Abstract
Members of the RecQ helicase family play critical roles in genome maintenance. There are five RecQ homologs in mammals, and defects in three of these (BLM, WRN, and RECQL4) give rise to cancer predisposition syndromes in humans. RECQL and RECQL5 have not been associated with a human disease. Here we show that deletion of Recql5 in mice results in cancer susceptibility. Recql5-deficient cells exhibit elevated frequencies of spontaneous DNA double-strand breaks and homologous recombination (HR) as scored using a reporter that harbors a direct repeat, and are prone to gross chromosomal rearrangements in response to replication stress. To understand how RECQL5 regulates HR, we use purified proteins to demonstrate that human RECQL5 binds the Rad51 recombinase and inhibits Rad51-mediated D-loop formation. By biochemical means and electron microscopy, we show that RECQL5 displaces Rad51 from single-stranded DNA (ssDNA) in a reaction that requires ATP hydrolysis and RPA. Together, our results identify RECQL5 as an important tumor suppressor that may act by preventing inappropriate HR events via Rad51 presynaptic filament disruption.
[Keywords: Recql5 helicase, DNA repair, homologous recombination, tumor suppressor, Rad51 recombinase]
Homologous recombination (HR) is a fundamental molecular process in all organisms. In meiosis, HR is necessary for the proper segregation of homologous chromosomes and generates genetic diversity through the shuffling of parental alleles (Paques and Haber 1999; Symington 2002). In mitotic cells, HR is an important pathway for repairing chromosomal breaks and gaps, and for restarting damaged or stalled DNA replication forks (Symington 2002; Wu and Hickson 2006). However, inappropriate or untimely HR events can have mutagenic and oncogenic consequences. For example, reciprocal exchanges (crossovers) between homologous chromosomes can lead to somatic loss of heterozygosity (LOH), while crossovers between nonhomologous chromosomes can result in translocation. Also, crossovers between repeated sequences on the same chromosome can result in deletions or inversions. For these reasons, specific mechanisms have evolved for regulating HR to minimize these potentially deleterious rearrangements (Sung and Klein 2006; Wu and Hickson 2006).
Genetic analyses in Saccharomyces cerevisiae indicate that two DNA helicases, Srs2 and Sgs1, function in different pathways to suppress crossover events in mitotic cells (Ira et al. 2003). Mutations in either Srs2 or Sgs1 result in a hyperrecombination phenotype (Aguilera and Klein 1988; Onoda et al. 2000; Liberi et al. 2005). In humans, mutations in BLM, which encodes the human ortholog of Sgs1, give rise to the rare hereditary disorder Bloom’s syndrome (Ellis et al. 1995). This disorder is marked by an elevated rate of sister chromatid exchange (SCE), increased chromosomal instability, and a high incidence of cancer (German 1993; Hickson 2003). BLM suppresses SCEs by acting in conjunction with the Type 1A topoisomerase, Topo IIIα, and a recently identified protein, BLAP75, to mediate the dissolution of double Holliday junctions (DHJ; a late HR intermediate), a process that yields solely noncrossover recombinants (Wu and Hickson 2003; Raynard et al. 2006; Sung and Klein 2006; Wu et al. 2006). Like its human counterpart, Sgs1 forms a complex with the Top3 topoisomerase (Gangloff et al. 1994; Bennett et al. 2000) and Rmi1 (the BLAP75 ortholog) (Chang et al. 2005; Mullen et al. 2005), suggesting that it might function to suppress SCEs by a similar mechanism.
Srs2 is a superfamily 1 helicase with similarities to the bacterial UvrD/Rep helicases. The mechanistic basis of the Srs2 function was elucidated by biochemical studies that revealed its ability to bind Rad51 and to dismantle the Rad51-ssDNA (single-stranded DNA) nucleoprotein filament, the key catalytic intermediate in recombination reactions (Krejci et al. 2003; Veaute et al. 2003; Sung and Klein 2006). To date, no apparent Srs2 ortholog has been identified in other eukaryotes, although the recently identified Fbh1 helicase shows some structural similarity to the Srs2 helicase family and studies in Schizosaccharomyces pombe suggest that Fbh1 plays a role in processing HR intermediates (Osman et al. 2005; Morishita et al. 2005). However, Fbh1-deficient DT40 cells show no prominent sensitivity to DNA damaging agents, and exhibit only a mild SCE phenotype (Kohzaki et al. 2007). This raises the question as to whether attenuation of HR by disruption of the Rad51 presynaptic filament represents a significant mechanism for HR regulation in higher eukaryotes.
Sgs1 and BLM are members of the RecQ family of DNA helicases. Sgs1 is the sole RecQ helicase in budding yeast. Interestingly, humans have a total of five RecQ helicase encoding genes (RECQL, BLM, WRN, RECQL4, and RECQL5). They share a conserved seven-motif helicase domain but are otherwise distinct from one another by their unique amino acid composition outside the helicase domain, suggesting that they have related but different roles (Hickson 2003). In addition to BLM, defects in both WRN and RECQL4 are also associated with heritable genome instability and cancer disorders (Hickson 2003). Therefore, while BLM likely represents the Sgs1 ortholog, the other RECQ-like helicases represent potential candidates as the functional equivalent of Srs2 in humans. We recently reported that mouse cells deficient in the RECQL5 homolog Recql5 exhibit an elevated level of SCEs, thus implicating this helicase in the regulation of HR (Hu et al. 2005). Importantly, deletion of both Recql5 and Blm further increases the SCE frequency, consistent with Recql5 acting to regulate SCEs in mitotic cells via a mechanism that is distinct from Blm, perhaps by functioning similarly to Srs2 to suppress the channeling of DNA lesions into HR.
In this paper, we show that deletion of Recql5 in mice results in increased susceptibility to cancer. Recql5-deficient cells exhibit elevated frequencies of spontaneous double-strand breaks (DSBs) and HR between direct repeats, and are prone to the accumulation of gross chromosomal rearrangements (GCRs) in response to replication stress. Moreover, by biochemical means, we provide a mechanistic basis by which RECQL5 functions in suppressing GCRs and tumorigenesis. Specifically, we show that human RECQL5 binds the Rad51 recombinase, and a catalytic quantity of this helicase inhibits Rad51-mediated D-loop formation markedly. Furthermore, we show that RECQL5 displaces Rad51 from ssDNA in a reaction that requires ATP hydrolysis by RECQL5 and is stimulated by the ssDNA-binding protein RPA. Taken together, our data provide compelling evidence that this unique member of the RecQ helicase family functions to minimize the propensity of oncogenic rearrangements by suppressing the accumulation of DSBs and attenuating HR by disrupting the Rad51 presynaptic filament.
