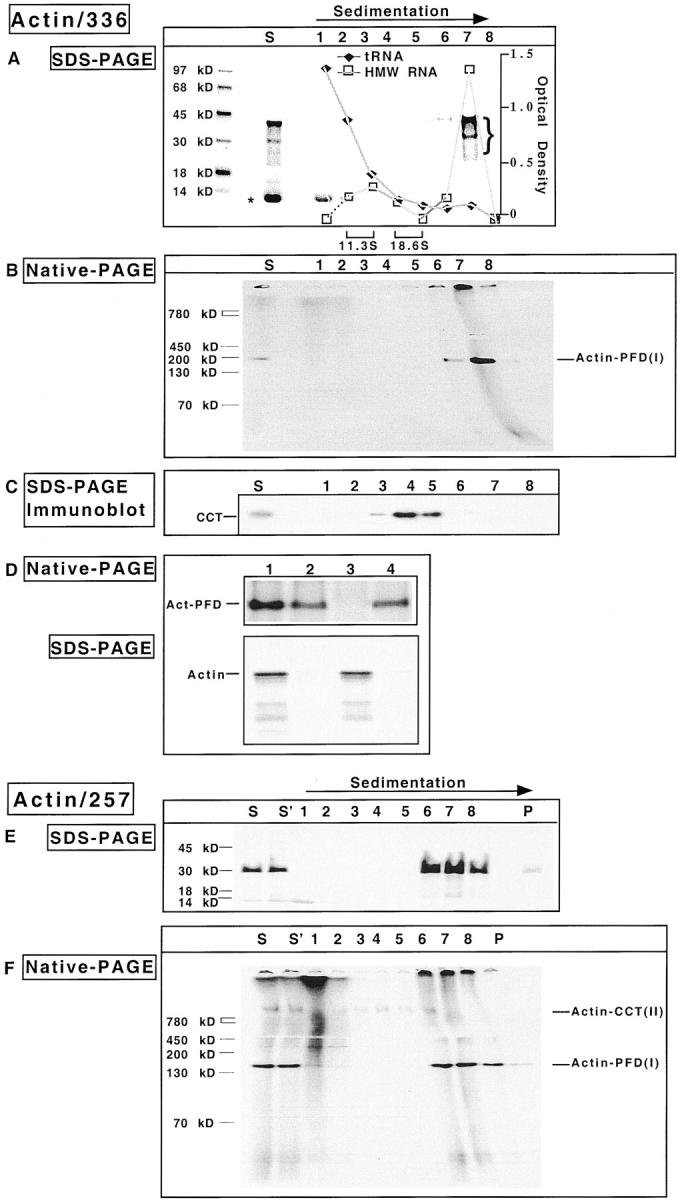
The actin–PFD complex forms cotranslationally. Reticulocyte lysate was programmed with mRNA encoding either a 336– or a 257–amino acid NH2-terminal fragment of actin. After 8 min, edeine and 7-MeGMP were added, and the reaction continued for an additional 9 min. Nascent chain–ribosome complexes were stabilized by addition of 0.5 mM cycloheximide, and polysomes isolated by gradient centrifugation. Aliquots from each fraction were incubated with puromycin to release nascent chains from the ribosome. (A and B) SDS-PAGE and native-PAGE analyses of the gradient fractions obtained from the 336–amino acid actin translation product. S denotes the starting material for the gradient. Shown are fluorograms of the gels with molecular mass markers on the left. The position of sedimentation of the standards catalase (11.3 S) and α2-macroglobulin (18.6 S) are indicated at the bottom of A. (C) The proteins in the starting material for the gradient and each of the gradient fractions were analyzed for their content of CCT by an immunoblot using anti-CCT antibodies. (D) The polysome-bound actin complex contains PFD. The purified polysomes shown in A and B (lane 7) were treated with puromycin and apyrase, and then incubated at 4°C for 30 min with immobilized preimmune antibody, immobilized PFD antibody, or DNase I coupled to Sepharose 4B. After clarification, the supernatant fractions from each of the reactions were analyzed by native-PAGE (top). The antibody beads were washed four times with PBS and the material retained by the beads was eluted by heating in Laemmli sample buffer, then analyzed by SDS-PAGE (bottom). The starting material is shown in lane 1; immobilized preimmune antibodies, lane 2; immobilized anti-PFD antibody, lane 3; immobilized DNase I, lane 4. The position of actin PFD is indicated on the left. (E and F) SDS-PAGE and native-PAGE analysis of the gradient fractions obtained from the 257–amino acid actin translation product, performed as in A and B. S denotes the starting material for the gradient which was immediately frozen while S′ denotes the starting material which was stored at 4°C during the centrifugation run.
