Abstract
The RNA-binding protein TIAR (related to TIA-1 [T-cell-restricted intracellular antigen 1]) was shown to associate with subsets of mRNAs bearing U-rich sequences in their 3′ untranslated regions. TIAR can function as a translational repressor, particularly in response to cytotoxic agents. Using unstressed colon cancer cells, collections of mRNAs associated with TIAR were isolated by immunoprecipitation (IP) of (TIAR-RNA) ribonucleoprotein (RNP) complexes, identified by microarray analysis, and used to elucidate a common signature motif present among TIAR target transcripts. The predicted TIAR motif was an unexpectedly cytosine-rich, 28- to 32-nucleotide-long element forming a stem and a loop of variable size with an additional side loop. The ability of TIAR to bind an RNA oligonucleotide with a representative C-rich TIAR motif sequence was verified in vitro using surface plasmon resonance. By this analysis, TIAR containing two or three RNA recognition domains (TIAR12 and TIAR123) showed low but significant binding to the C-rich sequence. In vivo, insertion of the C-rich motif into a heterologous reporter strongly suppressed its translation in cultured cells. Using this signature motif, an additional ∼2,209 UniGene targets were identified (2.0% of the total UniGene database). A subset of specific mRNAs were validated by RNP IP analysis. Interestingly, in response to treatment with short-wavelength UV light (UVC), a stress agent causing DNA damage, each of these target mRNAs bearing C-rich motifs dissociated from TIAR. In turn, expression of the encoded proteins was elevated in a TIAR-dependent manner. In sum, we report the identification of a C-rich signature motif present in TIAR target mRNAs whose association with TIAR decreases following exposure to a stress-causing agent.
Mammalian gene expression is extensively regulated at the posttranscriptional level, via mechanisms such as pre-mRNA splicing, transport, stability, and translation. Prominent among the posttranscriptional trans-acting factors that influence these processes are RNA-binding proteins (RBPs) that influence transcript splicing, localization, stability, and association with the translation machinery (11, 17, 42). Many mRNAs encoding stress-response, proliferative, immune, and developmental proteins comprise specific regulatory sequences in the untranslated regions (UTRs), often encompassing uridine- or adenine/uridine-rich stretches (hence termed “AREs”). AREs are bound by a specific subset of RBPs that influence the stability and translation of the ribonucleoprotein (RNP) complex. Many ARE-RBPs decrease the stability of target mRNAs, including AU-binding factor 1 (AUF1), tristetraprolin (TTP), K homology splicing-regulatory protein (KSRP), and the butyrate response factor 1 (BRF1) (7, 27, 34, 39, 46). Other ARE-RBPs, like the Hu proteins (HuR, HuB, HuC, and HuD), can stabilize target mRNAs instead (4, 6); Hu proteins have also been shown to modulate the translation of several target mRNAs, both enhancing (5, 24, 31) and inhibiting (9, 23, 33) protein synthesis. However, the best-studied ARE-RBPs functioning as translational inhibitors are the T-cell-restricted intracellular antigen 1 (TIA-1) and the TIA-1-related protein TIAR (1, 2, 14, 28, 32).
TIA-1 and TIAR contain three RNA-recognition motifs (RRMs) through which they bind mRNAs (10). In addition to participating in pre-mRNA splicing (12, 26, 38), TIA-1 and TIAR have been proposed to repress translation (1, 2, 32, 37). In unstressed cells, a preinitiation complex (comprising the eukaryotic translation initiation factor 1 [eIF-1], eIF-2, eIF-3, eIF-5, and the 40S ribosomal subunit) forms at the 5′ end of capped mRNAs. Following the recognition of an initiation codon, the 60S subunit assembles, displacing the eIFs and forming a functional ribosome to initiate translation. In cells exposed to damaging agents, phosphorylation of eIF-2α by a family of kinases (PKR, PERK, GCN2, and HRI) reduces the levels of functional preinitiation complex (recently reviewed in reference 16). Under these conditions, TIAR and TIA-1 have been postulated to function as translational repressors by associating with eIF-4F, eIF-3, and the 40S ribosomal subunit, to form nonfunctional preinitiation complexes (2). The self-aggregating properties of TIA-1 and TIAR were further proposed to facilitate the accumulation of the translationally inactive preinitiation complexes into discrete cytoplasmic foci called stress granules (SGs). Given the presence of RBPs implicated in the regulation of mRNA turnover (such as TTP and HuR) and translation (TIA proteins) at SGs, these foci are believed to function as dynamic sites of mRNA triage during stress, wherein the composition of mRNA RNP complexes and their subsequent engagement with the translation or degradation machineries are decided (20, 21).
While these mechanisms of TIA-1/TIAR action can lead to a general suppression of translation in the cell, they are believed to have a preferential effect upon specific subsets of bound mRNAs, such as ARE-containing mRNAs encoding tumor necrosis factor alpha (TNF-α), matrix metalloproteinase 13 (MMP-13), cyclooxygenase 2 (COX-2), and β2-adrenergic receptor (AR) (8, 14, 19, 37, 44). Accordingly, global searches have been undertaken to identify TIA-1/TIAR target mRNAs systematically. An earlier study in which pools of random RNA sequences were selected/amplified in vitro revealed that both TIA proteins recognized RNAs containing U-rich stretches (10). More recently, a genome-wide search for TIA-1 target mRNAs was carried out by immunoprecipitation (IP) of TIA-1 RNPs followed by the identification of bound transcripts using DNA microarrays (28). A computational analysis of the target mRNAs found by this approach led to the elucidation of a shared signature motif present among mRNAs which was also U rich. TIA-1 was shown to associate with target mRNAs bearing this motif in cells subjected to heat shock and to suppress their translation (28). Using a similar en masse approach, TIAR target mRNAs were found to include many mRNAs encoding translation factors and other proteins involved in translation (32). Further analysis of TIAR RNPs indicated that the association of TIAR with several target mRNAs increased following irradiation with short-wavelength UV light (UVC), thereby helping to suppress their translation (32).
Here, we sought to elucidate a shared motif among TIAR target mRNAs. The starting material was a collection of TIAR-bound transcripts that was isolated from untreated RKO cells (a human colon cancer line) using the RNP IP methodology and was identified by using a microarray (32). Computational analysis of this set of transcripts revealed a shared signature motif that was unexpectedly C rich. The ability of TIAR RRM domains to bind to a representative C-rich sequence was verified in vitro using surface plasmon resonance (SPR). Further validation of this interaction was obtained by studying the binding of mRNAs that were predicted to be TIAR targets because their 3′ untranslated regions (3′UTRs) contained at least one occurrence of the C-rich TIAR motif. Interestingly, the transcripts tested were found to dissociate from TIAR in response to UVC treatment, suggesting that this C-rich TIAR signature motif may occur within mRNAs whose binding to TIAR decreases following stress stimulation.
MATERIALS AND METHODS
Cell culture, treatment, and transfections.
Human colorectal carcinoma RKO cells were cultured in minimum essential medium (Invitrogen), and human cervical carcinoma HeLa cells in Dulbecco's modified essential medium, each supplemented with 10% fetal bovine serum and antibiotics. Where indicated, cells were irradiated with 25 J/m2 of short-wavelength UV light (UVC). Reporter plasmid pEGFP-GAPDH was constructed by inserting a 200 bp of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) 3′UTR (nucleotides 1111 to 1310 of GAPDH [RefSeq accession no. NM_002046]) after the stop codon of pEGFP-N1; reporter plasmid pEGFP-Motif was constructed by inserting the consensus C-rich motif sequence UUGCCACCUCCUGCUCCUGCCCAGACAG within the GAPDH 3′UTR from pEGFP-GAPDH (at nucleotide position 1211 of the above fragment); reporter plasmid pEGFP-APAF1 was constructed by subcloning the 3′UTR of APAF1 (nucleotide positions 5401 to 5825 of NM_013229, including the C-rich motif hit CTGCTCCCTCTTGTTTCTTACATATCAG) immediately after the enhanced green fluorescent protein (EGFP) stop codon. Subconfluent cells were transfected with TIAR-directed (AAGGGCTATTCATTTGTCAGA) or control (TTCTCCGAACGTGTCACGT) small interfering RNAs (siRNAs) (20 nM each) and 24 h later with reporter plasmids (0.2 μg) using Lipofectamine 2000. Cells were subsequently harvested for RNA and protein analyses.
