Abstract
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder of the gastrointestinal tract involving aberrant activation of innate and adaptive immune responses. We have used two complementary models of IBD to examine the roles of interleukin (IL)-12 family cytokines in bacterially induced intestinal inflammation. Our results clearly show that IL-23, but not IL-12, is essential for the induction of chronic intestinal inflammation mediated by innate or adaptive immune mechanisms. Depletion of IL-23 was associated with decreased proinflammatory responses in the intestine but had little impact on systemic T cell inflammatory responses. These results newly identify IL-23 as a driver of innate immune pathology in the intestine and suggest that selective targeting of IL-23 represents an attractive therapeutic approach in human IBD.
The maintenance of intestinal homeostasis is complex, involving interactions between the intestinal microflora, the epithelium, and the host immune system. A breakdown in the regulatory mechanisms that control homeostasis is thought to precipitate the dysregulated intestinal inflammation that characterizes human inflammatory bowel disease (IBD), including Crohn's disease (CD) (1, 2). Although the aetiopathogenesis of CD is not yet completely understood, it is widely held that chronic intestinal inflammation is driven by the proinflammatory cytokine IL-12, which promotes the development of pathogenic Th1 CD4+ effector cells (3, 4). This hypothesis was supported by results from several animal models of intestinal inflammation in which disease development could be blocked by treatment with monoclonal antibodies directed against the IL-12p40 subunit (5, 6) or by using other strategies to inhibit Th1 responses (7, 8). More recently, encouraging results were reported from initial clinical trials administering anti–IL-12p40 monoclonal antibodies to human CD patients (9).
IL-12 is a heterodimeric type I cytokine comprising the IL-12p40 subunit together with the IL-12p35 subunit. The recent discovery that the IL-12p40 subunit can also combine with a specific IL-23p19 subunit to form the closely related cytokine IL-23 (10) has led to a reappraisal of the roles of IL-12 and IL-23 in a variety of inflammatory disorders. Subsequent studies have clearly demonstrated that IL-23 is essential for driving several pathological reactions, including the autoimmune pathologies present in experimental autoimmune encephalomyelitis and collagen-induced arthritis (11–13). Like IL-12, IL-23 is primarily secreted by activated DCs, monocytes and macrophages, and transgenic mice constitutively overexpressing IL-23p19 develop fatal multiorgan inflammation (14). The proinflammatory activities of IL-23 have been partly ascribed to its ability to support the development of a novel subset of CD4+ inflammatory T cells known as Th17 cells (15–17). Th17 cells are characterized by their production of IL-17 (IL-17A), IL-6, and TNF-α and have been associated with the induction of autoimmune tissue inflammation (12, 18). Although the precise mechanism by which IL-23 promotes Th17 responses in vivo is still not completely understood, recent studies have shown that Th17 cell lineage commitment is driven by TGF-β and IL-6, whereas IL-23 appears to expand or maintain effector Th17 cell populations (19–23).
In addition to its effects on T cell responses, IL-23 also has potent effects on cells of the innate immune system, inducing the production of inflammatory cytokines, such as IL-1, IL-6, and TNF-α, by monocytes and macrophages (16, 24). The crucial role of innate immunity in intestinal inflammation was highlighted by the recent discovery that a substantial subgroup of human CD patients harbor mutations in the innate immune receptor NOD2, a cytoplasmic protein expressed by DCs, phagocytes, and some intestinal epithelial cells (25, 26). Although the mechanism by which NOD2 mutations predispose to the development of CD has not yet been ascertained, the finding that NOD2 recognizes a muropeptide motif derived from bacterial peptidoglycan (27) suggests that it may involve dysregulation of innate immune responses toward intestinal bacteria (28, 29). To study innate immune activation in intestinal pathology, we developed a model of T cell–independent intestinal inflammation triggered by infection with the pathogenic bacterium Helicobacter hepaticus. Infection of 129SvEvRAG−/− mice with H. hepaticus led to the development of chronic typhlocolitis mediated through activation and accumulation of innate immune cells, including granulocytes and monocytic cells (30, 31). Innate immune typhlocolitis was inhibited by treatment with anti–IL-12p40 monoclonal antibodies (30), indicating a requirement for IL-12 and/or IL-23 in disease. However, as bacteria-derived stimuli have been reported to induce secretion of both IL-12 and IL-23 by DCs and monocytes (16, 32), their relative roles in intestinal pathology remain undefined. Similarly, recent studies in CD patients (33, 34) reported elevated expression of both IL-12 and IL-23, highlighting this as a crucial issue for further study.
In this paper we have examined the role of IL-23 in two complimentary, well-characterized models of chronic intestinal inflammation. In addition to the H. hepaticus–triggered innate immune model, we also used the T cell transfer model of colitis in which adoptive transfer of naive CD4+ T cells into C57BL/6 RAG−/− recipients leads to severe colitis that is associated with expansion and accumulation of activated T cells and DCs in the intestinal lamina propria (35, 36). Our results reveal a crucial role for IL-23, but not IL-12, in intestinal inflammation mediated by sustained activation of either adaptive (CD4+ T cell–dependent) or innate immune mechanisms. The finding that microflora-induced IL-23 drives pathogenic innate and adaptive immune pathology locally in the intestine suggests that selective targeting of IL-23 may be an effective therapeutic approach in human IBD.
RESULTS
IL-23 is essential for innate intestinal inflammation
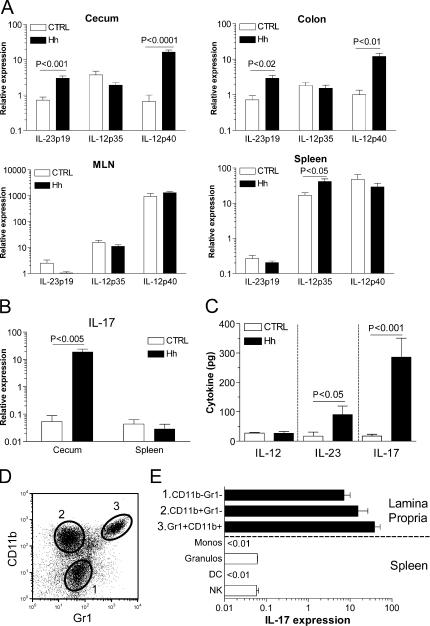
We initially examined whether H. hepaticus–induced innate immune typhlocolitis was associated with excessive production of IL-12 or IL-23 using real-time quantitative PCR (Q-PCR) to assay the expression of these cytokines in the normal and inflamed intestine. As illustrated in Fig. 1 A, significantly increased expression of IL-23p19 and IL-12p40 mRNA, but not of IL-12p35, was observed in the cecum and colon of H. hepaticus–infected (Hh +) 129SvEvRAG−/− mice. The increased expression of IL-23p19 was restricted to the intestine, as it was not observed in either the mesenteric LN (MLN) or spleen (Fig. 1 A).
Figure 1.
Increased expression of IL-23 and IL-17 in inflamed intestine. (A and B) RNA was purified from tissue samples isolated from H. hepaticus–infected (≥8 wk after infection) and control uninfected 129SvEvRAG−/− mice, and cytokine gene expression was assayed using Q-PCR. For each individual sample, cytokine gene expression was normalized relative to expression of HPRT (×104). Data shown represent mean relative expression levels (±SEM) from two independent experiments (n = 8–9 total mice per group). (C) Freshly isolated colon explants from H. hepaticus–infected and control uninfected 129SvEvRAG−/− mice were cultured overnight in complete RPMI, and cytokine release was measured using cytometric bead assay or ELISA. Results represent mean cytokine levels (±SEM; pg/100 mg colonic tissue) in samples pooled from three independent experiments (n = 9–12 total mice per group). (D and E) Spleen cells and LPLs (pooled from 6–12 mice) were isolated from H. hepaticus–infected 129SvEvRAG−/− mice and separated into the indicated subpopulations using FACS sorting. IL-17 gene expression was assayed using Q-PCR and normalized relative to expression of HPRT (×104). Data shown represent mean relative expression levels (±SEM) from two independent experiments.
