Abstract
Regulatory T cells (T reg cells) are a population of CD4+ T cells that limit immune responses. FoxP3 is a master control transcription factor for development and function of these cells, but its regulation is poorly understood. We have identified a T cell receptor–responsive enhancer in the FoxP3 first intron that is dependent on a cyclic-AMP response element binding protein (CREB)/activating transcription factor (ATF) site overlapping a CpG island. Methylation of this island inversely correlates with CREB binding and FoxP3 expression. Interestingly, transforming growth factor-β, which induces T reg cell formation, decreases methylation of the CpG island and increases FoxP3 expression. Similarly, inhibiting methylation with 5-azacytidine or knocking down the DNA methyltransferase Dnmt1 also induces FoxP3 expression. Conversely, methylation of the CpG island, which decreases CREB binding or expression of dominant-negative CREB, decreases FoxP3 gene expression. Thus, T cell receptor–induced FoxP3 expression in T reg cells is controlled both by sequence-specific binding of CREB/ATF and by DNA methylation of a CpG island.
Regulatory T cells (T reg cells) are a subpopulation of CD4+ T cells that are hyporesponsive to antigenic stimulation, but exert immunosuppressive effects to control the immune response (1). Several types of T reg cells can be identified based on cell surface markers or cytokine secretion. T reg cells develop in the thymus and express cell surface molecules, including the IL-2 receptor α chain (IL-2Rα), cytotoxic T-lymphocyte antigen-4, glucocorticoid-induced tumor necrosis factor receptor family–related gene, and lymphocyte activation antigen-3. Deficiency or dysfunction of T reg cells can augment tumor immunity, but predispose to autoimmunity; in contrast, an expansion of these cells can promote tolerance of transplanted tissues (1).
FoxP3 is expressed exclusively in T reg cells and regulates their development in the thymus and maintenance in the periphery (2). FoxP3 is mutated in the Scurfy mutant mouse and in patients with IPEX (immune dysregulation, polyendocrino pathy, enteropathy, X-linked) syndrome (3). FoxP3 belongs to the forkhead actor family, containing a highly conserved, C-terminal winged-helix/forkhead, DNA-binding domain, a C2H2 zinc finger, and a leucine zipper (3). The N-terminal domain of FoxP3 is unique to the FOXP subfamily, and it is critical for transcriptional repression (3). FoxP3 can interact with NF-AT and NF-κB to repress cytokine gene expression (4, 5).
In the thymus, intermediate affinity interactions between the TCR and MHC induce FoxP3 expression and promote the development of T reg cells (6). T reg cells can also be generated in the periphery in the context of suboptimal TCR stimulation (7). Production of human thymic stromal lymphopoietin, which is an IL-7–like cytokine that signals via thymic stromal-derived lymphopoietin receptor and IL-7Rα (8), by Hassall's corpuscles was reported to promote the generation of thymic T reg cells by activating dendritic cells (9). Mice lacking CD28 or its ligands, CD80 and CD86, have diminished T reg cells, suggesting a role for these accessory molecules in thymic selection of T reg cells and the maintenance of the peripheral T reg cell compartment (10). Nevertheless, the T reg cells seen in the absence of CD28 have normal levels of IL-2Rα and FoxP3 (11); thus, FoxP3 expression is not absolutely dependent on the CD28 pathway. IL-2 is important for T reg cell development, as shown by diminished FoxP3+ T reg cells in the thymus and peripheral lymphoid organs in Il2 (12), Il2ra (12), or Il2rb (13) knockout mice. Both IL-2 and transforming growth factor (TGF)-β can induce the conversion of CD4+CD25− into CD4+CD25+ T cells with elevated FoxP3 expression in vitro (14), which is consistent with the presence of both IL-2 and TGF-β response elements in the Il2ra gene (15–17).
Studies of FoxP3 regulation have revealed a proximal 5′ regulatory region that was reported to contribute to TCR-mediated regulation of the gene (18), whereas IL-2–induced FoxP3 expression has been attributed to intronic tandem GAS motifs that bind Stat5 in vitro (19). We now identify a potent new TCR response element in the FoxP3 gene and demonstrate DNA methylation-dependent control of FoxP3 gene expression.
RESULTS AND DISCUSSION
Identification of an enhancer-like TCR-response element in the FoxP3 gene
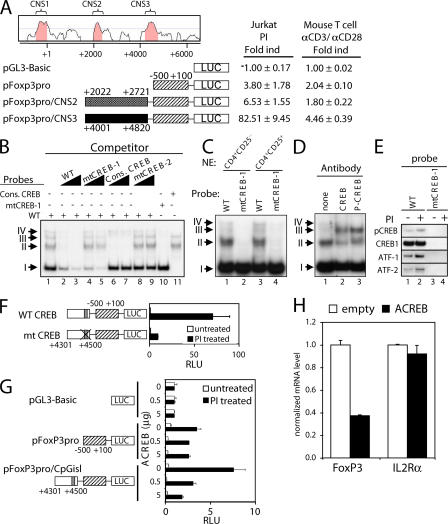
To delineate critical regulatory elements controlling FoxP3 gene expression, we used comparative genomics to identify conserved noncoding sequences (CNSs). Consistent with previous observations (18, 19), using the VISTA computer program, we identified three CNSs with >75% sequence identity between humans and mice. CNS1 is in the promoter region, whereas CNS2 and CNS3 are in the first intron (Fig. 1 A). The promoter region (CNS1; −500 to 100 region) was cloned 5′ to the luciferase reporter gene in pGL3-Basic and transfected into Jurkat T cells. It exhibited an approximately fourfold increase in activity in response to PMA + ionomycin (Fig. 1 A). When CNS2 was cloned upstream of CNS1, only a modest increase was observed, but CNS3 induced a 20-fold increase in activity, which is indicative of a potent enhancer (Fig. 1 A). When the same constructs were transfected into primary splenic T cells, followed by activation with anti-CD3 + anti-CD28, activity was lower than in Jurkat T cells, but the CNS3 region again exhibited enhancer-like activity (Fig. 1 A) that we delineated to the +4,301 to +4,500 region (bottom construct), with 5′ truncation to +4,351 (fourth construct) or 3′ truncation to +4,450 (seventh construct) significantly lowering activity (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20070109/DC1). These results underscore the potency of CNS3.
