Abstract
The role of CD4+ T cells in the control of persistent viral infections beyond the provision of cognate help remains unclear. We used polychromatic flow cytometry to evaluate the production of the cytokines interferon (IFN)-γ, tumor necrosis factor (TNF)-α, and interleukin (IL)-2, the chemokine macrophage inflammatory protein (MIP)-1β, and surface mobilization of the degranulation marker CD107a by CD4+ T cells in response to stimulation with cytomegalovirus (CMV)-specific major histocompatibility complex class II peptide epitopes. Surface expression of CD45RO, CD27, and CD57 on responding cells was used to classify CD4+ T cell maturation. The functional profile of virus-specific CD4+ T cells in chronic CMV infection was unique compared with that observed in other viral infections. Salient features of this profile were: (a) the simultaneous production of MIP-1β, TNF-α, and IFN-γ in the absence of IL-2; and (b) direct cytolytic activity associated with surface mobilization of CD107a and intracellular expression of perforin and granzymes. This polyfunctional profile was associated with a terminally differentiated phenotype that was not characterized by a distinct clonotypic composition. Thus, mature CMV-specific CD4+ T cells exhibit distinct functional properties reminiscent of antiviral CD8+ T lymphocytes.
Human CMV is a β herpes virus that establishes lifelong infection. Endothelial, renal epithelial, and pulmonary tissue as well as myeloid cells all contain latent CMV. Seroprevalence increases with age and reaches 30–70% in developed countries (1). Although serious disease can occur, it is rare. In most immunocompetent individuals, infection is asymptomatic (2).
The human immune system dedicates tremendous resources to the control of CMV. In CMV-seropositive healthy subjects >60 yr of age, the total number of CD8+ T cells is twice that in age-matched healthy CMV-seronegative subjects (3). The frequency of CD8+ T cells specific for individual CMV-derived peptide epitopes is often >1% of the total peripheral CD8+ T cell pool (4). CMV-specific CD4+ T cells are also frequent. It has been estimated that in 11% of healthy CMV-seropositive individuals, between 10 and 40% of the total peripheral CD4+ T cell pool is directed against CMV epitopes (5). Similar results have been reported in HIV-infected patients without apparent CMV disease (6). An exhaustive analysis of all 213 human CMV open reading frames found that a median of 10% of the total peripheral blood memory CD4+ T cells pooled from healthy CMV-seropositive individuals was directed against CMV epitopes (7). From these data, the frequency of CMV-specific CD4+ T cells appears to be much greater than required to ensure the production of antibodies and functional CD8+ T cells.
The occurrence of CMV disease during immunosuppressive treatments and AIDS demonstrates the importance of cellular immunity in the control of CMV infection. It is clear from many studies that CD8+ T cells play a role in the control of viral replication and may require CD4+ T cell help to achieve effective viral control (8–14). However, there is also increasing evidence that CD4+ T cells may also play a direct role in the control of CMV infection. In mice, salivary tissue is exempt from CD8+ T cell control of infection, and CMV-specific CD4+ T cells are required for the clearance of CMV from that tissue (15). In humans, reports have shown a correlation between the presence of CMV-specific CD4+ T cells and control of disease. Gamadia et al. (16) showed that CMV-specific CD4+ T cells are essential for protection against disease in primary infection, even in the presence of functional CMV-specific CD8+ T cells. Tu et al. (17) detected lower levels of CMV-specific CD4+ T cells after primary infection in children with persistent shedding of CMV compared with children who did not shed virus; this difference was observed even when the levels of CMV-specific CD8+ T cells were comparable to those present in seropositive children and adults who did not shed virus. However, the mechanism through which such CD4+ T cells exert their antiviral effect remains unclear.
Cytotoxic CD4+ T cells can be generated in vitro (18–24); however, most viruses infect a broad range of target cells in vivo that do not express MHC class II and therefore cannot be targeted by CD4+ T cells. Nevertheless, several authors have suggested recently that CD4+ T cells may have a direct effect on the control of viral infection in vivo. Suni et al. (25) reported a CD4+ CD8dim subset of T cells that can lyse whole CMV antigen-loaded EBV-transformed B lymphoblastoid cells (B-LCLs) ex vivo. Appay et al. (26) reported the presence of increased numbers of perforin-containing CD4+ T cells in HIV-infected individuals and showed that these cells can kill. Zaunders et al. (27) described Gag-specific CD4+ T cells in an HIV-infected long-term nonprogressor that express both granzymes A and B, as well as exhibited specific cytotoxicity after a brief time in culture. van Leeuwen et al. (28) have reported that CMV-specific ex vivo killing by CD4+ T cells in PBMCs isolated from a chronically infected individual is associated with a CD28− phenotype.
We hypothesized that CD4+ T cells found during chronic subclinical CMV infection may express a specific effector phenotype. To determine if CMV-specific CD4+ T cells had functional characteristics consistent with antiviral effector cells, we conducted a detailed characterization of CMV-specific CD4+ T cell function in human CMV infection using polychromatic flow cytometry. Specifically, within each MHC II–restricted antigen-specific response, we measured the production of IFN-γ and TNF-α, two cytokines often used to quantify CMV-specific CD4+ T cells (29); IL-2, a cytokine associated with helper function (30); macrophage inflammatory protein (MIP)-1β, a proinflammatory chemokine (31); and surface mobilization of CD107a, a marker of degranulation associated with cytolytic function (32). In conjunction, we classified CMV-specific CD4+ T cell responses based on the surface expression of CD45RO, a marker of memory T cells, CD27, which is irreversibly lost with prolonged exposure to antigen (26, 33), and CD57, a marker of terminal differentiation (34). Specific functional responses were also analyzed with respect to T cell clonotype composition. The results provide a detailed picture of CMV-specific CD4+ effector T cells and indicate a potential role for these cells in the control of CMV replication.
RESULTS
Measurement of functional responses to specific peptide epitopes
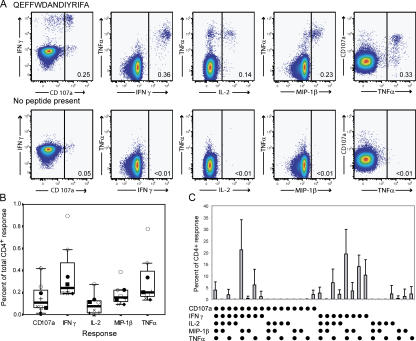
PBMCs from 34 CMV-seropositive healthy volunteers participating in the NIH research apheresis program were screened with five previously described CMV epitopes. Individuals with a frequency of IFN-γ peptide–specific CD4+ T cells of 0.2% or greater were studied further. Eight different CMV epitope-specific CD4+ T responses were found in seven subjects (Table I). Peptide-specific CD4+ T cell responses were polyfunctional. Stimulation with cognate epitopes resulted in the production of IFN-γ, TNF-α, and MIP-1β, as well as surface mobilization of CD107a (Fig. 1, A and B). In addition, IL-2 production was detected in seven of the eight responses. The functional hierarchy was dominated by production of IFN-γ and TNF-α, followed by MIP-1β and surface mobilization of CD107a. IL-2 production was the least common functional outcome (Fig. 1 B). In all control stimulations, backgrounds for each individual function were minimal (Fig. 1 A).
Table I.
Age and sex of CMV-seropositive subjects and total frequency of response to peptide epitopes
| Subject | Age (Sex) | Peptide (Class II restriction) | Location | Total response |
|---|---|---|---|---|
| 1 | 64 (M) | QEFFWDANDIYRIFA (DR52) | pp65281–295 | 0.88% |
| 2 | 55 (F) | LLQTGIHVRVSQPSL (DR15) | pp6541–55 | 0.64% |
| 3 | 58 (F) | LLQTGIHVRVSQPSL (DR15) | pp6541–55 | 0.28% |
| 4 | 44 (M) | PPWQAGILARNLVPMV (DR11) | pp65485–500 | 0.2% |
| IIKPGKISHIMLDVA (DR53) | pp65281–295 | 0.2% | ||
| 5 | 23 (M) | PQYSEHPTFTSQYRIQ (DR11) | pp65361–376 | 0.2% |
| 6 | 37 (M) | PQYSEHPTFTSQYRIQ (DR11) | pp65361–376 | 0.2% |
| 7 | 61 (M) | QEFFWDANDIYRIFA (DR52) | pp65281–295 | 0.39% |
Total response represents the sum of all CD4+ T cells showing surface mobilization of CD107, or production of IFN-γ, IL-2, MIP-1β, or TNF-α in response to stimulation with the peptide indicated.
Figure 1.