Results
Recql5-deficient mice are highly cancer prone
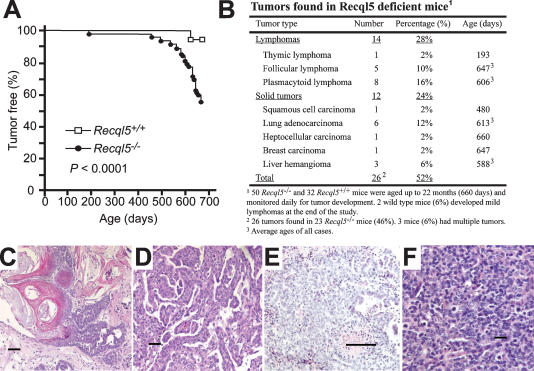
To examine whether Recql5 suppresses tumorigenesis, we generated large cohorts of Recql5 knockout mice and their wild-type littermates and monitored their phenotypes. No significant differences between the two groups were observed after 1 yr. However, as these animals aged further, a significantly higher percentage of Recql5 knockout mice developed cancer. Specifically, by 22 mo of age, 23 out of 50 (46%) of the knockout mice and only two out of 32 (6%) wild-type animals developed cancer (Fig. 1A–F). The only two cancer cases in the wild-type were both lymphomas. In mutant mice, approximately half of the cancer incidences were lymphomas, with late-onset follicular lymphoma, which is a common type of malignant lymphoma in humans, accounting for 36% (five out of 14) of the total lymphoma incidences. The rest of the cancers were solid tumors of different tissue origins. Intriguingly, lung adenocarcinomas predominated among them (six out 12, or 50%) (Fig. 1B,D). Three Recql5 knockout mice had two different types of primary cancers each (Fig. 1B). Interestingly, aside from the striking cancer susceptibility phenotype, Recql5 knockout mice were indistinguishable from their wild-type siblings. In particular, except for those animals that succumbed to cancer, Recql5 knockout mice had a lifespan that was not different from wild-type animals. Collectively, these data show that Recql5 is not required for normal embryonic development or for postnatal life in mice, but it has a potent tumor suppression function.
Figure 1.
Cancer susceptibility phenotype of Recql5−/− mice. (A) Cancer-free survival of wild-type (n = 32) and Recql5 knockout (n = 50) mice. The animals were monitored for up to 22 mo or until they succumbed to cancer. The two wild-type mice with cancer were identified when they were sacrificed at 22 mo of age. All tumor cases were determined based on the results of pathological analysis. (B) A table summarizing the cancers observed in Recql5−/− mice, including the types of cancers, the frequency for individual type of cancers, and their ages (or average ages) of on-set. (C–F) Images of hematoxylin and eosin (H&E) histology of four representative cancers found in Recql5−/− mice. (C) A well-differentiated squamous cell carcinoma with keratin formation and focal invasion. (D) A well-differentiated lung adenocarcinoma of the predominantly papillary type. (E) A breast carcinoma characteristic of malignant epithelial cells with poor gland formation by axillary mass histopathology. (F) A follicular lymphoma exhibiting characteristic mixture of small and medium-sized abnormal lymphocytes with features of centroblasts and centrocytes. Bars: C–E, 50 μm; F, 10 μm.
Recql5 deficiency is not associated with an elevated frequency of LOH
As Blm and Recql5 have nonredundant roles in suppressing SCEs in mitotic cells (Hu et al. 2005), our finding that Recql5 knockout mice are highly cancer prone suggests that misregulation of HR may be the underlying basis for the cancer phenotype in these animals. Mutations in both human BLM and mouse Blm result in elevated rates of LOH and engender cancer susceptibility (German 1993; Luo et al. 2000; Goss et al. 2002; Hickson 2003). Thus, we first examined whether the elevated SCEs in Recql5-deficient cells is also associated with an increased frequency of LOH. Consistent with a previous report (Luo et al. 2000), we found that Blm knockout embryonic stem (ES) cells exhibited a significant increase in LOH at the Rad50 locus compared with their wild-type counterparts (Supplementary Fig. S1A,B). In contrast, Recql5 knockout ES cells had similar frequencies of LOH as wild-type ES cells at both the Rad50 locus (Supplementary Fig. S1A,C) and the Recql5 locus (Supplementary Fig. S1D,E). This finding suggests that alteration in LOH is not the primary underlying mechanism of cancer susceptibility in Recql5 knockout mice. It also suggests that the main function of Recql5 with respect to suppressing SCEs does not lie at the dissolution of DHJ, a deduction that is consistent with our previous finding that Blm and Recql5 have nonredundant roles in suppressing SCEs in mitotic cells (Hu et al. 2005).
Recql5-deficient cells exhibit an increased frequency of Rad51 and γ-H2AX foci
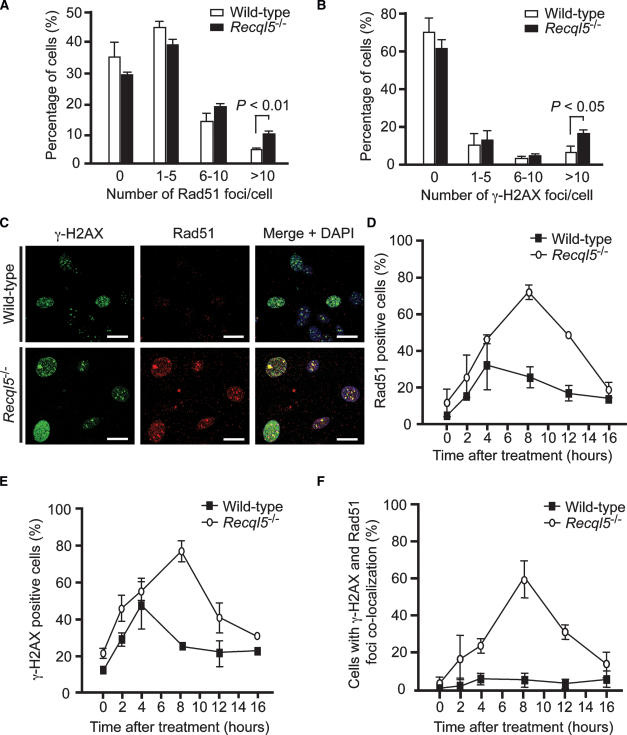
HR can also be regulated at an early step—for instance, during the assembly of presynaptic filaments of the Rad51 recombinase (Sung and Klein 2006). To determine whether Recql5 functions at this stage, we examined the effect of Recql5 ablation on the kinetics of HR induced by DSBs. We reasoned that if Recql5 suppresses SCEs by affecting the channeling of DSBs into HR, deletion of Recql5 should result in an elevated frequency of HR. First, γ-H2AX and Rad51 foci were used as indicators of DSB induction and HR activation, respectively, whereas the colocalization of γ-H2AX and Rad51 foci was used to monitor HR-mediated DSB repair (Paull et al. 2000; Rothkamm et al. 2003). We found that in mouse embryonic fibroblasts (MEFs) derived from Recql5 knockout mice, the percentage of cells with >10 Rad51 foci was significantly higher than wild-type MEFs (Fig. 2A; Supplementary Fig. S2A). Unexpectedly, Recql5-deficient MEFs also had elevated frequencies of cells with >10 spontaneous γ-H2AX foci (Fig. 2B). No significant difference in the frequency of S-phase cells or tetraploid cells between the wild-type and Recql5-deficient MEFs was seen by flow cytometry analysis (data not shown), ruling out the possibility that the increased percentages of cells with >10 γ−H2AX or Rad51 foci were due to differences in the cell cycle distributions. The majority of the spontaneous γ-H2AX foci were detected in PCNA positive cells (data not shown), suggesting that they arise during DNA replication. Consistent with this hypothesis, we found that after exposure to a sublethal dose of camptothecin (CPT), a potent S-phase-specific DSB inducer (Pommier et al. 2003), the proportion of cells with Rad51 foci increased dramatically in both mutant and wild-type MEFs, with the fraction of such cells being much higher in the mutant than wild-type (Fig. 2C,D). Importantly, the CPT-induced Rad51 foci persisted much longer in the mutant cells as compared with wild-type cells (Fig. 2D). The Recql5 mutant cells also exhibited more γ-H2AX foci and for a much longer length of time than wild-type cells (Fig. 2C,E). Moreover, γ-H2AX and Rad51 foci colocalization was rarely observed in wild-type MEFs, even after CPT treatment. In comparison, there were a significantly higher percentage of mutant cells with >10 γ-H2AX foci, of which at least 80% of the foci colocalized with Rad51. Parallel experiments using ES cells showed a similar trend (Supplementary Fig. S2B). In addition, Western blotting showed that chromatin-bound Rad51 was enriched in mutant ES cells compared with the wild-type control following CPT treatment (Supplementary Fig. S3).