Immunoblot analysis of the gradient fractions using anti-CCT mAbs revealed that the majority of the ribosome-bound nascent actin chains (fractions 6 and 7) was resolved from CCT (fractions 4 and 5) during the centrifugation (Fig. 5 C).
To verify that the 150-kD actin containing complex released from the isolated ribosomes contained PFD, an immunodepletion experiment was performed. Incubation of the puromycin-released complexes with immobilized anti– PFD 6 resulted in the complete depletion of the truncated actin chains as shown by native-PAGE analysis (Fig. 5 D, lane 3). Consistent with this observation was the capture of the labeled actin by the PFD antibody beads as determined by SDS-PAGE (bottom of Fig. 5 D, lane 3). Note that the immobilized preimmune antibodies or DNase I slightly reduced the amount of species I (top of Fig. 5 D, lanes 2 and 4). This likely is due to nonspecific binding, since the beads did not retain the radiolabeled actin chain after extensive washing (bottom of Fig. 5 D, lanes 2 and 4).
Similar results were obtained with the 257–amino acid actin truncation product (Fig. 5, E and F). Specifically, the nascent actin chains, upon their puromycin-mediated release from the polysomes, migrated in the native gel with an apparent mass of 150 kD. Again, this material could effectively be removed by preincubation with the PFD antibody, but not with antibodies specific for CCT (data not shown). We conclude that PFD interacts with nascent actin as the actin polypeptide is being synthesized on the ribosome.
Nascent Actin Chains Complexed with PFD Are in a Nonnative State
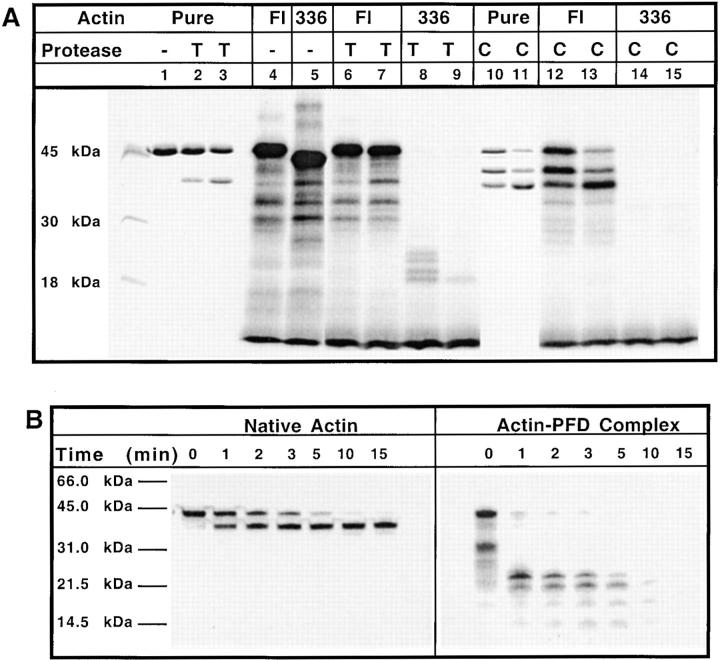
Previous work has shown that nascent chains bound to hsp70 are inaccessible to low levels of added protease unless the hsp70–nascent chain complex is first dissociated by incubation with ATP. In the absence of ATP, nascent chains are sequestered from the bulk solution (Hansen et al., 1994; Eggers et al., 1997). We found that the actin–PFD complex, present within the whole rabbit reticulocyte lysate, was insensitive to added ATP (data not shown). Moreover, protease protection experiments revealed that little or no protection was provided to nascent actin chains bound to PFD (Fig. 6 A). For example, under conditions where full-length actin exhibited relative resistance to added trypsin or chymotrypsin, the 336–amino acid translation product (which consists largely of actin molecules bound to PFD; see Fig. 2) was rapidly and completely digested (Fig. 6 A). In a second experiment, labeled full-length actin was denatured in 8 M urea, the target protein presented to purified PFD by sudden dilution, and the actin–PFD complex then isolated by gel filtration. The isolated actin–PFD complex, as well as native actin, was subjected to protease K treatment for varying times and the reaction products analyzed by SDS-PAGE (Fig. 6 B). The full-length actin molecules were only partially cleaved by the added protease. In contrast, actin bound to PFD was highly susceptible to the added protease. These results, together with our observation that actin–PFD complexes are unable to bind to DNase I, indicate that nascent actin chains bound to PFD are in a relatively solvent exposed, nonnative state.