Polyfunctional CMV-specific CD4+ T cell responses. (A) Representative plots from subject 7 showing functional profiles in response to cognate antigen (top) and background responses in the absence of cognate antigen (bottom). (B) Total frequency of individual peptide-specific CD4+ T cell functions. The frequencies of surface mobilization of CD107a and production of IFN-γ, IL-2, MIP-1β, and TNF-α are shown for subject 1 (◯), subject 2 (⋄), subject 3 (▪), subject 4 with peptide PPWQAGILARNLVPMV (×), subject 4 with peptide IIKPGKISHIMLDVA (+), subject 5 (♦), subject 6 (□), and subject 7 (•) in the total CD4+ population using box plots with the data for each individual superimposed. The line in the middle of the box represents the median, the top box represents the second quartile, and the box below represents the third quartile. (C) Frequency of normalized functional species in subjects shown in B. The mean percentage of the total CD4+ response for each of 31 functional species is shown ± SD.
Comparison of these responses between different individuals required normalization of the data (Table II). The total frequency of responding CD4+ T cells was set to 100%, and the contribution of each functional subgroup was expressed as a percentage of the total response (Fig. 1 C). Notably, cells that mobilized CD107a and produced IFN-γ, MIP-1β, and TNF-α were most frequent (mean 22 ± 14% of the total response). The next most frequent functional subgroup showed no evidence of surface mobilization of CD107a, but it produced IFN-γ, MIP-1β, and TNF-α (mean 20 ± 11% of the total response). Cells that produced IFN-γ and TNF-α only (mean 15 ± 6% of the total response) or IFN-γ alone (mean 12 ± 8% of the total response) were still prominent. Almost all responding cells produced IFN-γ and TNF-α. Interestingly, cells that produced IL-2 alone (2 ± 4% of the total response) were rare.
Table II.
Total CD4+ T cell response and percentage of the total response found in the naive, CD27+ memory, CD27− memory, CD27−CD45RO−, and terminally differentiated effector CD4+ T cell compartments
| Percentage total response mean ± SD | Total cells mean ± SD | |
|---|---|---|
| Naive (CD27+/CD45RO−) | 1 ± 1 | 23 ± 18 |
| CD27+ memory (CD27+/CD45RO+) | 15 ± 10 | 42 ± 10 |
| CD27− memory (CD27−/CD45RO+) | 77 ± 10 | 25 ± 12 |
| CD27−/CD45RO− | 1 ± 2 | 4 ± 3 |
| Terminally differentiated effector cells (CD57+/CD27−/CD45RO+) | 28 ± 18 | 16 ± 12 |
Values are given as mean ± SD.
Maturational phenotype of responding CD4+ T cells
We further characterized the CMV-specific CD4+ T cells by including maturational markers, allowing simultaneous measurement of function and maturational phenotype (Table II). Prolonged stimulation of CD27+CD45RO+ memory CD4+ T cells has been shown to result in the irreversible loss of CD27 expression in vitro (35). Other studies with multiple viral pathogens have suggested that loss of CD27 in vivo is accompanied by changes in CD4+ T cell function (30, 36–39). For the purposes of this study, we classified cells as naive (CD45RO−CD27+), CD27+ memory cells (CD45RO+CD27+), or CD27− memory (CD45RO+CD27−). In addition, surface expression of CD57 was used as a marker of terminally differentiated effector cells (34).
In our cohort, naive, CD27+ memory, CD27− memory, and CD27− CD45RO− cells accounted for 23 ± 18%, 42 ± 11%, 25 ± 12%, and 4 ± 3% of the peripheral blood CD4+ T cell pool, respectively (values expressed as mean ± SD; Table II). The vast majority of the CD4+ peptide–specific T cell responses were contained within the CD27− memory (77 ± 10%) and CD27+ memory (15 ± 10%) subsets (Table II). In addition, a substantial but widely variable percentage of the responding CD4+ T cells were terminally differentiated effector cells (28 ± 18%).
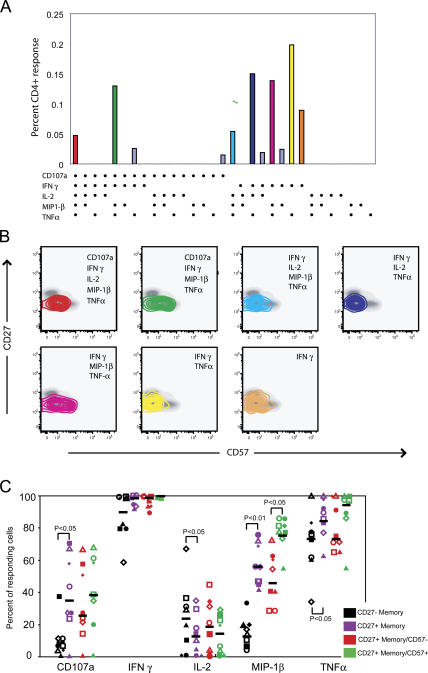
To explore the relationship between functional response and maturational phenotype in more detail, we mapped individual functional species to their respective phenotype. This is represented visually by mapping individual response profiles (Fig. 2 A) onto density plots showing CD27 versus CD57 expression (Fig. 2 B), a format that provides optimal resolution given that ∼95% of responding cells on average were CD45RO+. Inspection of these plots shows that MIP-1β–producing CD4+ cells are found at a greater frequency in terminally differentiated CD57+ CD4+ T cells than non– MIP-1β antigen-specific cells. This is most pronounced in IFN-γ–, MIP-1β–, and TNF-α–producing cells that also mobilize CD107a. Comparison of IFN-γ–, MIP-1β–, and TNF-α–producing cells also show that these cells have a higher frequency of CD57 positivity than cells that produce IFN-γ and TNF-α alone. Indeed, we found that antigen-specific CD4+ T cells that produced MIP-1β contained a significantly higher proportion of terminally differentiated cells compared with cells that lacked production of MIP-1β but were otherwise functionally identical (P < 0.05; not depicted). To describe further the relationship between function and maturational phenotype, we compared the frequency of each response in the different phenotypic subsets (Fig. 2 C). Memory CD4+ T cells that were CD27− were more likely than memory CD4+ T cells that were CD27+ to mobilize CD107a (P ≤ 0.05) and produce MIP-1β (P ≤ 0.01) and TNF-α (P ≤ 0.05). IL-2 was produced by a greater proportion of responding CD27+ memory CD4+ T cells than CD27− memory CD4+ T cells (P ≤ 0.05). MIP-1β production was the only function found to differ between terminally and nonterminally differentiated effector CD4+ T cells; it was increased in terminally differentiated cells (P ≤ 0.05).
Figure 2.
Mapping functional CMV-specific CD4+ T cell responses to maturational phenotype. (A) Individual responses for subject 1. The 31 possible response profiles are shown on the x axis, and the total frequency of each response profile is shown on the y axis. The dominant response profiles are color coded. (B) The maturational phenotypes of the CD4+ T cells expressing each of the dominant response profiles shown in A are overlaid on CD27 versus CD57 plots in which the phenotype of the total CD4+ T cell population is shown in gray. Responding cells are represented as 10% probability contour plots. (C) Functional responses for subject 1 (□), subject 2 (▪), subject 3 (⋄), subject 4 with peptide PPWQAGILARNLVPMV (◯), subject 4 with peptide IIKPGKISHIMLDVA (▵), subject 5 (▴), subject 6 (•), and subject 7 (♦) are shown with medians represented by a horizontal bar. To facilitate comparison between different CD4+ T cell subsets (CD45RO+CD27+ and CD45RO−CD27−; and CD45RO+CD27−CD57− and CD45RO+CD27−CD57+), data were normalized based on the total frequency of responding cells within each maturational subset.
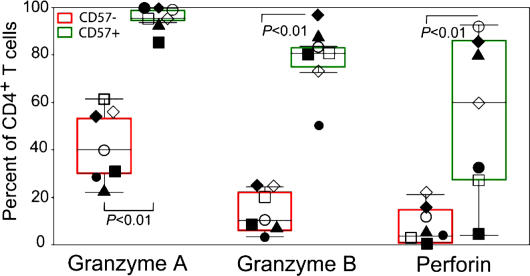
To study further the relationship between maturation and cytolytic function, we measured the presence of granzyme A, granzyme B, and perforin in CD27+ memory cells as well as in terminally and nonterminally differentiated CD27− memory CD4+ T cells in our cohort. Granzyme A was found frequently in CD27+ memory CD4+ T cells (13 ± 5%), but granzyme B and perforin were not (2 ± 1 and 1 ± 2%, respectively). These cytotoxic proteins were more commonly detected in CD27− memory CD4+ T cells (Fig. 3). The frequency of granzyme A (95 ± 5%), granzyme B (78 ± 15%), and perforin (53 ± 33%) in terminally differentiated CD27− CD4+ T cells was markedly higher than the frequency of granzyme A (41 ± 15%), granzyme B (14 ± 9%), and perforin (8 ± 9%) in nonterminally differentiated CD27− CD4+ T cells (P < 0.01%). Furthermore, in two subjects, the frequency of terminally differentiated CD27− memory CD4+ T cells containing granzyme A, granzyme B, and perforin was near 80%.