As increased IL-17 expression has been associated with several IL-23–driven autoimmune T cell–mediated pathologies (16), we analyzed whether increased IL-17 expression was also a feature of T cell–independent innate intestinal pathology. Somewhat unexpectedly, we observed a dramatic increase in expression of IL-17 in the inflamed cecum of Hh + 129SvEvRAG−/− mice (Fig. 1 B), which was again restricted to the intestine, as it was not increased in spleen (Fig. 1 B). To confirm that these differences in mRNA expression correlated with increased cytokine secretion, we cultured colon explants from Hh + or control uninfected 129SvEvRAG−/− mice overnight and quantified cytokine release into the medium. As shown in Fig. 1 C, significantly higher amounts of both IL-23 and IL-17 were produced by colon explants from Hh + 129SvEvRAG−/− mice, but there was no increase in IL-12 secretion. To identify the innate sources of IL-17 in the intestine, we used cell sorting to isolate different populations of lamina propria leukocytes (LPLs) from Hh + 129SvEvRAG−/− mice. Three major subpopulations could be readily identified within the LPLs: Gr1+CD11b+ (predominantly granulocytes), Gr1−CD11b+ (monocytic cells), and Gr1−CD11b− cells (Fig. 1 D). Q-PCR analysis also revealed that all leukocyte subpopulations isolated from the intestine expressed high levels of IL-17 mRNA (Fig. 1 E). In contrast, consistent with the results obtained using whole spleen samples (Fig. 1 B), leukocyte subpopulations isolated from the spleen of Hh + 129SvEvRAG−/− mice expressed only very low levels of IL-17, around 100–1,000-fold lower than their intestinal counterparts (Fig. 1 E). Collectively, these results indicated that increased local production of IL-23 and IL-17 correlated with innate intestinal pathology.
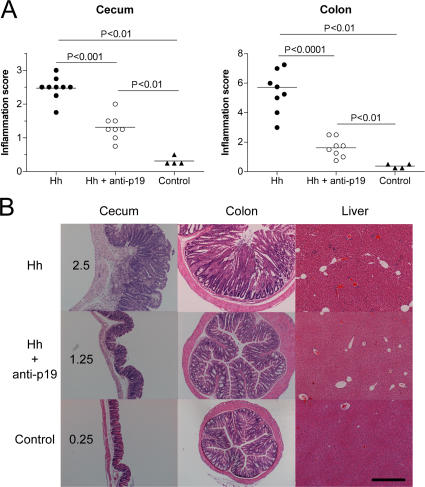
To formally assess the requirement for IL-23 in innate intestinal pathology, we used an anti–IL-23p19 monoclonal antibody to neutralize IL-23 in vivo. Although Hh + 129SvEvRAG−/− mice exhibited extensive inflammation in both the cecum and colon, treatment with anti-p19 throughout the course of the experiment resulted in highly attenuated intestinal pathology (Fig. 2 A). Anti-p19–treated mice had markedly reduced levels of inflammatory infiltrates and epithelial hyperplasia in the cecum and colon (Fig. 2 B). These results demonstrate that IL-23 plays an essential role in H. hepaticus–induced innate immune typhlocolitis.
Figure 2.
IL-23 drives innate immune typhlocolitis. (A) 129SvEvRAG−/− mice were infected with H. hepaticus and treated i.p. with 1 mg/wk of anti–IL-23p19 or isotype control antibody throughout the course of the experiment. Mice were killed 6–8 wk later, and pathology in the cecum and colon was assessed histologically. Each symbol represents a single animal, and the data shown represent combined results from two independent experiments (n = 4 total controls; and n = 8–9 total mice for experimental groups). Horizontal lines represent means. (B) Representative photomicrographs of tissue sections isolated from the mice outlined in A. Numbers indicate inflammation scores for the cecal sections shown. Bar, 500 μm.
IL-23 is essential for H. hepaticus–triggered systemic innate inflammatory responses
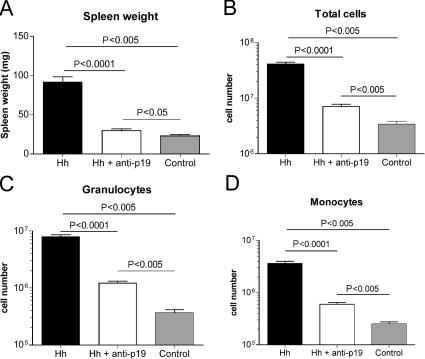
As Hh + 129SvEvRAG−/− mice also exhibit marked systemic inflammatory responses (30), we next examined the effect of anti–IL-23p19 treatment on these disease parameters. As shown in Fig. 3 (A and B), anti–IL-23p19 treatment significantly reduced H. hepaticus–induced splenomegaly in terms of both spleen weights and total spleen cell numbers. FACS analysis of spleen cells revealed that anti–IL-23p19 treatment significantly reduced the accumulation of granulocytes and monocytes that was found in Hh + 129SvEvRAG−/− mice (Fig. 3, C and D). Moreover, although scattered small inflammatory foci were present in the livers of Hh + 129SvEvRAG−/− mice, these were not observed in mice that received anti–IL-23p19 (Fig. 2 B). These results demonstrate that the systemic innate immune activation triggered by H. hepaticus infection is also dependent on IL-23.
Figure 3.
IL-23 drives systemic innate immune activation in H. hepaticus–infected 129SvEvRAG−/− mice. 129SvEvRAG−/− mice were infected with H. hepaticus and treated i.p. with 1 mg/wk of anti–IL-23p19 or isotype control antibody throughout the course of the experiment (6–8 wk). Spleen cell populations were enumerated using FACS analysis (reference 58). Graphs represent means ± SEM of (A) spleen weights, (B) spleen cell numbers, (C) granulocytes (FSCHiSSCHiGr1HiCD11bHiCD11c−), and (D) monocytes/macrophages (FSCHiSSCLoGr1IntCD11bHiCD11c−). Data shown represent combined results from two independent experiments (n = 4 total controls; and n = 8–9 total mice for experimental groups).
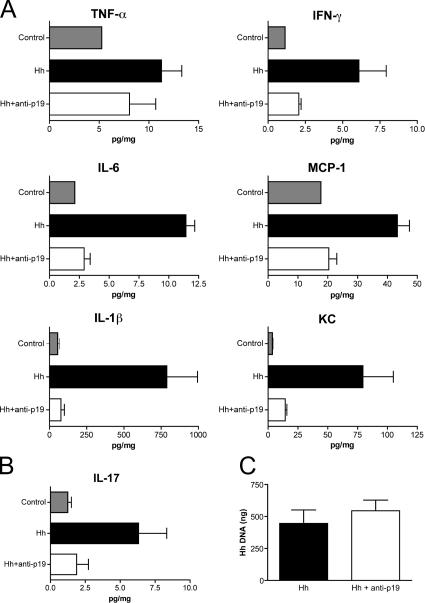
Anti–IL-23p19 treatment decreases proinflammatory cytokine production in the intestine
As the main producers of IL-23 are activated DCs and macrophages(16), we reasoned that high levels of IL-23 production might trigger a cascade of inflammatory cytokines that drives chronic intestinal inflammation. We therefore measured the levels of several proinflammatory cytokines in intestinal tissue homogenates. Although colon homogenates prepared from Hh + 129SvEvRAG−/− mice contained markedly elevated levels of IL-6, monocyte chemoattractant protein–1 (MCP-1), IFN-γ, TNF-α, mouse chemokine CXCL1 (KCs), and IL-1β, samples isolated from Hh + 129SvEvRAG−/− mice that had received anti–IL-23p19 expressed similar low levels of proinflammatory cytokines to uninfected controls (Fig. 4 A). Treatment of Hh + 129SvEvRAG−/− mice with anti–IL-23p19 also led to a similar reduction in IL-17 levels in the colon (Fig. 4 B), indicating that innate IL-17 production is also controlled by IL-23 in vivo. To confirm that anti–IL-23p19 was preventing intestinal pathology by inhibiting host innate immune activation and not through antibacterial effects, we used Q-PCR to assess H. hepaticus infection levels. As shown in Fig. 4 C, treatment with anti–IL-23p19 had no effect on the level of H. hepaticus colonization in 129SvEvRAG−/− mice. These results indicate that neutralization of IL-23 prevents intestinal inflammation by inhibiting innate immune responses and suggest that high production of IL-23 in the intestine triggers a proinflammatory cytokine cascade that mediates local and systemic pathology.