Figure 6.
Actin bound to PFD is nonnative. (A) Actin purified from rabbit muscle (pure), along with either in vitro translated [35S]methionine-labeled full-length actin (FL) or the 336–amino acid NH2-terminal fragment of actin (336) were each incubated at 23°C for 30 min, under control conditions (i.e., no added protease, indicated as in the panel, lanes 1, 4, and 5); with 0.15 mM of trypsin (T, lanes 2, 6, and 8) or 0.375 mM of trypsin (T, lanes 3, 7, and 9); with 0.6 mM of chymotrypsin (C, lanes 10, 12, and 14) or 1.5 mM of chymotrypsin (C, lanes 11, 13, and 15). Digestions were terminated by addition of 1 mM PMSF and the products were analyzed by SDS-PAGE. Shown is the Coomassie blue stained image of the purified actin, and a fluorograph of the gel containing the in vitro translated substrates. (B) Full-length [35S]methionine-labeled actin was synthesized in bacteria and then purified by anion exchange chromatography on MonoQ. A portion of the purified radiolabeled full-length recombinant actin was denatured by treatment with 8 M urea, and then was diluted out of urea in the presence of purified PFD. The recombinant actin–PFD complex was purified by gel filtration. Both the native actin (left) and the actin–PFD complex (right) then were incubated at 25°C with 50 nM proteinase K for the times indicated. The reactions were terminated by the addition of Laemmli sample buffer and the products resolved by SDS-PAGE. A fluorogram of the gel is shown.
A Limited Subset of Proteins Folds via the PFD-mediated Pathway
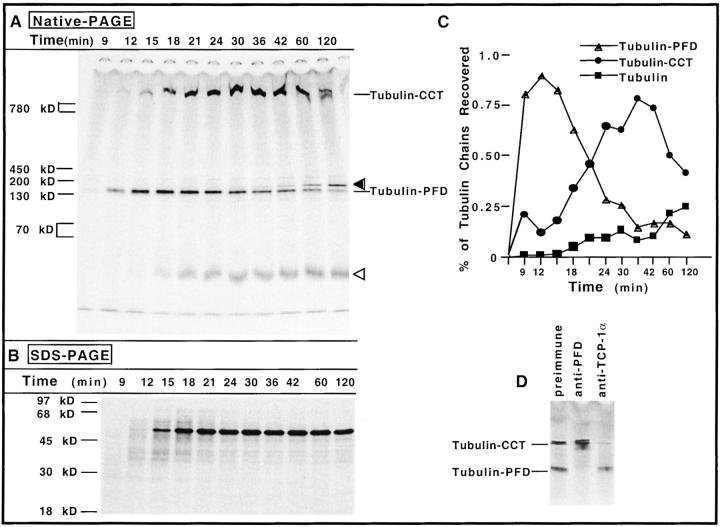
To determine whether nascent chains in general interact with PFD, we translated mRNAs encoding a number of other polypeptides and looked for their possible interaction with PFD. We first examined newly synthesized α- and β-tubulin because, like actin, their productive folding has been shown to be assisted by CCT (Frydman et al., 1992; Yaffe et al., 1992; Sternlicht et al., 1993; Lewis et al., 1996). In addition, urea-unfolded α- and β-tubulin have been shown to be targets for PFD (Vainberg et al., 1998). Finally, the yeast homologue of PFD, GimC, promotes the formation of functional α- and γ-tubulin in yeast (Geissler et al., 1998). Full-length β-tubulin mRNA was translated in reticulocyte lysate for 8 min, inhibitors of translational initiation were added, and the reactions were further incubated to allow for polypeptide chain elongation and completion. At various times, aliquots were removed and incubated with apyrase to deplete ATP, and with puromycin to release any unfinished chains. Analysis of the reaction products by SDS-PAGE revealed that little or none of the tubulin chains had completed their synthesis after 9 min of translation, that a small amount of full-length tubulin was generated by 12 min, and that the level of full-length molecules remained relatively unchanged after 18–21 min (Fig. 7 B). When analyzed by native-PAGE, two prominent tubulin containing intermediates were observed, one of ∼150 kD and the other of 800 kD (Fig. 7 A). Species I, which comigrated with the actin–PFD complex (data not shown), preceded the appearance of species II (Fig. 7, A and C).