Figure 3.
Frequency of granzyme A, granzyme B, and perforin in CD57− and CD57+ CD27− memory CD4+ T cells. The frequency of granzyme A, granzyme B, and perforin in CD57− and CD57+ CD27− memory CD4+ T cells is shown using box plots, with individual data values superimposed for subject 1 (□), subject 2 (▪), subject 3 (⋄), subject 4 (◯), subject 5 (▴), subject 6 (•), and subject 7 (♦).
Surface mobilization of CD107a by CD4+ T cells is associated with loss of cytolytic granules
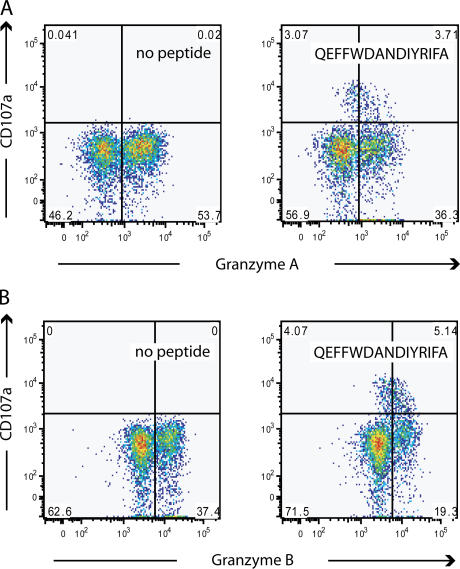
The high frequency of CD4+ T cells showing surface mobilization of CD107a was unexpected because surface mobilization of CD107a by CD8+ T cells identifies cells that can release cytolytic granules (32, 40, 41), a function not often associated with CD4+ T cells. For subject 1, one third of the response to the epitope QEFFWDANDIYRIFA was contained within the TCRVβ12 subset. This response accounted for 16% of the total TCRVβ12 subset. This allowed us to determine if loss of granzyme A occurred concurrently with surface mobilization of CD107a. After incubation of PBMCs with 2 μg/ml of peptide QEFFWDANDIYRIFA for 5 h, a population of CD107a+ granzyme A–low CD4+ T cells emerged. Cells with the brightest CD107a-associated fluorescence had the lowest granzyme A content (Fig. 4 A). Similar results were seen with granzyme B (Fig. 4 B). Concurrent with surface expression of CD107a, the population of CD4+ T cells that were CD107− and did not contain granzyme A or granzyme B increased by 11 and 9%, respectively, compared with CD4+ T cells from the same individual incubated without peptide. Unlike granzyme A, perforin was only present in 9% of CD4+ T cells from subject 1 and was almost undetectable in TCRVβ12+ CD4+ T cells. These data suggest that CD107a expression occurs concurrently with loss of granzyme A and granzyme B. These data also suggest that granzyme A and granzyme B are lost in these incubations without expression of CD107a. Whether this indicates that surface expression of CD107a does not identify all cells that degranulate, or if another mechanism is responsible for the loss of granzyme A and B in these cells, is not clear.
Figure 4.
Degranulation of epitope-specific CD4+ T cells. (A) Concurrent loss of granzyme A with surface mobilization of CD107a in response to peptide QEFFWDANDIYRIFA during a 5-h incubation (right) compared with the αCD28/49d control stimulation in the absence of peptide (left) from TCRVβ12+ CD4+ T cells from subject 1. (B) Concurrent loss of granzyme B with surface mobilization of CD107a in response to peptide QEFFWDANDIYRIFA during a 5-h incubation (right) compared with the αCD28/49d control stimulation in the absence of peptide (left) from TCRVβ12+ CD4+ T cells from subject 1.
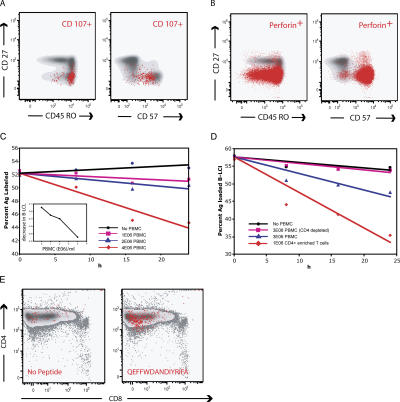
We next demonstrated that the CMV-specific CD4+ T cells were capable of specific target cell lysis ex vivo using PBMCs from subject 7. In this subject, 50% of the CD4+ T cells responding to the peptide epitope QEFFWDANIYRIFA showed evidence of surface mobilization of CD107a. 65% of the degranulating CD4+ T cells mapped to the terminally differentiated effector subset, and 85% of CD57+ CD4+ T cells contained perforin (Fig. 5, A and B). Autologous B-LCLs from subject 7 were stained with either CFSE or chloromethyl-benzoyl-aminotetramethyl-rhodamine (CMTMR). CFSE-stained B-LCLs were loaded with the target peptide QEFFWDANDIYRIFA, whereas CMTMR-stained B-LCLs were not loaded with peptide. PBMCs were added to an equal mixture of peptide-loaded CFSE- and CMTMR-stained B-LCLs that were not loaded with peptide. The percentage of CFSE- and CMTMR-stained B-LCLs was determined upon the mixing of B-LCLs with PBMCs and at 8, 16, and 24 h after mixing. Loss of antigen-loaded B-LCLs increased linearly with the number of PBMCs added (Fig. 5 C). Depletion of CD4+ T cells from PBMCs before the incubation of PBMCs with B-LCLs resulted in almost no loss of antigen-loaded B-LCLs during 24 h of incubation. The addition of 1E06 CD4+ T cells/ml, isolated by elutriation and then purified by positive selection on a MACS column to B-LCLs, resulted in a threefold increase in the rate of the killing of B-LCLs loaded with the peptide QEFFWDANDIYRIFA compared with that observed when 3E06 PBMCs/ml were incubated with antigen-loaded B-LCLs (Fig. 5 D). Parallel incubation in which 1E06 CD4+ T cells/ml were added to 30,000 unstained B-LCLs/ml loaded with the peptide QEFFWDANDIYRIFA resulted in production of IFN-γ of 0.2% of CD4+ T cells (Fig. 5 E). This response was entirely within the CD4+ T cell subset. Incubation of PBMCs with 30,000 B-LCLs that had not been loaded with peptide resulted in IFN-γ production by 0.03% of CD4+ T cells. Incubation of B-LCLs loaded with the PPWQAGILARNLVPMV, to which no response was detected, did not result in B-LCL killing. Similar rates of killing were observed whether peptide-loaded B-LCLs were stained with CMTMR or CFSE (not depicted). No CD8+ T cell response to the peptide QEFFWDANDIYRIFA was present in subject 7.
Figure 5.
Killing of antigen-loaded autologous B-LCLs by epitope-specific CD4+ T cells. (A) Mapping of CD4+ T cells from subject 7 that degranulate in response to peptide QEFFWDANDIYRIFA (red dots) to the CD45RO+CD27− CD4+ T cell compartment and the CD57+ compartment. (B) Mapping of perforin to the CD27−CD45RO+ and CD27−CD45RO− compartment and the CD57+ compartments. (C) Loss of antigen-loaded B-LCLs during incubation with PBMCs. Killing of B-LCLs was determined by loss of antigen-loaded, dye-stained B-LCLs as a percentage of the total amount of CFSE-stained antigen-loaded and CMTMR-stained unloaded B-LCLs in incubations containing none (•) and 1E06 PBMCs (▪), 2E06 PBMCs (♦), and 4E06 PBMCs/ml (▴). Lines are the least square fits of the data forced to intersect the y axis at the average of the initial percentage of CFSE-stained B-LCLs for all four incubations. Rate of decrease per million PBMCs is shown in the inset. (D) Killing of peptide-loaded B-LCLs in incubations containing none (•), 3E06 PBMCs after the depletion of CD4+ T cells (▪), 3E06 PBMCs (▴), and 1E06 CD4+ enriched PBMCs (♦). CD4-depleted PBMC CD4+ T cells contained >15× less CD4+ T cells compared with CD8+ T cells than nondepleted PBMCs. CD4+-enriched PBMC CD4+ T cells represented >99% of CD3+ cells. Both PBMCs and B-LCLs are from subject 7. Lines are the least square fits of the data forced to intersect the y axis at the average of the initial percentage of CFSE-stained B-LCLs for all four incubations. (E) Production of IFN-γ by CD4+-enriched T cells after a 6-h incubation with 30,000 B-LCLs either without peptide or loaded with peptide QEFFWDANDIYRIFA. IFN-γ–producing CD3+ T cells are overlaid on a histograph showing CD4+ and CD8+ T cells. These assays were performed without costimulation and run in parallel with the killing experiments shown in C.