Figure 2.
Detection of γ-H2AX and Rad51 focus formation in MEFs by immunostaining. Detection of spontaneous Rad51 (A) and γ-H2AX (B) foci in MEFs. (C) Representative images of cells with γ-H2AX, and/or Rad51 foci. (D–F) The accumulation of MEF cells with Rad51 foci (D), γ-H2AX foci (E), and cells with γ-H2AX and Rad51 foci colocalization (F) at various time points after exposure to 50 nM CPT. In each data set, only cells with >10 foci were scored as positive. Each data point represents the mean ± SEM from at least 500 nuclei examined in two independent experiments. Bars, 10 μm.
Recql5 suppresses DSB-induced HR
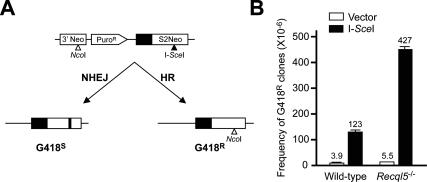
The finding that deletion of Recql5 leads to a dramatic increase in γ-H2AX foci-positive cells strongly suggests that Recql5 has a role in suppressing the accumulation of DSBs. Therefore, it seemed possible that the elevated Rad51 foci and Rad51/γ-H2AX colocalization in Recql5-deficient cells could be an indirect consequence of the increased accumulation of DSBs, rather than an inability to suppress HR. To distinguish between these two possibilities, we examined whether Recql5 deficiency affects the efficiency of HR-mediated repair of I-SceI-induced DSBs. An SCneo cassette for monitoring HR-mediated repair (Johnson et al. 1999) was introduced into the Rb locus in both wild-type and Recql5 knockout ES cells by gene targeting. Within this reporter system, a single DSB could be induced in the mutated version of a G418 resistance selection marker gene by the I-SceI endonuclease. The DSB triggers HR-mediated repair with a nonmutated fragment of the neomycin gene to generate G418-resistant clones (Fig. 3A; Johnson et al. 1999). Since Recql5 deletion does not seem to affect either HJ resolution (Supplementary Fig. S1) or nonhomologous end joining (Hu et al. 2005), differences in the proficiency of reconstructing a functional G418 resistance gene between wild-type and Recql5-deficient ES cells likely reflect differences in the channeling of DSBs into HR. The result showed that the frequency of G418-resistant colonies arising from Recql5-deficient ES cells was almost 3.5-fold higher than that from wild-type ES cells (Fig. 3B). Taken together, these data indicate that Recql5 deficiency is associated with increased DSB repair by HR.
Figure 3.
HR-mediated repair of I-SceI-induced DSBs in mouse ES cells. (A) Schematic illustration of the SCneo system for monitoring HR-mediated repair of I-SceI-induced DSBs. A single copy of the SCneo substrate was introduced into the Rb locus in both wild-type and Recql5 knockout ES cells by gene targeting. Repair of the DSB by HR results in the reconstitution of a functional Neo cassette and G418 resistance. (B) Recovery of G418-resistant colonies from cells containing the SCneo cassette. ES cells containing the SCneo cassette were cultured in medium containing G418 after transfection with either the expression vector for I-SceI or the corresponding empty vector by electroporation. G418-resistant colonies were scored at 10 d after transfection. The average values from three independent experiments are presented above the histograms.
Recql5 is required for genome stability in response to replicative stress
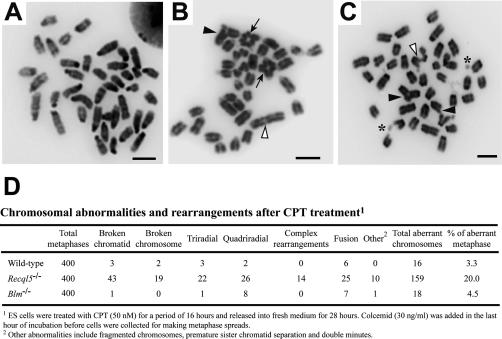
Both the accumulation of DSBs and unchecked inappropriate HR events that accompany Recql5 deletion can result in genome instability. To address this important issue, we examined metaphase chromosome spreads from both wild-type and Recql5-deficient ES cells. As reported previously, spontaneous GCRs are very rare in both wild-type and Recql5-deficient cells (Hu et al. 2005) and therefore difficult to quantify. Thus, to better assess the impact of Recql5 deficiency on GCRs, we compared the frequency of chromosomal aberrations between wild-type, Blm, and Recql5 mutant ES cells following exposure to a sublethal dose (50 nM) of CPT, a potent inducer of GCRs (Pommier et al. 2003). This treatment had only a modest impact on genome stability in the wild-type and Blm knockout ES cells, but it resulted in a dramatic increase in the incidence of chromosomal aberrations in the Recql5−/− mutant cells. The aberrations included chromatid and chromosome gaps, as well as multiradial structures (Fig. 4A–D). These data provide evidence that Recql5 deficiency elevates the susceptibility for GCRs.
Figure 4.
Effect of CPT treatment on chromosome stability in ES cells. (A) A metaphase spread from a wild-type cell. (B,C) Representative images of two metaphase spreads from CPT-treated Recql5 knockout ES cells. A number of abnormal features, including triradial (solid arrowheads), quadriradial (solid arrows), DNA fragments (asterisks), and end-to-end fusion (open arrowheads) are indicated. Bars, 5 μm. (D) Summary of the data from the metaphase spread analysis.
Human RECQL5 physically interacts with Rad51 and suppresses Rad51-mediated D-loop formation
To delineate how Recql5 regulates HR at the mechanistic level, we expressed human RECQL5 in Escherichia coli as a C-terminal fusion with a self-cleaving affinity tag composed of an Mxe-intein fragment and a chitin-binding domain (CBD) (Supplementary Fig. S4A; Garcia et al. 2004). RECQL5 was purified to near homogeneity using two chromatographic fractionation steps coupled with affinity chromatography on chitin beads (Supplementary Fig. S4B). Three independently purified RECQL5 preparations gave the same results in all the biochemical analyses.
Since human RECQL5 physically interacts with both Topo IIIα and its isoform, Topo IIIβ (Shimamoto et al. 2000), we first considered the possibility that RECQL5 helps mediate DHJ dissolution as has been shown for BLM (Wu and Hickson 2003). However, using a previously described DHJ dissolution assay (Wu and Hickson 2003), we could find no evidence of RECQL5 and Topo IIIα or Topo IIIβ being capable of dissolving the DHJ. The same negative result was obtained with the inclusion of BLAP75 in the reaction (data not shown).
