Abstract
The Bur1–Bur2 and Paf1 complexes function during transcription elongation and affect histone modifications. Here we describe new roles for Bur1–Bur2 and the Paf1 complex. We find that histone H3 K36 tri-methylation requires specific components of the Paf1 complex and that K36 tri-methylation is more strongly affected at the 5′ ends of genes in paf1Δ and bur2Δ strains in parallel with increased acetylation of histones H3 and H4. Interestingly, the 5′ increase in histone acetylation is independent of K36 methylation, and therefore is mechanistically distinct from the methylation-driven deacetylation that occurs at the 3′ ends of genes. Finally, Bur1–Bur2 and the Paf1 complex have a second methylation-independent function, since bur2Δ set2Δ and paf1Δ set2Δ double mutants display enhanced histone acetylation at the 3′ ends of genes and increased cryptic transcription initiation. These findings identify new functions for the Paf1 and Bur1–Bur2 complexes, provide evidence that histone modifications at the 5′ and 3′ ends of coding regions are regulated by distinct mechanisms, and reveal that the Bur1–Bur2 and Paf1 complexes repress cryptic transcription through a Set2-independent pathway.
Keywords: Bur2, histone methylation, Paf1, transcription, yeast
Introduction
RNA polymerase II (pol II)-directed transcription consists of distinct initiation, elongation, and termination steps, all of which occur in the context of a nucleosomal template (Hartzog et al, 2002; Li et al, 2007). Nucleosomes can block initiation and elongation by pol II in vitro (Li et al, 2007), but eukaryotes have evolved elaborate mechanisms to overcome nucleosome-mediated repression, including ATP-dependent nucleosome remodeling (Zofall et al, 2006) and post-translational histone modifications (Kouzarides, 2007; Li et al, 2007). We are still in the early stage of identifying and understanding the functional significance of histone modifications that occur as pol II progresses through a template.
Methylation of histone H3 on lysine 36 (K36) is a conserved modification in eukaryotes that is catalyzed by Set2 (Strahl et al, 2002). Set2 associates with the coding regions of transcribed genes through direct interaction with the elongating hyperphosphorylated form of pol II (Li et al, 2002; Xiao et al, 2003; Krogan et al, 2003b; Kizer et al, 2005). Consistent with the localization pattern of Set2, di-methylated and tri-methylated K36 are enriched throughout the coding regions of transcriptionally active genes (Xiao et al, 2003; Bannister et al, 2005; Morillon et al, 2005; Pokholok et al, 2005; Rao et al, 2005). Moreover, set2Δ causes a growth defect when combined with mutations in genes that encode elongation factors (Li et al, 2003; Krogan et al, 2003b). Current evidence indicates that Set2 and H3 K36 methylation perform inhibitory roles in transcription regulation: LexA–Set2 can repress transcription of a lacZ reporter that contains a LexA-binding site (Strahl et al, 2002), set2Δ or a histone H3 K36R substitution increases the expression of a UAS-less GAL4 mutant promoter (Landry et al, 2003) and restores the binding of pol II and TBP to promoters in strains lacking yeast FACT (Biswas et al, 2006), and SET2 represses cryptic transcription initiation within coding regions (Carrozza et al, 2005).
Histone acetylation affects transcription by leading to the recruitment of factors that further modify histones (Kouzarides, 2007) or by changing the electrostatic charge on histones and weakening histone–DNA interactions (Hong et al, 1993; Workman and Kingston, 1998). Histone acetylation across the Saccharomyces cerevisiae genome is not uniform, with promoter regions typically being acetylated more than the associated coding regions (Reid et al, 2004; Joshi and Struhl, 2005; Pokholok et al, 2005). The low level of acetylation in coding regions is maintained by a dynamic regulation between acetylation and deacetylation. Interestingly, at least part of this regulation is mediated by histone H3 K36 methylation, which can recruit the Rpd3S histone deacetylase (HDAC) complex through its chromodomain-containing Eaf3 subunit, resulting in histone deacetylation and repression of cryptic transcription initiation within the coding region (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005).