Figure 4.
IL-23 blockade reduces proinflammatory cytokine production in the intestine. 129SvEvRAG−/− mice were infected with H. hepaticus and treated i.p. with 1 mg/wk of anti–IL-23p19 or isotype control antibody throughout the course of the experiment (6–8 wk). (A and B) Cytokine concentrations in colon homogenates were measured and normalized to total protein content for each sample and are given as pg/mg total protein. Results represent mean cytokine levels (±SEM) from samples pooled from two similar experiments (n = 3–4 total samples per group). (C) Total amount of H. hepaticus DNA present in each cecal sample was determined using Q-PCR. Results represent mean H. hepaticus DNA levels (±SEM; n = 5 mice per group). H. hepaticus DNA was undetectable in cecal samples isolated from control uninfected mice.
IL-23 is essential for T cell–mediated intestinal inflammation but not systemic inflammatory responses
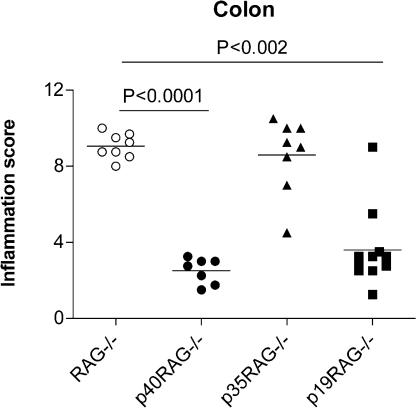
To determine whether IL-23 was a general mediator of intestinal inflammation, we next evaluated the roles of IL-12 and IL-23 in a well-established T cell–dependent model of IBD (35, 36). Thus, naive CD4+CD45RBhigh T cells isolated from C57BL/6 mice were adoptively transferred into age-matched cohorts of syngeneic RAG−/− recipient mice, p40−/−RAG−/− mice (lacking both IL-12 and IL-23), p35−/−RAG−/− mice (lacking only IL-12), or p19−/−RAG−/− mice (lacking only IL-23), and development of intestinal inflammation was monitored. As illustrated in Fig. 5, the severe colitis induced by naive CD4+CD45RBhigh T cell transfer into RAG−/− recipients was highly attenuated in p40−/−RAG−/− recipients, confirming that IL-12p40 was essential for disease. Strikingly, although p35−/−RAG−/− recipients developed colitis of similar severity to that found in control RAG−/− recipients, intestinal inflammation was highly attenuated in p19−/−RAG−/− recipients (Fig. 5). These results clearly indicate that IL-23, but not IL-12, is required for the development of T cell–mediated colitis.
Figure 5.
IL-23 drives T cell–mediated colitis. Cohorts of wild-type or IL-12/23–deficient RAG−/− mice were reconstituted with 4 × 105 CD4+CD45RBhigh T cells by i.p. injection. Mice were killed 6–8 wk later, and pathology in the colon was assessed histologically. Each symbol represents a single animal, and data shown represent combined results from two independent experiments (n = 7–11 total mice per group). Horizontal lines represent means.
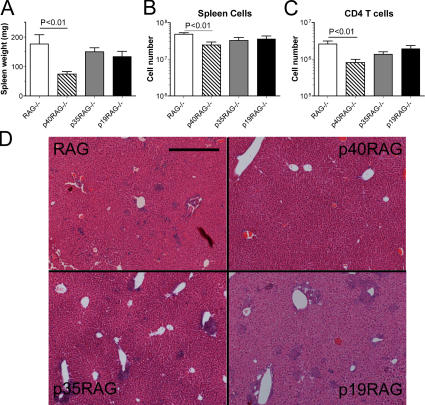
In addition to colitis, naive CD4+ T cell reconstitution of RAG−/− recipients also leads to systemic immune pathology, including splenomegaly and hepatic inflammation (35, 36). We therefore analyzed these parameters to examine the role of IL-23 in T cell–mediated systemic immune pathology. T cell–mediated colitis was accompanied by marked splenomegaly in RAG−/− recipients and, even though this was significantly decreased in p40−/−RAG−/− recipients, there was no significant decrease in splenomegaly in either p35−/−RAG−/− or p19−/−RAG−/− recipients (Fig. 6, A and B). FACS analysis confirmed that the adoptively transferred CD4+ T cells had reconstituted the spleens of all groups of RAG−/− mice and, even though this was reduced in p40−/−RAG−/− recipients, there was no significant reduction in CD4+ T cells in p35−/−RAG−/− or p19−/−RAG−/− recipients (Fig. 6 C). A similar pattern was observed with respect to liver pathology, with numerous prominent inflammatory foci present in RAG−/−, p35−/−RAG−/−, and p19−/−RAG−/− recipients but virtually absent in p40−/−RAG−/− recipients (Fig. 6 D). Collectively, these results show that IL-23 is not required for CD4+ T cell–mediated systemic inflammatory responses and further indicate that either IL-12 or IL-23 alone is able to facilitate CD4+ T cell–dependent inflammatory responses in the liver and spleen.
Figure 6.
T cell–mediated systemic inflammation does not require IL-12 or IL-23. Cohorts of wild-type or IL-12/23–deficient RAG−/− mice were reconstituted with 4 × 105 CD4+CD45RBhigh T cells by i.p. injection and killed 6–8 wk later. (A–C) Splenomegaly was assessed by measuring spleen weights (A) and spleen cell number (B), and CD4+ T cell reconstitution (C) was assessed using FACS analysis. Data represent means ± SEM from two similar experiments (n = 7–11 total mice per group). (D) Representative photomicrographs of liver sections isolated from the mice outlined earlier in this legend. Bar, 500 μm.
T cell–dependent intestinal proinflammatory cytokine production is attenuated in the absence of IL-23
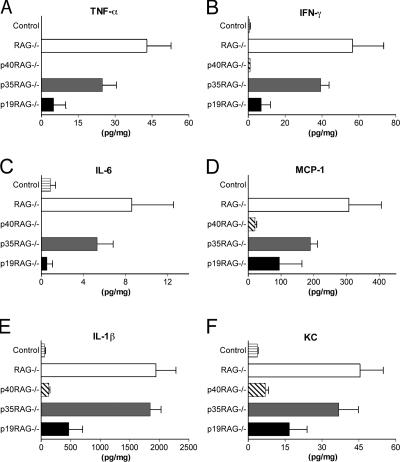
As the T cell transfer colitis model has been associated with increased local Th1 responses (8), we also examined the levels of several proinflammatory cytokines in intestinal tissue homogenates from the various groups of RAG−/− recipients. Although colon homogenates isolated from control RAG−/− recipients contained elevated levels of TNF-α, IFN-γ, IL-6, MCP-1, IL-1β, and KC, these levels were markedly decreased in p40−/−RAG−/− recipients (Fig. 7). As expected, p35−/−RAG−/− recipients with colitis expressed similarly elevated levels of proinflammatory cytokines as control RAG−/− recipients, whereas these were again attenuated in p19−/−RAG−/− recipients (Fig. 7). These results demonstrate that IL-23, but not IL-12, is required for the efficient expression of proinflammatory cytokine cascades in the intestine.
Figure 7.
IL-23 deficiency results in decreased levels of proinflammatory cytokine production in the intestine. Cohorts of wild-type or IL-12/23–deficient RAG−/− mice were reconstituted with 4 × 105 CD4+CD45RBhigh T cells by i.p. injection and killed 6–8 wk later. Cytokine concentrations in colon homogenates were measured and normalized to total protein content for each sample and are given as pg/mg total protein. Results represent mean cytokine levels (±SEM) from one of two similar experiments (n = 3–5 mice per group).