We then entertained the possibility that RECQL5 regulates the activity of the human Rad51 recombinase. To test this premise, we first examined whether RECQL5 is capable of associating with Rad51. Interestingly, we found that Rad51 can be coimmunoprecipitated with RECQL5 from U2OS cell extracts (Supplementary Fig. S5A). Likewise, Rad51 in HEK293 cell extracts bound to chitin beads coated with purified RECQL5-CBD protein, but not to control beads containing E. coli McrA-CBD protein (Supplementary Fig. S5B). This interaction is direct, as purified RECQL5 and Rad51 can be efficiently coimmunoprecipitated (Fig. 5A).
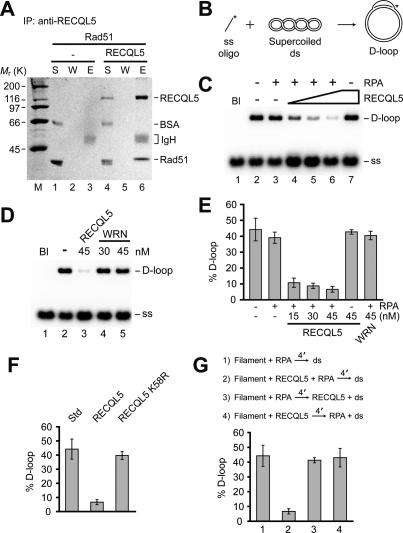
Figure 5.
Human RECQL5 binds Rad51 and inhibits the D-loop reaction. (A) Purified RECQL5 and Rad51 were mixed and subjected to immunoprecipitation with anti-RECQL5 antibodies. The reaction supernatant (S), wash (W), and eluate (E) were analyzed by SDS-PAGE. (IgH) Immunoglobulin heavy chains. (B) D-loop reaction scheme. (C,D) Rad51 K133R presynaptic filaments were incubated with RECQL5 or WRN and RPA (where indicated) before Hop2-Mnd1 and pBluescript form I DNA were incorporated. (Bl) Blank containing DNA substrates only. (E) The average values ± SEM from three or more independent experiments are plotted. (F) D-loop reactions with RPA and RECQL5 or RECQL5 K58R. (Std) Standard reaction with RPA but no RECQL5 or RECQL5 K58R. (G) Effects of order of addition of reaction components. Presynaptic filaments of Rad51 K133R were incubated with the rest of the components in the indicated orders. Hop2-Mnd1 was added together with the dsDNA to all the reactions. The concentration of RECQL5 and RECQL5 K58R was 45 nM. The average values ± SEM from three or more independent experiments are plotted in F and G.
A D-loop assay (Fig. 5B; Petukhova et al. 1998; Mazin et al. 2000) was used to test whether RECQL5 has an effect on the recombinase activity of Rad51. We initially used the Rad51 K133R protein, which binds ATP but is greatly attenuated for ATP hydrolysis, in the D-loop reaction. Since ATP hydrolysis prompts the dissociation of Rad51 from DNA, Rad51 K133R forms a highly stable presynaptic filament (Chi et al. 2006), and its use facilitated our analyses. Coimmunoprecipitation verified that RECQL5 has the same affinity for Rad51 K133R as Rad51 (data not shown). To enhance the efficiency of the D-loop reaction, the accessory factor Hop2-Mnd1 (Petukhova et al. 2005) was included. As expected, Rad51 K133R, and Hop2-Mnd1 catalyzed efficient D-loop formation both in the presence and absence of the ssDNA-binding factor, RPA (Fig. 5C, lanes 2,3). The addition of a catalytic quantity of RECQL5 with Rad51 K133R strongly inhibited D-loop formation when RPA was present (Fig. 5C,E). As little as 15 nM of RECQL5 suppressed the D-loop reaction by four- to fivefold. Interestingly, no significant inhibition by RECQL5 was observed upon the omission of RPA (Fig. 5C, lane 7). Exactly the same results—i.e., RPA-dependent inhibition of the D-loop reaction by RECQL5—were obtained when wild-type Rad51 was used (Supplementary Fig. S6A). To address the specificity of the reaction, we substituted RECQL5 with another human RecQ helicase, WRN, which was previously reported to directly interact with Rad51 (Otterlei et al. 2006). WRN, even at 45 nM, did not affect the D-loop reaction catalyzed by either Rad51 K133R (Fig. 5D,E) or Rad51 (Supplementary Fig. S6A, lanes 5,6). However, this specific WRN preparation was quite adept at dissociating a HJ substrate—in fact, much more so than RECQL5 (Supplementary Fig. S7A,B).
D-loop inhibitory action is dependent on ATP hydrolysis by RECQL5 and order of addition of proteins
We used the ATPase-defective RECQL5 K58R mutant (Garcia et al. 2004) to ask whether suppression of D-loop formation requires ATP hydrolysis by RECQL5. RECQL5 K58R retains the ability to interact with Rad51 and Rad51 K133R (data not shown), but has no effect on the D-loop reaction, indicating that the RECQL5 ATPase activity is required for inhibition (Fig. 5F). Rad51 forms a functional presynaptic filament in the presence of the nonhydrolyzable ATP analog AMP-PNP (Chi et al. 2006). The D-loop reaction catalyzed by Rad51 with AMP-PNP as the nucleotide cofactor is impervious to RECQL5 (data not shown), a result that further supports the conclusion that ATP hydrolysis by RECQL5 is needed for Rad51 inhibition.
A series of order-of-addition experiments were conducted to test the possibility that RECQL5 might use its helicase activity to dissociate the D-loop product. Interestingly, for RECQL5 to inhibit the D-loop reaction, it must be added with RPA during the assembly of the presynaptic filament (Fig. 5G; Supplementary Fig. S6B). This finding argues that RECQL5 acts not by unwinding the D-loop product, but by interfering with the presynaptic filament assembly process.
RECQL5 disrupts the Rad51 presynaptic filament
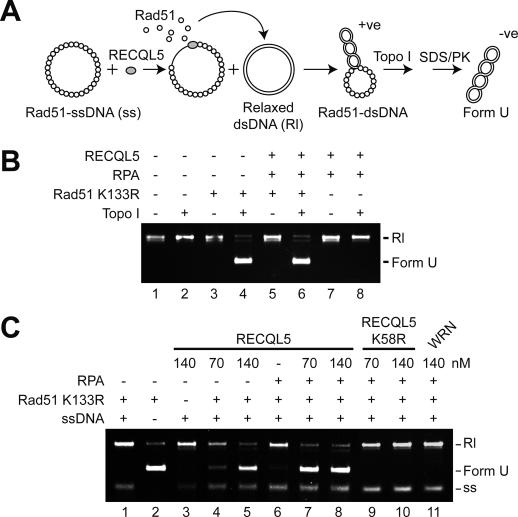
We employed a topoisomerase-linked assay previously used to characterize the presynaptic filament disruption function of Srs2 (Krejci et al. 2003) to enquire whether RECQL5 can catalyze the transfer of Rad51 K133R from ssDNA to a double-stranded DNA (dsDNA) trap. A preassembled presynaptic filament of Rad51 K133R was treated with RECQL5 with or without RPA and, after a brief incubation, an excess of topologically relaxed dsDNA was added to trap Rad51 K133R molecules displaced from the ssDNA. The binding of Rad51 K133R to topologically relaxed dsDNA induces lengthening of the DNA (Ogawa et al. 1993; Sung and Robberson 1995) that can be monitored as a change in the DNA linking number upon incubation with calf thymus topoisomerase I (Fig. 6A; Krejci et al. 2003). The product, Form U DNA, is well separated from the topologically relaxed substrate in an agarose gel (Fig. 6B; Krejci et al. 2003). Control experiments show that RPA and RECQL5 do not make Form U DNA (Fig. 6B, lane 8). The addition of a catalytic amount of RECQL5 resulted in the generation of Form U, indicating that it promotes the transfer of Rad51 K133R from the presynaptic filament to the dsDNA trap (Fig. 6C). The RECQL5-mediated release of Rad51 K133R from the ssDNA is enhanced by RPA (Fig. 6C, cf. lanes 7,8 and 4,5). In contrast, WRN is incapable of mediating the release of Rad51 K133R from the presynaptic filament, regardless of whether RPA is included (Fig. 6C, lane 11) or not (data not shown). The disruptive action of RECQL5 on the presynaptic filament is dependent on its ATPase activity, as the RECQL5 K58R mutant protein is completely inactive in this regard (Fig. 6C, lanes 9,10).