Figure 1.
Identification of a TCR response element in the FoxP3 gene. (A) A TCR-responsive enhancer in the CNS3 intronic region of the FoxP3 gene. Conserved noncoding regions are shown in the promoter and first intron regions. Jurkat cells were transfected with the indicated constructs. Transfected cells were either untreated or treated with PMA/ionomycin for 17 h. Results are representative of three or more independent experiments (shown as the mean ± the SD). (B and C) EMSAs with the indicated probes; competitor oligonucleotides were added where indicated. In B, nuclear extracts were obtained from anti-CD3/anti-CD28–activated mouse CD4+ splenic T cells. In C, nuclear extracts were obtained from either CD4+CD25− splenic T cells (lanes 1 and 2) or from natural CD4+CD25+ T reg cells (lanes 3 and 4). (D) EMSA in the presence of specific antibody using WT probes and nuclear extracts from anti-CD3/anti-CD28–activated mouse CD4+ splenic T cells. (E) DNA affinity purification, followed by Western blotting for phospho-CREB, CREB, ATF-1, and ATF-2. Nuclear extracts were from human Kit225 cells stimulated with PMA + ionomycin. (F) The CREB binding site in CNS3 is essential for PI-induced FoxP3 promoter activity. A mutation in the CREB binding site was introduced into FoxP3 reporter construct. Constructs were transfected into Jurkat cells, followed by no stimulation or stimulation with PI for 17 h. (G) Jurkat cells were transfected with the FoxP3 reporter construct plus either empty vector or 0.5 or 5 μg of the ACREB vector. Transfected cells were either untreated or treated with PMA/ionomycin for 17 h. (H) MT-2 cells were infected with the indicated lentiviruses, and puromycin-resistant cells were selected. Real-time PCR was used to analyze FoxP3 and IL-2Rα mRNA levels and normalized to the mRNA level of GAPDH. Results in F–H are representative of three or more independent experiments (shown as the mean ± the SD).
CREB/ATF is important for the activity of the TCR response element
The sequences of the FoxP3 promoter and intronic regions are shown in Fig. S2 (A and B, respectively; available at http://www.jem.org/cgi/content/full/jem.20070109/DC1). The promoter region contains sequence motifs that resemble NF-AT and AP-1 motifs and a GC box (Fig. S2 A) (18). The intronic region contains two GAS motifs (19); additionally, we identified a TGACGTCA putative cyclic-AMP response element binding protein (CREB)/activating transcription factor (ATF) motif with an overlapping YY1 motif (Fig. S2 B).
CREB and a family of ATFs are transcriptional activators (20), so we investigated if the putative FoxP3 CREB/ATF site is important for the activity of the intronic TCR response element. We evaluated CREB binding to this motif using electrophoretic mobility shift assays (EMSAs), with DNA probes spanning the CREB site at +4,470 (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20070109/DC1). For these studies, we used nuclear extracts from primary mouse splenic CD4+ T cells expanded with anti-CD3, anti-CD28, and IL-2 for 6 d. EMSAs revealed four complexes (denoted I, II, III, and IV; Fig. 1 B, lane 1) whose formation was inhibited by excess unlabeled probe (lanes 2 and 3). In contrast, an oligonucleotide containing a mutant CREB site (mtCREB-1) had little effect on the formation of complexes II and III, but still inhibited complex I (Fig. 1 B, lanes 4 and 5). Consistent with this, the formation of complexes II and III, but not of complex I, was inhibited by a consensus CREB binding site oligonucleotide (Fig. 1 B, lanes 6 and 7), whereas an oligonucleotide with a mutant CREB site (mtCREB-2) did not inhibit formation of any of the complexes (lanes 8 and 9). When EMSAs were performed with the mtCREB-1 probe, complexes II and III were not detected (Fig. 1 B, lane 10), and only complexes II and III were detected with a probe containing a consensus CREB binding site but lacking relevant surrounding sequences (lane 11). Similar CREB binding activity was observed using nuclear extracts from CD4+CD25+ natural T reg cells (Fig. 1 C, lanes 3 and 4 vs. 1 and 2). Antibodies specific for CREB or phospho-CREB supershifted complex II (Fig. 1 D), confirming CREB binding; because of the position of the supershifted band, the effect of the antibody on complex III could not be determined. ATF1 and ATF2 have similar binding specificity to CREB (20), and like CREB, both ATF1 and ATF2 also bind to a WT, but not mutant, probe (Fig. 1 E).
We next investigated the functional significance of the CREB/ATF sites. Mutation of the CREB site in the +4,301 to +4,500 TCR response element greatly decreased activity (Fig. 1 F). Moreover, cotransfection of 0.5 or 5 μg of a dominant-negative CREB construct (denoted “ACREB”) that can inhibit not only CREB but also ATF1 and ATF2 (21) diminished PI inducibility of the CNS3-containing construct in a dose-responsive fashion, whereas the empty expression vector had no effect (Fig. 1 G, bottom). To confirm the importance of CREB/ATF, we examined the ability of an ACREB-expressing lentivirus to lower FoxP3 expression in HLTV-I transformed MT-2 cells, which constitutively express FoxP3 (22). Compared with a control “empty” virus, the ACREB-expressing virus significantly decreased FoxP3 expression, but not IL-2Rα expression, which was used as an internal control (Fig. 1 H). We also observed that complex I contains YY1 (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20070109/DC1); however, mutation of the YY1 site did not affect activity, and knocking down YY1 expression with a shRNA construct also did not decrease FoxP3 expression (unpublished data). Thus, CREB/ATF, but not YY1, is a critical component of the TCR response element.