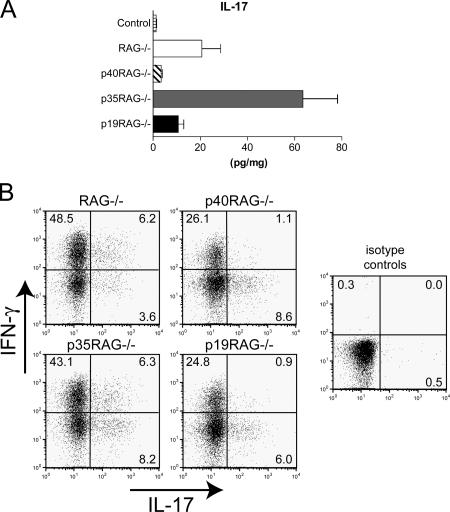
As IL-23 has been strongly associated with proinflammatory IL-17–secreting CD4+ T cells (Th17), we also examined the levels of IL-17 in the colon homogenates and assayed for Th17 cells using intracellular FACS analysis. The highest levels of IL-17 were detected in colon homogenates isolated from p35−/−RAG−/− recipients (Fig. 8 A). In contrast, only low levels of IL-17 were present in colon homogenates from p40−/−RAG−/− or p19−/−RAG−/− recipients (Fig. 8 A). CD4+ T cells isolated from the MLN and LPLs of RAG−/− or p35−/−RAG−/− recipients contained a high proportion of IFN-γ+ cells and a small population of IL-17+ cells (Fig. 8 B and not depicted). Interestingly, CD4+ T cells isolated from p40−/−RAG−/− or p19−/−RAG−/− recipients showed a marked decrease in the frequency of IFN-γ+ cells but still contained a small population of IL-17+ cells (Fig. 8 B). These results show that T cell–mediated colitis correlates with increased frequencies of IFN-γ–secreting Th1 cells in intestinal lymphoid tissue and increased levels of IL-17 in the colon, suggesting that both Th1 and Th17 cells may contribute to pathogenesis. They further show that neither IL-12 nor IL-23 is absolutely required for the differentiation of Th17 cells from naive CD4+ T cells in vivo.
Figure 8.
IL-12 or IL-23 are not required for differentiation of IL-17–secreting CD4+ T cells in vivo. Cohorts of wild-type or IL-12/23–deficient RAG−/− mice were reconstituted with 4 × 105 CD4+CD45RBhigh T cells by i.p. injection and killed 6–8 wk later. (A) IL-17 concentrations in colon homogenates were measured and normalized to total protein content for each sample and are given as pg/10 mg total protein. Results represent mean cytokine levels (±SEM) from one of two similar experiments (n = 3–5 mice per group). (B) Pooled MLN cells were restimulated for 4 h with PMA and ionomycin in the presence of Brefeldin A. IFN-γ– and IL-17–secreting cells were detected using intracellular FACS analysis. Results shown represent frequencies of cytokine-secreting cells among gated CD4+ cells and are representative of two similar experiments.
DISCUSSION
Until recently, it had been widely accepted that the chronic intestinal inflammation found in human CD patients, as well as in many animal models of IBD, was caused by IL-12–driven excessive CD4+ Th1 responses. However, the results presented in this paper clearly demonstrate that IL-23, but not IL-12, plays an essential role in the induction of chronic intestinal inflammation. Although the requirement for IL-23 in T cell–dependent colitis is consistent with previous findings on its ability to promote pathological T cell responses, the demonstration that it is also essential for typhlocolitis in Hh + 129RAG−/− mice highlights a novel role for IL-23 in innate immune pathology in vivo.
Attenuation of intestinal inflammation by blockade or genetic ablation of IL-23 was accompanied by decreased production of many proinflammatory cytokines, including TNF-α, IFN-γ, MCP-1, IL-6, IL-1β, and KC, several of which have been implicated in the pathogenesis of IBD. As IL-23 is produced rapidly by DCs and macrophages after exposure to pathogen-derived molecules (16, 32), the most straightforward interpretation of our data is that IL-23, produced in response to intestinal bacteria, triggers a proinflammatory cytokine cascade that, if left unchecked, can lead to the development of chronic intestinal inflammation. As IL-23R is also expressed by DCs and macrophage populations, it has been proposed that IL-23 secretion can drive an autocrine feedback loop that amplifies local expression of cytokines like IL-1β and TNF-α (37), which in turn stimulate release of additional proinflammatory mediators by stromal, epithelial, and endothelial cells.
Our experiments also highlight some novel aspects of the regulation of IL-17 expression in intestinal inflammation in vivo. To date, IL-23 has been proposed to trigger IL-17 production through the induction/expansion of a novel subset of CD4+ Th17 cells, which have been associated with autoimmune pathology (12, 13, 18, 38). However, our analysis of Hh + 129RAG−/− mice showed that, concomitant with the increased IL-23 expression, there was a striking increase in both IL-17 mRNA expression and protein release in inflamed intestine, indicating that IL-23 also induces the secretion of IL-17 by non–T cells in an inflammatory environment. FACS sorting revealed that several subpopulations of LPLs expressed high levels of IL-17, including granulocytes and monocytes. However, the accumulation of granulocytes and monocytes in the spleens of Hh + 129RAG−/− mice did not correlate with an increase in splenic IL-17 expression, suggesting that additional local signals in the inflamed intestine are required. A previous study noted that neutrophils may produce IL-17 in response to LPS, suggesting that bacteria may provide one such signal (39). Increased expression of IL-17 has been reported in the intestinal mucosa of IBD patients, and histological analysis suggested that both T cells and monocytes may act as sources of IL-17 in the inflamed gut (40). The IL-17R is widely expressed, and IL-17 binding promotes stromal, endothelial, and epithelial cells to secrete proinflammatory mediators that recruit neutrophils to sites of inflammation (41). Together with our previous finding that a prominent granulocytic infiltrate is characteristic of H. hepaticus–induced innate typhlocolitis (30), this supports a role for IL-17 in the induction of innate intestinal pathology and suggests that several cell types may contribute to IL-17 production in the gut.
A very recent study reported that the T cell–mediated colitis that develops in IL-10−/− mice or in RAG−/− recipients of IL-10−/− CD4+ T cells was also dependent on IL-23 (42). Although this was associated with increased development of pathogenic Th17 cells, anti–IL-17 treatment had little impact on colitis and had to be combined with anti–IL-6 treatment to attenuate disease (42). These results indicate that even though IL-23–driven Th17 responses may play an important role in colitis, they constitute one of several potential innate and adaptive immune mechanisms that can contribute to intestinal pathology. Support for this hypothesis was obtained in a parallel study (see Kullberg et al. [43] on p. 2485 of this issue) that examined the role of IL-23 in two models of H. hepaticus–triggered T cell–dependent colitis. Though again highlighting a critical role for IL-23, these studies additionally identified a pathogenic role for IFN-γ, indicating that Th1 and Th17 responses may synergize to elicit maximal pathology during bacterially induced colitis (43). Similarly, in our T cell–mediated colitis model the correlation between IL-17 and intestinal inflammation was not completely straightforward. Although elevated levels of IL-17 were found in colon homogenates from colitic RAG−/− and p35−/−RAG−/− recipients, disease severity correlated with high frequencies of IFN-γ–secreting T cells, whereas only low proportions of IL-17–secreting T cells were present. These results again suggest that both Th1 and Th17 cells contribute to IL-23–dependent colitis. In addition, our observations that small populations of IL-17–secreting T cells were present in both p40−/−RAG−/− and p19−/−RAG−/− recipients clearly indicate that neither IL-12 nor IL-23 is required for differentiation of IL-17–secreting T cells in vivo. These findings are in accord with those of a very recent study in which infection with the intestinal bacterial pathogen Citrobacter rodentium elicited potent Th17 responses in both wild-type and p19−/− mice (21). However, despite mounting strong Th17 responses, p19−/− mice had less colonic inflammation and failed to clear the C. rodentium infection, indicating that IL-23–driven inflammatory responses were an integral component of the protective response (21).