Figure 7.
The maturation pathway of β-tubulin involves both PFD and CCT. (A) Reticulocyte lysate was programmed with full-length mRNA encoding chicken β-tubulin in the presence of [35S]methionine. Edeine and 7-MeGMP were added after 8 min to block new initiation of translation. Aliquots of the reaction mixture were removed at various times (indicated at the top of the panel), the samples incubated with apyrase and puromycin, and analyzed by native-PAGE (top). On the right, the positions of migration of the tubulin–PFD complex and the β-tubulin–CCT complex are indicated. An open arrowhead marks the position of a fast migrating β-tubulin containing product (likely to be α/β-tubulin heterodimer, β-tubulin–cofactor A complex, or a mixture of the two). The filled arrowhead indicates the position of migration of a species that occurs very late in time, which likely is β-tubulin complexed with one of the several tubulin-specific chaperones that facilitate tubulin heterodimer formation. (B) Analysis of the same samples shown in A by SDS-PAGE. (C) Kinetics of appearance of species detected by native-PAGE in A was determined as described for Figs. 1 and 2. (D) Puromycin-released β-tubulin chains produced after 18 min of translation (shown in A) were incubated at 24°C for 15 min with preimmune serum, anti–PFD 6 serum, or purified anti–TCP-1α monoclonal IgG 23C, and the reaction products were analyzed by native-PAGE. The positions of tubulin bound to CCT and tubulin bound to PFD are indicated on the left.
The composition of the different β-tubulin intermediates was determined by examining the effects of antibodies specific for PFD and CCT upon the native-PAGE mobility of the complexes. When anti-PFD antiserum was incubated with the translation products, only the electrophoretic mobility of species I was affected, thus it contained PFD (Fig. 7 D). Species II represented full-length β-tubulin bound to CCT because the anti–TCP-1α antibody specifically altered its migration (Fig. 7 D). The species migrating slightly slower than the PFD-containing complex (indicated by an arrowhead in Fig. 7 A) probably represents β-tubulin complexed with one of the several tubulin-specific chaperones that facilitate tubulin heterodimer formation (Tian et al., 1996), while that species migrating near the bottom of the gel (indicated as tubulin) is probably α/β-tubulin heterodimer factor A–β-tubulin complex, or a mixture of the two (Tian et al., 1996).
Analysis of the process of α-tubulin maturation in reticulocyte lysate by the same methods yielded similar results: nascent α-tubulin was observed to first interact with PFD and was then found bound to CCT. The mobility of the α-tubulin complexes formed was also specifically altered by anti-PFD and anti-CCT antibodies (data not shown). We conclude that, like actin, both α- and β-tubulin interact with PFD before delivery to the CCT.
Neither PFD nor the CCT appeared to participate in the maturation pathway of four other proteins: cytosolic yeast invertase, green fluorescent protein, chloramphenicol acetyltransferase, or luciferase. A careful kinetic analysis of the different translation products via native-PAGE did not reveal any distinct intermediates of native mass similar to PFD or CCT during the maturation process (data not shown). Based on these observations we conclude that unlike actins or tubulins, maturation of these four proteins does not involve PFD or CCT.