IP assays.
IP of [TIAR-mRNA] complexes from RKO cell lysates was used to evaluate the association of endogenous TIAR with endogenous target mRNAs. The assay was performed essentially as described previously (29, 40), except that 100 million cells were used as starting material and lysate supernatants were precleared for 30 min at 4°C using 15 μg of immunoglobulin G (IgG) (Santa Cruz Biotech.) and 50 μl of protein-A Sepharose beads (Sigma) that had been previously swollen in NT2 buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40) supplemented with 5% bovine serum albumin. Beads (100 μl) were incubated (16 h at 4°C) with 30 μg of antibody (either goat IgG [Santa Cruz Biotechnology] or goat anti-TIAR [Santa Cruz Biotechnology]) and then for 2 h at 4°C with 1.5 mg of cell lysate. After extensive washes and digestion of proteins in the IP material (40), the RNA was extracted and used either for hybridization of cDNA arrays or for verification of individual TIAR target transcripts. The array analysis was previously reported (32). Briefly, RNA obtained after IP reactions using either an anti-TIAR antibody or IgG was reverse transcribed in the presence of [α-33P]dCTP and the radiolabeled product was used to hybridize cDNA arrays (Mammalian Gene Collection [MGC] arrays, containing ∼6,000 individual genes) employing previously reported methodologies (29, 40, 41). All of the data were analyzed using the Array Pro software (Media Cybernetics, Inc.) and then normalized by Z score transformation and used to calculate differences in signal intensities. Significant values were tested using a two-tailed Z test and P < 0.01. The data were calculated from three independent experiments. The complete cDNA array data are available from the authors. For the analysis of individual transcripts, RNA in the IP material was used in reverse transcription (RT) reactions followed by quantitative real-time PCR (qPCR) analysis to detect the presence of specific target mRNAs using gene-specific primer pairs (see the supplemental material). qPCR products were visualized after electrophoresis in 1% agarose gels stained with ethidium bromide to verify that single bands were amplified in each reaction.
Computational analysis to identify a TIAR signature motif.
Human UniGene records were first identified from the most strongly enriched TIAR targets derived from the array analysis using untreated RKO cells. The top 179 transcripts from which 3′UTRs were available served as the experimental data set (see Table S1 in the supplemental material) for the identification of the TIAR motif. Shared RNA motifs were elucidated from the the 3′UTR sequences; among the top candidate motifs, the motif with the highest statistical enrichment in the experimental 3′UTR data set was considered to be the best TIAR candidate motif (additional description in the supplemental material). The computational analysis was conducted as previously described (28) using the software RNAmotifPro (M. Zhan, unpublished). The motif logo was constructed using WebLogo (http://weblogo.berkeley.edu/). RNAplot was used to depict the secondary structure of the representative RNA motifs. The computation was performed using the NIH Biowulf computer farm. Both UniGene and RefSeq datasets were downloaded from NCBI.
Western blot analysis.
Whole-cell protein lysates (10 or 15 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene difluoride membranes, and used for Western blot analysis. Primary antibody incubations were performed using mouse monoclonal antibodies recognizing β-actin (Abcam) or c-Myc (BD Pharmingen) or using rabbit polyclonal antibodies recognizing Apaf-1 (Chemicon), eIF5a, PXN, or TCF3 (Santa Cruz Biotechnology). Following secondary antibody incubations, signals were visualized by enhanced chemiluminescence.
Plasmid construction and protein purification.
Constructs to express TIAR RRM123 (residues 1 to 283) and TIAR RRM12 (residues 1 to 208) (10) were transformed into Escherichia coli strain BL21(DE3), and the encoded proteins were expressed and purified as described previously (10). HuR RRM12 (residues 18 to 184) was cloned into pGEX-4T1, expressed in E. coli BL21(DE3), and purified according to previously established protocols (43). The proteins were further purified by size-exclusion and cation-exchange chromatography. The concentration of each protein was determined using the Bradford assay (Bio-Rad) and by A280 measurements using theoretical molar extinction coefficients (ProtParam). The extinction coefficients were validated for folded protein; A280 measurements were within 10% of measurements made in 6.0 M guanidium hydrochloride. The purity of each protein was confirmed by SDS-PAGE.
Biosensor analysis.
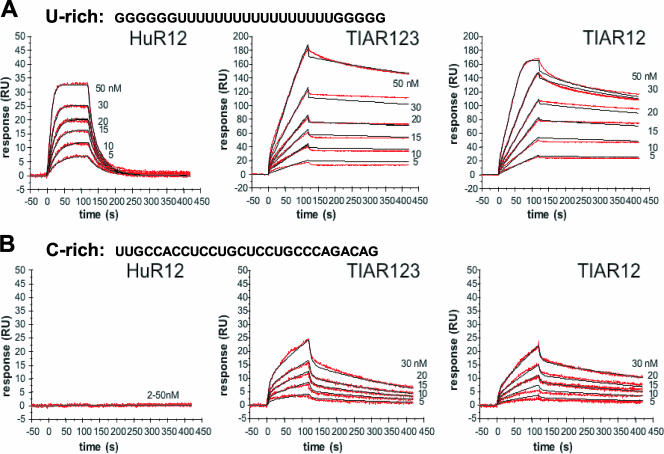
The dynamics of RNA-protein interactions were characterized by SPR using a BIACORE T100 instrument (Biacore Inc.). A U-rich RNA [containing a poly(U) stretch; 5′-GGGGGGUUUUUUUUUUUUUUUUUGGGGG-3′] and a C-rich RNA (5′-UUGCCACCUCCUGCUCCUGCCCAGACAG-3′) were chemically synthesized carrying a 5′-biotin tag (Dharmacon Research) to allow immobilization of the RNA onto streptavidin-coated sensor chips (series S sensor chip SA; Biacore, Inc.). RNAs were diluted to a final concentration of 1 μM in HBS buffer (10 mM HEPES [pH 7.4], 150 mM NaCl), followed by heating at 80°C for 10 min and cooling to room temperature. The sample was then diluted 500-fold in running buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM dithiothreitol, 0.025% surfactant P20 [Biacore, Inc.]) and injected over the sensor chip surface at 10 μl/min at 25°C to generate a 50-response unit (RU) RNA surface (for a low-density surface). Proteins were serially diluted in running buffer to the concentrations indicated in Fig. 3 and injected at 25°C at a flow rate of 50 μl/min for 2 min. Surface regeneration to remove any protein that remained bound after 3 min of dissociation was achieved using a 1-min injection of 2 M NaCl at 50 μl/min. Analysis of each protein concentration was done in duplicate, and samples were run in random order. Any background signal from a streptavidin-only reference flow cell was subtracted from every data set. Data were analyzed using a simple 1:1 Langmuir interaction model using the Biacore T100 evaluation software (Biacore, Inc.) to determine the affinities of the protein-RNA interactions.
FIG. 3.
Kinetic analysis of the interactions of TIAR12, TIAR123, and HuR12 proteins with U-rich and C-rich RNAs. The binding of TIAR123, TIAR12, and HuR12 to a U-rich or a C-rich RNA (28-mer each) is shown. Biotinylated RNA was captured on SA-coated sensor chips, and increasing concentrations of protein were injected over the surface. Injections were performed for 120 s (association phase), followed by a 300-s flow of running buffer to assess dissociation. The experiments were conducted in duplicate and showed good overlap. The red lines represent the binding responses for injections of protein analyte at specified concentrations (nM) over the RNA surface. The kinetic data were fit by a 1:1 Langmuir binding model which describes monovalent analyte binding to a single site on the immobilized ligand. Mass transport effects were not evident. The black curves superimposed on top of the sensorgrams represent the model fitted curves. The association and dissociation rate constants (Ka and Kd, respectively) were determined simultaneously as global fitting parameters from which KD was determined. The resulting parameter values are given in Table 2.