Interestingly, a study using the dextran sulfate sodium model of acute colitis reported that neutralization of IL-17 exacerbated intestinal inflammation, suggesting an inhibitory role for IL-17 in this disease (44). Furthermore, in vitro experiments using intestinal epithelial cell monolayers suggested that IL-17 could enhance mucosal barrier function (45). Several recent studies have clearly shown that TGF-β, in the presence of proinflammatory cytokines such as IL-6, induces the differentiation of Th17 cells (20–22). Conversely, in the absence of inflammatory mediators, TGF-β promotes the development of Foxp3+ T reg cells associated with suppression of inflammatory responses(21, 22, 46–49). These paradoxical observations illustrate the complexity of immune regulation in the intestine and indicate that understanding how interactions between pleiotropic factors such as IL-17 and TGF-β influence intestinal homeostasis presents an important future challenge.
As increasing scrutiny is given to the role of IL-23 in inflammatory responses, it is becoming apparent that it is a gross oversimplification to consider the IL-23–IL-17 and IL-12–IFN-γ pathways as two independent (and often mutually exclusive) axes of immune pathology. Instead, we favor the idea that IL-23 is a central conductor of a range of innate and adaptive inflammatory responses and that IL-23 itself may be regulated at several levels. Although bacterial stimuli may be a major inducer of IL-23 secretion, adaptive immune processes may also modulate its production. In support of this, we have recently observed that injection of agonistic anti-CD40 monoclonal antibody induces an IL-23–dependent acute inflammatory response in RAG−/− mice that was accompanied by intestinal inflammation (50). In contrast to most other models, anti-CD40–induced colitis was independent of the presence of a bacterial microflora (50), indicating that there is an alternative route of IL-23 induction. The anti-CD40 treatment most likely mimicked strong T cell activation, characterized by marked up-regulation of CD40L that can then signal through CD40 on APCs. Thus, during sustained immune responses, activated T cells may provide a positive feedback loop for inducing further production of IL-23, thereby perpetuating inflammation. It would also seem important to have means of inhibiting IL-23 production and, because T reg cells and IL-10 have been implicated in down-modulation of innate and adaptive inflammatory responses, these seem obvious candidates. In fact, macrophages isolated from IL-10−/− mice show elevated secretion of both IL-12 and IL-23 in response to bacterial stimuli (51).
Another important observation in these studies was that even though IL-23 was essential for local tissue inflammation in the intestine, it was not required for systemic inflammatory responses. This was clearly shown in the T cell transfer model of disease in which, even though colitis was highly attenuated in p19−/−RAG−/− mice, there was no inhibition of splenomegaly or in development of inflammatory foci in the liver. This suggests that IL-23 may be especially important for inflammatory responses within peripheral tissues, which is consistent with previous experimental observations in models of autoimmune inflammation in the brain (11) and joints (13) and in inflammation induced by bacterial pathogens in the lung (52) and intestine (21). Together with our results, these studies are consistent with the hypothesis that the natural function of IL-23 in host defense may be in coordinating inflammatory responses against bacterial infection in peripheral tissues, but that dysregulated expression of IL-23 may promote harmful immune pathology in these sites. This is consistent with the expression profile of IL-23 in Hh + RAG−/− mice, where up-regulated IL-23 was present only in the cecum and colon but not in peripheral lymphoid tissues. Our findings that splenomegaly and liver inflammation in Hh + RAG−/− mice were also attenuated after treatment with anti-p19 may reflect the sequential activation of local and systemic inflammatory responses in this model. In this case, we postulate that disease follows an “outside-in” sequence by which infection with the bacteria triggers local inflammation in the intestine, resulting in increased host cytokines and bacterial proinflammatory molecules reaching the systemic circulation. These in turn feed the systemic cytokine cascade that drives splenomegaly and liver pathology; therefore, preventing the initial inflammation in the intestine also shuts down the downstream systemic sequelae. In contrast, the T cell transfer model may represent an “inside-out” sequence in which naive T cell reconstitution is followed by a rapid T cell expansion in systemic lymphoid tissues. In the absence of T reg cells, this expansion proceeds in a dysregulated manner, allowing the excessive accumulation of both autoaggressive T cells that can mediate systemic inflammatory responses as well as bacterially reactive T cells that mediate intestinal inflammation. In this case, although IL-23 deficiency prevents the bacterially reactive T cells from causing colitis, it does not inhibit systemic T cell expansion and associated inflammation.
One additional point to note is that even though IL-23 is clearly a central mediator of intestinal inflammation, there may be additional IL-23–independent inflammatory pathways that also contribute to disease. This hypothesis is supported by our findings that although highly attenuated, some mild inflammation persisted in the anti-p19–treated Hh + 129RAG−/− mice, mainly in the cecum, where the highest levels of H. hepaticus colonization occur (30). This indicates that pathogenic bacteria can also activate alternative innate immune mechanisms that synergize with IL-23 to drive severe pathology. It is clear that excessive immune responses of almost any variety, Th1 cell, Th2 cell, or innate immunity, can mediate intestinal inflammation (1, 2).
The increasing clinical use of biological therapies such as infliximab (anti–TNF-α) in human IBD illustrates the potential benefits that may be derived through molecular analysis of immune pathogenesis. However, the long-term effects of such therapies are still unknown and, given the essential role of TNF-α in host defense, concerns have been voiced over possible increased incidences of infections, such as Mycobacteria, or tumors (53). Although some encouraging results have been obtained in initial clinical trials of anti–IL-12p40 in CD (9), the central role of IL-12 in resistance against many pathogenic infections (54) suggests that long-term administration may similarly depress systemic immune function. Agents that target IL-23 may have the advantage of selectively decreasing local immune responses in afflicted tissues while sparing systemic immune protective mechanisms, a highly desirable property for efficient therapeutic agents for IBD.
MATERIALS AND METHODS
Mice.
129SvEvRAG2−/− mice and wild-type B6, control B6RAG1−/−, and IL-12/23–deficient strains on a B6RAG1−/− background (IL-12p40−/−RAG−/−, IL-12p35−/−RAG−/−, and IL-23p19−/−RAG−/− mice) were bred and maintained under specific pathogen-free conditions in accredited animal facilities at the University of Oxford. Experiments were conducted in accordance with the UK Scientific Procedures Act of 1986. Mice were routinely screened for the presence of Helicobacter spp. and were >6 wk old when first used.
Bacteria.
H. hepaticus NCI-Frederick isolate 1A (strain 51449; American Type Culture Collection) was grown on blood agar plates containing trimethoprim, vancomycin, and polymixin B (all obtained from Oxoid) under microaerophilic conditions as previously described (30, 55). For H. hepaticus infections, bacterial viability was confirmed using fluorescent microscopy with a bacterial live/dead kit (BacLight; Invitrogen), and 129SvEvRAG2−/− mice were fed three times on alternate days with ∼5 × 107–2 × 108 CFU H. hepaticus.
Antibody treatment.
Anti–IL-23p19 monoclonal antibody (PAB1106) was produced and characterized as previously described (56). Antibody treatment was commenced on the day of the first inoculation with H. hepaticus, and mice received weekly i.p. injections of 1 mg anti–IL-23p19 or isotype control for the duration of the experiment.
Induction of colitis with naive CD4+CD45RBhigh T cells.
Naive CD4+CD45RBhigh T cells were isolated from spleens of C57BL/6 mice using FACS sorting as previously described (57). In brief, single cell suspensions were depleted of CD8+, MHC class II+, Mac-1+, and B220+ cells by negative selection using a panel of rat monoclonal antibodies, followed by sheep anti–rat–coated Dynabeads (Dynal). After staining with Cy-Chrome–conjugated anti-CD4, PE-conjugated anti-CD25, and FITC–anti-CD45RB (all obtained from BD Biosciences), naive CD4+CD25−CD45RBhigh T cells were purified (∼99%) by cell sorting with a cell sorter (MoFlo; DakoCytomation). Sex-matched control or IL-12/23–deficient B6RAG−/− mice received 4 × 105 CD4+CD45RBhigh T cells by i.p. injection, and development of intestinal inflammation was monitored as described in the next paragraph.