Figure 6.
Turnover of the presynaptic filament as revealed by Topoisomerase-linked DNA topology modification. (A) The reaction scheme. (PK) Proteinase K. (B) Topologically relaxed duplex DNA was incubated with the indicated proteins, with or without calf thymus topoisomerase I. Only Rad51 K133R makes form U DNA. (C) Rad51 K133R presynaptic filaments were assembled and then incubated with RECQL5, RECQL5 K58R, or WRN, with or without RPA. Topologically relaxed duplex DNA and topoisomerase were subsequently added to complete the reaction. Form U DNA marker was run in lane 2. (Rl) Topologically relaxed duplex DNA; (ss) ssDNA.
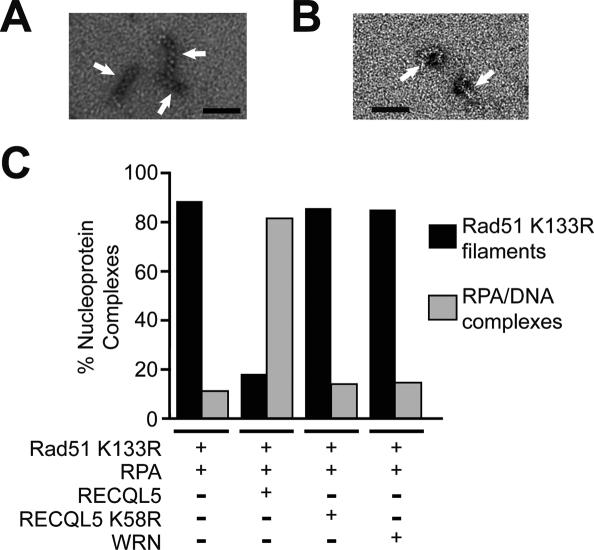
Electron microscopy was used to visualize disruption of the presynaptic filament by RECQL5. Incubation of Rad51 K133R with a 150-mer oligonucleotide and ATP produced abundant presynaptic filaments with distinctive striations (Fig. 7A). Nucleoprotein complexes of RPA with ssDNA were nondescript (Fig. 7B). The dissociation of the Rad51 K133R presynaptic filaments was quantified by determining the relative frequencies of filaments and RPA–ssDNA complexes. At least 2500 nucleoprotein complexes were counted for each reaction. In congruence with the biochemical data, incubation of the Rad51 K133R presynaptic filaments with RECQL5 in the presence of ATP led to its replacement by RPA–ssDNA complexes (Fig. 7C). As anticipated, the Rad51 K133R filaments were impervious to RECQL5 K58R and WRN (Fig. 7C). RECQL5, but not RECQL5 K58R or WRN, also catalyzed the dismantling of presynaptic filaments that comprise Rad51 protein (Supplementary Fig. S8A–C). Thus, the electron microscopic analyses provide clear validation that RECQL5 catalyzes disassembly of the Rad51 presynaptic filament in an ATPase-dependent fashion.
Figure 7.
Analysis of presynaptic filament dissociation by electron microscopy. (A) Presynaptic filaments of Rad51 K133R on the 150-mer ssDNA. (B) Nucleoprotein complexes of RPA and the 150-mer ssDNA. The black scale bar in A and B denotes a length of 50 nm. (C) The relative abundance of Rad51 K133R presynaptic filaments and RPA–ssDNA complexes in reactions that contained the indicated protein components. Over 2500 nucleoprotein complexes were counted in each case.
Discussion
Inappropriate or untimely HR events can have deleterious consequences. For example, the formation of crossovers can result in chromosome translocation, deletion, inversion, and LOH, which are potentially oncogenic in mammals; whereas inappropriate HR repair can lead to GCRs (Sung and Klein 2006; Wu and Hickson 2006). It is therefore not surprising that mammals have evolved multiple pathways for regulating HR-mediated repair in order to suppress potential oncogenic rearrangement in mitotic cells. The importance of this regulation is attested by several human cancer-prone disorders that are associated with aberrant or deregulated HR events (Thompson and Schild 2002). Recent studies have established that in yeast, HR is regulated by the Srs2 and Sgs1 helicases at the initiation and resolution stages, respectively (Sung and Klein 2006). In mammals, BLM appears to be the functional equivalent of yeast Sgs1. The relevance of BLM-dependent DHJ resolution in cancer avoidance has already been clearly established (German 1993; Luo et al. 2000; Goss et al. 2002).
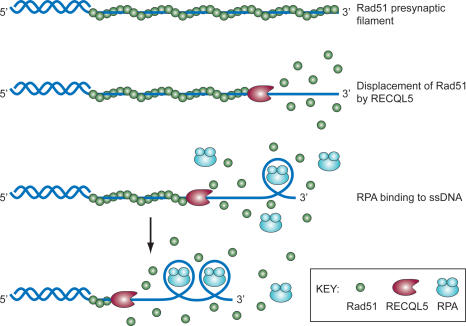
Recently, we demonstrated that Blm and Recql5 act in nonredundant pathways to suppress mitotic SCEs in mice (Hu et al. 2005). In this study, we provide new data showing that deletion of Recql5 in mice results in an increased susceptibility to cancer. Further, we show that Recql5-deficient cells exhibit elevated frequencies of spontaneous DSBs and HR between direct repeats, and are prone to GCRs in response to replication stress. We suggest that elevated spontaneous GCRs represent the primary underlying basis for the cancer susceptibility in Recql5 knockout mice. We also provide evidence that the GCR phenotype is due to a failure to regulate HR. Importantly, using biochemical approaches, we elucidate the mechanism by which RECQL5 regulates HR. Specifically, we demonstrate that human RECQL5 binds the Rad51 recombinase, and a catalytic quantity of this helicase inhibits Rad51-mediated D-loop formation markedly. Furthermore, we show that RECQL5 displaces Rad51 from ssDNA in a reaction that requires ATP hydrolysis by RECQL5. This reaction is stimulated by the ssDNA-binding protein RPA, probably due to the sequestering of ssDNA by RPA after Rad51 removal to prevent Rad51 renucleation on the DNA (Fig. 8). In contrast, the related WRN helicase is incapable of such a feat, even though it is considerably more active than RECQL5 in HJ unwinding. Similarly, the RECQL helicase unwinds the HJ much more efficiently than RECQL5 but is incapable of Rad51 removal from ssDNA (our unpublished data).
Figure 8.
Model depicting the action mechanism of RECQL5 on Rad51 presynaptic filaments. RECQL5 utilizes the free energy from ATP hydrolysis to catalyze the dismantling of the Rad51 presynaptic filament. The ssDNA generated as a result of Rad51 removal is immediately occupied by RPA to prevent the reloading of Rad51.