To ensure that the CD4+ T cell killing shown with subject 7 was not an anomaly, we screened PBMCs from 10 HIV-infected individuals coinfected with CMV for their responses to overlapping pp65 peptides. None of these subjects had evidence of CMV end organ disease. We identified two individuals in whom the majority of the CD4+ T cells that mobilized CD107a in response to stimulation with pp65 were highly associated with CD57 positivity. Peptide epitopes were identified using pp65 matrices constructed from 138 15 mers overlapping by 11 (42). The responses of these CD4+ pp65-specific T cells were studied further. The first subject was a 53-yr-old male with a CD4 count of 656 and a viral load of 553 copies/ml who had not taken antiretroviral drugs for >4 yr. In this individual, 0.45% of the CD4+ T cell population responded to pp65489–503, AGILARNLVPMVATV. Two different assays performed on two different days with different cell preparations showed CD4+ T cells killing. In one assay that contained 1.6 E06 CD4+ T cells and 30,000 peptide-loaded autologous B-LCLs, peptide-loaded B-LCLs were decreased by 13%. In the second incubation that contained 1.0 E06 PBMCs and 30,000 autologous peptide-loaded B-LCLs, peptide-loaded B-LCLs decreased by 11%. The second HIV-infected subject was a 43 yr old on antiretroviral therapy with a CD4+ T cell count of 612 and a viral load of <50 copies/ml. In this individual, 0.34% of the CD4+ T cell population responded to the peptide pp65505–520, GQNLKYQEFFWDAND. In this individual, killing assays were performed three times with three different cell preparations on three different days. Two incubations contained 2.0E06 CD4+ T cells per 30,000 peptide-loaded autolgous B-LCLs, and one incubation contained 1E06 CD4+ T cells per 30,000 peptide-loaded autolgous B-LCLs. In the two incubations containing 2E06 CD4+ T cells per 30,000 autologous peptide-loaded B-LCLs, decreases in peptide-loaded B-LCLs of 13.2 and 5.2% were observed during a 24-h incubation. In the incubation containing 1E06 CD4+ T cells/ml, an 8.8% decrease in antigen-loaded B-LCLs was observed.
We also tried to show expression of CD95L by peptide-specific CD4+ T cells in subject 7. Although we were able to see CD95L expression with staphylococcal enterotoxin B stimulation, we could not see evidence of surface mobilization CD95L in response to stimulation with the peptide QEFFWDANDIYRIFA.
Comparison of functional response profiles in different viral infections
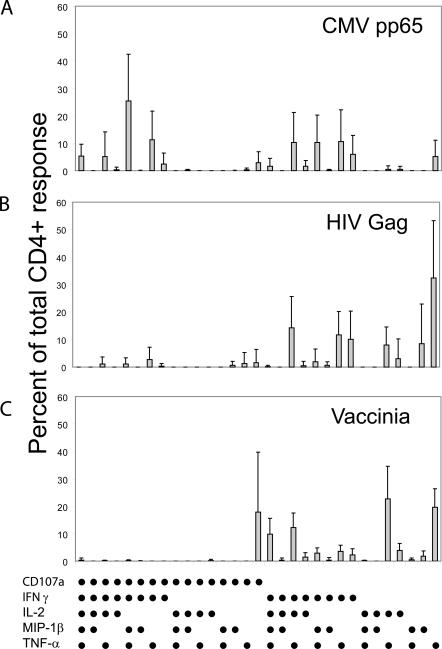
Virus-specific CD4+ T cell responses have been reported to exhibit different functional responses in different infections (30, 38, 39, 43, 44). We therefore conducted similar functional profiling studies of responding CD4+ T cells in HIV-infected long-term nonprogressors and individuals infected with vaccinia virus during a smallpox vaccine trial (Fig. 6). Multifunctional responses, including the production of MIP-1β and surface expression of CD107a, were substantially more frequent in CD4+ T cell populations specific for CMV pp65 than in those specific for either HIV-1 Gag in long-term nonprogressors or vaccinia virus in vaccine recipients.
Figure 6.
Functional response patterns for CD4+ T cells with different viral specificities. PBMCs from (A) subjects with chronic CMV stimulated with overlapping pp65 peptides (n = 5); (B) HIV-infected long-term nonprogressors stimulated with overlapping Gag peptides (n = 11); and (C) smallpox vaccinees stimulated with whole vaccinia virus 1 mo after vaccination (n = 6). In each panel, the 31 possible response profiles are shown on the x axis, and the percentage of the total response is shown on the y axis. The mean percentage of the total CD4+ response for each of 31 functional species is shown ± SD.
Clonotype analysis of degranulating CD4+ T cells
We next asked whether terminally differentiated degranulating CD4+ T cells represent a clonally restricted subset of the total CD4+ T cell response to a given epitope. To accomplish this, we needed to sort live, unpermeabilized CD4+ T cells that did and did not degranulate (45). Although not routinely used in this study, surface mobilization of CD154 identifies almost all activated CD4+ T cells (46). Our data support these findings in CMV-specific CD4+ T cells. Using CMV-derived peptides, we found that surface expression of CD154 in our cohort occurred concomitantly with IFN-γ production (not depicted) and was limited almost exclusively to the CD27+ and CD27− memory CD4+ T cell pools. Therefore, by using surface mobilization of CD154 and CD107a, we were able to sort responding CD4+ T cells into degranulating and nondegranulating populations suitable for clonotypic analysis.
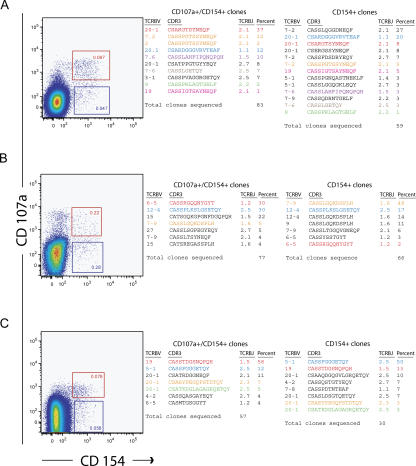
After stimulation with cognate peptide, CD107a+CD154+ (degranulating) and CD107a−CD154+ (nondegranulating) CD4+ T cells were sorted and subjected to clonotype analysis (Fig. 7). In each case, the sorted CD4+ T cell population was oligoclonal; the three dominant CD107a+CD154+ clonotypes accounted for the majority of the degranulating population (subject 7, 61%; subject 1, 82%; subject 2, 81%). Although the clonotypes sequenced for the degranulating and nondegranulating responses were similar, the frequency of clonotypes was different in these subsets. Clonotypes that were represented more frequently within the degranulating CD4+ T cell population occurred less frequently in the nondegranulating subset. The frequency of clonotypes determined by molecular analysis was substantiated by flow cytometric studies with TCRVβ mAbs in two subjects (1 and 7), thereby validating the quantitative nature of the RT-PCR method in this setting (not depicted). In sum, these data are consistent with the acquisition of lytic function by terminally differentiated effector memory phenotypes that are unrelated to clonotypic composition.
Figure 7.
Clonotypic composition of peptide-responsive CD4+CD107a+CD154+ and CD4+CD154+ T cell populations. Functional responses to cognate antigen stimulation and sort gates are shown in the left panels for subject 7 (A), subject 1 (B), and subject 2 (C). The corresponding TCRBV usage, CDR3 amino acid sequence, and TCRBJ usage of the depicted CD4+ T cell populations are shown in the right panels with the percentage frequency of each clonotype and the total number of clones sequenced.
DISCUSSION
The role of CD4+ T cells in B cell maturation, immunoglobin class-switching, and licensing of antigen-presenting cells to promote the expansion of functional CD8+ cytotoxic T lymphocytes is well established. Less is known about the role of CD4+ T cells in the direct control of infection. Here, we demonstrate that highly differentiated CD4+ T cells, specific for CMV pp65, mediate antiviral effector functions. Specifically, we find that: (a) CD27− memory CD4+ T cells can degranulate in response to cognate antigen; (b) a greater proportion of CD4+ T cells contains granzyme A, granzyme B, and perforin as they mature; (c) CD4+ T cells from an individual in which degranulation occurred in a subset of cells with a high frequency of perforin killed target cells bearing the cognate CMV pp65-derived MHC class II–restricted epitope; and (d) the frequency of CD4+ T cells that degranulate and produce MIP-1β increases with maturational status. In addition, we sorted activated CD4+ T cells into degranulating and nondegranulating populations and showed that degranulating CD4+ T cells were not clonotypically unique.
Although our data show epitope-specific killing of B-LCLs by CD4+ T cells from subject 7, the subject in whom the majority of the surface mobilization of CD107a occurred in a population of CD4+ T cells with a high frequency of perforin expression, our data do not show a direct link between surface mobilization of CD107a, perforin, and granzyme content and killing. In fact, it is possible that perforin-independent pathways may be involved in CD4+ T cell killing that we have shown in three individuals. Although we looked for other pathways and could not provide evidence that they were operative, it is still possible that the expression of FAS ligand was below our ability to detect, or a TNF-α–dependent mechanism was responsible for the observed killing.
