Abstract
Expression of the type II collagen gene (human COL2A1, mouse Col2a1) heralds the differentiation of chondrocytes. It is also expressed in progenitor cells of some nonchondrogenic tissues during embryogenesis. DNA sequences in the 5′ flanking region and intron 1 are known to control tissue-specific expression in vitro, but the regulation of COL2A1 expression in vivo is not clearly understood. We have tested the regulatory activity of DNA sequences from COL2A1 on the expression of a lacZ reporter gene in transgenic mice. We have found that type II collagen characteristic expression of the transgene requires the enhancer activity of a 309-bp fragment (+2,388 to +2,696) in intron 1 in conjunction with 6.1-kb 5′ sequences. Different regulatory elements were found in the 1.6-kb region (+701 to +2,387) of intron 1 which only needs 90-bp 5′ sequences for tissue-specific expression in different components of the developing cartilaginous skeleton. Distinct positive and negative regulatory elements act together to control tissue-specific transgene expression in the developing midbrain neuroepithelium. Positive elements affecting expression in the midbrain were found in the region from −90 to −1,500 and from +701 to +2,387, whereas negatively acting elements were detected in the regions from −1,500 to −6,100 and +2,388 to +2,855.
During embryogenesis, the selective and combinatorial expression of genes signifies the developmental program and phenotype of differentiated cells. The activation and repression of gene expression is usually controlled at the level of transcription by multiple positive and negative regulatory elements (Darnell, 1982; Gutman et al., 1994).
Expression of the type II collagen gene is a hallmark of chondrogenesis in vertebrates. In chickens, rodents, and humans, the type II collagen gene is transcribed at high levels by cells of the chondrogenic lineage (Kosher et al., 1986; Cheah et al., 1991; Ng et al., 1993; Lui et al., 1995). In Xenopus, avian, and mouse embryos (Von der Mark et al., 1976; Kravis and Upholt, 1985; Kosher et al., 1986; Thorogood et al., 1986; Swalla et al., 1988; Kosher and Solursh, 1989; Cheah et al., 1991; Su et al., 1991; Wood et al., 1991; Ng et al., 1993), expression of the type II collagen gene is not restricted to chondrogenic cells. In humans, COL2A1 mRNAs are also widely expressed in tissues that are not involved in cartilage formation, such as the notochord, neural retina, heart, fetal brain, and the epithelium of the otic vesicle (Sandell, 1994; Lui et al., 1995; see Table I).
Table I.
Expression Pattern of COL2A1 in Human, cosHcol2 Transgene in Mouse, Col2a1 in Mouse, and COL2A1-lacZ in Transgenic Mice
| Gene | Age/dpc | Tg | lacZ | Ch (Ap) | Sm/Ch (Ax) | Ba1 | Ba2 | Ov* | Fn/Sn | Br | No | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | COL2A1 in | 27–29 | — | — | N | + | + | + | +‡ | + | + | + | ||||||||||||
| human fetuses | 42–44 | — | — | + | + | N | N | +‡ | − | − | − | |||||||||||||
| b | cosHcol2 in | 9.5 | 1 | — | N | + | + | + | + | + | + | + | ||||||||||||
| mouse | 14.5 | 1 | — | + | + | N | N | + | + | + | + | |||||||||||||
| c | mouse Col2a1 | 8.5 | — | — | N | + | N | N | N | N | + | + | ||||||||||||
| 9.5 | — | — | N | + | + | + | + | + | + | + | ||||||||||||||
| 13.5 | — | — | + | + | N | N | + | + | + | + | ||||||||||||||
| d | COL2A1-lacZ | |||||||||||||||||||||||
| pKL9 | 9.5 | 19 | 5 | N | 1 | 4 | 2 | 4 | 3 | — | 4 | |||||||||||||
| 13.5 | 10 | 4 | 4 | 4 | N | N | 4 | 4 | 1§ | 4 | ||||||||||||||
| pKL8 | 9.5 | 4 | — | N | — | — | — | — | — | — | — | |||||||||||||
| 12.5 | 16 | — | — | — | N | N | — | — | — | — | ||||||||||||||
| pKL80.3 | 8.5 | 2 | 2 | N | 2 | N | N | N | N | — | 2 | |||||||||||||
| 9.5 | 21 | 7 | N | 4 | 6 | 3 | 6 | 1 | 1§ | 7 | ||||||||||||||
| 14.5 | 13 | 10 | 10 | 10 | N | N | 5 | — | 2§ | 7 | ||||||||||||||
| pKL10 | 9.5 | 12 | 4 | N | 1 | 2 | 3 | 2 | 1 | 3§ | 3 | |||||||||||||
| 13.5 | 22 | 7 | 7 | 7 | N | N | 7 | 7 | — | 7 | ||||||||||||||
| pKL10R | 8.5 | 4 | 3 | N | — | N | N | N | N | — | 3 | |||||||||||||
| 9.5 | 6 | 4 | N | 2 | 4 | 4 | 4 | 3 | 3§ | 4 | ||||||||||||||
| 13.5 | 3 | 3 | 3 | 3 | N | N | 3 | 3 | — | 3 | ||||||||||||||
| pKL12 | 9.5 | 20 | 5 | N | — | 5 | 3 | 3 | 1 | — | 2 | |||||||||||||
| 14.5 | 1 | 1 | 1 | 1 | N | N | 1 | 1 | — | 1 | ||||||||||||||
| 15.5 | 4 | 3 | 3 | 3 | N | N | 1 | 1 | — | 1 | ||||||||||||||
| pKL10X | 9.5 | 18 | 6 | N | 4 | 4 | — | 5 | 3 | 3§ | 1 | |||||||||||||
| 14.5 | 5 | 4 | — | 4 | N | N | 4 | 4 | — | 4 | ||||||||||||||
| pKL12X | 9.5 | 8 | 5 | N | 5 | 5 | 1 | 5 | 5 | 5§ | 1 | |||||||||||||
| 13.5 | 11 | 8 | — | 6 | N | N | 8 | 7 | 6§ | — | ||||||||||||||
| pKL10RX | 9.5 | 11 | 9 | N | 2 | 1 | 1 | — | 2 | — | — | |||||||||||||
| 14.5 | 15 | 9 | 9 | — | N | N | — | 2 | — | — |
Tg, number of transgenic embryos detected by PCR for lacZ sequence; lacZ, number of transgenic embryos expressing the lacZ transgene; dpc, days post coitum; Ch, chondrocytes; Sm, somites; Ap, appendicular skeleton; Ax, axial skeleton; Ba, branchial arch; Ov, otic vesicle; Fn, frontonasal mesenchyme; Sn, snout; Br, brain; He, heart; No, notochord; Nd, node; N, tissues not present.
Ventral portion.
Full expression.
Expressed in midbrain only. For the expression of COL2A1 gene from Lui et al., (1995): +, expressed; −, not expressed.
The complex expression pattern of type II collagen in chondrogenic and nonchondrogenic tissues during vertebrate development implies intricate regulation. Several cis-regulatory elements important for chondrogenic expression of the type II collagen gene have been identified for the rat and mouse genes by assaying the activity of transfected reporter genes in cultured chondrocytes. Such studies identified sequences in the first intron of the rat and mouse type II collagen genes that bind chondrocyte-specific proteins (Horton et al., 1987; Wang et al., 1991; Lefebvre et al., 1996) and are important for enhancer activity (Horton et al., 1987; Wang et al., 1991; Mukhopadhyay et al., 1995; Krebsbach et al., 1996; Lefebvre et al., 1996). The 5′ flanking region of the type II collagen gene contains domains that repress transcription in cultured nonchondrocytic cells in vitro (Savagner et al., 1990). Synergistic action between the rat type II collagen gene promoter and first intron has also been shown (Savagner et al., 1995). In the human COL2A1, a helix-loop-helix recognition sequence, analogous to the intron 1 decamer enhancer sequence of the rat gene, has been found to be critical for maintaining basal activity of the promoter (Seghatoleslami et al., 1994).