Discussion
Here we have shown that the cytoskeletal proteins, actin, and α- and β-tubulin undergo a defined series of interactions with at least two chaperones during the course of maturation. In the case of β-actin, cotranslational interaction of the nascent actin chain with PFD occurs following synthesis of the first 145 amino acids. Once engaged, the actin–PFD complex persists until the completion of actin synthesis. Actin molecules bound to PFD are nonnative and accessible to added proteases. Upon completion of synthesis, full-length actin molecules are transferred to CCT where folding to the native state is thought to occur (Fig. 8). A similar maturation pathway applies in the case of both α- and β-tubulin, although interaction of nascent β-tubulin with PFD occurs later than that observed for actin (i.e., after ∼250 amino acids have been synthesized; data not shown). Consequently, as in the case of actin, we suspect that once bound to the nascent chain, PFD remains engaged until the completion of tubulin synthesis, probably until transfer to CCT has occurred.
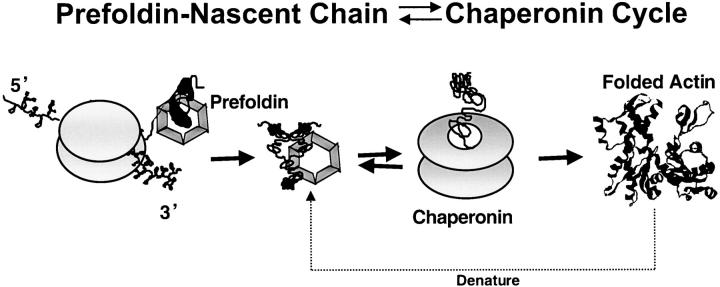
Figure 8.
Cytoskeletal proteins, actin, and tubulin interact with PFD and then with CCT during the course of their maturation in vivo. Data presented here indicate that at least a portion of actin, as well as α- and β-tubulin, interacts with the PFD chaperone while still bound to the ribosome. Following release from the ribosome, the full-length newly synthesized actin and tubulins are transferred over to the CCT for completion of folding to the native state. It remains possible that the actin or tubulin molecules that fail to reach the native state upon release from the chaperonin rebind to PFD and perhaps are transferred to the chaperonin again to allow for a new round of folding. Finally, full-length native actin, which is denatured, likely interacts with PFD before being transferred over to the CCT (Vainberg et al., 1998). It remains possible that other pathways of actin maturation may also exist in the living cell.
Support for the aforementioned pathway of actin and tubulin maturation follows from a number of observations. First, kinetic analysis of translation products derived from full-length actin shows that the newly synthesized protein first accumulated in a complex with PFD. Over time the newly synthesized actin molecules were then found in a complex with CCT, before their appearance as monomeric actin (Fig. 1). Only the monomeric actin appeared properly folded as evidenced by its ability to interact with DNase I (Fig. 3). Second, translation of a truncated actin mRNA resulted in the majority of the puromycin-released actin becoming complexed with PFD; very little of the truncated actin molecules was found in the chaperonin complex (Fig. 2). To rule out the possibility that the truncated actin chains released prematurely from the ribosome were simply interacting with PFD molecules present within the reticulocyte lysate, polysomes containing nascent actin were isolated. Subsequent release of the nascent chains and analysis by native-PAGE revealed actin in a complex with PFD, but not with the CCT (Fig. 4). The most likely interpretation of these data is that PFD was associated with polysomes based on its interaction with nascent actin chains. Analysis of the various actin intermediates by native-PAGE followed by SDS-PAGE (Fig. 2) demonstrates that nascent, as well as full-length, actin could be found complexed with PFD. Relatively low levels of the puromycin-released nascent actin chains could be found in a complex with CCT: rather, the full-length actin molecules appeared to be the preferential binding partner for CCT. Similar results were found for α- and β-tubulin (Fig. 7).
Taken together, our results lead us to conclude that during the course of their maturation, these cytoskeletal proteins interact first with PFD and, upon completion of their synthesis, are transferred over to the chaperonin complex for their folding to the native state. We cannot rule out that other pathways might also feature in the maturation of these proteins. Frydman and Hartl described the cotranslational interaction of nascent actin with the hsp70 chaperone, as well as with CCT (1996). While we also observed an interaction of newly synthesized actin with CCT, this interaction appeared to occur primarily after release of the actin chain from the ribosome. It remains possible that in the absence of PFD actin and tubulin maturation may still occur, but presumably with slower kinetics and reduced fidelity. Indeed, this is precisely the case in yeast strains carrying mutations in the different PFD subunits (Geissler et al., 1998; Vainberg et al., 1998; Siegers et al., 1999).