RESULTS
Sequence and structure of the predicted TIAR motif.
A collection of mRNAs that were TIAR targets was identified using human colorectal carcinoma RKO cells following IP under conditions that preserved the pools of mRNAs bound to TIAR (32). The RNA in the IP materials (associated with TIAR or bound in a nonspecific fashion in the IgG IP samples) was then extracted and reverse transcribed, and the resulting products were hybridized to human cDNA arrays (http://www.grc.nia.nih.gov/branches/rrb/dna/dna.htm#, MGC arrays). Three hundred array spots (∼3.1% of the total spots on the array) had Z ratios of >1.00 when comparing the signals in TIAR IP arrays with those in IgG IP arrays and were thus deemed to represent specific TIAR-associated transcripts. Among them, the 179 transcripts for which full-length mRNAs were available (the experimental data set) were selected for further analysis. A subset of transcripts from the experimental data set are listed in Table 1. The complete experimental data set is provided in Table S1 in the supplemental material, and numerous target transcripts were validated elsewhere (32).
TABLE 1.
Top TIAR target mRNAs identified on MGC arrays after RNP IPa
| Name | Symbol | RefSeq accession no. | Unigene no. | Z ratio |
|---|---|---|---|---|
| Nucleolar protein family A3 (H/ACA small nucleolar RNPs) | NOLA3 | NM_018648 | Hs#S2294033 | 3.38 |
| ADP-ribosylation factor 3 | ARF3 | NM_001659 | Hs#S1726280 | 3.13 |
| Metallothionein 1H | MT1H | NM_005951 | Hs#S3219010 | 3.06 |
| Small nuclear ribonucleoprotein D3 polypeptide 18 kDa | SNRPD3 | NM_004175 | Hs#S1729293 | 2.90 |
| Metallothionein 1L | MT1L | NM_002450 | Hs#S1727187 | 2.88 |
| Phospholipid scramblase 3 | PLSCR3 | NM_020360 | Hs#S2294612 | 2.66 |
| Serine hydroxymethyltransferase 2 (mitochondrial) | SHMT2 | NM_005412 | Hs#S1730829 | 2.61 |
| Cystatin B (stefin B) | CSTB | NM_000100 | Hs#S1730514 | 2.52 |
| F11 receptor | JAM1 | NM_144503 | Hs#S4554561 | 2.42 |
| Laminin, alpha 5 | LAMA5 | NM_005560 | Hs#S3619103 | 2.40 |
| Adenylosuccinate lyase | ADSL | NM_000026 | Hs#S1728269 | 2.26 |
| H3 histone, family 3B (H3.3B) | H3F3B | NM_005324 | Hs#S1730616 | 2.26 |
| 3-Hydroxybutyrate dehydrogenase (heart, mitochondrial) | BDH | NM_004051 | Hs#S4001852 | 2.24 |
| SWI/SNF related, matrix associated, regulator of chromatin | SMARCE1 | NM_003079 | Hs#S1730834 | 2.23 |
| Long-chain fatty-acyl elongase | LCE | NM_024090 | Hs#S3355556 | 2.16 |
| H2A histone family, member Y2 | H2AFY2 | NM_018649 | Hs#S2294022 | 2.16 |
| Non-SMC (structural maintenance of chromosomes) 1 | NSE1 | NM_145080 | Hs#S4554546 | 2.14 |
| CHK1 checkpoint homolog (Schizosaccharomyces pombe) | CHEK1 | NM_001274 | Hs#S1726468 | 2.13 |
| Transforming growth factor beta 1-induced transcript 1 | TGFB1I1 | NM_015927 | Hs#S2140275 | 2.13 |
| MAD1 mitotic arrest-deficient-like 1 (yeast) | MAD1L1 | NM_003550 | Hs#S1731867 | 2.12 |
| Amplified in osteosarcoma | OS-9 | NM_006812 | Hs#S1731315 | 2.12 |
| Fibrinogen-like 1 | FGL1 | NM_004467 | Hs#S1732393 | 2.11 |
| Cytochrome c oxidase subunit IV isoform 1 | COX4I1 | NM_001861 | Hs#S1730504 | 2.10 |
| Mevalonate (diphospho) decarboxylase | MVD | NM_002461 | Hs#S1731983 | 2.10 |
| Ubiquitin-conjugating enzyme E2C | UBE2C | NM_007019 | Hs#S1731533 | 2.06 |
| Wiskott-Aldrich syndrome protein-interacting protein | WASPIP | NM_003387 | Hs#S1728188 | 2.02 |
| Dihydropyrimidine dehydrogenase | DPYD | NM_000110 | Hs#S1728556 | 1.98 |
| Small nuclear ribonucleoprotein polypeptide F | SNRPF | NM_003095 | Hs#S1727894 | 1.96 |
| Ribosomal protein L5 | RPL5 | NM_000969 | Hs#S1727704 | 1.94 |
| Malic enzyme 2, NAD+-dependent, mitochondrial | ME2 | NM_002396 | Hs#S1727151 | 1.92 |
| G protein-coupled receptor 56 | GPR56 | NM_005682 | Hs#S1729809 | 1.91 |
| COX10 homolog, cytochrome c oxidase assembly protein | COX10 | NM_001303 | Hs#S1732347 | 1.91 |
Whole-cell lysates prepared from untreated RKO cells were used for IP assays by employing either IgG or anti-TIAR antibodies. RNA was subsequently extracted from the RNP complexes present in the IP material and was reverse transcribed; the resulting radiolabeled molecules were used to hybridize a cDNA array (32). The most enriched transcripts found in association with TIAR (TIAR IP material compared with IgG IP material) are listed. The top 179 enriched transcripts with complete 3′UTR sequences, the experimental dataset (see Table S1 in the supplemental material), were used to derive the TIAR motif. The Z ratio column reflects the differences in signal intensity when comparing TIAR IP with IgG IP array signals (32). Transcripts were deemed TIAR targets if Z ratios are >1.
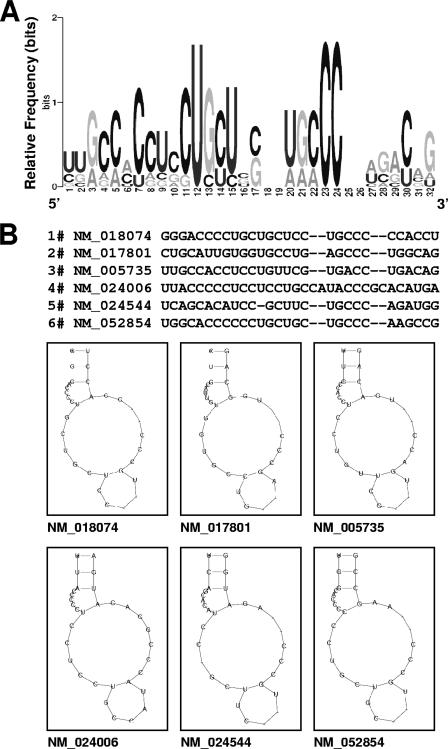
The RNA sequences of the experimental data set were subjected to computational analysis (Materials and Methods) to identify TIAR motifs, based on both primary RNA sequences and secondary structures. Of the 100 possible candidate motifs initially derived from the experimental data set, one motif comprising 28 to 32 nucleotides had the highest relative number of hits in the experimental data set compared with the entire UniGene database (as described previously in [28] and in the supplemental material). The sequence alignment, motif logo (graphic representation of the relative frequency of nucleotides at each position), and examples of the secondary structures of this putative TIAR are shown in Fig. 1A and B. The motif was found to be predominantly C rich (46%); U, G, and A nucleotides were significantly less abundant (21%, 18%, and 14%, respectively [Fig. 1A]). The sequence of this motif was unexpected, given our previous finding using SELEX analysis that suggested that TIAR bound to U-rich sequences, although these were often flanked by C residues (10). Figure 1B depicts six examples of the C-rich TIAR motif, with the corresponding mRNAs indicated below.