The importance of sequence elements for tissue-specific expression of genes is often inferred from cotransfection assays in cultured cells. Although such studies are valuable, differing results may be obtained depending on whether transfection is stable or transient. It is also not possible to identify elements important for stage specificity of expression by this approach. Experiments using transgenic animals to recapitulate the normal pattern of gene expression have shown that in certain cases, such as elastase and α1(I) collagen genes that DNA sequences identified as essential for gene regulation in transfected cells may not have any functional role in vivo (Swift et al., 1989; Sokolov et al., 1995).
The regulatory activity of some type II collagen gene sequences has been examined in vivo by transgenesis. The first intron of the mouse type II collagen gene (Col2a1) is essential for cartilage-specific expression of an Escherichia coli β-galactosidase (lacZ) reporter gene in transgenic mice (Metsäranta et al., 1995). Sequences within a 182-bp fragment of the mouse Col2a1 first intron were also shown to direct chondrogenic expression of a lacZ reporter gene when present as tandem copies (Mukhopadhyay et al., 1995; Zhou et al., 1995). However, some sequences shown to be important for enhancer activity of the first intron in cell transfection assays were found to be redundant for chondrocyte-specific expression (Mukhopadhyay et al., 1995). These studies on the regulation of the Col2a1 gene have concentrated mainly on defining minimal sequences important for chondrocyte-specific enhancer activity and on expression in the chondrogenic lineage from 10.5 days post coitum (dpc)1 in the mouse. The elements necessary for expression of Col2a1 before 10.5 dpc and in nonchondrogenic tissues in the mouse have not been determined. Furthermore, the transgenic studies performed so far have not revealed any activity of the silencing (repressor) elements that have been identified by cell transfection studies (Savagner et al., 1990; Seghatoleslami et al., 1994).
The sequence elements directing human type II collagen gene expression are not well defined. We have previously shown that a 30-kbp genomic fragment containing the human COL2A1 gene can be expressed in chondrogenic tissues in the mouse (Lovell Badge et al., 1987). More recent studies on the COL2A1 gene have shown that a combination of 1.8-kbp 5′ flanking DNA with a 3.0-kbp first intron or 4.5-kbp upstream sequence with 1.8-kbp of intron 1 DNA, was sufficient for chondrogenic-specific lacZ reporter gene expression in transgenic mice (Seghatoleslami et al., 1994; Cheah et al., 1995). These studies concentrated on later stages and on the chondrogenic lineage; there is little information on the cis-regulatory elements governing COL2A1 transcription in vivo or the ability of these sequences to direct expression at early stages of embryogenesis and in nonchondrogenic tissues. We have assayed such elements in the 5′ flanking and intron 1 regions of COL2A1 expressed in transgenic mice.
A prerequisite for analysis of the regulation of the human COL2A1 gene in mice is that the developmental pattern of expression can be recapitulated in the transgenic animal. We have previously shown very similar patterns of expression of the endogenous human and mouse type II collagen genes in nonchondrogenic and prechondrogenic cells from stages equivalent to 9.5 dpc in mouse embryos (Ng et al., 1993; Lui et al., 1995). To provide a starting point for the identification of regulatory elements controlling appropriate COL2A1 transcription in this study, we compared expression of a genomic clone containing COL2A1 in transgenic mice with that of the endogenous mouse Col2a1 gene. In addition, we have studied the regulation of COL2A1 expression in transgenic mice during early organogenesis and fetal stages.
In the present study we identified a 309-bp fragment within the first intron of human COL2A1 which is capable of directing reporter transgene (COL2A1-lacZ) expression in prechondrogenic and chondrogenic sites as well as in a subset of nonchondrogenic tissues (such as node, neural crest cells, and notochord), characteristic of type II collagen. These sequences were sufficient for correct temporal regulation of gene expression. We show that the 5′ flanking DNA and the first intron need to work synergistically to direct the expression of the transgene in tissues that normally express Col2a1. We present evidence that positive and negative tissue-specific regulatory elements in the first intron and the 5′ flanking region of the COL2A1 gene are important for directing expression in the developing midbrain neuroepithelium.
Materials and Methods
Transgenes
Cosmid clone cosHcol2 contained the complete human COL2A1 structural gene with a 9.1-kbp 5′ flanking and a 1.7-kbp 3′ flanking DNA (gift of E. Solomon, Imperial Cancer Research Fund, London, UK).
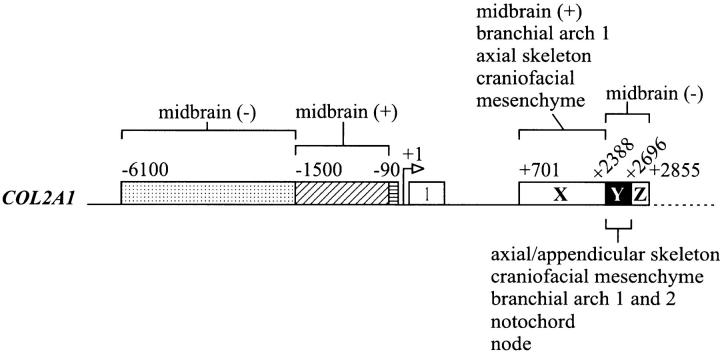
Reporter gene constructs were created in pPOLYIII I vector by fusing various lengths of 5′-flanking promoter sequences to a 125-bp 5′ untranslated region of human COL2A1 gene and an E. coli β-galactosidase (lacZ) reporter gene cassette with an initiation translation codon and polyadenylation signal, followed by various lengths of the first intron of the human COL2A1 gene. The COL2A1 content in the different constructs were as shown in Fig. 1. For ease of reference, the intron 1 sequences analyzed in this study have been subdivided into three regions, designated X, Y and Z, the coordinates of which are +701 to +2,387, +2,388 to +2,696, and +2,697 to +2,855, respectively (see Figs. 1 and 6).
Figure 1.
Schematic representation of constructs used to generate transgenic mice in this study. Each construct contains a lacZ cassette flanked on the 5′ side by upstream sequences from the following genes: COL2A1 (hatched box); human β-globin gene promoter (cross-striped box); mouse Col10a1 5′ flanking DNA (stippled box) and on the 3′ side by COL2A1 first intron sequences (black or white boxes) or the SV-40 enhancer (vertically striped box). Numbers indicate the positions of the nucleotide in the sequence relative to the start site of transcription in the COL2A1 gene. For ease of reference, the intron 1 sequences analyzed have been subdivided into three regions, designated X, Y, and Z (refer to Materials and Methods). ATG, translation start site; pA, polyadenylation signal.
Figure 6.
A summary of the organization of promoter and enhancer elements of the human COL2A1 gene as revealed by in vivo reporter activity in transgenic mouse embryos. The 1.6-kbp intron 1 (+701 to +2,387) sequence (region X) contains the enhancer elements that are required, in conjunction with a minimal 90-bp fragment of the 5′ flanking region of the COL2A1 gene, to direct prechondrogenic and chondrogenic and some nonchondrogenic expression of the reporter. Expression of the reporter is found in the midbrain, first branchial arch, craniofacial mesenchyme, and axial skeleton. Region Y of intron 1 (+2,388 to +2,696) in conjunction with 6.1-kbp 5′ flanking sequence confers similar prechondrogenic and chondrogenic enhancer activity. With this combination of regulatory elements, the transgene is expressed widely in the chondrogenic tissues (axial and appendicular skeleton and craniofacial mesenchyme), branchial arch 1 and 2, notochord, and node. Midbrain-positive elements may be present in the sequences from −90 to −1,500 and from +701 to +2,387 (region X). On the other hand, midbrain-negative elements may be present in the sequences from −1,500 to −6,100 and +2,388 to +2,855 (region Y+Z). Our study also shows that chondrogenic expression of the reporter can be achieved only by using homologous 5′ flanking and intron 1 sequences of the COL2A1 gene and substituting either one with heterologous regulatory elements results in nonspecific reporter expression.