Assessment of intestinal inflammation.
Mice were killed when symptoms of clinical disease (weight loss or diarrhea) became apparent in control groups, usually 6–8 wk after initiation of experiments. Samples of liver, cecum, and proximal, mid-, and distal colon were immediately fixed in buffered 10% formalin. 4–5 μm of paraffin-embedded sections was stained with hematoxylin and eosin, and inflammation was assessed as previously described (30, 57). Each sample was graded semiquantitatively from 0 to 4, and typical features of each grade are as follows: 0 = normal; 1 = mild epithelial hyperplasia; 2 = pronounced hyperplasia with substantial inflammatory infiltrates; 3 = severe hyperplasia and infiltration with marked decrease in goblet cells; and 4 = severe hyperplasia, severe transmural inflammation, ulceration, crypt abscesses, and severe depletion of goblet cells. Ceca and colons were assessed separately, and three separate sections from each sample were examined. The total colonic score was obtained by adding the individual scores from the sections of proximal, mid-, and distal colon.
Flow cytometry.
Aliquots of 1–5 × 105 cells were stained in FACS buffer (HBSS, 0.1% BSA, 5 mM EDTA; both obtained from Sigma-Aldrich) using the following panel of monoclonal antibody to mouse cell surface molecules (all obtained from BD Biosciences): biotinylated anti–NK cells (DX5), biotinylated anti-Gr1 (Ly6G), allophycocyanin-conjugated anti-CD11b, PE–anti-CD11c, PerCP-conjugated anti-CD4, and PE–anti–mouse TCRβ. Biotinylated antibodies were detected using PE-, allophycocyanin-, or FITC-conjugated streptavidin (all obtained from BD Biosciences). Samples were first incubated for 10 min with Fc block (eBioscience) to block any nonspecific binding, and subsequent staining steps were performed for 20 min on ice, followed by washing with FACS buffer. Samples were fixed in FACS buffer containing 2% paraformaldehyde (Biolab) acquired using a FACSCalibur (Becton Dickinson) and analyzed with software (FlowJo; Tree Star, Inc.). For intracellular cytokine analysis, cells were incubated in complete RPMI 1640 (10% heat-inactivated FCS, 2 mM l-glutamine, 100 U/ml penicillin and streptomycin [obtained from Invitrogen], and 0.05 mM 2-mercaptoethanol [obtained from Sigma-Aldrich]) containing 0.1 μM PMA and 1 μM ionomycin (both obtained from Sigma-Aldrich), plus 10 μg/ml Brefeldin A (Sigma-Aldrich), for 4 h at 37°C in a humidified incubator with 5% CO2. Cell surface staining was performed as described previously in this paper, and, after fixing, the samples were incubated in permeabilization buffer (PBS, 0.1% BSA, 0.5% saponin; Sigma-Aldrich) containing allophycocyanin–anti–mouse IFN-γ, PE–anti–mouse IL-17 or appropriate isotype controls, allophycocyanin–rat IgG1, and PE–rat IgG2b (all obtained from BD Biosciences) for 30 min at room temperature, washed in permeabilization buffer, and analyzed as described in this paper.
Isolation of leukocyte subpopulations.
Spleen cells and LPLs were isolated from 6–12 pooled Hh +129SvEvRAG2−/− mice as previously described (30). Cells were stained with fluorescent antibodies, and leukocyte subpopulations were purified using FACS sorting as outlined previously in this paper. Cells were sorted into distinct subpopulations on the basis of size (forward scatter [FSC]) and granularity (side scatter [SSC]) in combination with surface marker expression (58). Characteristics of subpopulations were as follows: granulocytes, FSCHiSSCHiGr1HiCD11bHiCD11c−; monocytes, FSCHiSSCLoGr1IntCD11bHiCD11c−; DCs, FSCHiSSCLoGr1−CD11cHi; and NK cells, FSCLoSSCLoDX-5Hi.
Quantitation of cytokine gene expression using real-time PCR.
RNA was purified from frozen tissue samples or sorted cells using RNAeasy kits (QIAGEN). Homogenization was performed using a Polytron Homogenizer (Kinematica AG). RNA purity and quantification was determined using a Nanodrop spectrophotometer (Nanodrop Technologies). cDNA synthesis was performed using a reverse transcriptase kit (Superscript III) with Oligo dT (both obtained from Invitrogen). Q-PCR reactions were performed using the following primers, together with FAM/TAMRA- or VIC/TAMRA-labeled probes: HPRT primers, 5′-GACCGGTCCCGTCATGC-3′ and 5′-TCATAACCTGGTTCATCATCGC-3′, and probe, 5′-ACCCGCAGTCCCAGCGTCGTC-3′; IL-23p19 primers, 5′-AGCGGGACATATGAATCTACTAAGAGA-3′ and 5′-GTCCTAGTAGGGAGGTGTGAAGTTG-3′, and probe, 5′-CCAGTTCTGCTTGCAAAGGATCCGC-3′; IL-12p35 primers, 5′-TACTAGAGAGACTTCTTCCACAACAAGAG-3′ and 5′-TCTGGTACATCTTCAAGTCCTCATAGA-3′, and probe, 5′-AGACGTCTTTGATGATGACCCTGTGCCT-3′; IL-12p40 primers, 5′-GACCATCACTGTCAAAGAGTTTCTAGAT-3′and 5′-AGGAAAGTCTTGTTTTTGAAATTTTTTAA-3′, and probe, 5′-CCACTCACATCTGCTGCTCCACAAGAAG-3′; and IL-17A primers, 5′-GCTCCAGAAGGCCCTCAG-3′ and 5′-CTTTCCCTCCGCATTGACA-3′, and probe, 5′-ACCTCAACCGTTCCACGTCACCCTG-3′.
cDNA samples were assayed in triplicate using a detection system (Chromo4; GRI), and cytokine gene expression levels for each individual sample were normalized relative to HPRT using ΔCt calculations (59).
Quantitation of cytokine protein levels in intestinal tissues.
In initial experiments, small pieces of colon (∼5 mm of mid-colon) were isolated and rinsed in HBSS/BSA and weighed. Colon explants were cultured overnight in 24-well tissue culture plates (Costar) in 500 μl complete RPMI 1640 at 37°C in an atmosphere containing 5% CO2. After centrifugation at 10,000 g to pellet debris, culture supernatants were transferred to fresh tubes and stored at −20°C. Cytokine concentrations were measured using specific sandwich ELISAs (IL-23, R&D Systems; IL-17, Bio-Rad Laboratories) and were normalized to the weight of the colon explant.
For more comprehensive analysis, frozen intestinal tissue samples were homogenized in PBS containing a cocktail of protease inhibitors (Protease Inhibitor Cocktail Tablets; Roche) using a Polytron Homogenizer. After centrifugation at 10,000 g to pellet debris, concentrations of a panel of proinflammatory cytokines in supernatants were measured either using the cytometric bead assay (BD Biosciences) or the Luminex 100 assay (Bio-Rad Laboratories). In all cases, cytokine levels were normalized to the total protein level in each sample, as measured using the Bradford assay (Bio- Rad Laboratories).
Quantitation of H. hepaticus using real-time PCR.
DNA was purified from cecal contents taken from H. hepaticus–infected mice using the DNA Stool kit (QIAGEN). H. hepaticus DNA was determined using a Q-PCR method based on the cdtB gene and performed with a Chromo4 detection system, as previously described (30, 60).
Statistics.
The nonparametric Mann-Whitney test was used for comparing pathology scores and Q-PCR data, and an unpaired t test was used to examine spleen weights and cell counts. Differences were considered statistically significant when P < 0.05.