FIG. 1.
Sequence and structure of the predicted TIAR motif, as identified among TIAR-bound transcripts. (A) Probability matrix (graphic logo) of the TIAR motif indicating the relative frequency of finding each residue at each position within the motif, as elucidated from the array-derived experimental data set. (B) Secondary structure of six representative examples of the TIAR motif in specific mRNAs; the corresponding RefSeq accession numbers names are shown.
Both HuR and TIAR proteins bind U-rich RNA.
The unanticipated identification of a C-rich RNA motif for TIAR target transcripts prompted us to investigate the relative binding affinity of TIAR to a U-rich [a poly(U) stretch comprising 17 U residues flanked by G residues] RNA compared with this C-rich RNA in vitro. We prepared recombinant TIAR proteins comprising two or three of its N-terminal RRM domains (TIAR12 and TIAR123, respectively), as previously described (10). We also prepared a recombinant protein comprising the N-terminal two RRMs of HuR (HuR12) to serve as a positive control for binding to the U-rich RNA and negative control for binding to the C-rich RNA (Fig. 2).
FIG. 2.
Recombinant proteins used in in vitro binding assays. The construction of plasmids to express recombinant proteins comprising RRM1 and RRM2 or all three RRMs (HuR12, TIAR123, and TIAR12) was previously described (10). Proteins were expressed in bacteria and purified to homogeneity (details in Materials and Methods). Std., protein standard, with molecular mass (kDa) indicated.
We investigated the ability of these proteins to bind to the U-rich 28-mer RNA using SPR. This methodology allowed not only a qualitative indication of binding but also a comparative measure of on rates, off rates, and overall affinities to the RNA tethered to a chip via its 5′ end. The sensorgrams in Fig. 3 (top) show the binding of a range of concentrations of HuR12, TIAR123, and TIAR12 when injected across the U-rich RNA-coated chip. The on rates, off rates and overall affinities (equilibrium dissociation constant [KD]) for each protein, as approximated using a simple Langmuir binding model, are listed in Table 2. As shown, all three proteins bound the U-rich RNA. HuR12 bound with a KD in the nM range, with both high on and off rates. This finding is in keeping with previous SPR studies of HuD proteins binding to AU-rich sequences, wherein HuD12 bound with a KD of 5.4 nM to a 38-mer and a KD of 15.3 nM to a 13-mer (35, 36).
TABLE 2.
Kinetic and affinity constants for the interactions of TIAR123, TIAR12, and HuR12 proteins with U-rich and C-rich RNAsa
| Protein | RNA | Ka (1/M−1 s−1) | Kd (1/s) | KD (Kd/Ka [nM]) |
|---|---|---|---|---|
| HuR12 | U rich | (1.29 ± 0.01) × 108 | 4.35 ± 0.03 | 33.73 ± 0.50 |
| TIAR123 | U rich | (1.58 ± 0.02) × 106 | (1.56 ± 0.01) × 10−3 | 0.99 ± 0.02 |
| C rich | (2.72 ± 0.03) × 104 | (36.78 ± 0.12) × 10−4 | 135.2 ± 1.93 | |
| TIAR12 | U rich | (4.10 ± 0.11) × 106 | (2.83 ± 0.08) × 10−3 | 0.69 ± 0.04 |
| C rich | (3.48 ± 0.04) × 104 | (19.64 ± 0.06) × 10−4 | 56.39 ± 0.79 |
The association and dissociation rate constants (Ka and Kd) were determined as global fitting parameters for a 1:1 binding model. The equilibrium dissociation constant, KD, was determined as Kd/Ka.
The TIAR proteins also bound to the U-rich RNA with a KD in the nM range (TIAR123 KD, ∼1 nM; TIAR12 KD, ∼0.7 nM). These proteins showed slower on and off rates compared with HuR12, reflecting an intrinsically different and potentially more complex process of association and dissociation with the RNA. These findings are also reflected in the imperfect fit of the data by a simple 1:1 Langmuir interaction model and suggest that the KD values may only be accurate to within an order of magnitude. These affinities are in keeping with those measured previously using nitrocellulose filter-binding assays, which showed binding affinities for poly(U) of 8 nM, 20 nM, and 40 nM by TIAR, TIAR123, and TIAR12, respectively (10). The similar binding properties of TIAR12 compared with TIAR123 also suggest, as previously noted (10), that the primary poly(U) binding contact is made by the first two RRMs of TIAR.
TIAR, but not HuR, binds the C-rich RNA motif.
To test if TIAR was able to bind to the newly identified C-rich RNA motif, an RNA sequence was designed by choosing the most frequent nucleotide present at each position within the probability matrix (Fig. 1A). SPR was used to examine the binding kinetics of recombinant HuR and TIAR proteins to this sequence in comparison with the binding seen for the U-rich sequence. The sensorgrams in Fig. 3 (bottom) show the binding of HuR12, TIAR123, and TIAR12 when injected across the C-rich RNA-coated chip. There was no evidence of an interaction between HuR12 and the C-rich RNA, consistent with the known specificity of HuR for U- and AU-rich sequences and demonstrating that the experimental conditions of SPR employed here did not allow the occurrence of nonspecific interactions. In contrast, TIAR proteins were able to bind the C-rich RNA, confirming that this is a bona fide, novel TIAR target sequence. On rates, off rates, and overall affinities (KD) are indicated in Table 2. TIAR123 and TIAR12 bound the C-rich RNA with nM affinities (∼135 and ∼56 nM, respectively). These values represent significant binding affinities, although they are approximately 100-fold weaker than TIAR protein binding to the U-rich RNA.
Functional analysis of the C-rich motif using heterologous reporter assays.
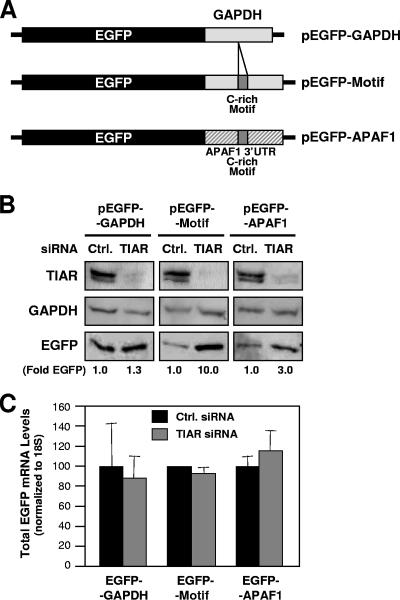
The functional role of the C-rich motif present in a given transcript was first tested by using reporter vectors in cells that expressed different levels of TIAR. Depicted in Fig. 4A are three plasmids that were engineered to express the EGFP reporter from chimeric mRNAs that also contained either the GAPDH 3′UTR (pEGFP-GAPDH), the GAPDH 3′UTR with one embedded copy of the consensus C-rich motif (pEGFP-Motif), or the APAF1 3′UTR (one of the TIAR target transcripts), including one hit of the C-rich motif (pEGFP-APAF1). HeLa cells were transfected with siRNAs that effectively silenced the endogenous TIAR (Fig. 4B) and were subsequently transfected with the EGFP reporter constructs. As shown in Fig. 4B, silencing of TIAR potently elevated (∼10-fold) EGFP expression from the EGFP reporter bearing the C-rich motif (pEGFP-Motif transfection group) and also induced EGFP expression (approximately threefold) from the EGFP reporter bearing the APAF1 3′UTR and its C-rich motif hit (pEGFP-APAF1 transfection group), but it did not significantly increase EGFP levels in cells transfected with pEGFP-GAPDH. These changes in EGFP expression did not arise from changes in the levels of the corresponding transcripts, as EGFP mRNA was essentially unchanged in the various transfection groups (Fig. 4C). These data strongly support the view that TIAR suppressed the translation of a transcript bearing the C-rich motif, thereby reducing the expression levels of the encoded protein.