Production of Transgenic Mice
Transgenic mice harboring the various COL2A1 constructs were generated by pronuclear injection of one-cell zygotes of F1 (CBA × C57BL6) hybrid mice as described (Hogan et al., 1986). Transgenic fetuses were collected at different stages of development and tested for integration of the transgene by PCR. PCR was performed on genomic DNA using oligonucleotide primers for the lacZ gene 5′-CGTAATAGCGAAGAGGCCCG-3′ (nucleotides [nt] 112–130; translation start = 1); and 5′-TCCAACGCAGCACCATCACC-3′ (complementary to nt 558–577). PCR reactions, containing 2.5 U Taq polymerase (Life Technologies, Inc., Gaithersburg, MD), were denatured at 95°C for 3 min, followed by 30 cycles of 95°C for 1 min, denaturing, 55°C for 1 min, annealing, and then 72°C for 1 min of elongation.
Section and Whole Mount In Situ Hybridization
In situ hybridization to sections was carried out as described previously (Cheah et al., 1991; Nonchev et al., 1996). Two hybridization probes used were human COL2A1-specific (KH14) and mouse Col2a1-specific (KH15). KH14 and KH15 probes each contained 70 bp of relatively less conserved 3′ untranslated sequence from equivalent regions of the human and mouse type II collagen genes corresponding to +31,102 to +31,171 (COL2A1) and +28,624 to +28,693 (Col2a1), i.e., KH14 is the human homologue of KH15. These probes were so designed because they contained short stretches of nonconserved bases and were species-specific when sections were treated with RNase posthybridization, and then washed at high stringency. Autoradiography and photography of sections were carried out as described previously (Cheah et al., 1991).
Whole mount in situ hybridization on 8.0–9.0 dpc CBA mouse embryos was carried out as described by Wilkinson (1992) using single-stranded, digoxgenin-UTP (Boehringer Mannheim, Mannheim, Germany)–labeled antisense riboprobes generated from pNJ61 (Ng et al., 1993), which contains exon 1–5B of Col2a1.
X-Gal Staining and Histochemistry
Mouse embryos or fetuses of various stages were harvested in PBS, pH 7.4, and then fixed for 15–60 min, according to their size, at 4°C in 4% paraformaldehyde in PBS. They were assayed for expression of lacZ by X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining essentially as described in Yee and Rigby (1993). After staining, fetuses were embedded in paraffin. 6-μm sections were counterstained with eosin.
Results
Human COL2A1 Transgene Is Expressed Like the Endogenous Col2a1 Gene
The expression of 45.8-kbp transgene, cosHcol2, containing the whole COL2A1 gene, 9.1-kbp 5′ and 1.7-kbp 3′ flanking sequence was examined by in situ hybridization using riboprobes specific for the human transgene (KH14) and the mouse gene (KH15). The COL2A1 transgene and the mouse Col2a1 gene were coexpressed in the prechondrogenic mesenchyme and chondrocytes (Fig. 2, a, c, and e; Table I, COL2A1; Fig. 2, b, d, and f; Table I, Col2a1). COL2A1 was also expressed in the nonchondrogenic tissues that express the Col2a1 gene, such as the brain (Fig. 2 a) and otic vesicle (Fig. 2 c). However, COL2A1 transgene was not expressed in the mouse heart (Fig. 2 c) which normally expresses the Col2a1 gene strongly. No ectopic expression of the transgene was found. COL2A1 riboprobe did not display any nonspecific hybridization in nontransgenic mouse embryo (Fig. 2 g).
Figure 2.
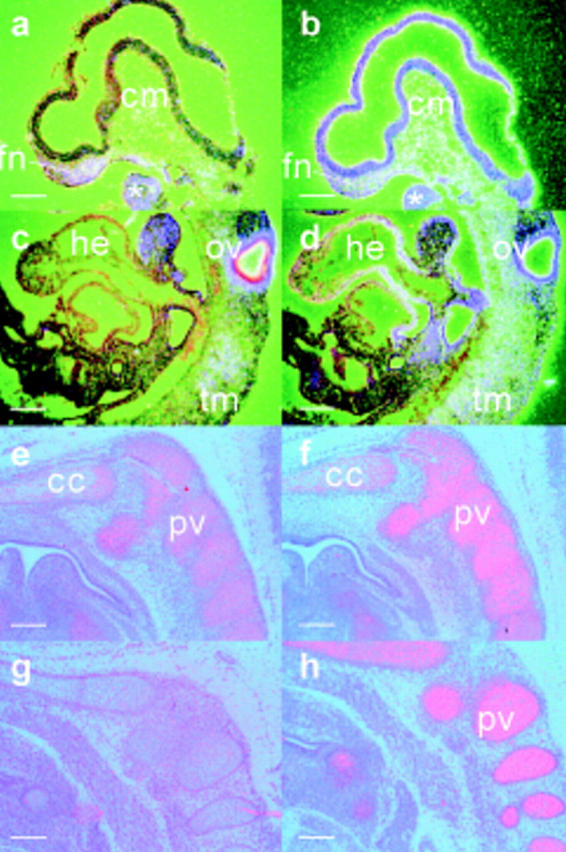
A comparison of the expression of the human COL2A1 (a and c, 9.5-dpc; e and g, 14.5-dpc embryo) and mouse Col2a1 (b and d, 9.5-dpc; f and h, 14.5-dpc embryo) genes in transgenic (a–f) and nontransgenic (g and h) embryos. The transgenic embryos are heterozygous for a transgenic locus containing a 9.1-kbp 5′ and a 1.7-kbp 3′ flanking sequences and the COL2A1 gene (cosHcol2 construct). In the transgenic embryos, transcripts of the cosHcol2 transgene are found in the first branchial arch (asterisk), frontonasal (fn) and cranial paraxial mesenchyme (cm) (a), the otic vesicle (ov) and prechondrogenic trunk mesenchyme (tm) (c), the prevertebrae (pv) and the chondrocranial cartilage (cc) (e). These tissues in the transgenic mice also express the endogenous mouse Col2a1 (b, d, and f). The cosHcol2 transgene is not expressed in the heart tissues (he) that express the endogenous Col2a1 gene (compare c with d). In the nontransgenic embryo, no COL2A1 mRNA is detected in the chondrogenic mesenchyme (g) and expression of the endogenous Col2a1 transcript is shown in (h). Bar, 200 μm.