Acknowledgments
We would like to acknowledge Nigel Rust for assistance with cell sorting, Liz Darley and Richard Stillion for histology, Janine Coombes for critical reading of the manuscript, and the staff at the University of Oxford for excellent animal care.
This work was supported by grants from the Wellcome Trust to K.J. Maloy (Career Development Fellowship) and F. Powrie (Senior Fellowship), and by a grant from a Marie Curie European Union Research Training Network program (MRTN-CT-2004-005632 to F. Powrie and K.J. Maloy).
The authors declare that they have no competing financial interests.
Abbreviations used: CD, Crohn's disease; IBD, inflammatory bowel disease; FSC, forward scatter; KC, mouse chemokine CXCL1; LPL, lamina propria leukocyte; MCP-1, monocyte chemoattractant protein–1; MLN, mesenteric LN; Q-PCR, quantitative PCR; SSC, side scatter.
References
- 1.Macdonald, T.T., and G. Monteleone. 2005. Immunity, inflammation, and allergy in the gut. Science. 307:1920–1925. [DOI] [PubMed] [Google Scholar]
- 2.Elson, C.O., Y. Cong, V.J. McCracken, R.A. Dimmitt, R.G. Lorenz, and C.T. Weaver. 2005. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol. Rev. 206:260–276. [DOI] [PubMed] [Google Scholar]
- 3.Strober, W., I.J. Fuss, and R.S. Blumberg. 2002. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 20:495–549. [DOI] [PubMed] [Google Scholar]
- 4.Bouma, G., and W. Strober. 2003. The immunological and genetic basis of inflammatory bowel disease. Nat. Rev. Immunol. 3:521–533. [DOI] [PubMed] [Google Scholar]
- 5.Neurath, M.F., I. Fuss, B.L. Kelsall, E. Stuber, and W. Strober. 1995. Antibodies to interleukin-12 abrogate established experimental colitis in mice. J. Exp. Med. 182:1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson, S.J., S. Shah, M. Comiskey, Y.P. de Jong, B. Wang, E. Mizoguchi, A.K. Bhan, and C. Terhorst. 1998. T cell–mediated pathology in two models of experimental colitis depends predominantly on the interleukin-12–signal transducer and activator of transcription (Stat)–4 pathway but is not conditional on interferon γ expression by T cells. J. Exp. Med. 187:1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullberg, M.C., A.G. Rothfuchs, D. Jankovic, P. Caspar, T.A. Wynn, P.L. Gorelick, A.W. Cheever, and A. Sher. 2001. Helicobacter hepaticus-induced colitis in interleukin-10-deficient mice: cytokine requirements for the induction and maintenance of intestinal inflammation. Infect. Immun. 69:4232–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powrie, F., M.W. Leach, S. Mauze, S. Menon, L.B. Caddle, and R.L. Coffman. 1994. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1:553–562. [DOI] [PubMed] [Google Scholar]
- 9.Mannon, P.J., I.J. Fuss, L. Mayer, C.O. Elson, W.J. Sandborn, D. Present, B. Dolin, N. Goodman, C. Groden, R.L. Hornung, et al. 2004. Anti-interleukin-12 antibody for active Crohn's disease. N. Engl. J. Med. 351:2069–2079. [DOI] [PubMed] [Google Scholar]
- 10.Oppmann, B., R. Lesley, B. Blom, J.C. Timans, Y. Xu, B. Hunte, F. Vega, N. Yu, J. Wang, K. Singh, et al. 2000. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 13:715–725. [DOI] [PubMed] [Google Scholar]
- 11.Cua, D.J., J. Sherlock, Y. Chen, C.A. Murphy, B. Joyce, B. Seymour, L. Lucian, W. To, S. Kwan, T. Churakova, et al. 2003. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 421:744–748. [DOI] [PubMed] [Google Scholar]
- 12.Langrish, C.L., Y. Chen, W.M. Blumenschein, J. Mattson, B. Basham, J.D. Sedgwick, T. McClanahan, R.A. Kastelein, and D.J. Cua. 2005. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy, C.A., C.L. Langrish, Y. Chen, W. Blumenschein, T. McClanahan, R.A. Kastelein, J.D. Sedgwick, and D.J. Cua. 2003. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J. Exp. Med. 198:1951–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiekowski, M.T., M.W. Leach, E.W. Evans, L. Sullivan, S.C. Chen, G. Vassileva, J.F. Bazan, D.M. Gorman, R.A. Kastelein, S. Narula, and S.A. Lira. 2001. Ubiquitous transgenic expression of the IL-23 subunit p19 induces multiorgan inflammation, runting, infertility, and premature death. J. Immunol. 166:7563–7570. [DOI] [PubMed] [Google Scholar]
- 15.Bettelli, E., and V.K. Kuchroo. 2005. IL-12– and IL-23–induced T helper cell subsets: birds of the same feather flock together. J. Exp. Med. 201:169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenzie, B.S., R.A. Kastelein, and D.J. Cua. 2006. Understanding the IL-23-IL-17 immune pathway. Trends Immunol. 27:17–23. [DOI] [PubMed] [Google Scholar]
- 17.Harrington, L.E., R.D. Hatton, P.R. Mangan, H. Turner, T.L. Murphy, K.M. Murphy, and C.T. Weaver. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6:1123–1132. [DOI] [PubMed] [Google Scholar]
- 18.Park, H., Z. Li, X.O. Yang, S.H. Chang, R. Nurieva, Y.H. Wang, Y. Wang, L. Hood, Z. Zhu, Q. Tian, and C. Dong. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6:1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwal, S., N. Ghilardi, M.H. Xie, F.J. de Sauvage, and A.L. Gurney. 2003. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 278:1910–1914. [DOI] [PubMed] [Google Scholar]
- 20.Veldhoen, M., R.J. Hocking, C.J. Atkins, R.M. Locksley, and B. Stockinger. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. [DOI] [PubMed] [Google Scholar]
- 21.Mangan, P.R., L.E. Harrington, D.B. O'Quinn, W.S. Helms, D.C. Bullard, C.O. Elson, R.D. Hatton, S.M. Wahl, T.R. Schoeb, and C.T. Weaver. 2006. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 441:231–234. [DOI] [PubMed] [Google Scholar]
- 22.Bettelli, E., Y. Carrier, W. Gao, T. Korn, T.B. Strom, M. Oukka, H.L. Weiner, and V.K. Kuchroo. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. [DOI] [PubMed] [Google Scholar]
- 23.Cua, D.J., and R.A. Kastelein. 2006. TGF-beta, a ‘double agent’ in the immune pathology war. Nat. Immunol. 7:557–559. [DOI] [PubMed] [Google Scholar]
- 24.Puccetti, P., M.L. Belladonna, and U. Grohmann. 2002. Effects of IL-12 and IL-23 on antigen-presenting cells at the interface between innate and adaptive immunity. Crit. Rev. Immunol. 22:373–390. [PubMed] [Google Scholar]
- 25.Hugot, J.P., M. Chamaillard, H. Zouali, S. Lesage, J.P. Cezard, J. Belaiche, S. Almer, C. Tysk, C.A. O'Morain, M. Gassull, et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 411:599–603. [DOI] [PubMed] [Google Scholar]
- 26.Ogura, Y., D.K. Bonen, N. Inohara, D.L. Nicolae, F.F. Chen, R. Ramos, H. Britton, T. Moran, R. Karaliuskas, R.H. Duerr, et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 411:603–606. [DOI] [PubMed] [Google Scholar]
- 27.Girardin, S.E., I.G. Boneca, J. Viala, M. Chamaillard, A. Labigne, G. Thomas, D.J. Philpott, and P.J. Sansonetti. 2003. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278:8869–8872. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi, K.S., M. Chamaillard, Y. Ogura, O. Henegariu, N. Inohara, G. Nunez, and R.A. Flavell. 2005. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 307:731–734. [DOI] [PubMed] [Google Scholar]
- 29.Maeda, S., L.C. Hsu, H. Liu, L.A. Bankston, M. Iimura, M.F. Kagnoff, L. Eckmann, and M. Karin. 2005. Nod2 mutation in Crohn's disease potentiates NF-kappaB activity and IL-1beta processing. Science. 307:734–738. [DOI] [PubMed] [Google Scholar]