FIG. 4.
Functional assessment of the C-rich motif using a heterologous reporter. (A) Plasmid pEGFP-GAPDH contains a 200-bp fragment of the GAPDH 3′UTR after the EGFP coding region; plasmid pEGFP-APAF1 contains nucleotide positions 5401 to 5825 of APAF1 (NM_013229), including one hit of the APAF1 3′UTR C-rich motif (positions 5599 to 5626); plasmid pEGFP-Motif contains a 28-bp insert (the consensus C-rich motif) within the GAPDH 3′UTR (Materials and Methods). (B) Western blot analysis of TIAR, loading control GAPDH, and reporter protein EGFP expression levels in RKO cells that had been transfected 48 h earlier with either control (Ctrl.) or TIAR-directed siRNAs. Signals (representative of five independent experiments) were quantified by densitometry and shown as EGFP levels (fold) in TIAR siRNA compared with control siRNA. (C) The levels of chimeric EGFP-GAPDH, EGFP-APAF1, and EGFP-Motif mRNAs were measured in each transfection group (B) by RT-qPCR analysis. Data are shown as the means and standard errors of the mean of three independent experiments.
Identification of novel TIAR target transcripts containing the C-rich TIAR motif.
We used the C-rich TIAR motif to query transcript databases for additional putative TIAR target mRNAs. A total of 2,209 transcripts were identified from the UniGene database (2.0% of the complete database); a subset of transcripts is presented in Table 3, along with the positions of the individual TIAR motif hits within the 3′UTR. Each motif hit was assigned a score (in parentheses), a value that reflects the degree to which each particular motif matches the TIAR motif model (Fig. 1A).
TABLE 3.
Putative TIAR target mRNAs bearing the C-rich signature motif in the 3′UTRa
| Position(s) in 3′UTRb | Name | Symbol | Unigene no. |
|---|---|---|---|
| 1445-1472 (0.58), 2180-2207 (3.38), 2630-2659 (1.56) | Sideroflexin 5 | SFXN5 | Hs#S4546027 |
| 1087-1115 (0.85), 1430-1457 (2.60), 1461-1488 (1.50), 1585-1612 (0.24) | Ras-related protein Rab-40C | RAB40C | Hs#S4044974 |
| 190-217 (3.95) | Ribosomal protein L34 | RPL34 | Hs#S3940107 |
| 250-277 (3.60) | Calcium-dependent protein kinase IG | CAMK1G | Hs#S3619658 |
| 206-235 (1.30) | Homolog of mouse LGP1 | LGP1 | Hs#S3619636 |
| 172-200 (1.14) | Myosin, heavy polypeptide 9, non-muscle | MYH9 | Hs#S3220052 |
| 1392-1419 (3.55) | Microtubule-associated proteins 1A/1B light-chain 3B | MAP1LC3B | Hs#S3219949 |
| 770-797 (3.12) | Matrix metalloproteinase 25 | MMP25 | Hs#S3219838 |
| 683-710 (1.53) | Junctophilin 3 | JPH3 | Hs#S3219743 |
| 135-164 (0.89), 556-583 (2.38) | Phospholysine phospho-His inorganic pyrophosphate phosphatase | LHPP | Hs#S3219695 |
| 62-89 (3.63) | Prostate and breast cancer overexpressed 1 | PBOV1 | Hs#S3219375 |
| 140-167 (1.16) | Potassium inwardly-rectifying channel, J11 | KCNJ11 | Hs#S3218881 |
| 828-853 (2.14) | Solute carrier family 2 (facilitated glucose transporter), member 6 | SLC2A6 | Hs#S2294542 |
| 284-308 (2.73) | Sphingosine kinase 2 | SPHK2 | Hs#S2294536 |
| 2014-2041 (3.10) | Ring finger protein 144 | RNF144 | Hs#S2139432 |
| 1472-1501 (0.04), 2649-2677 (0.59), 3189-3216 (2.93) | Protein phosphatase 1F | PPM1F | Hs#S2139385 |
| 1308-1335 (3.66) | Apoptotic protease-activating factor | APAF1 | Hs#S2138799 |
| 1839-1866 (1.31) | ATPase, (Na+)/K+-transporting, beta 4 polypeptide | ATP1B4 | Hs#S1824340 |
| 431-459 (3.68) | Poly(A) binding protein, nuclear 1 | PABPN1 | Hs#S1732404 |
| 318-345 (4.38) | Interleukin-24 | IL-24 | Hs#S1732294 |
| 194-222 (1.53) | Growth factor, augmenter of liver regeneration (ERV1 homolog) | GFER | Hs#S1732174 |
| 975-1002 (3.31) | Mitogen-activated protein kinase kinase kinase 14 | MAP3K14 | Hs#S1731892 |
| 287-314 (4.38) | Polyamine-modulated factor 1 | PMF1 | Hs#S1731724 |
| 1013-1040 (3.21) | Huntingtin-interacting protein 1-related | HIP1R | Hs#S15639035 |
| 67-96 (0.38), 276-303 (3.80), 347-372 (0.24), 511-538 (2.12), 745-771 (1.33) | Histamine receptor H3 | HRH3 | Hs#S1731688 |
| 13-40 (1.34) | Neuro-oncological ventral antigen 2 | NOVA2 | Hs#S1731549 |
| 109-136 (3.34), 227-254 (0.17), 956-983 (2.38), 2107-2134 (0.23) | SRY (sex-determining region Y)-box 12 | SOX12 | Hs#S1731519 |
| 1554-1583 (4.23), 2449-2476 (0.27) | Calcium-dependent protein kinase 2 | CAMKK2 | Hs#S1731273 |
| 1218-1246 (1.31), 1960-1985 (0.80) | NK3 transcription factor related, locus 1 (Drosophila) | NKX3-1 | Hs#S1730286 |
| 101-128 (1.05), 859-886 (3.75) | Signal-regulatory protein beta 1 | SIRPB1 | Hs#S1730130 |
| 240-267 (1.12) | Brain-specific angiogenesis inhibitor 1 | BAI1 | Hs#S1729737 |
| 311-337 (0.99), 625-653 (0.78), 4921-4948 (0.95), 5654-5681 (2.12), 5818-5845 (1.25) | Methyl CpG binding protein 2 | MECP2 | Hs#S1729484 |
| 809-836 (3.48) | Vanin 1 | VNN1 | Hs#S1729359 |
| 51-78 (5.63), 660-689 (1.89), 785-812 (0.14), 841-868 (0.21), 3296-3326 (0.02), 3504-3531 (0.06) | Synaptogyrin 1 | SYNGR1 | Hs#S1729312 |
| 1501-1528 (2.31) | Eukaryotic translation termination factor 1 | ETF1 | Hs#S1729236 |
| 155-182 (3.63) | Natural killer cell transcript 4 | NK4 | Hs#S1729141 |
| 357-384 (0.21), 773-801 (1.95), 1094-1125 (0.04) | Paired box gene 2, transcript variant c | PAX2 | Hs#S1728525 |
| 330-358 (0.76), 2112-2139 (0.62), 2211-2240 (2.46) | Major histocompatibility complex class II transactivator | MHC2TA | Hs#S1728490 |
| 55-82 (2.12) | Alpha-l-iduronidase precursor | IDUA | Hs#S1728450 |
| 52-82 (3.79) | Tu translation elongation factor | TUFM | Hs#S1728121 |
| 663-690 (2.37) | Transforming growth factor, beta receptor III | TGFBR3 | Hs#S1728020 |
| 290-317 (1.16) | Syntrophin, alpha 1 | SNTA1 | Hs#S1727897 |
| 962-988 (1.34) | Regulator of G-protein signaling 16 | RGS16 | Hs#S1727647 |
| 16-43 (3.16) | RAB3B, member RAS oncogene family | RAB3B | Hs#S1727591 |
| 525-552 (0.73), 999-1027 (0.27), 1322-1351 (1.05) | Paxillin | PXN | Hs#S1727580 |
| 1734-1761 (1.44), 2017-2042 (1.56) | Oligophrenin 1 | OPHN1 | Hs#S1727255 |
| 464-491 (1.12) | Eukaryotic translation initiation factor 5 | EIF5A | Hs#S1726700 |
| 782-810 (1.95) | C-terminal PDZ domain ligand of neuronal NOS | CAPON | Hs#S15898574 |
| 32-59 (3.12) | Transcription factor 3 | TCF3 | Hs#S11062708 |
Shown is a partial list of genes bearing the C-rich signature motif in the 3′UTR of the corresponding transcripts. (The complete list of predicted target transcripts is available from the authors.)