Identifying the cis-Acting Elements for COL2A1-specific Expression
To test the activity of the cis-acting elements that may direct lineage-specific expression of the COL2A1 gene, a COL2A1-lacZ transgene (Fig. 1, pKL9), containing the 6.1-kbp (−1 to −6,100) 5′ flanking sequence, 125-bp 5′ untranslated region and 2.1-kbp intron 1 (+701 to +2,855, regions X, Y, and Z, refer to Materials and Methods) was made. The lacZ gene was expressed in the first and second branchial arches, otic vesicle, frontonasal mesenchyme, and the notochord in the 9.5-dpc embryo (Fig. 3, c–e; Table I). At 13.5 dpc, lacZ activity was detected in the craniofacial chondrogenic tissues (e.g., the frontonasal, occipital, parietal, and facial mesenchyme), the axial (prevertebrae and ribs) and appendicular skeleton (Fig. 4, a–e, limb cartilages). Expression of the pKL9 transgene at both ages was very similar to that of cosHcol2 (Fig. 2, a, c and e) and Col2a1 (Figs. 2 b, d, f and 3 b; Table I). These observations suggest that the regulatory elements of pKL9 are sufficient for driving the COL2A1-specific expression in transgenic mice. Some necessary regulatory elements may, however, be missing in pKL9 since at 9.5 dpc lacZ transgene expression was absent from the dorsal half of the otic vesicle and the brain (Fig. 3, c–e). A construct (pKL8) containing only the 6.1-kbp 5′ flanking region but without the intron 1 sequence linked to the reporter gene did not express in all the transgenic embryos (Table I). Some critical tissue-specific regulatory elements are therefore present within the first intron of the gene.
Figure 3.
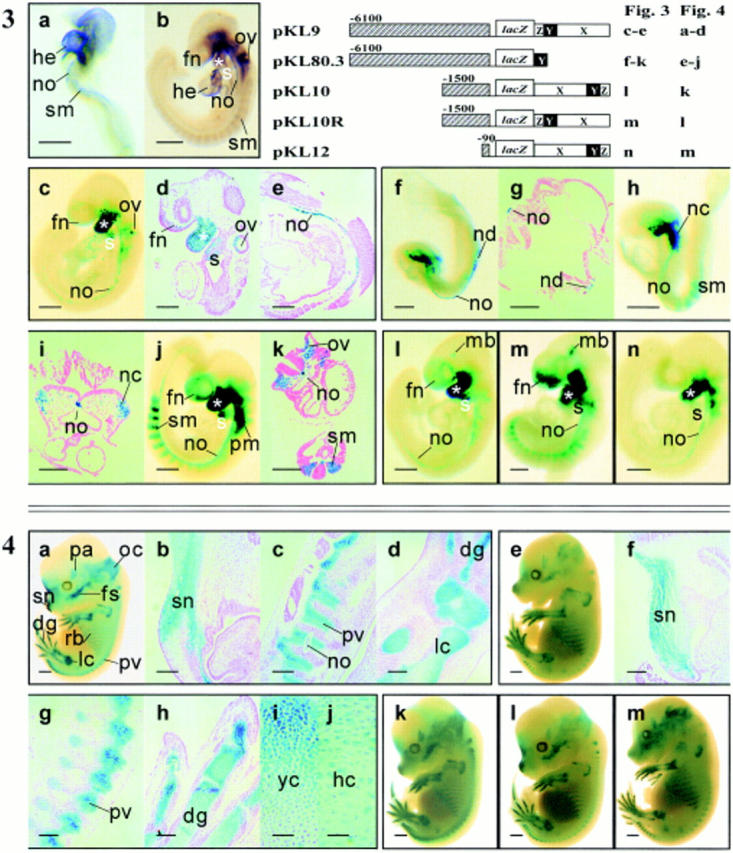
The delineation of minimal COL2A1 sequences required for expression of the lacZ reporter in prechondrogenic tissues. Col2a1 is expressed in the heart (he), notochord (no), and somites (sm) of (a) 8.5- and (b) 9.5-dpc embryos. The tissue-specific pattern of Col2a1 expression is emulated by that of the COL2A1-lacZ transgene with a 6.1-kbp 5′ flanking and regions X, Y, and Z of intron 1 in the pKL9 transgenic embryo (c–e). The transgene is expressed in the frontonasal mesenchyme (fn), otic vesicle (ov), first (asterisk) and second (s) branchial arches (c and d) and notochord (no) (c and e). Reduction of the first intron to region Y in pKL80.3 embryos still allows the full expression of the reporter gene in prechondrogenic tissues both in 8.5- (f–i) and 9.5-dpc (j and k) embryos and mimics the expression of Col2a1 gene (a, 8.5 dpc; b, 9.5 dpc). Transgene expression is found in the node (nd) and notochord (no) of the three-somite stage embryo but not in the embryonic heart (f and g). In the eight-somite stage pKL80.3 embryo, transgene expression is found in the notochord (no), neural crest cells (nc) (h and i), and somites (sm) (h). At 9.5 dpc, pKL80.3 is expressed in a pattern (j and k) similar to that of the endogenous Col2a1 (b). In addition, it is found to express in the paraxial mesenchyme (pm) (j). Reduction of the 5′ sequence of pKL9 to 1.5 kbp (l, pKL10; m, pKL10R) and 90 bp (n, pKL12) allows the full expression of the reporter gene, suggesting that the −1 to−90 fragment of the COL2A1 gene acts like a minimal promoter in conjunction with regions X, Y, and Z of intron 1. The intron 1 sequence functions as an enhancer element and gives the same reporter gene expression when placed in either orientation (compare l with m). It is interesting to find transgene expression in the midbrain (mb) of pKL10 and pKL10R (l and m). Bars: (a–c, h, j, and l–n) 500 μm; (d and e) 200 μm; (g, i, and k) 100 μm.Figure 4. Delineation of the minimal COL2A1 sequences needed to direct expression of the transgene in chondrogenic tissues. pKL9 embryo at 13.5 dpc (a) shows characteristic chondrogenic expression in the snout (sn), digits (dg), limb (lc), ribs (rb), prevertebrae (pv), facial skeleton (fs) and parietal (pa), and occipital (oc) mesenchyme. Histological sections of pKL9 13.5-dpc embryos show reporter gene expression in the condensation of the nasal mesenchyme (sn) (b), prevertebrae (pv), notochord (no) (c), and mesenchymal condensation of the zygopods (lc), and digits (dg) of the limb (d). Deletion of the intron 1 to region Y in pKL80.3 does not affect expression of the transgene in chondrogenic tissues (e–j), indicating that region Y of intron 1, in conjunction with 6.1-kbp 5′ flanking sequence, contains sufficient cis-acting tissue-specific elements to direct chondrogenic expression. Histological sections of pKL80.3 14.5-dpc embryos show similar lacZ expression in chondrogenic tissues (f–h) as in pKL9 embryos (b–d). Transgene expression in the hypertrophic chondrocytes (hc) (j) is weaker than that in young chondrocytes (yc) (i), which mimics the endogenous Col2a1 expression pattern. Reduction of the 5′ flanking sequences to 1.5 kbp in pKL10 (k) and pKL10R (l) and 90 bp in pKL12 (m) also has no effect, indicating that only 90-bp 5′ flanking sequence is enough to give chondrogenic expression in conjunction with regions X, Y, and Z of intron 1. Bars: (a, e, and k–m) 500 μm; (b–d and f–h) 200 μm; (i and j) 50 μm.
DNA Modules for COL2A1 Expression during Early Organogenesis
In the mouse, Col2a1 is expressed early at 8.0–8.5 dpc. A distinct craniocaudal gradient of Col2a1 mRNA expression was observed in the paraxial mesoderm (Fig. 3 a). In addition, the midline notochord displayed a uniform expression throughout its length (Fig. 3 a). A 309-bp fragment (region Y) of the first intron of the COL2A1 gene (84.1% homologous to the minimal enhancer of the rat type II collagen gene [Krebsbach et al., 1996]) when linked in normal orientation to the lacZ gene and 6.1-kbp 5′ flanking COL2A1 sequence (pKL80.3), could direct transgene expression like that of endogenous Col2a1 in early organogenesis stage mouse embryos. β-galactosidase activity was found in the paraxial mesoderm and was expressed strongly in the node and notochord at 8.5 dpc (Fig. 3, f–i; Table I). At 9.5 dpc, pKL80.3 transgene was expressed in the first and second branchial arch and the paraxial mesoderm (Fig. 3, j and k), like that of the endogenous Col2a1 (Fig. 3 b). pKL80.3 expression was stronger in the newly formed somites than in the more mature somites (Fig. 3 j). These findings indicate that region Y, together with 6.1-kbp 5′ flanking DNA, was sufficient to drive lacZ expression in tissues that normally express mouse Col2a1 during early organogenesis (Table I).