Although we cannot say with certainty what the mechanism of CMV antigen-specific killing is, we have shown ample evidence of ex vivo killing. We have shown killing with three different individuals using three different peptide epitopes. In each case killing was shown with repeated assays. In subject 7, we demonstrated killing twice with purified CD4+ T cells and five different times with concentrations of PBMCs ranging from 1 to 4E06/ml. In the two individuals infected with HIV, CMV epitope-specific killing was shown in two and three different assays, respectively. In all of our assays, the ratio of peptide-loaded B-LCLs to nonloaded B-LCLs was determined at either three or four different time points. In addition, to assure that the observed killing was not artifactual, we loaded autologous B-LCLs with an irrelevant peptide and failed to show killing using PBMCs from subject 7. In almost all cases, and at least once for each person in which killing was demonstrated, we ran parallel control incubations using the same antigen-loaded and -unloaded stained autologous B-LCLs that were used in the killing assays. In these assays no effector cells were added. This assured that there was not a significant difference in the rate of growth between B-LCLs caused by either the peptide used or by staining with CMTMR or CSFE. In all of the above-mentioned incubations, when incubations were performed under the appropriate circumstances, killing was observed. We did not observe killing in all cases. We tried to show killing using cells from subjects 1 and 6. In subject 1, using 1E06 CD4+ T cells prepared by either positive or negative selection, we failed to show killing in two different assays. In subject 6, using either 1E06 PBMCs or 1E06 CD4+ T cells prepared by negative selection, we also failed to see any evidence of killing in two different assays.
Several reports have tried to describe functional capacity using changes in the expression of surface markers (26, 39, 47, 48). These reports have used a combination of reversible and irreversible surface markers. Many of these markers have well-established functional roles themselves. We followed three easily measured surface markers: CD45RO, the isoform of CD45 associated with the transition from naive to memory cell, CD27, a costimulatory marker irreversibly lost with exposure to cognate epitope, and CD57, a marker of replicative incompetence. As others have suggested, our data show frequent production of IFN-γ, and TNF-α, and decreased IL-2 frequencies as CD4+ T cells mature (49, 50). Similarly we find that CD4+ T cells show an increased frequency of degranulation and MIP-1β production with maturation. Nonetheless, ascribing all functionality to maturation appears to be an over simplification. The amount of perforin present in CD4+ T cells varies greatly in CD57+ T cells. Similarly, it seems clear from the data presented in Fig. 6 that degranulation occurs at different frequencies in response to different pathogens, even with similar levels of expression of CD27 and CD57. Whether these differences are due to different milieus at initial encounter of cognate epitope, or to changes that occur during chronic stimulation, remains to be determined.
The delay in the development of cytotoxic function and MIP-1β production to the stage at which CD4+ T cells acquire an effector phenotype is attractive teleologically. Such a delay in the development of these functions would serve to protect MHC class II–presenting cells in lymphoid tissue from damage during periods when CD4+ T cell help is required to develop and maintain an effective immune response. At the same time, effector CD4+ T cells could contribute to the control of pathogens in peripheral tissues. Indeed, in many ways the functional progression that we demonstrate here during CMV-specific CD4+ T cell differentiation is similar to that seen in CD8+ CTL development (37), even though effector CD4+ T cells in the periphery should still be able to supply help through the CD154–CD40 pathway. The persistent expression of CD154/40L, even in cells that are CD57+, suggests the importance of this ligand in CD4+ T cell function. Although much speculation is possible, we can certainly conclude that the pattern of response described here is more complicated than has been previously suggested for most virus-specific CD4+ T cells (51, 52).
The clonotypic data indicate a more varied CD4+ clonal response to CMV than suggested previously by others (53). Rather than one dominant clonotype, we see a more balanced response with at least three or four prevalent clonotypes in each epitope-specific CD4+ T cell population. There are several possible reasons for this difference in the apparent number of antigen-responsive clones. First, sorting antigen-responsive CD4+ T cells by CD107a and CD154 readouts may identify lower frequency clones than previously reported. It is also possible that the selection of subjects with a high response frequency to a specific epitope could select for more polyclonal MHC class II–restricted CD4+ T cell populations.
Overall, our data indicate that as pp65-specific CD4+ T cell clonotypes progress from CD27+ memory to a terminally differentiated effector memory phenotype, they acquire greater effector, and specifically cytotoxic, potential. The role of these CD4+ T cells in providing help is unclear. The relatively low frequency of IL-2–producing CD4+ T cells compared with IFN-γ, TNF-α, and MIP-1β production suggests that the primary role of CMV pp65-specific CD4+ T cells during chronic infection is not one of supplying CD4+ T cell help. The role of the CD40L–CD95 pathway in these cells remains unclear. Our findings do not apply to all CD4+ T cells specific to every virus (Fig. 6) and may not necessarily apply to other CMV proteins. However, our data support a model where certain unique antiviral effector functions of virus-specific CD4+ T cells are elaborated after an appropriate maturational state has been achieved. It remains to be determined whether the expression of these functions is dependent on key characteristics of the pathogen to which the CD4+ T cells are targeted, or the immunologic milieu in which those T cells become initially or subsequently stimulated during the course of the infection.
MATERIALS AND METHODS
Subjects.
PBMCs were obtained from CMV-seropositive healthy volunteers participating in the NIH research apheresis program (Table I) and from healthy HIV-infected volunteers participating in the vaccine research centers (VRC) apheresis protocol or by venapucture from HIV-infected volunteers participating in the VRC positive and negative protocol. When elutriated lymphocytes were prepared, mononuclear cells prepared by apheresis were further purified by elutriation using a Gambro Elutra. Lymphocytes prepared in this manner were routinely >85% lymphocytes. Signed informed consent approved by the NIH Institutional Review Board was obtained in each case. PBMCs were obtained by standard Ficoll-Hypaque (GE Healthcare) density gradient centrifugation and cryopreserved in FCS (Invitrogen) containing 10% DMSO (Fisher Scientific) using a Cryomed Freezer (model 7454; Thermo Electron Corporation). All PBMC preparations were screened for responses to five previously identified CMV-specific MHC class II–restricted epitopes (54, 55). Subjects with CD4+ T cell responses to a single peptide >0.2%, as determined by intracellular production of IFN-γ, were used for further study.
Peptides.
Peptides representing CMV-derived epitopes (54, 55) were synthesized and purified by Bio-Synthesis Inc. and were >80% pure by high performance chromatography (Table II). 138 15 mers overlapping by 11 spanning the entire pp65 protein were obtained from JPT Peptide Technology and were >70% pure. All peptides were dissolved in DMSO and stored at −20°C. The final concentration of each peptide in PBMC assays was 2 μg/ml.
Antibodies.
Directly conjugated mAbs specific for the molecules listed were obtained from the following: IL-2-allophycocyanin (APC), CD3-Cy7APC, CD20-APC, CD107a-FITC, CD95-PE, CD154-PE, IFN-γ–FITC, MIP-1β–PE, TNF-α–PECy7, perforin-FITC, granzyme A–PE, and granzyme A-FITC from BD Biosciences; CD45RO–Texas red–PE (TRPE) and TCRVβ12-PE from Beckman Coulter; and granzyme B-APC and CD4-PECy5.5 from Caltag. The following antibodies were conjugated in our laboratory according to standard protocols (http://drmr.com/abcon/index.html): CD4–Cascade blue, CD8–quantum dot (QD) 655, CD27–Cascade blue, CD14-PECy5, CD19-PECy5, CD57-QD545, and CD107a–Alexa 680. Unconjugated mAbs were obtained from BD Biosciences, Cascade blue and Alexa 680 were obtained from Invitrogen; Cy5 was obtained from GE Healthcare, and quantum dots were obtained from Quantum Dot Corporation.
Cell stimulation and staining.