The relative positions of the C-rich TIAR motif within each transcript and the corresponding scores (in parentheses) are indicated.
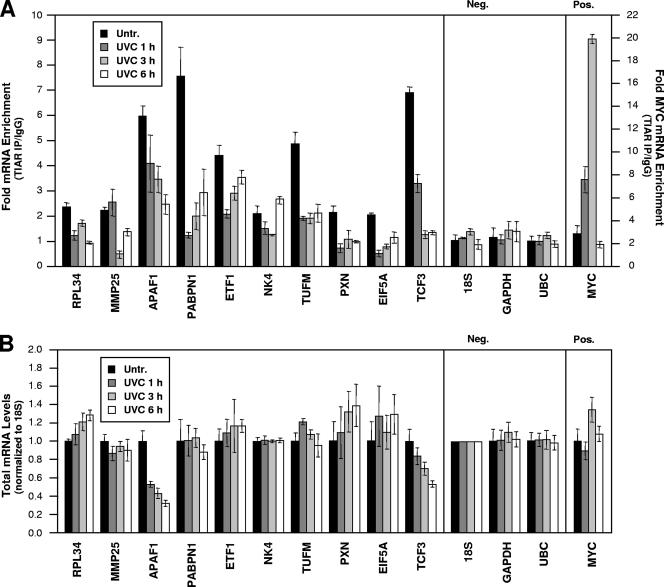
To test whether the C-rich motif could be used to identify TIAR target mRNAs computationally, we monitored the presence of putative target mRNAs in immunoprecipitated TIAR-RNP complexes. Using gene-specific primers, we evaluated the abundance of 12 predicted and randomly selected TIAR target mRNAs by RT followed by qPCR. Indeed, all 12 transcripts (10 shown in Fig. 5A [CAMKK2 and HIP1R not shown]) appeared to be bona fide targets of TIAR, as they were found to be enriched in TIAR IP material compared with IgG IP material (time zero); the degrees of enrichment ranged from just over twofold for RPL34 and MMP25 mRNAs to greater than sevenfold for PABPN1 mRNA. With the same experimental approach, we further sought to determine whether the abundance of such putative RNP complexes would be affected by treatment with short-wavelength UV light (UVC), a genotoxic agent that was previously found to increase binding of TIAR to a subset of mRNAs (32). To our surprise, however, treatment with 25 J/m2 UVC significantly reduced the association of these mRNAs with TIAR, as their abundance in the TIAR IP material was markedly diminished following exposure to UVC (with few exceptions, such as NK4 at 6 h after UVC). Importantly, negative controls 18S rRNA and housekeeping GAPDH and UBC mRNAs showed negligible enrichment in TIAR IPs relative to IgG IPs; these non-TIAR target RNAs bound the beads and IP reagents in a nonspecific fashion and served to monitor the even input of the samples (Fig. 5A, Neg.). In addition, the binding of MYC mRNA was tested as a positive control, since TIAR-MYC mRNA complexes were previously shown to increase following UVC irradiation (32); in keeping with these earlier findings, MYC mRNA showed strikingly increased binding to TIAR at 1 and 3 h after UVC treatment (Fig. 5A, Pos.).
FIG. 5.
Analysis of the binding and whole-cell levels of predicted TIAR target mRNAs in untreated and UVC-irradiated cells. (A) The association of endogenous TIAR with endogenous putative target mRNAs was tested using lysates prepared from RKO cells that were either left without treatment (Untr. [0 h]) or were irradiated with 25 J/m2 UVC and collected 1, 3, or 6 h afterwards. Anti-TIAR or IgG antibodies were used in IP reactions followed by the analysis of predicted target transcripts by RT-qPCR analysis of the IP material. Neg., negative control transcripts 18S rRNA and housekeeping GAPDH and UBC mRNAs; Pos., positive control MYC mRNA, a known TIAR target (32). Numbers on the right y axis indicate MYC mRNA enrichment (TIAR/IgG). (B) Total RNA was extracted from cells that were processed as described for panel A. The whole-cell levels of each mRNA were calculated and normalized to 18S rRNA levels. Data show means and standard deviations (A and B).
The reductions in RNPs comprising TIAR and mRNAs bearing the C-rich motif did not simply reflect changes in the total levels of these cellular mRNAs for two reasons. First, by the RNP IP assay, global reductions in mRNA are reflected in binding both in the IgG IP and the TIAR IP, so by measuring mRNA enrichment, such differences would already be accounted for. Second, individual testing of each of the transcripts in whole-cell RNA preparations showed that UVC elicited modest or no changes in the levels of most mRNAs, as shown in Fig. 5B; only whole-cell APAF1 and TCF3 mRNA levels were reduced to about one-third and one-half of their original abundance, respectively.
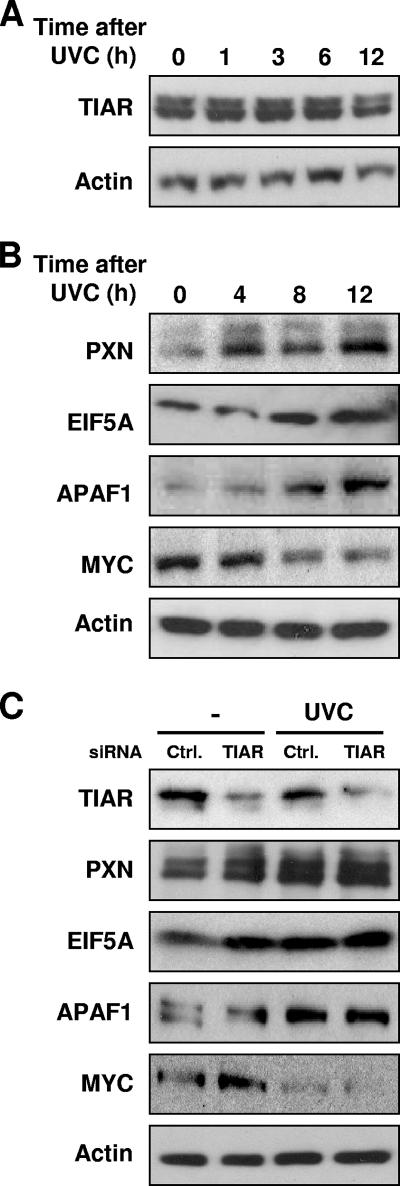
Finally, we tested the expression levels of proteins encoded by four TIAR target mRNAs. Three of the proteins encoded by dissociation target mRNAs (PXN, EIF5A, and APAF1) were readily detectable (TCF3 could not be detected with the antibodies available). While the levels of TIAR itself remained unchanged following UVC irradiation (Fig. 6A), the levels of PXN, EIF5A, and APAF1 were markedly elevated following UVC irradiation, in agreement with the view that TIAR repressed their expression (Fig. 6B). Of note, the steady-state levels of these mRNAs either remained unchanged (PXN and EIF5A) or were actually reduced (APAF1), as shown in Fig. 5B. As a positive control, MYC expression was tested, as UVC stress was previously shown to increase TIAR-MYC mRNA association and to decrease MYC translation and protein levels (Fig. 5A) (32); here, MYC protein abundance was also reduced potently following UVC irradiation (Fig. 6B). Evidence that TIAR contributed to these changes in protein expression was obtained through silencing experiments. As shown by Western blot analysis in Fig. 6C, PXN, EIF5A, and APAF1 abundance increased in RKO cells that expressed reduced TIAR levels (by TIAR siRNA transfection). Combined UVC irradiation and TIAR silencing did not further elevate the expression of these proteins, supporting the view that both interventions shared common mechanisms of action.