Inverse correlation of FoxP3 expression and methylation of the intronic CpG island
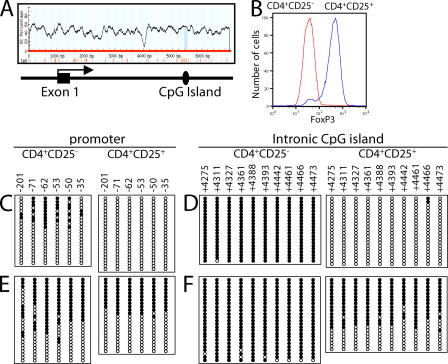
Because of the lineage- and tissue-specific expression of the FoxP3 gene and because CREB binding can be inhibited by CpG methylation (23), we hypothesized that DNA methylation might be an important control mechanism for FoxP3 expression. Strikingly, analysis of the FoxP3 genomic locus with the MethPrimer computer program (24) revealed a single putative CpG island in the +4,393 to +4,506 region (Fig. 2 A) of CNS3. To evaluate the methylation status of CpG sequences in the promoter and CNS3 regions, we isolated FoxP3−CD4+CD25− naive splenic T cells and FoxP3+CD4+CD25+ regulatory T cells (Fig. 2 B). For the −250 to +1 promoter region, we sequenced the sense strand of ≥20 individual DNA clones and found that in male mice 10–45% of the CpG sites were methylated in CD4+CD25− cells, whereas none were methylated in CD4+CD25+ T reg cells (Fig. 2 C and Fig. S5 A, available at http://www.jem.org/cgi/content/full/jem.20070109/DC1). Thus, even though the FoxP3 gene is not expressed in CD4+CD25− cells, less than half of the CpG sites in the promoter region were methylated in these cells. We also analyzed the CpG sites in the +4,201 to +4,500 intronic CpG island. In CD4+CD25+ T reg cells from male mice, almost no methylation of these sites was found, similar to the promoter region, but in CD4+CD25− T cells, the intronic CpG sites were almost completely methylated (Fig. 2 D and Fig. S5 B), suggesting that methylation of sites in the CpG island inhibits FoxP3 expression in CD4+CD25− T cells.
Figure 2.
DNA methylation status of CpG sites in the promoter and intronic CpG island region of the FoxP3 gene. (A) Identification of a CpG island in the first intron that corresponds to the TCR response element that binds CREB. (B) Expression of FoxP3 in CD4+CD25− versus CD4+CD25+ cells. (C–F) The DNA methylation status for these regions was determined from male (C and D) or female (E and F) C57BL/6 mice by bisulfite sequencing analysis. Each line represents one DNA strand; open circle, unmethylated CpGs; filled circle, methylated CpGs.
We also examined the DNA methylation status of the FoxP3 promoter and CpG island regions in female mice (Fig. 2, E and F; and Fig. S5, A and B). In the promoter region, female mice exhibited greater methylation in CD4+CD25− T cells and ∼50% methylation in CD4+CD25+ T reg cells (Fig. 2 E). In the intronic CpG island, they exhibited essentially complete methylation in the CD4+CD25− T cells, but only 45–68% methylation in the CD4+CD25+ T reg cells (Fig. 2 F). Because the FoxP3 gene is located on chromosome X, the difference in methylation of the intronic CpG island region in male versus female T reg cells may result from the methylation status of the inactivated X chromosome. Thus, FoxP3 expression inversely correlates with the methylation status of the intronic CpG island.
Like the splenic T reg cells, thymic CD4+CD25+ T reg cells from C57BL/6 mice exhibited little methylation in the promoter region (Fig. S6 A, far right, available at http://www.jem.org/cgi/content/full/jem.20070109/DC1) and ∼70% demethylation in the intronic CpG island region (Fig. S6 B, far right). However, cell populations that do not express FoxP3, including CD4−CD8− DN, CD4+CD8+ DP, CD4+CD8−CD25−, and CD4−CD8+CD25− SP thymocytes, had almost complete methylation of the intronic CpG island (Fig. S6, B [first 4 panels] and C), even though they were only partially methylated in the promoter region, further inversely correlating FoxP3 expression and methylation of the intronic CpG island.
To determine if the demethylated status in T reg cells was specific for the FoxP3 gene or a more general phenomenon, we examined the methylation status of the genes encoding IL-2 and IFN-γ (Fig. S7, A and B, available at http://www.jem.org/cgi/content/full/jem.20070109/DC1), focusing on sites whose methylation is known to be important for the regulation of these genes, either before versus after T cell activation (for the Il2 gene) (25, 26) or in Th1 versus Th2 cells (for the Ifng gene) (27). Unlike FoxP3, neither of these genes is expressed in T reg cells. The methylation status for the 5′ regulatory regions for both genes was similar in CD4+CD25− or CD4+CD25+ cell populations (Fig. S7, A–C), which is consistent with neither Il2 nor Ifng genes being expressed in the cell populations, thus further supporting the specificity of our findings for the differential methylation of the FoxP3 gene in these two populations.