Cell stimulation and staining were performed using a modification of the method described by Betts et al. (32). Purified PBMCs were thawed, resuspended at 2 × 106 cells/ml in complete RPMI media (RPMI 1640 supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin G, 100 μg/ml streptomycin sulfate, and 1.7 mM sodium glutamamine; R10), and rested for 2 h at 37°C in the presence of 10 U/ml DNase I (Roche Diagnostics). Cells were then washed with R10 and readjusted to 2 × 106 cells/ml. Costimulatory antibodies (αCD28 and αCD49d; 1 μg/ml final concentration; Becton Dickinson), monensin (0.7 μg/ml final concentration; BD Biosciences), brefeldin A (10 μg/ml final concentration; Sigma-Aldrich), αCD107a–Alexa 680 (pretitered volume), and in some cases αCD154-PE (pretitered volume) were added to cells, which were then transferred in 1-ml aliquots to polystyrene tubes containing 5 μl of each peptide or peptide mix. A negative control containing PBMCs from the same individual but with no added peptide was included for each assay. Cells were incubated for 5.5 h at 37°C. For Vaccinia virus–specific responses, 2 × 106 PBMCs were infected with Vaccinia virus (multiplicity of infection = 1) in 200 μl R10 for 1 h at 37°C. R10 was then added to adjust the total volume to 1 ml, and the cells were incubated for an additional 3 h before the addition of brefeldin A, monensin, and αCD107a–Alexa 680. Cells were then mixed and incubated at 37°C for an additional 5 h. Negative control tubes, without the addition of vaccinia virus, were included for all samples. After incubation, cells were washed once with PBS containing 1% bovine serum albumin and 0.1% sodium azide, and surface stained with the appropriate directly conjugated antibodies for 20 min in the dark at 4°C. The cells were then washed again and permeabilized using the cytofix/cytoperm kit (BD Biosciences) according to the manufacturer's instructions. After intracellular staining for CD3, CD4, IFN-γ, MIP-1β, IL-2, and TNF-α, the cells were washed one final time and fixed in PBS containing 1% paraformaldehyde. Fixed cells were stored at 4°C until the time of collection.
Maximal responses were obtained between 5 1/2 and 8 h. During this time, no change in the ratio of responses was observed with the increases in all functions being proportional to each other. Although total CD107a responses continued to increase between 5 1/2 and 8 h, mean florescence started to decrease during this time. To maximize the accuracy of CD107a gating, an incubation time of 5 1/2 h was used for all multicolor flow experiments.
In incubations in which cells were purified by either positive or negative selection, purification was performed using MACS methodology as recommended by the manufacturer (Miltenyi Biotec). CD4+ T cells purified by positive selection were >99.5% pure based on total CD3+ expression. In all cases, cells were allowed to rest for at least 2 h before assay.
Flow cytometric analysis.
Cells were analyzed with either a modified flow cytometer (LSRII; BD Immunocytometry Systems) equipped for the detection of 17 fluorescent parameters, or with a FACSCalibur (BD Immunocytometry). For four-color flow cytometry, between 100,000 and 250,000 events were collected from each sample, and for polychromatic flow cytometry, between 500,000 and 1,000,000 total events were collected from each sample. Electronic compensation was conducted with antibody capture beads (BD Biosciences) stained separately with individual mAbs used in the test samples. Data analysis was performed using FlowJo version 6.0 (TreeStar). For polychromatic analysis, initial gating of each sample set used a forward scatter area versus a forward scatter height plot to gate out cell aggregates. The cells were then gated through a forward scatter area versus a side scatter height plot to isolate small lymphocytes. After this, CD14+ and CD19+ cells were removed from the analysis to reduce background staining with αCD107a. Finally, cells were gated through a CD3 versus Cascade blue plot to remove dead cells (Cascade blue bright staining can be used as a surrogate marker of viability; unpublished data). Data are reported after background correction using costimulated cells from the same individuals in the absence of peptide for comparison. Importantly, nonspecific background becomes extremely low when combinations of functions are being examined, nearly always reaching zero events when three or more functions are being examined simultaneously. The background tends to be higher for single-positive responses, particularly for CD107a. Cut-off for a positive responses was 10 events.
CD95L staining.
CD95 staining was performed using a coculture method similar to that used for CD107. PBMCs were incubated with and without peptide in R10 containing 10 ng/ml Galardin (Sigma-Aldrich) and costimulatory antibodies (αCD28 and αCD49d; 1 μg/ml final concentration for 5 h, and either or both fluorescently labeled CD107a and CD95L). Cells were washed and surface stained for CD3 and CD4. When IFN-γ was stained, cells were permeabilized and stained as described previously (56).
Killing assays.
The killing assay used was a modification of the previously published VITAL assay (57, 58). EBV-transformed B-LCLs were prepared using standard procedures (59) and cryopreserved until needed. B-LCLs were stained with either CFSE (34) or CMTMR (57), as described previously. Stained cells were then added to either R10 medium or R10 supplemented with 2 μg/ml of the peptide epitope of interest. After incubation for 1 h, cells were counted and washed twice with R10. The B-LCLs were then mixed to contain equal numbers of peptide-loaded CFSE-stained B-LCLs and unloaded control CMTMR-stained B-LCLs, or equal numbers of peptide-loaded CMTMR-stained B-LCLs and unloaded control CFSE-stained B-LCLs. Next, 300,000 of these mixed cells were washed one final time and combined with PBMCs or CD4+ T cells in a total volume of 5 ml. 1-ml aliquots were added to 4.5 ml polypropylene tubes. Final concentrations of antigen-loaded B-LCLs and unloaded control B-LCLs were 30,000/ml each in all assays. PBMCs and CD4+ T cells concentrations varied as noted in Results. Tubes were then placed in a 37°C, 5% CO2 incubator. Incubation mixtures were processed at 0, 8, 16, and 24 h. For processing, cells were washed once in PBS containing 1% bovine serum albumin and 0.1% sodium azide, and stained for 30 min with αCD20-APC at 4°C. Cells were then washed again, fixed with 1% paraformaldehyde, and analyzed using a FACSCalibur flow cytometer (BD Immunocytometry).
Cell sorting.
PBMCs were prepared and rested as described above, and then incubated for 5 h in R10 supplemented with αCD28 and αCD49d (each at 1 μg/ml final concentration), monensin (0.7 μg/ml final concentration), αCD107a–Alexa 680 (pretitered volume), αCD154-PE (pretitered volume), and 2 μg/ml of peptide in a modification of the method described by Chattopadhyay et al. (46) and Betts et al. (32). Cells were then washed once more with R10, and propidium iodide (Sigma-Aldrich) was added to a final concentration of 5 μg/ml as a viability marked immediately before sorting. Cells were sorted at 25 pounds per square inch using a modified FACS DIVA (Becton Dickinson). Instrument set-up was performed according to the manufacturer's instructions. Electronic compensation was conducted with antibody capture beads (BD Biosciences) stained separately with the individual mAbs used in the test samples. Small CD3+, CD4+, CD14−, and CD19− lymphocytes that were CD154+/CD107a+ or CD154+/CD107a− were collected separately and frozen in RNAlater (Ambion) for clonotype analysis.
Clonotype analysis.
Thawed cells were lysed and subjected to mRNA extraction (Oligotex kit; QIAGEN). A template switch–anchored RT-PCR using a 3′ TCRB constant region primer (5′-GCTTCTGATGGCTCAAACACAGCGACCTC-3′) was then performed as described previously (45, 60). Amplified products were ligated into pGEM-T Easy vector (Promega) and cloned by transformation of competent DH5α Escherichia coli. Selected colonies were amplified by PCR using standard M13 primers and then sequenced from an insert-specific primer using fluorescent dye terminator chemistry (Applied Biosystems). A minimum of 50 clones was generated and analyzed per sample. Pseudogenes and “nonfunctional” sequences that could not be resolved after inspection of the individual chromatograms were discarded from the analysis. Nucleotide comparisons were used to establish clonal identity. All sequences that were only found once in each cell population were disregarded. Data analysis was performed using Sequencher Version 4.2 (Gene Codes Corporation). The International Immunogenetics nomenclature system is used throughout this work (61).
Statistical analysis.
Comparison between groups was performed using a criterion of significance of P ≤ 0.05. All statistical tests were conducted using SPSS for Windows, and all means are reported ± the SD. Pair-wise comparisons were made using a Wilcoxon signed rank statistic.
Online supplemental material.
Figs. S1 and S2 show CMV epitope–specific CD4+ T cell killing assays for two different individuals, both infected with HIV. In addition, the mapping of CD4+ T cells with respect to surface expression of CD27, CD45RO, and CD57 is shown for CD4+ T cells that surface mobilize CD107a in response to stimulation with cognate epitope. Similar figures are shown for perforin containing CD4+ T cells. Figs. S1 and S2 are available at http://www.jem.org/cgi/content/full/jem.20052246/DC1.
Supplemental Material
Acknowledgments
All DIVA analyses were performed by David Ambrozak. We also wish to acknowledge David Ambrozak for assistance with polychromatic flow analysis.
D.A. Price is a Medical Research Council (UK) Senior Clinical Fellow. This research was supported by the Intramural Research Program of the NIH, NIAID.
The authors have no conflicting financial interests.
Abbreviations used: B-LCL, B lymphoblastoid cell; CMTMR, chloromethyl-benzoyl-aminotetramethyl-rhodamine; MIP, macrophage inflammatory protein.