Identification of Minimal 5′ Flanking Sequence for COL2A1 Expression
The 6.1-kbp 5′ flanking sequence was systematically examined in conjunction with 2.1 kbp of the intron 1 sequence (regions X, Y, and Z) to determine the minimal upstream DNA required to regulate appropriate lacZ expression. The 5′ flanking sequence was reduced to 1.5 kbp (−1 to −1,500) (pKL10, and pKL10 which differs by the orientation of the intron 1 sequence, refer to Fig. 1) and also to 90 bp (−1 to −90, pKL12, refer to Fig. 1). None of these constructs showed any significantly different expression patterns from one another (Fig. 3, l–n; Table I) or that of pKL9 (Fig. 3, c–e). Constructs pKL9 and pKL80.3 were expressed in chondrogenic tissues (Fig. 4, a–d and e–j; Table I) and were appropriately downregulated in the hypertrophic chondrocytes like the endogenous Col2a1 (Fig. 4, i and j; Ng et al., 1993; Lui et al., 1995). pKL10, pKL10R, and pKL12 transgenes were expressed in the same prechondrogenic and chondrogenic tissue types as pKL9 and pKL80.3 (Table I). Reduction of the 5′ flanking sequence of pKL9 to 1500 (−1 to −1,500, pKL10 and pKL10R) or 90 bp (−1 to −90, pKL12) did not seem to affect the expression of the transgene (Fig. 4, k–m at 13.5 dpc). Transgene expression was found in the notochord when the transgene was regulated by 6.1-kbp, 1.5-kbp, or 90-bp upstream sequence in combination with 2.1-kbp first intron (Fig. 3, c and e, pKL9; Fig. 3 l, pKL10; Fig. 3 m, pKL10R; Fig. 3 n, pKL12). Similar notochord expression was also achieved with region Y and 6.1-kbp 5′ flanking DNA (Fig. 3, f–k, pKL80.3). Therefore, the 5′ flanking sequence −1 to −90 with 2.1-kbp intron 1 sequence is sufficient for COL2A1 expression, and 6.1 kbp is required if only region Y of intron 1 is present.
Separate Regulatory Elements Direct Expression in Different Components of the Cartilaginous Skeleton
The pKL10 construct was modified by deleting region Y+Z from the intron 1 fragment (refer to Fig. 1, pKL10X). This transgene was expressed in sites that normally would express Col2a1 (Fig. 5, a–e; e.g., craniofacial mesenchyme, paraxial mesoderm, and prevertebrae), except the notochord, the second branchial arch (Fig. 5, a and b, 9.5 dpc), and the digits of the limbs (Fig. 5, c and f, 14.5 dpc). Transgene expression was generally patchy (Fig. 5 a) and reduced in limbs, digits, the upper trunk, vertebral cartilage, and rib cage (Fig. 5 c). A similar expression pattern was observed (Fig. 5, g–l) for construct pKL12X containing the minimal 90-bp (−1 to −90) 5′ flanking sequence and region X (refer to Fig. 1). Together, these results suggest that at least two separable modules of DNA are present in intron 1. Region X may contain regulatory elements for the first branchial arch, facial skeleton, and lower trunk vertebrae. Indeed, deletion of region X from pKL10R (Fig. 1, pKL10RX) led to loss of expression in most tissues except the limbs and digits (Fig. 5, m–q; Table I). Region Y+Z may contain regulatory elements for regulating COL2A1 expression in the second branchial arch, notochord, upper trunk (axial), and limb (appendicular) cartilage.
Figure 5.
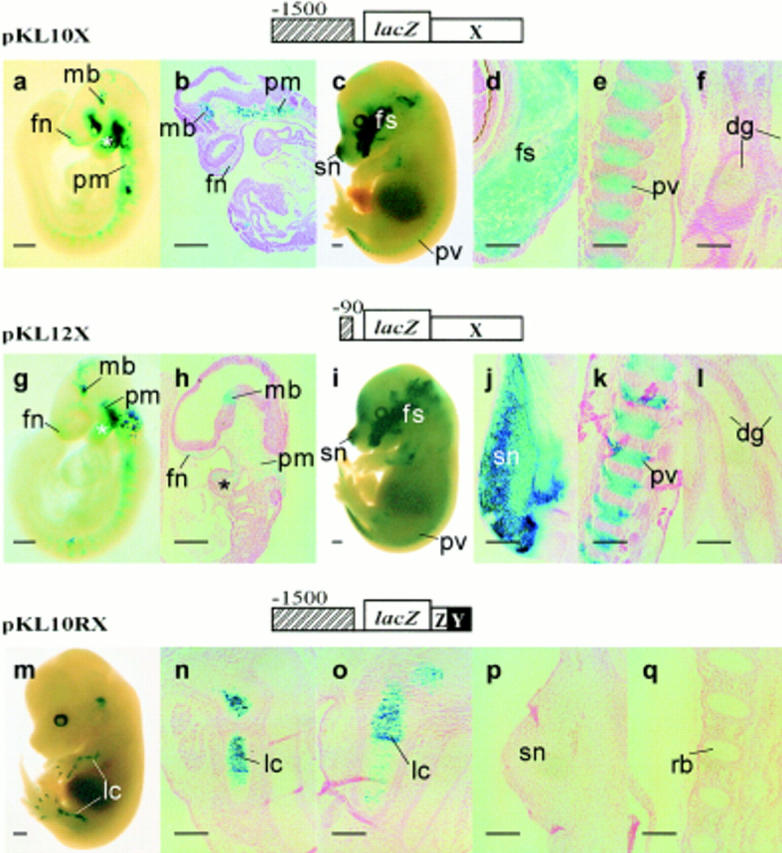
Demonstration of separable regulatory modules in the first intron. Deletion of regions Y and Z from pKL10 and pKL12 causes restricted and patchy expression of the reporter in the paraxial mesoderm (pm), the first branchial arch mesenchyme (asterisk), and the mesenchyme in the midbrain (mb) and frontonasal (fn) regions of the 9.5-dpc pKL10X (a and b) and pKL12X (g and h) embryos. lacZ expression is found in the snout (sn), facial skeleton (fs), and the prevertebrae (pv) but not in the cartilage of the digits (dg) of the 14.5-dpc pKL10X transgenic fetus (c–f) and that of the 15.5-dpc pKL12X transgenic fetus (i–l). Deletion of region X from pKL10R (refer to Fig. 1, pKL10RX) leads to the loss of expression in most tissues except the digits and limbs (lc) (m–o). No expression in the snout (sn) (p) or ribs (rb) (q) is found. Bars: (a, c, g, i, and m) 500 μm; (b, d–f, h, j–l, and n–q) 200 μm.
Positive and Negative Regulatory Elements for Expression in the Ventral Midbrain
The lacZ reporter was expressed in the neuroepithelium of the ventral midbrain for constructs containing 1.5-kbp (−1 to −1,500) 5′ fragment and regions X+Y+Z (Fig. 3, l and m; Table I, pKL10 and pKL10R). In contrast, midbrain expression was absent for constructs containing additional sequences from −1,500 to −6,100 (Fig. 3, c and d, pKL9) or only 90-bp 5′ fragment (Fig. 3 n, −1 to −90 pKL12). This suggests that negative elements that suppress midbrain expression may be localized in the sequence between −1,500 and −6,100, and positive elements may be found between −90 and −1,500.