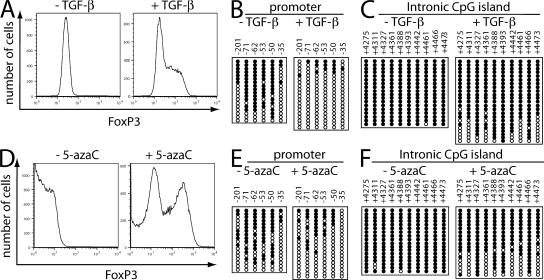
TGF-β–induced FoxP3 expression correlated with hypomethylation of the CpG intronic island
In addition to naturally occurring thymus-derived T reg cells, TGF-β can induce FoxP3 expression in splenic CD4+CD25− T cells and their differentiation into T reg cells. We thus investigated the effect of TGF-β on methylation of the FoxP3 locus. Naive CD4+CD25− splenic T cells from male C57BL/6 mice were purified by fluorescence-activated cell sorting, activated with anti-CD3 + anti-CD28, and expanded with 100 U/ml IL-2. FoxP3 expression was induced by treatment with TGF-β (Fig. 3 A). After 6 d in culture, the methylation status was assayed. In CD4+CD25− T cells not treated with TGF-β, which do not express FoxP3, the promoter region was more methylated than in naive CD4+CD25− T cells (see the CpG sites at −71, −62, −53, and −50; Fig. 3 B, left, vs. Fig. 2 C, left). When the cells were cultured with TGF-β, which induces FoxP3 expression, the promoter region was less methylated than in naive CD4+CD25− cells (Fig. 3 B, right, vs. Fig. 2 C, left). The intronic CpG island remained fully methylated in the absence of TGF-β (Fig. 3 C, left, vs. Fig. 2 D, left), but the CpG sites at +4,275, +4,311, +4,361, +4,461, and +4,473 exhibited partial demethylation after treatment with TGF-β (Fig. 3 C, right). Thus, TGF-β induced partial demethylation at the promoter and intronic CpG island, correlating with increased FoxP3 expression (Fig. S8 A, available at http://www.jem.org/cgi/content/full/jem.20070109/DC1).
Figure 3.
Expression of the FoxP3 gene in TGF-β–stimulated T reg cells correlates with DNA demethylation, and treatment with 5-azacytidine stimulates FoxP3 expression in non–T reg cells. (A–C) CD4+CD25− cells from male C57BL/6 mice isolated by FACS sorting were stimulated with 5 μg/ml of plate-bound anti-CD3 + 0.5 μg/ml anti- CD28 + 100 U/ml IL-2 in the absence or presence of 5 ng/ml TGF-β for 6 d. (A) The cells were stained with APC-CD4, PerCP-Cy5.5-CD25, and PE-FoxP3. Graphs are shown for FoxP3 expression, gated for live cells. (B and C) The methylation status of CpG sites in the promoter region (B) or the intronic CpG island region (C) was determined by bisulfite sequencing analysis. (D–F) CD4+CD25− cells from male C57BL/6 mice were isolated by FACS sorting and stimulated with 5 μg/ml anti-CD3, 0.5 μg/ml anti-CD28, and 100 U/ml IL-2 for 6 d, and 5-azacytidine was added for days 4–6. (D) The cells were stained with APC-CD4, PerCP-Cy5.5-CD25, and PE-FoxP3. Graphs are shown for FoxP3 expression, gated for live cells. (E and F) The methylation status of CpG sites in the promoter region (E) or the intronic CpG island region (F) was determined by bisulfite sequencing analysis. Each line represents a DNA strand; open circle, unmethylated CpGs; filled circle, methylated CpGs.
Treatment with 5-azacytidine induces FoxP3 expression
To further test whether DNA methylation repressed FoxP3 expression, we used 5-azacytidine, which is a cytosine nucleoside analogue that inhibits DNA methyltransferase and which was previously shown to augment FoxP3 expression in human NK cells (19). CD4+CD25− cells were stimulated with anti-CD3 + anti-CD28 and IL-2 for 6 d. When cells were treated with 5-azacytidine from day 4 to 6, FoxP3 expression increased (Fig. 3 D), and DNA methylation in the promoter (Fig. 3 E) and intronic CpG island (Fig. 3 F) significantly decreased (Fig. S8 B), supporting the hypothesis that DNA methylation prevents FoxP3 expression in non–FoxP3-producing CD4+CD25− T cells. A similar trend in methylation was seen after treatment with TGF-β (Fig. 3, A–C) or 5-azacytidine (Fig. 3, D–F).
We also studied the effect of 5-azacytidine in mouse cytotoxic CTLL-2 CD8+ T cells. 5-azacytidine markedly increased FoxP3 expression (Fig. S9 A, available at http://www.jem.org/cgi/content/full/jem.20070109/DC1), and the promoter and intronic CpG island region, which were highly methylated in the untreated cells, became substantially less methylated (Fig. S9, B–D), again, correlating hypomethylation with FoxP3 expression, even in a CD8+ cytotoxic T cell line where FoxP3 is not normally expressed.