Our combined genetic, cell biological, and biochemical data provide compelling evidence that RECQL5/Recql5 functions as the mammalian equivalent of yeast Srs2 in suppressing inappropriate HR events. Interestingly, our data also indicate that in the absence of Recql5, cells accumulate excessive DSBs despite having an elevated HR capacity. This observation suggests that this helicase plays an important role in mitigating the lesions associated with damaged or collapsed replication forks. If a replication fork does collapse, RECQL5/Recql5 then functions to suppress the HR-mediated repair of the one-ended DNA break associated with the collapsed fork.
Recent studies show that RECQL5 forms nuclear foci that colocalize with the DNA replication machinery in S-phase nuclei (Kanagaraj et al. 2006). Moreover, the RECQL5 foci persist at sites of stalled replication forks upon exposure of cells to UV irradiation. RECQL5 physically interacts with the DNA polymerase processivity factor PCNA (Kanagaraj et al. 2006). This interaction could serve to target RECQL5 to stalled replication forks where it may act to suppress spurious HR events, by channeling the stalled or damaged replication forks into a nonrecombinational repair pathway. We note that such a postulated means of targeting RECQL5 would be analogous to the PCNA-mediated recruitment of the Srs2 helicase to DNA replication forks (Papouli et al. 2005; Pfander et al. 2005).
In an accompanying paper, Bugreev et al. (2007) provide evidence that the BLM helicase can also disrupt presynaptic filaments of Rad51. In aggregate, the published information and the new findings indicate that in both the mouse and humans, BLM/Blm and RECQL5/Recql5 function via two distinct pathways to regulate the HR process at both early and late stages to prevent excessive crossovers and other inappropriate HR events. Specifically, RECQL5/Recql5 suppresses HR via Rad51 presynaptic filament disruption that serves to channel DNA lesions into the appropriate alternative mechanism(s), whereas BLM/Blm, in addition to providing a presynaptic filament disruptive function (Bugreev et al. 2007), acts to resolve a late HR intermediate in favor of gene conversions (Wu and Hickson 2003). Importantly, both mechanisms are required for tumor suppression.
Materials and methods
See the Supplemental Material for details of protein purification, cell culture, and DNA substrates.
Establishment and culture of mouse ES cells and MEFs
Culture and genetic manipulation of mouse ES cells was performed as described (Hu et al. 2005). The detailed information for individual genetically modified ES cell lines used in this study is described in the Supplemental Material. Primary MEF cultures were derived from 13.5-d-post-coitus (13.5-dpc) embryos of various genotypes, as described (Hu et al. 2005; Mann et al. 2005), except that a low oxygen condition (3% O2, 5% CO2, and 92% N2) was used to reduce oxidative damage-induced stress (Parrinello et al. 2003). All MEF cells used in experiments were under passage number 4.
Mouse and pathological analysis of tumors
Recql5+/+ and Recql5−/− mice were generated by crossing between Recql5+/− mice in a mixed genetic background of 129Sv/Ev and C57BL/6. Mice were maintained in a standard transgenic mouse facility and monitored daily. Individual mice were autopsied, and potential lesions were recorded and fixed in 10% phosphate-buffered formalin. After the fixation, an experienced pathologist examined each animal again. All lesions identified were processed and subjected to standard histological and pathological analyses.
Immunofluorescence
Anti-γ-H2AX (1:200, Upstate Biotechnology; 1:100, Trevigen), anti-Rad51 (1:100, Santa Cruz Biotechnology), and anti-PCNA (1:200, Santa Cruz Biotechnology) antibodies were used in immunofluorescence experiments. Passage 3 MEFs were seeded on glass coverslips for 24 h before drug treatment. After treatment, coverslips were washed briefly in PBS and processed by treating in 0.5% Triton X-100 in PBS (pH 7.1) for 5 min, followed by fixation with 3.7% paraformaldehyde (pH 7.1) for 10 min. Coverslips were incubated with primary antibodies overnight at 4°C and then with Alexa Fluor-conjugated secondary antibodies (Molecular Probes) for 1 h at room temperature, and then were counterstained with DAPI (Vector Laboratories). Images were captured using a confocal microscope (TCS SP2, UV/spectral confocal laser scanner, Leica) and analyzed using Adobe Photoshop (Adobe Systems).
Repair of I-SceI-induced DSBs in mouse ES cells
The SCneo substrate was introduced into the Rb locus in both wild-type and Recql5 knockout ES cells by gene targeting essentially as described (Stark and Jasin 2003), except that the Hyg selection marker was replaced by a Puro marker and puromycin was used to select for transfected cells. After the correctly targeted ES cell lines were obtained, individual cell lines containing the SCneo cassette were transfected with either the expression vector for I-SceI or the corresponding empty vector by electroporation. The transfected cells were divided equally into several plates and then cultured in medium with or without 180 μg/mL G418. The number of colonies from each plate was scored at 10 d after transfection. For each data point, the average frequency of recovering G418-resistant colonies was calculated by dividing the number of G418-resistant colonies by the estimated total number of cells plated after adjusting to the corresponding plating efficiencies. Each experiment was repeated three times.
Chromosome and mitotic abnormalities
Conventional cytogenetic protocols were used for analyzing chromosomal abnormalities as described (Hu et al. 2005; Mann et al. 2005).
RECQL5–Rad51 coimmunoprecipitation
All the incubation steps were performed at 4°C. Purified RECQL5 or RECQL5 K58R and Rad51 or Rad51 K133R, 4 μg each, were incubated in 30 μL of K buffer (20 mM KH2PO4 at pH 7.4, 10% glycerol, 1 mM DTT, 0.5 mM EDTA, 0.01% Igepal) containing 150 mM KCl and 100 μg mL−1 BSA for 10 min, and then rabbit polyclonal anti-RECQL5 antibody (0.1 μg) (Kanagaraj et al. 2006) was added, followed by a 30-min incubation. To capture RECQL5 and associated Rad51, the reactions were incubated for 30 min with 10 μL of Protein-G-coupled magnetic beads (Dynal Biotech) with frequent agitation. The magnetic beads and bound proteins were isolated using a magnet and, after washing the beads twice with 15 μL of K buffer containing 150 mM KCl, the bound proteins were eluted with 30 μL of SDS-PAGE sample loading buffer. The supernatant (S) that contained unbound proteins, the wash (W), and the SDS eluate (E), 10 μL each, were subject to 10% SDS-PAGE and staining with Coomassie blue to visualize proteins.
D-loop reaction
Buffer R (25 mM Tris-HCl at pH 7.5, 2 mM ATP, 1 mM MgCl2, 50 mM KCl, 1 mM DTT, 100 μg mL−1 BSA, containing an ATP-regenerating system consisting of 20 mM creatine phosphate and 20 μg mL−1 creatine kinase) was used for the D-loop reactions, and all the incubation steps were performed at 37°C. Rad51 K133R (1 μM) was incubated with the 5′-end-labeled 90-mer oligonucleotide D1 (3 μM nucleotides) in 11 μL of buffer R for 5 min, followed by the incorporation of Hop2-Mnd1 (300 nM) in 0.75 μL of K buffer and a 1-min incubation. The reaction was initiated by adding pBluescript replicative form I DNA (50 μM base pairs) in 0.75 μL of water. The reactions were terminated after 6 min by the addition of 0.8 μL each of 10% SDS and proteinase K (10 mg mL−1). Following a 3-min incubation, the reaction mixtures were resolved in 0.9% agarose gels in TAE buffer (40 mM Tris acetate at pH 7.4, 0.5 mM EDTA). The gels were dried and subjected to phosphorimaging analysis. When present, RPA (135 nM) and RECQL5 (15–45 nM), WRN (30 and 45 nM), or RECQL5 K58R (45 nM) were added to the preassembled Rad51 filament, followed by a 4-min incubation before the incorporation of Hop2-Mnd1 and replicative form I DNA.