Although pKL12 was not expressed in the midbrain of the transgenic embryo, the removal of region Y+Z from pKL12 (i.e., pKL12X) has resulted in the reexpression of the lacZ reporter in the midbrain (Fig. 5, g and h; Table I d). Exchanging region X for Y+Z (pKL10RX) resulted in loss of midbrain expression (Table I). These findings therefore suggest that negative regulatory elements controlling midbrain expression may be localized in region Y+Z. These elements may act in an orientation-independent manner, since midbrain expression was similar for pKL10 and pKL10R (refer to Fig. 3, l and m). Deletion of the same region (Y+Z) from pKL10 (i.e., pKL10X), which was expressed in the midbrain, has no effect on the expression of the reporter (compare Fig. 3 l with Fig. 5, a and b). Therefore, it suggests that the positive elements found between −90 and −1,500 could overcome the negative effect of region Y+Z.
Tissue-specific Expression Requires Both COL2A1 5′ and Intron 1 Sequences
To investigate whether region Y needs to interact specifically with the COL2A1 promoter, we replaced the 6.1-kbp 5′ flanking sequence of pKL80.3 with heterologous promoter fragments from the human β-globin and mouse type X collagen (Col10a1) genes (refer to Fig. 1, pKL21 and pWP16 constructs). The human β-globin promoter fragment has been shown to cooperate with tissue-specific elements from heterologous genes, such as myogenin (Yee and Rigby, 1993) and Hoxa2 (Nonchev et al., 1996), to drive tissue-specific lacZ expression in 9.5-dpc transgenic mice embryos. The Col10a1 promoter fragment can direct lacZ expression in developing bone of 14.5-dpc transgenic mouse fetuses (Poon, W.S., and K.S.E. Cheah, unpublished data).
The β-globin promoter (pKL21) was unable to direct transgene expression in the Col2a1 pattern despite the presence of region Y of intron 1 (seven expressing embryos showing low and nonspecific expression, data not shown). Furthermore, type X collagen promoter was also unable to substitute for the COL2A1 5′ flanking sequence and no expression was seen in seven transgenic embryos. The specific requirement for intron 1 sequence is further demonstrated by the inappropriate expression of pKL28 construct, in which the intron 1 fragment was replaced by the SV-40 enhancer (refer to Fig. 1). Only one of six transgenic embryos expressed lacZ and was not in a pattern characteristic of the Col2a1 (pKL9/pKL80.3) pattern (data not shown).
Discussion
Different Combinations of DNA Modules Confer Specific Expression of the COL2A1 Gene
In this study, we have identified DNA elements that regulate the expression of the COL2A1 gene by assaying the expression of COL2A1-lacZ transgenes in mouse embryos. We found that the 6.1-kbp upstream region and the first intron of COL2A1 contain regulatory elements required for transcription in tissues that normally express the endogenous mouse type II collagen gene. The first intron operates in an orientation-independent manner. These results agree with other studies using cell transfection assays and transgenic mice, which demonstrate an enhancer role of the first intron of type II collagen genes (Horton et al., 1987; Wang et al., 1991; Seghatoleslami et al., 1994; Metsäranta et al., 1995; Mukhopadhyay et al., 1995; Zhou et al., 1995; Krebsbach et al., 1996; Lefebvre et al., 1996).
In our experiments, neither the first intron nor the COL2A1 5′ flanking DNA could be substituted by enhancer or promoters of unrelated genes. These results are consistent with the report that the promoter and first intron of the rat type II collagen gene act synergistically (Savagner et al., 1995), but contrast with another report showing that mouse Col2a1 intron 1 sequences linked to the human β-globin or adenovirus major late promoters could direct transcription in chondrocytes (Mukhopadhyay et al., 1995; Zhou et al., 1995). These contradictory findings could be related to differences on the strength of the promoter–enhancer interaction and the proximity of the promoter to sequences in the first intron, which may affect the efficiency of transcription. In the constructs tested in the present study, the first intron sequences are separated from the promoter sequence by at least 3 kbp of lacZ DNA. This may hamper the formation of the correct promoter-enhancer loop, and thus diminishes the activity of the heterologous promoters. Furthermore, in other studies, multiple intron 1 sequences are placed immediately upstream of heterologous promoters. This arrangement may facilitate the stabilization of promoter–intron interactions allowing the expression of the transgene (Mukhopadhyay et al., 1995; Zhou et al., 1995).
The requirement for proper physical configuration and/ or differing combination of integrity of regulatory elements for transgene expression has also been shown in the present study. Appropriate expression of the lacZ transgene is achieved by a combination of either the 6.1-kbp 5′ flanking fragment and region Y, or a 90-bp 5′ flanking sequence and the 2.1-kbp intron 1 sequence containing X+Y+Z. Therefore, regions X and Z may contain some elements that interact with the short (90-bp) 5′ flanking sequence. The deletion of sequences −1,500 to −6,100 resulted in the loss of lacZ expression. Both combinations of regulatory elements were equally effective for correct tissue-specific expression, despite sequence differences between modules. The results are consistent with the notion that sufficient lengths of specific combinations of the 5′ flanking sequence and intron 1 of COL2A1 are essential to achieve the appropriate conformation for establishing the necessary heteromeric complex to activate transcription.
COL2A1 Activity in the Midbrain Is Regulated by Positive and Negative Regulatory Elements
Transgene expression in the midbrain is found to be modulated by a combination of positive and negative elements. DNA sequences in the upstream region −1,500 to −6,100 and region Y+Z of intron 1 seem to exert a suppressive activity, whereas the sequences in the −1 to −1,500 region and region X are required for the expression of the COL2A1 transgene (Fig. 6). The data also suggest that expression of COL2A1 in different skeletal elements may be mediated by different combinations of regulatory elements. For example, 1.5-kbp with region Z+Y gave limb expression only (Fig. 5 m) but 1.5-kbp region X gave expression in the axial skeleton but not in the limbs (Fig. 5 c). Recently, cis-acting elements that control α2(XI) collagen gene expression in different cartilaginous components of the skeleton (Tsumaki and Kimura, 1995) and separable DNA modules for tissue and site-specific expression of the α1(I) (Sokolov et al., 1995; Rossert et al., 1995, 1996) and α1(VI) collagen genes (Braghetta et al., 1996) have been described. The differential impact of selective enhancer and repressor elements of the COL2A1 gene on lacZ reporter activity in the brain suggests lineage-specific or region-specific deployment of regulatory elements may be a common mechanism of gene regulation.
Some Regulatory Motifs Identified by Cell Transfection Assays Have No Function In Vivo
Region Y contains the analogous sequences to those in the rodent type II collagen genes shown to be important for enhancer activity in transfected chondrocytes (Wang et al., 1991; Krebsbach et al., 1996; Lefebvre et al., 1996) but not all the motifs identified were present in this region. An AT-rich element previously shown to be important for the enhancer activity and for transcriptional regulation of the Col2a1 gene (Krebsbach et al., 1996) was absent in region Y, suggesting that this sequence is not essential for tissue-specific expression of COL2A1 in vivo.