References
- 1.Pass, R.F. 1985. Epidemiology and transmission of cytomegalovirus. J. Infect. Dis. 152:243–248. [DOI] [PubMed] [Google Scholar]
- 2.Sissons, J.G., and A.J. Carmichael. 2002. Clinical aspects and management of cytomegalovirus infection. J. Infect. 44:78–83. [DOI] [PubMed] [Google Scholar]
- 3.Looney, R.J., A. Falsey, D. Campbell, A. Torres, J. Kolassa, C. Brower, R. McCann, M. Menegus, K. McCormick, M. Frampton, et al. 1999. Role of cytomegalovirus in the T cell changes seen in elderly individuals. Clin. Immunol. 90:213–219. [DOI] [PubMed] [Google Scholar]
- 4.Moss, P., and N. Khan. 2004. CD8(+) T-cell immunity to cytomegalovirus. Hum. Immunol. 65:456–464. [DOI] [PubMed] [Google Scholar]
- 5.Sester, M., U. Sester, B. Gartner, B. Kubuschok, M. Girndt, A. Meyerhans, and H. Kohler. 2002. Sustained high frequencies of specific CD4 T cells restricted to a single persistent virus. J. Virol. 76:3748–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komanduri, K.V., S.M. Donahoe, W.J. Moretto, D.K. Schmidt, G. Gillespie, G.S. Ogg, M. Roederer, D.F. Nixon, and J.M. McCune. 2001. Direct measurement of CD4+ and CD8+ T-cell responses to CMV in HIV-1-infected subjects. Virology. 279:459–470. [DOI] [PubMed] [Google Scholar]
- 7.Sylwester, A.W., B.L. Mitchell, J.B. Edgar, C. Taormina, C. Pelte, F. Ruchti, P.R. Sleath, K.H. Grabstein, N.A. Hosken, F. Kern, et al. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202:673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riddell, S.R., K.S. Watanabe, J.M. Goodrich, C.R. Li, M.E. Agha, and P.D. Greenberg. 1992. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 257:238–241. [DOI] [PubMed] [Google Scholar]
- 9.Walter, E.A., P.D. Greenberg, M.J. Gilbert, R.J. Finch, K.S. Watanabe, E.D. Thomas, and S.R. Riddell. 1995. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N. Engl. J. Med. 333:1038–1044. [DOI] [PubMed] [Google Scholar]
- 10.Einsele, H., E. Roosnek, N. Rufer, C. Sinzger, S. Riegler, J. Loffler, U. Grigoleit, A. Moris, H.G. Rammensee, L. Kanz, et al. 2002. Infusion of cytomegalovirus (CMV)-specific T cells for the treatment of CMV infection not responding to antiviral chemotherapy. Blood. 99:3916–3922. [DOI] [PubMed] [Google Scholar]
- 11.Bunde, T., A. Kirchner, B. Hoffmeister, D. Habedank, R. Hetzer, G. Cherepnev, S. Proesch, P. Reinke, H.D. Volk, H. Lehmkuhl, and F. Kern. 2005. Protection from cytomegalovirus after transplantation is correlated with immediate early 1–specific CD8 T cells. J. Exp. Med. 201:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gratama, J.W., J.W. van Esser, C.H. Lamers, C. Tournay, B. Lowenberg, R.L. Bolhuis, and J.J. Cornelissen. 2001. Tetramer-based quantification of cytomegalovirus (CMV)-specific CD8+ T lymphocytes in T-cell-depleted stem cell grafts and after transplantation may identify patients at risk for progressive CMV infection. Blood. 98:1358–1364. [DOI] [PubMed] [Google Scholar]
- 13.Reusser, P., G. Cathomas, R. Attenhofer, M. Tamm, and G. Thiel. 1999. Cytomegalovirus (CMV)-specific T cell immunity after renal transplantation mediates protection from CMV disease by limiting the systemic virus load. J. Infect. Dis. 180:247–253. [DOI] [PubMed] [Google Scholar]
- 14.Sacre, K., G. Carcelain, N. Cassoux, A.M. Fillet, D. Costagliola, D. Vittecoq, D. Salmon, Z. Amoura, C. Katlama, and B. Autran. 2005. Repertoire, diversity, and differentiation of specific CD8 T cells are associated with immune protection against human cytomegalovirus disease. J. Exp. Med. 201:1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucin, P., I. Pavic, B. Polic, S. Jonjic, and U.H. Koszinowski. 1992. Gamma interferon-dependent clearance of cytomegalovirus infection in salivary glands. J. Virol. 66:1977–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamadia, L.E., E.B. Remmerswaal, J.F. Weel, F. Bemelman, R.A. van Lier, and I.J. Ten Berge. 2003. Primary immune responses to human CMV: a critical role for IFN-gamma-producing CD4+ T cells in protection against CMV disease. Blood. 101:2686–2692. [DOI] [PubMed] [Google Scholar]
- 17.Tu, W., S. Chen, M. Sharp, C. Dekker, A.M. Manganello, E.C. Tongson, H.T. Maecker, T.H. Holmes, Z. Wang, G. Kemble, et al. 2004. Persistent and selective deficiency of CD4+ T cell immunity to cytomegalovirus in immunocompetent young children. J. Immunol. 172:3260–3267. [DOI] [PubMed] [Google Scholar]
- 18.Norris, P.J., and E.S. Rosenberg. 2001. Cellular immune response to human immunodeficiency virus. AIDS. 15:S16–S21. [DOI] [PubMed] [Google Scholar]
- 19.Littaua, R.A., M.B. Oldstone, A. Takeda, and F.A. Ennis. 1992. A CD4+ cytotoxic T-lymphocyte clone to a conserved epitope on human immunodeficiency virus type 1 p24: cytotoxic activity and secretion of interleukin-2 and interleukin-6. J. Virol. 66:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanna, R., and S.R. Burrows. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu. Rev. Microbiol. 54:19–48. [DOI] [PubMed] [Google Scholar]
- 21.Bickham, K., C. Munz, M.L. Tsang, M. Larsson, J.F. Fonteneau, N. Bhardwaj, and R. Steinman. 2001. EBNA1-specific CD4+ T cells in healthy carriers of Epstein-Barr virus are primarily Th1 in function. J. Clin. Invest. 107:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, Z., A. Vafai, J. Lee, R. Mahalingam, and A.R. Hayward. 1992. Specific lysis of targets expressing varicella-zoster virus gpI or gpIV by CD4+ human T-cell clones. J. Virol. 66:2664–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahon, B.P., K. Katrak, A. Nomoto, A.J. Macadam, P.D. Minor, and K.H. Mills. 1995. Poliovirus-specific CD4+ Th1 clones with both cytotoxic and helper activity mediate protective humoral immunity against a lethal poliovirus infection in transgenic mice expressing the human poliovirus receptor. J. Exp. Med. 181:1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaye, A., A.F. Magnusen, A.D. Sadiq, T. Corrah, and H.C. Whittle. 1998. Ex vivo analysis of cytotoxic T lymphocytes to measles antigens during infection and after vaccination in Gambian children. J. Clin. Invest. 102:1969–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suni, M.A., S.A. Ghanekar, D.W. Houck, H.T. Maecker, S.B. Wormsley, L.J. Picker, R.B. Moss, and V.C. Maino. 2001. CD4(+)CD8(dim) T lymphocytes exhibit enhanced cytokine expression, proliferation and cytotoxic activity in response to HCMV and HIV-1 antigens. Eur. J. Immunol. 31:2512–2520. [DOI] [PubMed] [Google Scholar]
- 26.Appay, V., J.J. Zaunders, L. Papagno, J. Sutton, A. Jaramillo, A. Waters, P. Easterbrook, P. Grey, D. Smith, A.J. McMichael, et al. 2002. Characterization of CD4(+) CTLs ex vivo. J. Immunol. 168:5954–5958. [DOI] [PubMed] [Google Scholar]
- 27.Zaunders, J.J., W.B. Dyer, B. Wang, M.L. Munier, M. Miranda-Saksena, R. Newton, J. Moore, C.R. Mackay, D.A. Cooper, N.K. Saksena, and A.D. Kelleher. 2004. Identification of circulating antigen-specific CD4+ T lymphocytes with a CCR5+, cytotoxic phenotype in an HIV-1 long-term nonprogressor and in CMV infection. Blood. 103:2238–2247. [DOI] [PubMed] [Google Scholar]
- 28.van Leeuwen, E.M., E.B. Remmerswaal, M.H. Heemskerk, I.J. Ten Berge, and R.A. van Lier. 2006. Strong selection of virus-specific cytotoxic CD4+ T cell clones during primary human cytomegalovirus infection. Blood. 108:3121–3127. [DOI] [PubMed] [Google Scholar]