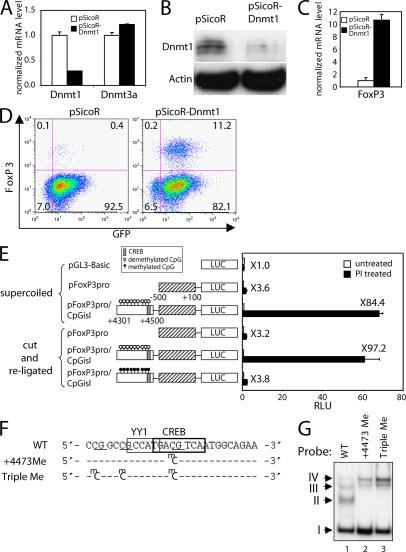
Decreasing Dnmt1 induces FoxP3 expression
The major DNA methyltransferase responsible for maintaining DNA methylation is Dnmt1 (28). We further assessed the role of DNA methylation for FoxP3 gene expression in vivo by using a lentiviral vector and RNA interference to decrease Dnmt1 expression. CTLL-2 cells that were GFP+ (indicative of successful viral transduction) were sorted by FACS. The expression of Dnmt1 mRNA (Fig. 4 A) and protein (Fig. 4 B) was markedly decreased in CTLL-2 cells infected with lentivirus containing shRNA against Dnmt1, whereas Dnmt3a mRNA was unaffected (Fig. 4 A). Strikingly, FoxP3 mRNA (Fig. 4 C) and protein (Fig. 4 D) were increased in Dnmt1-diminished cells within 4 d after infection, with 15–20% of GFP-positive cells expressing FoxP3 by day 7 (Fig. 4 D), indicating an essential role for Dnmt1 in repressing FoxP3 gene expression.
Figure 4.
Knockdown of Dnmt1 induces FoxP3 expression. (A–D) CTLL-2 cells were infected with the indicated lentiviruses, and GFP-positive cells were sorted. (A) Real-time PCR was performed to analyze the mRNA levels of Dnmt1 and Dnmt3a and normalized with the 18S rRNA level. (B) Whole-cell lysates were separated with SDS-PAGE and were subjected to Western blotting using antibodies to Dnmt1 and actin. (C) Real-time PCR was performed to analyze the mRNA levels of FoxP3 expression and normalized with the 18S rRNA level. Results in A and C are representative of at least three independent experiments (shown as the mean ± the SD). (D) Dot plot showing the expression of FoxP3 in CTLL-2 cells infected with the indicated lentiviruses. (E–G) Essential role of CREB and its methylation for FoxP3 expression. (E) Inhibition of the FoxP3 promoter activity by methylation at CpG sites in the intronic CpG island. Jurkat cells were transfected with the indicated constructs. Transfected cells were either untreated or treated with PMA/ionomycin for 17 h. Results in E are representative experiments (shown as the mean ± the SD). (F) Sequence of DNA probes used in EMSAs in G. (G) EMSA using a WT probe or probes methylated at the indicated CpGs and nuclear extracts from mouse splenic CD4+ T cells expanded with anti-CD3, anti-CD28, and IL-2 for 6 d.
DNA methylation of the CpG island inhibits FoxP3 promoter activity and CREB binding
The ability of 5-azacytidine and the Dnmt1 shRNA to activate FoxP3 gene expression could be indirect or direct. We thus investigated if DNA methylation of the CpG island could inhibit CNS3-mediated reporter activity and CREB DNA-binding activity. The +4,301 to +4,500 region was either not methylated or was methylated using SssI (CpG) methylase and S-adenosylmethionine and inserted into unmethylated pFoxP3Pro. A transient reporter assay showed an almost complete reduction in luciferase activity after methylation of the intronic CpG island (Fig. 4 E, sixth construct), but not with the unmethylated CpG island (fifth construct), indicating that DNA methylation can directly inhibit its transcriptional activating potential. This is consistent with a direct effect of 5-azacytidine and the Dnmt1 shRNA.
We identified CREB/ATF as a regulator of FoxP3 expression and provided data that methylation of the CpG island in CNS3 can inhibit FoxP3 promoter activity. To directly study the effect of DNA methylation on CREB binding activity, we performed EMSAs using DNA probes spanning the FoxP3 +4,473 CREB motifs that were either unmethylated, methylated only at the +4,473 CpG, or simultaneously methylated at the +4,461, +4,466, and +4,473 CpGs (Fig. 4 F). As in Fig. 1 B, EMSAs performed with the unmethylated oligonucleotide probe showed complexes I–IV (Fig. 4 G, lane 1). In contrast, the same sequence methylated at only the +4,473 position or at all three sites abolished formation of CREB complexes II and III, whereas complexes I and IV remained and a new complex appeared (Fig. 4 G, lanes 2 and 3). Thus, methylation of the CpG site at +4,473 or all three CpG sites inhibit in vitro binding of CREB, but not YY1.
The FoxP3 promoter and CNS3 regions have open chromatin structures
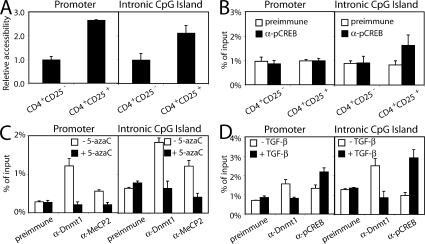
We next investigated whether the FoxP3 locus exists in an open or closed chromatin conformation using a restriction enzyme accessibility (REA) assay. As compared with naive CD4+CD25− T cells, higher REA was observed in CD4+CD25+ T cells at both the promoter and CNS3 (Fig. 5 A) regions, suggesting that an open chromatin structure in these regions facilitates transcription of the FoxP3 gene and further establishing the importance of CNS3 and CREB/ATF in FoxP3 gene regulation. When we examined phospho-CREB binding in vivo by chromatin immunoprecipitation (ChIP) assay, we did not observe binding to the promoter region in either CD4+CD25− or CD4+CD25+ cells (Fig. 5 B); however, in vivo phospho-CREB binding was observed in the CNS3 intronic CpG island region, but only in T reg cells (Fig. 5 B), which is consistent with the vital role of CREB for FoxP3 expression. Having shown that methylation could inhibit reporter activity of CNS3, as well as in vitro binding of CREB/ATF, we sought to clarify the basis for FoxP3 methylation, examining the interaction of DNA methyltransferase and methyl-CpG–binding proteins at the FoxP3 locus. MeCP2, like other methyl-CpG–binding proteins, can bind sequences containing methylated cytosines and repress transcription (28). Chromatin from CTLL-2 cells that were untreated or treated with 5-azacytidine for 3 d was precipitated with antibodies to Dnmt1 or MeCP2, and the precipitated DNA was amplified using primer sets for the promoter or CpG island region of the FoxP3 gene. This ChIP analysis revealed Dnmt1 and MeCP2 binding to both the promoter and the intronic CpG island (Fig. 5 C) regions in untreated CTLL-2 cells, with a significant reduction after treatment with 5-azacytidine (Fig. 5 C), correlating with the lower methylation seen after this treatment. Moreover, when primary mouse CD4+ T cells were cultured with anti-CD3, anti-CD28, and IL-2, the addition of TGF-β resulted in decreased Dnmt1 binding to the FoxP3 locus in vivo, as evaluated by ChIP. In contrast, in vivo binding of phospho-CREB was increased in TGF-β–treated cells (Fig. 5 D), again, inversely correlating DNA methylation with factor binding in vivo.