The 90-bp upstream region of COL2A1 is highly conserved among human, mouse, and rat and contains many potential transcription factor–binding motifs, including a potential Sp1-binding site (−75 to −80) (Vikkula et al., 1992). In the rat Col2a1 gene, a potential Sp1-binding site outside this 90-bp region (−114 to −119) interacts with the first intron enhancer via a complex consisting of Sp1 factor and a zinc finger protein, CIIZFP (Savagner et al., 1995). Our results showing appropriate transgene expression in the absence of this Sp1-binding site (construct pKL12) suggest either that the site is redundant or the other potential Sp1-binding site (−74 to −80) or yet undefined elements within the 90-bp sequence are sufficient for gene expression. The increased specific binding activity to Sp1 motifs found for nuclear extract prepared from non- Col2a1–expressing cells such as fibroblasts and dedifferentiated chondrocytes suggest that the Sp1-binding site may mediate negative regulation of the gene (Dharmavaram et al., 1997).
The rat type II collagen gene contains two sequence elements at positions −421 to −436 and −657 to −666 which behave as silencers in cell transfection assays (Savagner et al., 1990). This region is not well conserved between human and rat type II collagen gene (Vikkula et al., 1992) and the silencing activity has not been tested in vivo. Omitting the −90 to −1,500 region of COL2A1 did not change the tissue-specificity of transgene expression, suggesting that the putative silencer elements in the 5′ flanking DNA do not play any role in the regulation of COL2A1 activity in transgenic mice.
Molecular Mechanisms for Regulation of COL2A1 Expression: A Model of the Organization of COL2A1 Lineage-specific Regulatory Elements
The main structural and functional features of the DNA modules regulating lineage-specific expression of human COL2A1 (Fig. 6) are: (a) early organogenesis and chondrogenic expression are regulated by the interaction of elements in the upstream (−1 to −6,100) region and the 309-bp region Y of the first intron and of elements in −1 to −90 region and region X of the first intron; (b) expression in the midbrain tissue is mediated by interactions between positive elements in the −1 to −1,500 region and the negative elements in −1,500 to −6,100 and region Y+Z of the first intron; (c) critical regulatory elements for gene expression in the notochord are found in the region −1 to −1,500 and region Y of the first intron.
Such an interplay of positive and negative regulation to achieve defined expression patterns has been found for many developmentally regulated genes (Ip and Hemavathy, 1997). Transcription factors such as Bicoid, Dorsal, snail, and Elf-1 regulate patterning in the Drosophila embryo by restricting gene expression to specific sites (Novina and Roy, 1996; Ip and Hemavathy, 1997). We and others have recently shown that SOX9 directly regulates the mouse and human type II collagen genes via SRY/SOX binding motifs found in the Y region of the first intron (Bell et al., 1997; Lefebvre et al., 1997). However, the expression of SOX9 does not always transactivate the Col2a1 gene, especially in nonchondrogenic tissues, suggesting that other factors are also required (Bell et al., 1997). Future challenges lie in identifying those additional factors which bind to and interact with the various activating/repressing DNA modules in vivo, to direct appropriate tissue-specific expression of COL2A1 during key differentiation events in organogenesis and chondrogenesis.
Acknowledgments
We thank S. Zhou (Children's Medical Research Institute) for technical assistance with histology and P. Rowe (Children's Medical Research Institute), P. Koopman (Centre for Molecular and Cellular Biology, Australia), and J. Marsh (Clinical Trials Centre, University of Hong Kong, Hong Kong) for critical reading of the manuscript.
This work was supported by grants from the Research Grants Council (Hong Kong) (HKU356/94M), the Croucher Foundation (Hong Kong) and the Arthritis and Rheumatism Council (UK) (CO523).
Abbreviations used in this paper
- dpc
day(s) post coitum
- nt
nucleotide(s)
Footnotes
Address all correspondence to Professor Kathryn S.E. Cheah, Department of Biochemistry, The University of Hong Kong, Sassoon Road, Hong Kong. Tel.: (852) 2819-9240 or 2819-9170. Fax: (852) 2855-1254. E-mail: hrmbdkc@hkumd1.hku.hk
References
- Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, Cheah KS. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- Braghetta P, Fabbro C, Piccolo S, Marvulli D, Bonaldo P, Volpin D, Bressan GM. Distinct regions control transcriptional activation of the alpha 1(VI) collagen promoter in different tissues of transgenic mice. J Cell Biol. 1996;135:1163–1177. doi: 10.1083/jcb.135.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah KS, Lau ET, Au PK, Tam PP. Expression of the mouse alpha 1(II) collagen gene is not restricted to cartilage during development. Development (Camb) 1991;111:945–953. doi: 10.1242/dev.111.4.945. [DOI] [PubMed] [Google Scholar]
- Cheah KS, Levy A, Trainor PA, Wai AW, Kuffner T, So CL, Leung KK, Lovell RH, Badge, Tam PP. Human COL2A1-directed SV40 T antigen expression in transgenic and chimeric mice results in abnormal skeletal development. J Cell Biol. 1995;128:223–237. doi: 10.1083/jcb.128.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE. Variety in the level of gene control in eukaryotic cells. Nature. 1982;297:365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Dharmavaram RM, Liu G, Mowers SD, Jimenez SA. Detection and characterization of Sp1 binding activity in human chondrocytes and its alterations during chondrocyte dedifferentiation. J Biol Chem. 1997;272:26918–26925. doi: 10.1074/jbc.272.43.26918. [DOI] [PubMed] [Google Scholar]
- Gutman A, Gilthorpe J, Rigby PW. Multiple positive and negative regulatory elements in the promoter of the mouse homeobox gene Hoxb-4. Mol Cell Biol. 1994;14:8143–8154. doi: 10.1128/mcb.14.12.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, B., F. Costantini, and E. Lacy. 1986. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 157–173.