Through binding cotranslationally to its target proteins, which are in a relatively unfolded state, PFD may prevent misfolding/aggregation during synthesis or after release from the ribosome. In this context, binding by PFD may be a prerequisite for efficient substrate presentation to the CCT. We cannot rule out the possibility that PFD also participates in the rebinding of actin or tubulin molecules that are released from the CCT in a nonnative state. Recently, it has been reported that the yeast PFD homologue, GimC, acts primarily in a complex with the CCT during the folding of actin and perhaps also after release of actin from the chaperonin (Siegers et al., 1999). Our studies add to this complex picture by showing that PFD (and presumably GimC) act, at least in part, by binding cotranslationally to the actin chain and subsequently presenting the full-length actin (and tubulin) polypeptide to the chaperonin complex.
In vivo, there is a five- to eightfold decrease in the rate of actin folding in GimC (PFD) knockout strains of yeast (Siegers et al., 1999). The authors suggest that GimC (PFD) prevents the release of nonnative actin folding intermediates from the chaperonin, and thereby accelerate their folding. Alternatively, it has been suggested that if the actin released from CCT has not yet reached the final native state, then the interaction of actin with PFD (GimC) ensures that the substrate will be retargeted to CCT (Vainberg et al., 1998). The data presented here may help to explain the slower appearance of folded actin observed in yeast GimC mutants (Siegers et al., 1999): the absence of a functional GimC (PFD) chaperone may reduce the efficiency of transfer of the newly synthesized actin chain from the ribosome to CCT.
Exactly what property (sequence) of nascent actin or tubulin dictates their interaction with PFD remains to be determined. Interestingly, several amino acids (present within amino acids 103–133 of actin), which have emerged from the ribosome when actin first binds to PFD, are destined to pack into the native actin structure against residues found in the extreme COOH terminus (Kabsch et al., 1990). Perhaps PFD helps to prevent misfolding of residues 103–133 until the COOH-terminal region of actin has been synthesized.
Finally, it is important to note that so far we have been unable to demonstrate any significant interaction of other nascent/newly synthesized proteins with PFD or CCT. Hence, we suspect that like the chaperonin with which it interacts (Vainberg et al., 1998), PFD appears to have a relatively narrow range of target proteins. However, the family of proteins that possess the actin fold (i.e., potential PFD substrates) is growing. Members of the actin-related protein family function in diverse essential processes, such as transcriptional regulation (Fryer and Archer, 1998; Peterson et al., 1998), orientation of the mitotic spindle, nuclear migration (Melki et al., 1993; Muhua et al., 1994), and membrane polarity and endocytosis (Moreau et al., 1996). Therefore, it will be interesting to see whether these various relatives of actin similarly require the PFD–CCT pathway for their proper maturation.
Acknowledgments
We thank G. Tian for help with protease sensitivity experiments and S. Lewis and V. Lingappa for critical comments on the manuscript.
This work was supported by the National Institutes of Health (GM33551 to W.J. Welch).