Figure 5.
REA assay with RsaI at the promoter and CNS3 CpG island regions of the FoxP3 locus. (A) The data were quantitated using a two-step nested real-time PCR strategy and expressed as a ratio of digestion at the FoxP3 promoter or CNS3 to digestion at the Actin gene locus. (B) ChIP at the promoter and CNS3 CpG island regions of the FoxP3 locus revealing phospho-CREB binding only to the CpG island region in CD4+CD25+ T reg cells. (C) CTLL-2 cells cultured in the presence of 100 U/ml IL-2 for 6 d were incubated with 5-azacytidine for the last 3 d, and ChIP assays were performed with preimmune serum or antibodies to MeCP2 or Dnmt1. (D) CD4+CD25− cells from male C57BL/6 mice isolated by FACS sorting were stimulated with 5 μg/ml anti-CD3, 0.5 μg/ml anti-CD28, and 100 U/ml IL-2 in the absence or presence of 5 ng/ml TGF-β for 6 d. ChIP assays were performed with preimmune serum or antibodies to Dnmt1 or phospho-CREB. Results are representative of three or more independent experiments (shown as the mean ± the SD).
Conclusion and model
We found a new TCR response element in the first intron of the FoxP3 gene that is located within a CNS. This CNS contains a CpG island that we have shown is regulated by CREB. Epigenetic modifications have been implicated in gene regulation during embryonic development, genomic imprinting, and X chromosome inactivation (28). In mammals, epigenetic regulation is mediated by changes in chromatin structure, resulting from either histone modification or DNA methylation. We have shown that DNA methylation can influence long-lasting changes in FoxP3 expression during T reg cell development.
Fig. S10 (available at http://www.jem.org/cgi/content/full/jem.20070109/DC1) shows a model focusing on the intronic CNS3 region that integrates the contribution of sequence-specific transcription factors, including CREB (based on this study) and Stat5 proteins (19), as well as DNA methylation in the regulation of the FoxP3 gene. In T reg cells, IL-2 and TCR promote the recruitment of Stat5 and CREB, thus driving FoxP3 transcription, whereas in non–T reg cells, these critical sites are methylated, inhibiting transcription factor binding, and thus preventing FoxP3 transcription. Overall, our studies substantially extend our knowledge of FoxP3 regulation, with our discovery of a novel CREB-dependent TCR response element that is regulated by methylation to control expression of a gene that exhibits exquisite lineage-restricted expression.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were obtained from The Jackson Laboratory. All experiments were performed under protocols approved by the National Heart, Lung, and Blood Institute (NHLBI) Animal Use and Care Committee and followed the National Institutes of Health guidelines for Using Animals in Intramural Research. CD4+CD25− and CD4+CD25+ T cells were purified from the spleens of C57BL/6 mice by FACS. The purity of each cell population was >98%.
Antibodies, cell staining, and flow cytometry.
Conjugated antibodies were purchased from BD Biosciences and eBioscience. A FACSort (Becton Dickinson) was used for flow cytometry, and data were analyzed with FlowJo software. For intracellular FoxP3 flow cytometry, cells were stained using a standard protocol according to the manufacturer's instructions (eBioscience).
Plasmid constructs.
The −500 to +100 region of the FoxP3 gene was PCR amplified and cloned into pGL3-Basic to yield pFoxP3pro. The +2,022 to +2,721 and +4,001 to +4,820 regions were PCR amplified and inserted into pFoxP3pro to yield pFoxP3pro/CNS2 and pFoxP3pro/CNS3. The CpG region between +4,301 and +4,500 was similarly inserted into FoxP3Pro to yield FoxP3Pro/CpGisl. The QuikChange Site-Directed Mutagenesis kit (Stratagene) was used to mutate the CREB site (+4,473) in FoxP3Pro/CpGisl. The dominant-negative CREB construct (ACREB) was provided by J.-M. Park (Harvard Medical School, Boston, MA) and subcloned into pLentilox RSV vector, which has a puromycin marker. The plasmids encoding shRNA against Dnmt1 were provided by T. Jacks (Massachussetts Insitute of Technology, Cambridge, MA) (29).
Lentiviral transduction.
293T cells (8 × 106) were seeded onto 150-mm dishes, and 16 h later they were transiently transfected with 1 μg of lentiviral vector and 3 μg of each packaging vector (Invitrogen) by using Effectene (QIAGEN). Supernatants were collected 48 h after transfection, filtered through a 0.45-μM filter, and centrifuged at 25,000 rpm for 2 h. The viral pellet was resuspended in RPMI medium. MT-2 or CTLL-2 cells (106) were seeded into 24-well plates, and 6 μg/ml of polybrene and virus were added at a 5:1 (virus/cell) multiplicity of infection. Cells were spin infected for 90 min at 2,500 rpm and cultured at 37°C.