- 29.Waldrop, S.L., C.J. Pitcher, D.M. Peterson, V.C. Maino, and L.J. Picker. 1997. Determination of antigen-specific memory/effector CD4+ T cell frequencies by flow cytometry: evidence for a novel, antigen-specific homeostatic mechanism in HIV-associated immunodeficiency. J. Clin. Invest. 99:1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harari, A., S. Petitpierre, F. Vallelian, and G. Pantaleo. 2004. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood. 103:966–972. [DOI] [PubMed] [Google Scholar]
- 31.Maurer, M., and E. von Stebut. 2004. Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol. 36:1882–1886. [DOI] [PubMed] [Google Scholar]
- 32.Betts, M.R., D.A. Price, J.M. Brenchley, K. Lore, F.J. Guenaga, A. Smed-Sorensen, D.R. Ambrozak, S.A. Migueles, M. Connors, M. Roederer, et al. 2004. The functional profile of primary human antiviral CD8+ T cell effector activity is dictated by cognate peptide concentration. J. Immunol. 172:6407–6417. [DOI] [PubMed] [Google Scholar]
- 33.Baars, P.A., M.M. Maurice, M. Rep, B. Hooibrink, and R.A. van Lier. 1995. Heterogeneity of the circulating human CD4+ T cell population. Further evidence that the CD4+CD45RA−CD27− T cell subset contains specialized primed T cells. J. Immunol. 154:17–25. [PubMed] [Google Scholar]
- 34.Brenchley, J.M., N.J. Karandikar, M.R. Betts, D.R. Ambrozak, B.J. Hill, L.E. Crotty, J.P. Casazza, J. Kuruppu, S.A. Migueles, M. Connors, et al. 2003. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 101:2711–2720. [DOI] [PubMed] [Google Scholar]
- 35.Hintzen, R.Q., R. de Jong, S.M. Lens, M. Brouwer, P. Baars, and R.A. van Lier. 1993. Regulation of CD27 expression on subsets of mature T-lymphocytes. J. Immunol. 151:2426–2435. [PubMed] [Google Scholar]
- 36.Gamadia, L.E., R.J. Rentenaar, R.A. van Lier, and I.J. ten Berge. 2004. Properties of CD4(+) T cells in human cytomegalovirus infection. Hum. Immunol. 65:486–492. [DOI] [PubMed] [Google Scholar]
- 37.Appay, V., and S.L. Rowland-Jones. 2004. Lessons from the study of T-cell differentiation in persistent human virus infection. Semin. Immunol. 16:205–212. [DOI] [PubMed] [Google Scholar]
- 38.Yue, F.Y., C.M. Kovacs, R.C. Dimayuga, P. Parks, and M.A. Ostrowski. 2004. HIV-1-specific memory CD4+ T cells are phenotypically less mature than cytomegalovirus-specific memory CD4+ T cells. J. Immunol. 172:2476–2486. [DOI] [PubMed] [Google Scholar]
- 39.Amyes, E., C. Hatton, D. Montamat-Sicotte, N. Gudgeon, A.B. Rickinson, A.J. McMichael, and M.F. Callan. 2003. Characterization of the CD4+ T cell response to Epstein-Barr virus during primary and persistent infection. J. Exp. Med. 198:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Betts, M.R., J.M. Brenchley, D.A. Price, S.C. De Rosa, D.C. Douek, M. Roederer, and R.A. Koup. 2003. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods. 281:65–78. [DOI] [PubMed] [Google Scholar]
- 41.Betts, M.R., and R.A. Koup. 2004. Detection of T-cell degranulation: CD107a and b. Methods Cell Biol. 75:497–512. [DOI] [PubMed] [Google Scholar]
- 42.Kern, F., N. Faulhaber, C. Frommel, E. Khatamzas, S. Prosch, C. Schonemann, I. Kretzschmar, R. Volkmer-Engert, H.D. Volk, and P. Reinke. 2000. Analysis of CD8 T cell reactivity to cytomegalovirus using protein-spanning pools of overlapping pentadecapeptides. Eur. J. Immunol. 30:1676–1682. [DOI] [PubMed] [Google Scholar]
- 43.Lucas, M., C.L. Day, J.R. Wyer, S.L. Cunliffe, A. Loughry, A.J. McMichael, and P. Klenerman. 2004. Ex vivo phenotype and frequency of influenza virus-specific CD4 memory T cells. J. Virol. 78:7284–7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Day, C.L., N.P. Seth, M. Lucas, H. Appel, L. Gauthier, G.M. Lauer, G.K. Robbins, Z.M. Szczepiorkowski, D.R. Casson, R.T. Chung, et al. 2003. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Invest. 112:831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Douek, D.C., M.R. Betts, J.M. Brenchley, B.J. Hill, D.R. Ambrozak, K.L. Ngai, N.J. Karandikar, J.P. Casazza, and R.A. Koup. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 168:3099–3104. [DOI] [PubMed] [Google Scholar]
- 46.Chattopadhyay, P.K., J. Yu, and M. Roederer. 2005. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat. Med. 11:1113–1117. [DOI] [PubMed] [Google Scholar]
- 47.Hamann, D., P.A. Baars, M.H. Rep, B. Hooibrink, S.R. Kerkhof-Garde, M.R. Klein, and R.A. van Lier. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 186:1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712. [DOI] [PubMed] [Google Scholar]
- 49.Amyes, E., A.J. McMichael, and M.F. Callan. 2005. Human CD4+ T cells are predominantly distributed among six phenotypically and functionally distinct subsets. J. Immunol. 175:5765–5773. [DOI] [PubMed] [Google Scholar]
- 50.Harari, A., F. Vallelian, and G. Pantaleo. 2004. Phenotypic heterogeneity of antigen-specific CD4 T cells under different conditions of antigen persistence and antigen load. Eur. J. Immunol. 34:3525–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seder, R.A., and R. Ahmed. 2003. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 4:835–842. [DOI] [PubMed] [Google Scholar]
- 52.Harari, A., G.P. Rizzardi, K. Ellefsen, D. Ciuffreda, P. Champagne, P.A. Bart, D. Kaufmann, A. Telenti, R. Sahli, G. Tambussi, et al. 2002. Analysis of HIV-1- and CMV-specific memory CD4 T-cell responses during primary and chronic infection. Blood. 100:1381–1387. [DOI] [PubMed] [Google Scholar]
- 53.Bitmansour, A.D., S.L. Waldrop, C.J. Pitcher, E. Khatamzas, F. Kern, V.C. Maino, and L.J. Picker. 2001. Clonotypic structure of the human CD4+ memory T cell response to cytomegalovirus. J. Immunol. 167:1151–1163. [DOI] [PubMed] [Google Scholar]
- 54.Kern, F., T. Bunde, N. Faulhaber, F. Kiecker, E. Khatamzas, I.M. Rudawski, A. Pruss, J.W. Gratama, R. Volkmer-Engert, R. Ewert, et al. 2002. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. J. Infect. Dis. 185:1709–1716. [DOI] [PubMed] [Google Scholar]
- 55.Khattab, B.A., W. Lindenmaier, R. Frank, and H. Link. 1997. Three T-cell epitopes within the C-terminal 265 amino acids of the matrix protein pp65 of human cytomegalovirus recognized by human lymphocytes. J. Med. Virol. 52:68–76. [DOI] [PubMed] [Google Scholar]
- 56.Betts, M.R., J.P. Casazza, and R.A. Koup. 2001. Monitoring HIV-specific CD8+ T cell responses by intracellular cytokine production. Immunol. Lett. 79:117–125. [DOI] [PubMed] [Google Scholar]
- 57.Ritchie, D.S., I.F. Hermans, J.M. Lumsden, C.B. Scanga, J.M. Roberts, J. Yang, R.A. Kemp, and F. Ronchese. 2000. Dendritic cell elimination as an assay of cytotoxic T lymphocyte activity in vivo. J. Immunol. Methods. 246:109–117. [DOI] [PubMed] [Google Scholar]
- 58.Hermans, I.F., J.D. Silk, J. Yang, M.J. Palmowski, U. Gileadi, C. McCarthy, M. Salio, F. Ronchese, and V. Cerundolo. 2004. The VITAL assay: a versatile fluorometric technique for assessing CTL- and NKT-mediated cytotoxicity against multiple targets in vitro and in vivo. J. Immunol. Methods. 285:25–40. [DOI] [PubMed] [Google Scholar]
- 59.Tosato, G. 2005. Genearation of Epstein-Barr Virus (EBV)-immortalized B cell lines. In Current Protocols in Immunology. J.E. Coligan, B. Bierer, D.H. Marguiles, and E.M. Shevach, editors. John Wiley and Sons, Hoboken, NJ. 7.22.21–23. [DOI] [PubMed]
- 60.Price, D.A., S.M. West, M.R. Betts, L.E. Ruff, J.M. Brenchley, D.R. Ambrozak, Y. Edghill-Smith, M.J. Kuroda, D. Bogdan, K. Kunstman, et al. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 21:793–803. [DOI] [PubMed] [Google Scholar]
- 61.Lefranc, M.P., C. Pommie, M. Ruiz, V. Giudicelli, E. Foulquier, L. Truong, V. Thouvenin-Contet, and G. Lefranc. 2003. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 27:55–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.