Abbreviations used in this paper
- 7-MeGMP
7-methylguanosine monophosphate
- CCT
cytosolic chaperonin
- PFD
prefoldin
References
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Beckmann RP, Mizzen LA, Welch WJ. Interactions of hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Blattler DP, Garner F, Van Slyke K, Bradley A. Quantitative electrophoresis in polyacrylamide gels of 2–40% J Chromatogr. 1972;64:147–155. [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Eggers DK, Welch WJ, Hansen WJ. Complexes between nascent polypeptides and their molecular chaperones in the cytosol of mammalian cells. Mol Biol Cell. 1997;8:1559–1573. doi: 10.1091/mbc.8.8.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr GW, Scharl EC, Schumacher RJ, Sondek S, Horwich AL. Chaperonin-mediated folding in the eukaryotic cytosol proceeds through rounds of release of native and nonnative forms. Cell. 1997;89:927–937. doi: 10.1016/s0092-8674(00)80278-0. [DOI] [PubMed] [Google Scholar]
- Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–1501. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO (Eur Mol Biol Organ) J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Gao Y, Vainberg IE, Chow RL, Cowan NJ. Two cofactors and cytoplasmic chaperonin are required for the folding of alpha- and beta-tubulin. Mol Cell Biol. 1993;13:2478–2485. doi: 10.1128/mcb.13.4.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO (Eur Mol Biol Organ) J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen W, Garcia PD, Walter P. In vitro protein translocation across the yeast endoplasmic reticulum: ATP-dependent posttranslational translocation of the prepro-alpha-factor. Cell. 1986;45:397–406. doi: 10.1016/0092-8674(86)90325-9. [DOI] [PubMed] [Google Scholar]
- Hansen WJ, Lingappa VR, Welch WJ. Complex environment of nascent polypeptide chains. J Biol Chem. 1994;269:26610–26613. [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Jackson R, Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Craig EA. Protein folding in vivo: unraveling complex pathways. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin: DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg PA, Melton DA. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazarides E, Lindberg U. Actin is the naturally occurring inhibitor of deoxyribonuclease I. Proc Natl Acad Sci USA. 1974;71:4742–4746. doi: 10.1073/pnas.71.12.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WM, Galbraith RM. The extracellular actin-scavenger system and actin toxicity. N Engl J Med. 1992;326:1335–1341. doi: 10.1056/NEJM199205143262006. [DOI] [PubMed] [Google Scholar]
- Lewis SA, Tian G, Vainberg IE, Cowan NJ. Chaperonin-mediated folding of actin and tubulin. J Cell Biol. 1996;132:1–4. doi: 10.1083/jcb.132.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis VA, Hynes GM, Zheng D, Saibil H, Willison K. T-complex polypeptide-1 is a subunit of a heteromeric particle in the eukaryotic cytosol. Nature. 1992;358:249–252. doi: 10.1038/358249a0. [DOI] [PubMed] [Google Scholar]
- Melki R, Vainberg IE, Chow RL, Cowan NJ. Chaperonin-mediated folding of vertebrate actin-related protein and gamma-tubulin. J Cell Biol. 1993;122:1301–1310. doi: 10.1083/jcb.122.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Madania A, Martin RP, Winson B. The Saccharomyces cerevisiaeactin-related protein Arp2 is involved in the actin cytoskeleton. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kD heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU. Protein folding in the cytosol: chaperonin dependent and independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- Ngsee JK, Hansen W, Walter P, Smith M. Cassette mutagenic analysis of the yeast invertase signal peptide: effects on protein translocation. Mol Cell Biol. 1989;9:3400–3410. doi: 10.1128/mcb.9.8.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Zhao Y, Chait BT. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J Biol Chem. 1998;273:23641–23644. doi: 10.1074/jbc.273.37.23641. [DOI] [PubMed] [Google Scholar]
- Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. The molecular chaperone Ssb from Saccharomyces cerevisiaeis a component of the ribosome– nascent chain complex. EMBO (Eur Mol Biol Organ) J. 1998;14:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponte P, Ng SY, Engel J, Gunning P, Kedes L. Evolutionary conservation in the untranslated regions of actin mRNAs: DNA sequence of a human beta-actin cDNA. Nucleic Acids Res. 1984;12:1687–1696. doi: 10.1093/nar/12.3.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Siegers K, Waldmann T, Leroux M, Grein K, Shevchenko A, Schiebel E, Hartl FU. Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO (Eur Mol Biol Organ) J. 1999;18:75–84. doi: 10.1093/emboj/18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht H, Farr GW, Sternlicht ML, Driscoll JK, Willison K, Yaffe MB. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc Natl Acad Sci USA. 1993;90:9422–9426. doi: 10.1073/pnas.90.20.9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded β-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Peng I. Deletion of amino acids from the COOH-terminal end of actin. Cell Motil Cytoskelet. 1995;32:163–172. doi: 10.1002/cm.970320302. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H. Tcp1 complex is a molecular chaperone in tubulin biosynthesis. Nature. 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]