Topoisomerase I-linked DNA topology modification
Buffer R was used for the reactions and all the incubation steps were performed at 37°C. Rad51 K133R (1.5 μM) was incubated for 5 min with pBluescript (−) strand (6 μM nucleotides) in 10.5 μL of buffer R. RECQL5, RECQL5 K58R, or WRN (70 and 140 nM each) and RPA (150 nM) were each added in 0.5 μL of K buffer, followed by a 4-min incubation. Topologically relaxed ϕX174 (7 μM base pairs) in 0.5 μL of water and 2.5 U calf thymus topoisomerase I (Invitrogen) were then incorporated to complete the reaction. The reaction mixtures were incubated for 6 min, terminated, and analyzed as above.
Electron microscopy
Buffer R was used for the reactions and all the incubation steps were performed at 37°C. Rad51 K133R (4 μM) was incubated with 150-mer ssDNA (12 μM nucleotides) for 5 min in 11.5 μL of buffer R, followed by addition of RPA (0.55 μM) in 0.5 μL of K buffer and either buffer, RECQL5, RECQL5 K58R, or WRN (90 nM each) in 0.5 μL of K buffer. After a 4-min incubation, the reactions were diluted eightfold with buffer, and a 4-μL aliquot was applied to 400-mesh grids coated with carbon film that was glow-discharged in air. After staining for 30 sec with 2% uranyl acetate, the samples were examined in a Tecnai 12 Biotwin electron microscope (FEI Company) equipped with a tungsten filament at 100 keV. Digital images were captured with a Morada (Olympus Soft Imaging Solutions) charge-coupled device camera at a nominal magnification of ×87,000.
Acknowledgments
We are grateful to Daniel Camerini-Otero for the Hop2-Mnd1 expression plasmid and to Alessandro Vindigni for the recombinant RECQL baculovirus. This study was supported by grants RO1 CA110415, RO1 CA88939, P20 CA103736, RO1 ES015632, and RO1 ES015252 from the US National Institutes of Health, the Searle Scholar Program (01-E-109), and the Swiss National Science Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1609107
References
- Aguilera A., Klein H.L., Klein H.L. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.J., Noirot-Gros M.F., Wang J.C., Noirot-Gros M.F., Wang J.C., Wang J.C. Interaction between yeast Sgs1 helicase and DNA Topoisomerase III. J. Biol. Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Yu X., Egelman E.H., Mazin A.V., Yu X., Egelman E.H., Mazin A.V., Egelman E.H., Mazin A.V., Mazin A.V. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes & Dev. 2007 doi: 10.1101/gad.1609007. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Bellaoui M., Zhang C., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G.A., Boone C., Brown G.W., Bellaoui M., Zhang C., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G.A., Boone C., Brown G.W., Zhang C., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G.A., Boone C., Brown G.W., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G.A., Boone C., Brown G.W., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G.A., Boone C., Brown G.W., Delgado-Cruzata L., Rothstein R., Freyer G.A., Boone C., Brown G.W., Rothstein R., Freyer G.A., Boone C., Brown G.W., Freyer G.A., Boone C., Brown G.W., Boone C., Brown G.W., Brown G.W. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P., Van Komen S., Sehorn M.G., Sigurdsson S., Sung P., Van Komen S., Sehorn M.G., Sigurdsson S., Sung P., Sehorn M.G., Sigurdsson S., Sung P., Sigurdsson S., Sung P., Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst.) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J., Lennon D.J., Ciocci S., Proytcheva M., German J., Ciocci S., Proytcheva M., German J., Proytcheva M., German J., German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald J.P., Bendixen C., Arthur L., Rothstein R., McDonald J.P., Bendixen C., Arthur L., Rothstein R., Bendixen C., Arthur L., Rothstein R., Arthur L., Rothstein R., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P.L., Liu Y., Jiricny J., West S.C., Janscak P., Liu Y., Jiricny J., West S.C., Janscak P., Jiricny J., West S.C., Janscak P., West S.C., Janscak P., Janscak P. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom syndrome: A Mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. [PubMed] [Google Scholar]
- Goss K.H., Risinger M.A., Kordich J.J., Sanz M.M., Straughen J.E., Slovek L.E., Capobianco A.J., German J., Boivin G.P., Groden J., Risinger M.A., Kordich J.J., Sanz M.M., Straughen J.E., Slovek L.E., Capobianco A.J., German J., Boivin G.P., Groden J., Kordich J.J., Sanz M.M., Straughen J.E., Slovek L.E., Capobianco A.J., German J., Boivin G.P., Groden J., Sanz M.M., Straughen J.E., Slovek L.E., Capobianco A.J., German J., Boivin G.P., Groden J., Straughen J.E., Slovek L.E., Capobianco A.J., German J., Boivin G.P., Groden J., Slovek L.E., Capobianco A.J., German J., Boivin G.P., Groden J., Capobianco A.J., German J., Boivin G.P., Groden J., German J., Boivin G.P., Groden J., Boivin G.P., Groden J., Groden J. Enhanced tumor formation in mice heterozygous for Blm mutation. Science. 2002;297:2051–2053. doi: 10.1126/science.1074340. [DOI] [PubMed] [Google Scholar]
- Hickson I.D. RecQ helicases: Caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- Hu Y., Lu X., Barnes E., Yan M., Lou H., Luo G., Lu X., Barnes E., Yan M., Lou H., Luo G., Barnes E., Yan M., Lou H., Luo G., Yan M., Lou H., Luo G., Lou H., Luo G., Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell. Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J.E., Malkova A., Liberi G., Foiani M., Haber J.E., Liberi G., Foiani M., Haber J.E., Foiani M., Haber J.E., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.D., Liu N., Jasin M., Liu N., Jasin M., Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- Kanagaraj R., Saydam N., Garcia P.L., Zheng L., Janscak P., Saydam N., Garcia P.L., Zheng L., Janscak P., Garcia P.L., Zheng L., Janscak P., Zheng L., Janscak P., Janscak P. Human RECQ5β helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzaki M., Hatanaka A., Sonoda E., Yamazoe M., Kikuchi K., Vu Trung N., Szuts D., Sale J.E., Shinagawa H., Watanabe M., Hatanaka A., Sonoda E., Yamazoe M., Kikuchi K., Vu Trung N., Szuts D., Sale J.E., Shinagawa H., Watanabe M., Sonoda E., Yamazoe M., Kikuchi K., Vu Trung N., Szuts D., Sale J.E., Shinagawa H., Watanabe M., Yamazoe M., Kikuchi K., Vu Trung N., Szuts D., Sale J.E., Shinagawa H., Watanabe M., Kikuchi K., Vu Trung N., Szuts D., Sale J.E., Shinagawa H., Watanabe M., Vu Trung N., Szuts D., Sale J.E., Shinagawa H., Watanabe M., Szuts D., Sale J.E., Shinagawa H., Watanabe M., Sale J.E., Shinagawa H., Watanabe M., Shinagawa H., Watanabe M., Watanabe M., et al. Cooperative roles of vertebrate Fbh1 and Blm DNA helicases in avoidance of crossovers during recombination initiated by replication fork collapse. Mol. Cell. Biol. 2007;27:2812–2820. doi: 10.1128/MCB.02043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P., Reddy M.S., Klein H., Ellenberger T., Sung P., Klein H., Ellenberger T., Sung P., Ellenberger T., Sung P., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J.E., Foiani M., Lopes M., Pellicioli A., Haber J.E., Foiani M., Pellicioli A., Haber J.E., Foiani M., Haber J.E., Foiani M., Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes & Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Santoro I.M., McDaniel L.D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., Bradley A., Santoro I.M., McDaniel L.D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., Bradley A., McDaniel L.D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., Bradley A., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., Bradley A., Mills M., Youssoufian H., Vogel H., Schultz R.A., Bradley A., Youssoufian H., Vogel H., Schultz R.A., Bradley A., Vogel H., Schultz R.A., Bradley A., Schultz R.A., Bradley A., Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- Mann M.B., Hodges C.A., Barnes E., Vogel H., Hassold T.J., Luo G., Hodges C.A., Barnes E., Vogel H., Hassold T.J., Luo G., Barnes E., Vogel H., Hassold T.J., Luo G., Vogel H., Hassold T.J., Luo G., Hassold T.J., Luo G., Luo G. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum. Mol. Genet. 2005;14:813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- Mazin A.V., Zaitseva E., Sung P., Kowalczykowski S.C., Zaitseva E., Sung P., Kowalczykowski S.C., Sung P., Kowalczykowski S.C., Kowalczykowski S.C. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T., Furukawa F., Sakaguchi C., Toda T., Carr A.M., Iwasaki H., Shinagawa H., Furukawa F., Sakaguchi C., Toda T., Carr A.M., Iwasaki H., Shinagawa H., Sakaguchi C., Toda T., Carr A.M., Iwasaki H., Shinagawa H., Toda T., Carr A.M., Iwasaki H., Shinagawa H., Carr A.M., Iwasaki H., Shinagawa H., Iwasaki H., Shinagawa H., Shinagawa H. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol. Cell. Biol. 2005;25:8074–8083. doi: 10.1128/MCB.25.18.8074-8083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J.R., Nallaseth F.S., Lan Y.Q., Slagle C.E., Brill S.J., Nallaseth F.S., Lan Y.Q., Slagle C.E., Brill S.J., Lan Y.Q., Slagle C.E., Brill S.J., Slagle C.E., Brill S.J., Brill S.J. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1–Top3 complex. Mol. Cell. Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Yu X., Shinohara A., Egelman E.H., Yu X., Shinohara A., Egelman E.H., Shinohara A., Egelman E.H., Egelman E.H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Onoda F., Seki M., Miyajima A., Enomoto T., Seki M., Miyajima A., Enomoto T., Miyajima A., Enomoto T., Enomoto T. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom’s syndrome gene. Mutat. Res. 2000;459:203–209. doi: 10.1016/s0921-8777(99)00071-3. [DOI] [PubMed] [Google Scholar]
- Osman F., Dixon J., Barr A.R., Whitby M.C., Dixon J., Barr A.R., Whitby M.C., Barr A.R., Whitby M.C., Whitby M.C. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol. Cell. Biol. 2005;25:8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei M., Bruheim P., Ahn B., Bussen W., Karmakar P., Baynton K., Bohr V.A., Bruheim P., Ahn B., Bussen W., Karmakar P., Baynton K., Bohr V.A., Ahn B., Bussen W., Karmakar P., Baynton K., Bohr V.A., Bussen W., Karmakar P., Baynton K., Bohr V.A., Karmakar P., Baynton K., Bohr V.A., Baynton K., Bohr V.A., Bohr V.A. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J. Cell Sci. 2006;119:5137–5146. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- Papouli E., Chen S., Davies A.A., Huttner D., Krejci L., Sung P., Ulrich H.D., Chen S., Davies A.A., Huttner D., Krejci L., Sung P., Ulrich H.D., Davies A.A., Huttner D., Krejci L., Sung P., Ulrich H.D., Huttner D., Krejci L., Sung P., Ulrich H.D., Krejci L., Sung P., Ulrich H.D., Sung P., Ulrich H.D., Ulrich H.D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Paques F., Haber J.E., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S., Campisi J., Samper E., Krtolica A., Goldstein J., Melov S., Campisi J., Krtolica A., Goldstein J., Melov S., Campisi J., Goldstein J., Melov S., Campisi J., Melov S., Campisi J., Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M., Kirchgessner C.U., Gellert M., Bonner W.M., Gellert M., Bonner W.M., Bonner W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Petukhova G., Stratton S., Sung P., Stratton S., Sung P., Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Petukhova G.V., Pezza R.J., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D., Pezza R.J., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D., Ploquin M., Masson J.Y., Camerini-Otero R.D., Masson J.Y., Camerini-Otero R.D., Camerini-Otero R.D. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- Pfander B., Moldovan G.L., Sacher M., Hoege C., Jentsch S., Moldovan G.L., Sacher M., Hoege C., Jentsch S., Sacher M., Hoege C., Jentsch S., Hoege C., Jentsch S., Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Redon C., Rao V.A., Seiler J.A., Sordet O., Takemura H., Antony S., Meng L., Liao Z., Kohlhagen G., Redon C., Rao V.A., Seiler J.A., Sordet O., Takemura H., Antony S., Meng L., Liao Z., Kohlhagen G., Rao V.A., Seiler J.A., Sordet O., Takemura H., Antony S., Meng L., Liao Z., Kohlhagen G., Seiler J.A., Sordet O., Takemura H., Antony S., Meng L., Liao Z., Kohlhagen G., Sordet O., Takemura H., Antony S., Meng L., Liao Z., Kohlhagen G., Takemura H., Antony S., Meng L., Liao Z., Kohlhagen G., Antony S., Meng L., Liao Z., Kohlhagen G., Meng L., Liao Z., Kohlhagen G., Liao Z., Kohlhagen G., Kohlhagen G., et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Raynard S., Bussen W., Sung P., Bussen W., Sung P., Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIα, and BLAP75. J. Biol. Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Rothkamm K., Kruger I., Thompson L.H., Lobrich M., Kruger I., Thompson L.H., Lobrich M., Thompson L.H., Lobrich M., Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto A., Nishikawa K., Kitao S., Furuichi Y., Nishikawa K., Kitao S., Furuichi Y., Kitao S., Furuichi Y., Furuichi Y. Human RecQ5β, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3α and 3β. Nucleic Acids Res. 2000;28:1647–1655. doi: 10.1093/nar/28.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark J.M., Jasin M., Jasin M. Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 2003;23:733–743. doi: 10.1128/MCB.23.2.733-743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Klein H., Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Sung P., Robberson D.L., Robberson D.L. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Symington L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L.H., Schild D., Schild D. Recombinational DNA repair and human disease. Mutat. Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F., Kowalczykowski S.C., Le Cam E., Fabre F., Le Cam E., Fabre F., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D., Hickson I.D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D., Hickson I.D. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- Wu L., Bachrati C.Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Bachrati C.Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Chang M., Wang W., Li L., Brown G.W., Hickson I.D., Wang W., Li L., Brown G.W., Hickson I.D., Li L., Brown G.W., Hickson I.D., Brown G.W., Hickson I.D., Hickson I.D. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]