- 30.Maloy, K.J., L. Salaun, R. Cahill, G. Dougan, N.J. Saunders, and F. Powrie. 2003. CD4+CD25+ T reg cells suppress innate immune pathology through cytokine-dependent mechanisms. J. Exp. Med. 197:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erdman, S.E., T. Poutahidis, M. Tomczak, A.B. Rogers, K. Cormier, B. Plank, B.H. Horwitz, and J.G. Fox. 2003. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am. J. Pathol. 162:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker, C., S. Wirtz, M. Blessing, J. Pirhonen, D. Strand, O. Bechthold, J. Frick, P.R. Galle, I. Autenrieth, and M.F. Neurath. 2003. Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells. J. Clin. Invest. 112:693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt, C., T. Giese, B. Ludwig, I. Mueller-Molaian, T. Marth, S. Zeuzem, S.C. Meuer, and A. Stallmach. 2005. Expression of interleukin-12-related cytokine transcripts in inflammatory bowel disease: elevated interleukin-23p19 and interleukin-27p28 in Crohn's disease but not in ulcerative colitis. Inflamm. Bowel Dis. 11:16–23. [DOI] [PubMed] [Google Scholar]
- 34.Fuss, I.J., C. Becker, Z. Yang, C. Groden, R.L. Hornung, F. Heller, M.F. Neurath, W. Strober, and P.J. Mannon. 2006. Both IL-12p70 and IL-23 are synthesized during active Crohn's disease and are down-regulated by treatment with anti-IL-12 p40 monoclonal antibody. Inflamm. Bowel Dis. 12:9–15. [DOI] [PubMed] [Google Scholar]
- 35.Powrie, F., M.W. Leach, S. Mauze, L.B. Caddle, and R.L. Coffman. 1993. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int. Immunol. 5:1461–1471. [DOI] [PubMed] [Google Scholar]
- 36.Leach, M.W., A.G. Bean, S. Mauze, R.L. Coffman, and F. Powrie. 1996. Inflammatory bowel disease in C.B-17 scid mice reconstituted with the CD45RBhigh subset of CD4+ T cells. Am. J. Pathol. 148:1503–1515. [PMC free article] [PubMed] [Google Scholar]
- 37.Langrish, C.L., B.S. McKenzie, N.J. Wilson, R. de Waal Malefyt, R.A. Kastelein, and D.J. Cua. 2004. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol. Rev. 202:96–105. [DOI] [PubMed] [Google Scholar]
- 38.Nakae, S., S. Saijo, R. Horai, K. Sudo, S. Mori, and Y. Iwakura. 2003. IL-17 production from activated T cells is required for the spontaneous development of destructive arthritis in mice deficient in IL-1 receptor antagonist. Proc. Natl. Acad. Sci. USA. 100:5986–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferretti, S., O. Bonneau, G.R. Dubois, C.E. Jones, and A. Trifilieff. 2003. IL-17, produced by lymphocytes and neutrophils, is necessary for lipopolysaccharide-induced airway neutrophilia: IL-15 as a possible trigger. J. Immunol. 170:2106–2112. [DOI] [PubMed] [Google Scholar]
- 40.Fujino, S., A. Andoh, S. Bamba, A. Ogawa, K. Hata, Y. Araki, T. Bamba, and Y. Fujiyama. 2003. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 52:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolls, J.K., and A. Linden. 2004. Interleukin-17 family members and inflammation. Immunity. 21:467–476. [DOI] [PubMed] [Google Scholar]
- 42.Yen, D., J. Cheung, H. Scheerens, F. Poulet, T. McClanahan, B. McKenzie, M.A. Kleinschek, A. Owyang, J. Mattson, W. Blumenschein, et al. 2006. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Invest. 116:1310–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kullberg, M.C., D. Jankovic, C.G. Feng, S. Hue, P.L. Gorelick, B.S. McKenzie, D.J. Cua, F. Powrie, A.W. Cheever, K.J. Maloy, and A. Sher. 2006. IL-23 is a key mediator of Helicobacter hepaticus–induced T cell–dependent colitis. J. Exp. Med. 203:2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogawa, A., A. Andoh, Y. Araki, T. Bamba, and Y. Fujiyama. 2004. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin. Immunol. 110:55–62. [DOI] [PubMed] [Google Scholar]
- 45.Kinugasa, T., T. Sakaguchi, X. Gu, and H.C. Reinecker. 2000. Claudins regulate the intestinal barrier in response to immune mediators. Gastroenterology. 118:1001–1011. [DOI] [PubMed] [Google Scholar]
- 46.Chen, W., W. Jin, N. Hardegen, K.J. Lei, L. Li, N. Marinos, G. McGrady, and S.M. Wahl. 2003. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fantini, M.C., C. Becker, G. Monteleone, F. Pallone, P.R. Galle, and M.F. Neurath. 2004. Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25− T cells through Foxp3 induction and down-regulation of Smad7. J. Immunol. 172:5149–5153. [DOI] [PubMed] [Google Scholar]
- 48.Wan, Y.Y., and R.A. Flavell. 2005. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc. Natl. Acad. Sci. USA. 102:5126–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marie, J.C., J.J. Letterio, M. Gavin, and A.Y. Rudensky. 2005. TGF-β1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J. Exp. Med. 201:1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhlig, H., B.S. McKenzie, S. Hue, C. Thompson, R. Stepankova, N.J. Robinson, H. Tlaskalova-Hogenova, D.J. Cua, and F. Powrie. 2006. Differential acitvity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 25:309–318. [DOI] [PubMed] [Google Scholar]
- 51.Kamada, N., T. Hisamatsu, S. Okamoto, T. Sato, K. Matsuoka, K. Arai, T. Nakai, A. Hasegawa, N. Inoue, N. Watanabe, et al. 2005. Abnormally differentiated subsets of intestinal macrophage play a key role in Th1-dominant chronic colitis through excess production of IL-12 and IL-23 in response to bacteria. J. Immunol. 175:6900–6908. [DOI] [PubMed] [Google Scholar]
- 52.Happel, K.I., P.J. Dubin, M. Zheng, N. Ghilardi, C. Lockhart, L.J. Quinton, A.R. Odden, J.E. Shellito, G.J. Bagby, S. Nelson, and J.K. Kolls. 2005. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 202:761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamilton, C.D. 2004. Infectious complications of treatment with biologic agents. Curr. Opin. Rheumatol. 16:393–398. [DOI] [PubMed] [Google Scholar]
- 54.Trinchieri, G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133–146. [DOI] [PubMed] [Google Scholar]
- 55.Young, V.B., K.A. Knox, J.S. Pratt, J.S. Cortez, L.S. Mansfield, A.B. Rogers, J.G. Fox, and D.B. Schauer. 2004. In vitro and in vivo characterization of Helicobacter hepaticus cytolethal distending toxin mutants. Infect. Immun. 72:2521–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen, Y., C.L. Langrish, B. McKenzie, B. Joyce-Shaikh, J.S. Stumhofer, T. McClanahan, W. Blumenschein, T. Churakovsa, J. Low, L. Presta, et al. 2006. Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J. Clin. Invest. 116:1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Read, S., V. Malmstrom, and F. Powrie. 2000. Cytotoxic T lymphocyte–associated antigen 4 plays an essential role in the function of CD25+CD4+ regulatory cells that control intestinal inflammation. J. Exp. Med. 192:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lagasse, E., and I.L. Weissman. 1996. Flow cytometric identification of murine neutrophils and monocytes. J. Immunol. Methods. 197:139–150. [DOI] [PubMed] [Google Scholar]
- 59.Pfaffl, M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ge, Z., D.A. White, M.T. Whary, and J.G. Fox. 2001. Fluorogenic PCR-based quantitative detection of a murine pathogen, Helicobacter hepaticus. J. Clin. Microbiol. 39:2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]