- Horton W, Miyashita T, Kohno K, Hassell JR, Yamada Y. Identification of a phenotype specific enhancer in the first intron of the rat type II collagen gene. Proc Natl Acad Sci USA. 1987;84:8864–8868. doi: 10.1073/pnas.84.24.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip YT, Hemavathy K. Drosophiladevelopment: delimiting patterns by repression. Curr Biol. 1997;7:216–218. doi: 10.1016/s0960-9822(06)00104-7. [DOI] [PubMed] [Google Scholar]
- Kosher RA, Solursh M. Widespread distribution of type II collagen during embryonic chick development. Dev Biol. 1989;131:558–566. doi: 10.1016/s0012-1606(89)80026-0. [DOI] [PubMed] [Google Scholar]
- Kosher RA, Kulyk WM, Gay SW. Collagen gene expression during limb bud differentiation. J Cell Biol. 1986;102:1151–1156. doi: 10.1083/jcb.102.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravis D, Upholt WB. Quantitation of type II procollagen mRNA levels during chick limb cartilage differentiation. Dev Biol. 1985;108:164–172. doi: 10.1016/0012-1606(85)90018-1. [DOI] [PubMed] [Google Scholar]
- Krebsbach PH, Nakata K, Bernier SM, Hatano O, Miyashita T, Rhodes CS, Yamada Y. Identification of a minimum enhancer sequence for the type II collagen gene reveals several core sequence motifs in common with the link protein. J Biol Chem. 1996;271:4298–4303. doi: 10.1074/jbc.271.8.4298. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Zhou G, Mukhopadhyay K, Smith CN, Zhang Z, Eberspaecher H, Zhou X, Sinha S, Maity SN, De Crombrugghe B. An 18-base-pair sequence in the mouse proα1(II) collagen gene is sufficient for expression in cartilage and binds nuclear proteins that are selectively expressed in chondrocytes. Mol Cell Biol. 1996;16:4512–4523. doi: 10.1128/mcb.16.8.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Huang W, Harley VR, Goodfellow PN, De Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the proα1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell Badge, R.H., A. Bygrave, A. Bradley, E. Robertson, R. Tilly, and K.S. Cheah. Tissue-specific expression of the human type II collagen gene in mice. Proc Natl Acad Sci USA. 1987;84:2803–2807. doi: 10.1073/pnas.84.9.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui VCH, Ng LJ, Nicholls J, Tam PPL, Cheah KSE. Tissue-specific and differential expression of alternatively spliced α1(II) collagen mRNAs in early human embryos. Dev Dynamics. 1995;203:198–211. doi: 10.1002/aja.1002030208. [DOI] [PubMed] [Google Scholar]
- Metsäranta M, Garofalo S, Smith C, Niederreither K, De Crombrugghe B, Vuorio E. Developmental expression of a type II collagen/β-galactosidase fusion gene in transgenic mice. Dev Dynamics. 1995;204:202–210. doi: 10.1002/aja.1002040211. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay K, Lefebvre V, Zhou G, Garofalo S, Kimura JH, De Crombrugghe B. Use of a new rat chondrosarcoma cell line to delineate a 119-base pair chondrocyte-specific enhancer element and to define active promoter segments in the mouse pro-α1(II) collagen gene. J Biol Chem. 1995;270:27711–27719. doi: 10.1074/jbc.270.46.27711. [DOI] [PubMed] [Google Scholar]
- Ng LJ, Tam PPL, Cheah KSE. Preferential expression of alternatively spliced mRNAs encoding type II procollagen with a cysteine-rich amino-propeptide in differentiating cartilage and nonchondrogenic tissues during early mouse development. Dev Biol. 1993;159:403–417. doi: 10.1006/dbio.1993.1251. [DOI] [PubMed] [Google Scholar]
- Nonchev S, Vesque C, Maconochie M, Seitanidou T, Ariza L, McNaughton, Frain M, Marshall H, Sham MH, Krumlauf R, Charnay P. Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development (Camb) 1996;122:543–554. doi: 10.1242/dev.122.2.543. [DOI] [PubMed] [Google Scholar]
- Novina CD, Roy AL. Core promoters and transcriptional control. Trends Genet. 1996;12:351–355. [PubMed] [Google Scholar]
- Rossert J, Eberspaecher H, De Crombrugghe B. Separate cis-acting DNA elements of the mouse pro-α1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J Cell Biol. 1995;129:1421–1432. doi: 10.1083/jcb.129.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossert JA, Chen SS, Eberspaecher H, Smith CN, De Crombrugghe B. Identification of a minimal sequence of the mouse pro-alpha 1(I) collagen promoter that confers high-level osteoblast expression in transgenic mice and that binds a protein selectively present in osteoblasts. Proc Natl Acad Sci USA. 1996;93:1027–1031. doi: 10.1073/pnas.93.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LJ. In situ expression of collagen and proteoglycan genes in notochord and during skeletal development and growth. Microsc Res Tech. 1994;28:470–482. doi: 10.1002/jemt.1070280603. [DOI] [PubMed] [Google Scholar]
- Savagner P, Miyashita T, Yamada Y. Two silencers regulate the tissue-specific expression of the collagen II gene. J Biol Chem. 1990;265:6669–6674. [PubMed] [Google Scholar]
- Savagner P, Krebsbach PH, Hatano O, Miyashita T, Liebman J, Yamada Y. Collagen II promoter and enhancer interact synergistically through Sp1 and distinct nuclear factors. DNA Cell Biol. 1995;14:501–510. doi: 10.1089/dna.1995.14.501. [DOI] [PubMed] [Google Scholar]
- Seghatoleslami MR, Lichtler AC, Upholt WB, Kosher RA, Clark SH, Mack K, Rowe DW. Differential regulation of COL2A1 expression in developing and mature chondrocytes. Matrix Biol. 1994;14:753–764. doi: 10.1016/s0945-053x(05)80018-6. [DOI] [PubMed] [Google Scholar]
- Sokolov BP, Ala-Kokko L, Dhulipala R, Arita M, Khillan JS, Prockop DJ. Tissue-specific expression of the gene for type I procollagen (COL1A1) in transgenic mice. Only 476 base pairs of the promoter are required if collagen genes are used as reporters. J Biol Chem. 1995;270:9622–9629. doi: 10.1074/jbc.270.16.9622. [DOI] [PubMed] [Google Scholar]
- Su M, Suzuki HR, Bieker JJ, Solursh M, Ramirez F. Expression of two non-allelic type II procollagen genes during Xenopus laevisembryogenesis is characterized by stage-specific production of alternatively spliced transcripts. J Cell Biol. 1991;115:565–575. doi: 10.1083/jcb.115.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swalla BJ, Upholt WB, Solursh M. Analysis of type II collagen RNA localization in chick wing buds by in situ hybridization. Dev Biol. 1988;125:51–58. doi: 10.1016/0012-1606(88)90057-7. [DOI] [PubMed] [Google Scholar]
- Swift GH, Kruse F, MacDonald RJ, Hammer RE. Differential requirements for cell-specific elastase I enhancer domains in transfected cells and transgenic mice. Genes Dev. 1989;3:687–696. doi: 10.1101/gad.3.5.687. [DOI] [PubMed] [Google Scholar]
- Thorogood P, Bee J, Von der Mark K. Transient expression of collagen type II at epitheliomesenchymal interfaces during morphogenesis of the cartilaginous neurocranium. Dev Biol. 1986;116:497–509. doi: 10.1016/0012-1606(86)90150-8. [DOI] [PubMed] [Google Scholar]
- Tsumaki N, Kimura T. Differential expression of an acidic domain in the amino-terminal propeptide of mouse pro-α2(XI) collagen by complex alternative splicing. J Biol Chem. 1995;270:2372–2378. doi: 10.1074/jbc.270.5.2372. [DOI] [PubMed] [Google Scholar]
- Vikkula M, Metsäranta M, Syvänen A-C, Ala-Kokko L, Vuorio E, Peltonen L. Structural analysis of the regulatory elements of the type-II procollagen gene. Conservation of promoter and first intron sequences between human and mouse. Biochem J. 1992;285:287–294. doi: 10.1042/bj2850287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von der Mark H, Von der Mark K, Gay S. Study of differential collagen synthesis during development of the chick embryo by immunofluorescence. I. Preparation of type I and type II specific antibodies and their applications to early stages of the chick embryo. Dev Biol. 1976;48:237–249. doi: 10.1016/0012-1606(76)90088-9. [DOI] [PubMed] [Google Scholar]
- Wang L, Balakir R, Horton WE. Identification of a cis-acting sequence in the collagen II enhancer required for chondrocyte expression and the binding of a chondrocyte nuclear factor. J Biol Chem. 1991;266:19878–19881. [PubMed] [Google Scholar]
- Wilkinson, D.G. 1992. Whole mount in situ hybridization of vertebrate embryos. In In Situ Hybridization: A Practical Approach. D.G. Wilkinson, editor. IRL Press at Oxford University Press, New York. 75–83.
- Wood A, Ashurst D, Corbett A, Thorogood P. The transient expression of type II collagen at tissue interfaces during mammalian craniofacial development. Development (Camb) 1991;111:955–968. doi: 10.1242/dev.111.4.955. [DOI] [PubMed] [Google Scholar]
- Yee S-P, Rigby PWJ. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- Zhou G, Garofalo S, Mukhopadhyay K, Lefebvre V, Smith CN, Eberspaecher H, De Crombrugghe B. A 182 bp fragment of the mouse proα1(II) collagen gene is sufficient to direct chondrocyte expression in transgenic mice. J Cell Sci. 1995;108:3677–3684. doi: 10.1242/jcs.108.12.3677. [DOI] [PubMed] [Google Scholar]