FIG. 6.
Expression levels of proteins encoded by predicted TIAR target mRNAs in untreated and UVC-irradiated RKO cells. (A) Western blot analysis of TIAR expression levels in whole-cell lysates (15 μg per lane) that were prepared at the times indicated after UVC irradiation. (B) Western blot analysis of the expression levels of PXN, EIF5A, and APAF1 (encoded by TIAR dissociation target mRNAs), positive control MYC, and loading control β-actin. Whole-cell lysates (10 μg per lane) were prepared at the times shown following UVC irradiation. Shown are representative Western blots from three independent experiments. (C) Western blot analysis of the expression of TIAR, PXN, EIF5A, APAF1, MYC, and β-actin in cells that were transfected with either control (Ctrl.) or TIAR siRNAs. Forty-eight hours later, cells were left untreated or treated with UVC and collected after an additional 8 h. Data are representative of three independent experiments.
DISCUSSION
A C-rich motif is bound by TIAR RRMs in vitro and functions to suppress translation in the presence of TIAR in vivo.
We report the identification of a C-rich signature motif present in TIAR target mRNAs. The C-rich motif was unexpectedly distinct from the U-rich target sequence we previously reported for TIAR-bound mRNAs (10). In keeping with these earlier findings, TIAR proteins containing two or three RRM domains (TIAR12 and TIAR123, respectively) bound with high affinity to a U-rich sequence containing a stretch of 17 uracils; however, they also bound, albeit with 50- to 100-fold-less affinity, to the C-rich motif, as measured by SPR (Fig. 3). TIAR123 and TIAR12 bound to the C-rich sequence with KDs in the nanomolar range, representing a significant binding affinity of TIAR for the C-rich motif. These results demonstrate that TIAR binding to RNA may not be restricted to a single class of target motif. Besides serving as a target sequence for TIAR, the C-rich motif had a significant functional role, as revealed by using a chimeric reporter construct that comprised the EGFP coding region linked to the GAPDH 3′UTR. Insertion of the consensus C-rich motif in the GAPDH 3′UTR strongly reduced the expression of the heterologous reporter EGFP (Fig. 4). The C-rich sequence (the consensus motif or the motif hit within the APAF1 3′UTR) specifically suppressed the translation of the reporter construct without affecting reporter mRNA levels; this regulation was TIAR dependent, since silencing of TIAR restored reporter protein production.
Endogenous TIAR forms complexes with endogenous mRNAs.
The collection of UniGene transcripts that were predicted to be TIAR targets based on the presence of at least one hit of the C-rich motif was then studied. (A subset of predicted target transcripts is listed in Table 2.) Following TIAR RNP IP analysis and RT-qPCR identification of individual mRNAs present in TIAR RNP complexes, all of the transcripts chosen randomly among this collection were found to be enriched in the TIAR RNP IP material relative to IgG IP; no predicted targets were found not to be enriched (Fig. 5). These observations reveal that in addition to the binding of recombinant purified TIAR to the C-rich motif in vitro, endogenous TIAR was also found in association with endogenous mRNAs bearing the C-rich motif. These results underscore the usefulness of the C-rich motif in predicting TIAR target mRNAs.
It was interesting to discover that UVC irradiation triggered a reduction in binding of TIAR to all of the mRNA targets bearing the C motif (Fig. 5A). This finding suggested that the C-rich signature motif identifies mRNAs which will dissociate from TIAR following UVC treatment and possibly other forms of cell damage. Along with the reduced association of TIAR with C-rich target transcripts (PXN, EIF5A, and APAF1), we documented a sizeable increase in the expression of the encoded proteins following UVC irradiation (Fig. 6). In support of the notion that TIAR contributed to this increase, silencing of TIAR alone similarly elevated the expression levels of these proteins, an effect that recapitulated the regulation of the chimeric reporters (Fig. 4). At this time, the magnitude or kinetics of TIAR-mRNA association/dissociation do not appear to correlate with either score values or the number of motif hits on a given target transcript (not shown), although further analysis of these parameters is warranted. As these studies move forward, it will also be interesting to examine whether the coordinate regulation of subsets of C-rich motif-bearing mRNAs elicits a particular phenotype. This task will be complex, since >2,000 transcripts bearing the C-rich motif have been identified (Table 3 [complete list available from the authors]) and the encoded proteins participate in a broad range of cellular functions. We will first focus our attention on C-rich motif-bearing targets implicated in the stress response, particularly those that influence cell survival and proliferation.
The reduced binding of TIAR to these target transcripts was in contrast to the results obtained earlier when testing other TIAR target transcripts, particularly those encoding translation-modulatory proteins such as translation regulatory factors EIF4A1, EIF4E2, and EEF1B2, as well as MYC (which transcriptionally upregulates the expression of translation regulatory proteins) (32). In that investigation, binding of TIAR to target mRNAs increased following UVC irradiation, in turn causing a reduction in the expression of those translation regulators and contributing to an overall reduction in protein biosynthesis following UVC treatment. None of the TIAR-bound mRNAs encoding translational regulators was found to have the C-rich motif, in agreement with the idea that the C-rich motif is present in a different subset of mRNAs. Based on these observations, we postulate the existence of a different signature motif on TIAR target mRNAs that would instead signal increased association after cellular stress. Our efforts to identify such a signature motif have led to the preliminary identification of a shared sequence containing a stretch of U residues (see Fig. S1 in the supplemental material) flanked by C residues, similar to what was found by SELEX analysis (10). Studies are under way to test if this U-rich motif is a bona fide recognition sequence for TIAR. Should the U-rich motif be validated by testing recombinant and endogenous target transcripts, then further investigation will assess whether binding of TIAR to mRNAs bearing the U-rich motif increases following exposure to stimuli such as UVC and will examine its influence upon the expression of mRNAs in which it is present.
Multiple RNA target motifs for a given RBP?
If indeed the C-rich RNA sequence represents a “TIAR dissociation motif” which signals decreased mRNA association with TIAR after stress, it is conceivable that multiple motifs exist which can direct either increased or decreased TIAR binding to mRNAs after cellular damage. Moreover, signature motifs may exist to guide binding of TIAR (or for that matter, also other RBPs), depending on cellular energy levels, tissue type, proliferation status, subcellular locale, etc. A comprehensive elucidation of the group of TIAR motifs and their functional characteristics will require the development of more sophisticated analysis methods. The TIAR binding properties studied here are also likely to be influenced by UVC-triggered changes, including alterations in its subcellular localization (e.g., stress granules) (2); posttranslational modifications (e.g., by phosphorylation) (18); interactions with chaperones such as heat shock protein 70 (HSP70) (15); or interactions with other proteins that alter its RNA-binding properties, as recently reported for SRC-3, a nuclear transcriptional activator that also functions as a cytoplasmic activator of TIA-1/TIAR (3, 45). Whether UVC affects any of these potential modulators of TIAR activity remains to be addressed experimentally. Other regulatory schemes that merit consideration also await further testing. For example, microRNAs or other RBPs could preferentially facilitate or hinder the binding of TIAR to a target mRNA in cells exposed to stress agents; notably among these, RBPs with preference for C-rich RNA motifs (such as hnRNP K and PCBP1 to -4/αCP1 to -4 [reviewed in references 13 and 30]) might be anticipated to compete for binding, particularly since they have been implicated in regulating the translation of target transcripts and their function is modulated by stress (reviewed in reference 30).