Transient transfections and luciferase assays.
Jurkat cells were transiently transfected using DEAE-dextran. Transient transfections of normal mouse T cells were performed by electroporation (30).
EMSA.
EMSAs were performed as previously described (16). For supershifting assays, nuclear extracts were preincubated for 10 min with CREB and phospho-CREB (Millipore). The oligonucleotide probes are shown in the Supplemental materials and methods (available at http://www.jem.org/cgi/content/full/jem.20070109/DC1) and in Fig. 4 F.
Quantitative RT-PCR.
Total RNA was isolated from cells using RNeasy (QIAGEN). First-strand cDNAs were made using the Omniscript RT kit (QIAGEN). Quantitative real-time PCR was performed on a 7900H sequence detection system (Applied Biosystems). TaqMan Gene Expression Assays (Applied Biosystems) were used to measure the mRNA levels of human FOXP3, human IL-2Rα, human GAPDH, mouse Dnmt1, mouse Dnmt3a, and mouse FOXP3.
Methylation analysis.
Genomic DNA was purified with the Wizard genomic DNA purification kit (Promega). Methylation analysis was performed by bisulfite conversion of genomic DNA using the MethylDetector kit (Active Motif). The primer sequences to amplify the FoxP3 promoter region and the FoxP3 CpG island region are listed in the Supplemental materials and methods. The PCR product was cloned using the TOPO TA Cloning kit (Invitrogen).
Plasmid methylation.
For plasmid methylation, the +4,301 to +4,500 CpG island region was amplified by PCR, and 5 μg of PCR product was incubated with 80 U of SssI methylase and 640 μM S-adenosylmethionine (New England Biolabs) for 2 h at 37°C. DNA methylation was verified using AatII, which is a methylation-sensitive restriction enzyme. The pGL3-Basic vector containing the FoxP3 promoter region was cut with KpnI and SacI and ligated with an equimolar concentration of the methylated or unmethylated CpG island region at 16°C for 2 h. DNA was purified using the Qia-Quick method (QIAGEN), and 4 μg of ligated DNA was transfected into Jurkat cells using DEAE-Dextran.
REA assay.
REA assays were performed as previously described (17). The sequences of primers and probes used for the PCR are listed in the Supplemental materials and methods.
ChIP.
ChIP assays were performed as previously described (17). The primers and Taqman probe sequences for the FoxP3 promoter and the intronic CpG island are listed in the Supplemental materials and methods.
Online supplemental material.
Fig. S1 shows the delineation of CNS3 region as a TCR response element. Fig. S2 shows the sequence of the promoter and CNS3. Fig. S3 shows sequence of the EMSA probes. Fig. S4 shows that complex I contains YY1. Fig. S5 shows a summary of the data in Fig. 2 (C–F). Fig. S6 shows the DNA methylation status of CpG sites in the promoter and intronic CpG island region of the FoxP3 gene in thymocytes. Fig. S7 shows the DNA methylation status of CpG sites in the IL-2 and IFN-γ genes. Fig. S8 shows the summary of the data in Fig. 3 (B, C, E, and F). Fig. S9 shows that treatment with 5-azacytidine stimulates FoxP3 expression in CTLL-2 cells. Fig. S10 shows the schematic of FoxP3 gene regulation in T reg and non–T reg cells. The Supplemental materials and methods show the sequence of primer and probe used in the experiment. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20070109/DC1.
Supplemental Material
Acknowledgments
We thank Drs. Keji Zhao (NHLBI), Rosanne Spolski (NHLBI), Hyok Joon Kwon (NHLBI), Jian-Xin Lin (NHLBI), Kuan-Teh Jeang (National Institute of Allergy and Infectious Disease), Eui-Cheol Shin (National Institute of Diabetes and Digestive and Kidney Disease), Kye-Yoon Park (National Institute on Neurological Disorders and Stroke), and Karl Pfeifer (National Institute of Child Health and Human Development) for critical comments and valuable discussions; Dr. Jin Mo Park (Harvard Medical School) for the ACREB construct; Dr. Tyler Jacks (Massachussetts Institute of Technology) for the Dnmt1 shRNA construct; and Dr. J. Philip McCoy (NHLBI) and Leigh Samsel (NHLBI) for FACS sorting.
This research was supported by the Intramural Research Program, NHLBI, at the National Institutes of Health.
The authors have no conflicting financial interests.
Note added in proof. While this manuscript was under review, Floess et al. also reported the role of DNA methylation at CNS3 on FoxP3 expression (Floess, S., J. Freyer, C. Siewert, U. Baron, S. Olek, J. Polansky, K. Schlawe, H.D. Chang, T. Bopp, E. Schmitt, S. Klein-Hessling, E. Serfling, A. Hamann, J. Huehn. 2007. PLoS Biol. 5:e38).