Individual examples of RBPs dissociating from a target mRNA in response to a stimulus have been reported: for instance, TIAR was recently shown to dissociate from GADD45α mRNA following exposure to the alkylating agent methylmethane sulfonate (25). To our knowledge, however, this is the first study to identify an RNA motif that defines a subset of mRNAs which dissociate from an RBP in response to a stimulus. Broadly speaking, our findings illustrate the existence of an additional layer of posttranscriptional gene regulation for a given mRNA, in agreement with the “RNA regulon” model, whereby collections of mRNAs are coordinately regulated at the posttranscriptional level by specific RBPs (recently reviewed in reference 22). In addition to the interplay between trans factors (RBPs and microRNAs) acting upon the transcript, the influence of the subcellular environment, and the conditions of cellular growth at a given time, the specific mRNA sequence can also dictate the dynamic association of RBPs and hence the composition of the RNP and its posttranscriptional fate.
Supplementary Material
Acknowledgments
We thank K. G. Becker and the NIA Array Facility for providing cDNA arrays for analysis and E. Cummings for invaluable assistance with protein preparation.
This research was supported by the Intramural Research Program of the NIA-IRP, NIH (Y.K., M.Z., R.P., K.M.M., H.L., and M.G.); grant NIH AI33600 (P.A. and N.K.); an Australian Research Council Fellowship (J.A.W.); and a Monash University postgraduate scholarship (H.S.K.).
Footnotes
Published ahead of print on 6 August 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 115:3227-3234. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2002. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, P., and N. Kedersha. 2007. On again, off again: the SRC-3 transcriptional coactivator moonlights as a translational corepressor. Mol. Cell 25:796-797. [DOI] [PubMed] [Google Scholar]
- 4.Antic, D., and J. D. Keene. 1997. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am. J. Hum. Genet. 61:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antic, D., N. Lu, and J. D. Keene. 1999. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 13:449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell Mol. Life Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 8.Cok, S. J., S. J. Acton, and A. R. Morrison. 2003. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J. Biol. Chem. 278:36157-36162. [DOI] [PubMed] [Google Scholar]
- 9.Colegrove-Otero, L. J., A. Devaux, and N. Standart. 2005. The Xenopus ELAV protein ElrB represses Vg1 mRNA translation during oogenesis. Mol. Cell. Biol. 25:9028-9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dember, L. M., N. D. Kim, K. Q. Liu, and P. Anderson. 1996. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 271:2783-2788. [DOI] [PubMed] [Google Scholar]
- 11.Derrigo, M., A. Cestelli, G. Savettieri, and I. Di Liegro. 2000. RNA-protein interactions in the control of stability and localization of messenger RNA. Int. J. Mol. Med. 5:111-123. [PubMed] [Google Scholar]
- 12.Forch, P., and J. Valcarcel. 2001. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis 6:463-468. [DOI] [PubMed] [Google Scholar]
- 13.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gueydan, C., L. Droogmans, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem. 274:2322-2326. [DOI] [PubMed] [Google Scholar]
- 15.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holcik, M., and N. Sonenberg. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6:318-327. [DOI] [PubMed] [Google Scholar]
- 17.Hollams, E. M., K. M. Giles, A. M. Thomson, and P. J. Leedman. 2002. mRNA stability and the control of gene expression: implications for human disease. Neurochem. Res. 27:957-980. [DOI] [PubMed] [Google Scholar]
- 18.Izquierdo, J. M., and J. Valcarcel. 2007. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J. Biol. Chem. 282:1539-1543. [DOI] [PubMed] [Google Scholar]
- 19.Kandasamy, K., K. Joseph, K. Subramaniam, J. R. Raymond, and B. G. Tholanikunnel. 2005. Translational control of beta2-adrenergic receptor mRNA by T-cell-restricted intracellular antigen-related protein. J. Biol. Chem. 280:1931-1943. [DOI] [PubMed] [Google Scholar]
- 20.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 21.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fitzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keene, J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8:533-543. [DOI] [PubMed] [Google Scholar]
- 23.Kullmann, M., U. Gopfert, B. Siewe, and L. Hengst. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 16:3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal, A., T. Kawai, X. Yang, K. Mazan-Mamczarz, and M. Gorospe. 2005. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 24:1852-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lal, A., K. Abdelmohsen, R. Pullmann, T. Kawai, S. Galban, X. Yang, G. Brewer, and M. Gorospe. 2006. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol. Cell 22:117-128. [DOI] [PubMed] [Google Scholar]
- 26.Le Guiner, C., F. Lejeune, D. Galiana, L. Kister, R. Breathnach, J. Stevenin, and F. Del Gatto-Konczak. 2001. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 276:40638-40646. [DOI] [PubMed] [Google Scholar]
- 27.Loflin, P., C. Y. Chen, and A.-B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López de Silanes, I., S. Galbán, J. L. Martindale, X. Yang, K. Mazan-Mamczarz, F. E. Indig, G. Falco, M. Zhan, and M. Gorospe. 2005. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 25:9520-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López de Silanes, I., M. Zhan, A. Lal, X. Yang, and M. Gorospe. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 101:2987-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makeyev, A. V., and S. A. Liebhaber. 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazan-Mamczarz, K., S. Galban, I. López de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 100:8354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazan-Mamczarz, K., A. Lal, J. L. Martindale, T. Kawai, and M. Gorospe. 2006. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 26:2716-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng, Z., P. H. King, L. B. Nabors, N. L. Jackson, C. Y. Chen, P. D. Emanuel, and S. W. Blume. 2005. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 33:2962-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min, H., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023-1036. [DOI] [PubMed] [Google Scholar]
- 35.Park, S., D. G. Myszka, M. Yu, S. J. Littler, and I. A. Laird-Offringa. 2000. HuD RNA recognition motifs play distinct roles in the formation of a stable complex with AU-rich RNA. Mol. Cell. Biol. 20:4765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park-Lee, S., S. Kim, and I. A. Laird-Offringa. 2003. Characterization of the interaction between neuronal RNA-binding protein HuD and AU-rich RNA. J. Biol. Chem. 278:39801-39808. [DOI] [PubMed] [Google Scholar]
- 37.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shukla, S., W. P. Dirksen, K. M. Joyce, C. Le Guiner-Blanvillain, R. Breathnach, and S. A. Fisher. 2004. TIA proteins are necessary but not sufficient for the tissue-specific splicing of the myosin phosphatase targeting subunit 1. J. Biol. Chem. 279:13668-13676. [DOI] [PubMed] [Google Scholar]
- 39.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenenbaum, S. A., P. J. Lager, C. C. Carson, and J. D. Keene. 2002. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods 26:191-198. [DOI] [PubMed] [Google Scholar]
- 41.Vawter, M. P., T. Barrett, C. Cheadle, B. P. Sokolov, W. H. Wood III, D. M. Donovan, M. Webster, W. J. Freed, and K. G. Becker. 2001. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res. Bull. 55:641-650. [DOI] [PubMed] [Google Scholar]
- 42.Wilkie, G. S., K. S. Dickson, and N. K. Gray. 2003. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 28:182-188. [DOI] [PubMed] [Google Scholar]
- 43.Yeap, B. B., D. C. Voon, J. P. Vivian, R. K. McCulloch, A. M. Thomson, K. M. Giles, M. F. Czyzyk-Krzeska, H. Furneaux, M. C. Wilce, J. A. Wilce, and P. J. Leedman. 2002. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J. Biol. Chem. 277:27183-27192. [DOI] [PubMed] [Google Scholar]
- 44.Yu, Q., S. J. Cok, C. Zeng, and A. R. Morrison. 2003. Translational repression of human matrix metalloproteinase-13 by an alternatively spliced form of T-cell-restricted intracellular antigen-related protein (TIAR). J. Biol. Chem. 278:1579-1584. [DOI] [PubMed] [Google Scholar]
- 45.Yu, C., B. York, S. Wang, Q. Feng, J. Xu, and B. W. O'Malley. 2007. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol. Cell 25:765-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, W., B. J. Wagner, K. Ehrenman, A. W. Schaefer, C. T. DeMaria, D. Crater, K. DeHaven, L. Long, and G. Brewer. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.