References
- 1.Sakaguchi, S., M. Ono, R. Setoguchi, H. Yagi, S. Hori, Z. Fehervari, J. Shimizu, T. Takahashi, and T. Nomura. 2006. Foxp3CD25CD4 natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol. Rev. 212:8–27. [DOI] [PubMed] [Google Scholar]
- 2.Fontenot, J.D., M.A. Gavin, and A.Y. Rudensky. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4:330–336. [DOI] [PubMed] [Google Scholar]
- 3.Ziegler, S.F. 2006. FOXP3: of mice and men. Annu. Rev. Immunol. 24:209–226. [DOI] [PubMed] [Google Scholar]
- 4.Bettelli, E., M. Dastrange, and M. Oukka. 2005. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc. Natl. Acad. Sci. USA. 102:5138–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu, Y., M. Borde, V. Heissmeyer, M. Feuerer, A.D. Lapan, J.C. Stroud, D.L. Bates, L. Guo, A. Han, S.F. Ziegler, et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 126:375–387. [DOI] [PubMed] [Google Scholar]
- 6.Maloy, K.J., and F. Powrie. 2001. Regulatory T cells in the control of immune pathology. Nat. Immunol. 2:816–822. [DOI] [PubMed] [Google Scholar]
- 7.Kretschmer, K., I. Apostolou, D. Hawiger, K. Khazaie, M.C. Nussenzweig, and H. von Boehmer. 2005. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 6:1219–1227. [DOI] [PubMed] [Google Scholar]
- 8.Pandey, A., K. Ozaki, H. Baumann, S.D. Levin, A. Puel, A.G. Farr, S.F. Ziegler, W.J. Leonard, and H.F. Lodish. 2000. Cloning of a receptor subunit required for signaling by thymic stromal lymphopoietin. Nat. Immunol. 1:59–64. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe, N., Y.H. Wang, H.K. Lee, T. Ito, Y.H. Wang, W. Cao, and Y.J. Liu. 2005. Hassall's corpuscles instruct dendritic cells to induce CD4+CD25+ regulatory T cells in human thymus. Nature. 436:1181–1185. [DOI] [PubMed] [Google Scholar]
- 10.Salomon, B., D.J. Lenschow, L. Rhee, N. Ashourian, B. Singh, A. Sharpe, and J.A. Bluestone. 2000. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 12:431–440. [DOI] [PubMed] [Google Scholar]
- 11.Sansom, D.M., and L.S. Walker. 2006. The role of CD28 and cytotoxic T-lymphocyte antigen-4 (CTLA-4) in regulatory T-cell biology. Immunol. Rev. 212:131–148. [DOI] [PubMed] [Google Scholar]
- 12.Fontenot, J.D., J.P. Rasmussen, M.A. Gavin, and A.Y. Rudensky. 2005. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6:1142–1151. [DOI] [PubMed] [Google Scholar]
- 13.Malek, T.R., A. Yu, V. Vincek, P. Scibelli, and L. Kong. 2002. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 17:167–178. [DOI] [PubMed] [Google Scholar]
- 14.Chen, W., W. Jin, N. Hardegen, K.J. Lei, L. Li, N. Marinos, G. McGrady, and S.M. Wahl. 2003. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, H.P., J. Imbert, and W.J. Leonard. 2006. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 17:349–366. [DOI] [PubMed] [Google Scholar]
- 16.Kim, H.P., J. Kelly, and W.J. Leonard. 2001. The basis for IL-2-induced IL-2 receptor alpha chain gene regulation: importance of two widely separated IL-2 response elements. Immunity. 15:159–172. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H.P., B.G. Kim, J. Letterio, and W.J. Leonard. 2005. Smad-dependent cooperative regulation of interleukin 2 receptor alpha chain gene expression by T cell receptor and transforming growth factor-beta. J. Biol. Chem. 280:34042–34047. [DOI] [PubMed] [Google Scholar]
- 18.Mantel, P.Y., N. Ouaked, B. Ruckert, C. Karagiannidis, R. Welz, K. Blaser, and C.B. Schmidt-Weber. 2006. Molecular mechanisms underlying FOXP3 induction in human T cells. J. Immunol. 176:3593–3602. [DOI] [PubMed] [Google Scholar]
- 19.Zorn, E., E.A. Nelson, M. Mohseni, F. Porcheray, H. Kim, D. Litsa, R. Bellucci, E. Raderschall, C. Canning, R.J. Soiffer, et al. 2006. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 108:1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2:599–609. [DOI] [PubMed] [Google Scholar]
- 21.Ahn, S., M. Olive, S. Aggarwal, D. Krylov, D.D. Ginty, and C. Vinson. 1998. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol. 18:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, S., N. Ishii, S. Ine, S. Ikeda, T. Fujimura, L.C. Ndhlovu, P. Soroosh, K. Tada, H. Harigae, J. Kameoka, et al. 2006. Regulatory T cell-like activity of Foxp3+ adult T cell leukemia cells. Int. Immunol. 18:269–277. [DOI] [PubMed] [Google Scholar]
- 23.Iguchi-Ariga, S.M., and W. Schaffner. 1989. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 3:612–619. [DOI] [PubMed] [Google Scholar]
- 24.Li, L.C., and R. Dahiya. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 18:1427–1431. [DOI] [PubMed] [Google Scholar]
- 25.Bruniquel, D., and R.H. Schwartz. 2003. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat. Immunol. 4:235–240. [DOI] [PubMed] [Google Scholar]
- 26.Thomas, R.M., L. Gao, and A.D. Wells. 2005. Signals from CD28 induce stable epigenetic modification of the IL-2 promoter. J. Immunol. 174:4639–4646. [DOI] [PubMed] [Google Scholar]
- 27.Jones, B., and J. Chen. 2006. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 25:2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaenisch, R., and A. Bird. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33:245–254. [DOI] [PubMed] [Google Scholar]
- 29.Ventura, A., A. Meissner, C.P. Dillon, M. McManus, P.A. Sharp, L. Van Parijs, R. Jaenisch, and T. Jacks. 2004. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. USA. 101:10380–10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, H.P., and W.J. Leonard. 2002. The basis for TCR-mediated regulation of the IL-2 receptor alpha chain gene: role of widely separated regulatory elements. EMBO J. 21:3051–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.