Recent studies have found that elongation factors such as Bur1–Bur2 and the Paf1 complex are required for specific histone modifications. BUR1 and BUR2 encode the catalytic and regulatory subunits of a cyclin-dependent protein kinase that is essential for normal growth and has a general role in transcription elongation (Yao et al, 2000; Keogh et al, 2003). The yeast Paf1 complex, which consists of Paf1, Ctr9, Cdc73, Rtf1, and Leo1 subunits (Mueller and Jaehning, 2002; Squazzo et al, 2002; Mueller et al, 2004), has been implicated in transcription elongation based on physical association with pol II and the elongation factors Spt4–Spt5 and yFACT, its localization to the ORFs of active genes in vivo, and mutant sensitivity to 6-azauracil and mycophenolic acid (Krogan et al, 2002; Mueller and Jaehning, 2002; Squazzo et al, 2002). The Rtf1, Paf1, and Ctr9 subunits of the Paf1 complex are required for histone H2B mono-ubiquitylation and histone H3 K4 di- and tri-methylation on actively transcribed genes (Wood et al, 2003; Krogan et al, 2003a; Ng et al, 2003b; Xiao et al, 2005). Proper recruitment of the Paf1 complex is dependent on Bur1–Bur2, and therefore mutations in BUR1 and BUR2 also cause defects in histone H2B mono-ubiquitylation and H3 K4 methylation (Laribee et al, 2005; Wood et al, 2005; Qiu et al, 2006). We recently showed that Bur1–Bur2 is also required for tri-methylation of histone H3 at K36 (Chu et al, 2006). The role of the Paf1 complex in histone H3 K36 methylation is unclear, however, as conflicting results have been reported (Ng et al, 2003a; Krogan et al, 2003b). Effects of Bur1–Bur2 and the Paf1 complex on histone acetylation have not been studied.
Although the precise mechanisms by which Bur1–Bur2 and the Paf1 complex influence elongation are not fully elaborated, it remains important to define and investigate their shared and unshared roles in regulating histone modifications and transcription. To gain more insight into these issues, we examined the methylation of H3 K36 in strains deleted for Paf1 complex subunits and the functional consequences of Bur1–Bur2 and Paf1 complex defects on histone acetylation and cryptic transcription. Our results indicate that the Bur1 and Paf1 complexes regulate histone acetylation and repress cryptic transcription by a mechanism that is distinct from the mechanism that regulates H3 K36 tri-methylation.
Results
Mutations in the Paf1 complex affect the histone H3 K36 methylation state
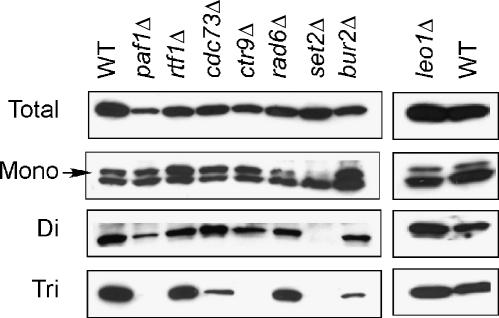
To identify factors that are required for the transition from the di-methylated to the tri-methylated state of histone H3 K36, we screened mutations in genes that have been functionally linked to histone modifications or transcription elongation. In particular, we focused on the multifunctional Paf1 complex, which is comprised of Paf1, Ctr9, Cdc73, Leo1, and Rtf1 (Krogan et al, 2002; Mueller and Jaehning, 2002; Squazzo et al, 2002; Mueller et al, 2004). Using antibodies specific for distinct methylated forms of H3 K36 in western blots of yeast cell lysates, we observed a strong reduction of H3 K36 tri-methylation in paf1Δ and ctr9Δ strains, a moderate reduction in the cdc73Δ strain, and no detectable K36 methylation defect in rtf1Δ and leo1Δ strains (Figure 1). The defect was restricted to tri-methylated H3 K36, as effects on mono- or di-methylated H3 K36 levels were either modest or absent. The dependence of K36 tri-methylation on only a subset of the Paf1 complex members was not due to allelic differences, since null alleles were used for each gene. Specific subunits of the Paf1 complex are therefore required for normal levels of histone H3 K36 tri-methylation.
Figure 1.
A subset of Paf1 complex mutations selectively affect histone H3 K36 tri-methylation. Whole-cell extracts were prepared from yeast strains with the genotypes shown at the top, and western blots were performed using antibodies specific for total H3, mono-, di-, or tri-methylated K36. The arrow indicates the position of mono-methylated H3 migrating slightly above a background band.
Extensive genetic interactions between the Paf1 complex, Set2, the pol II CTD, and the Bur1–Bur2 kinase
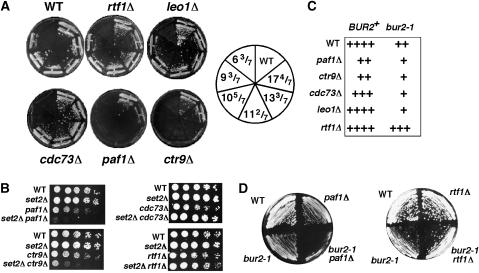
Because mutations in a subset of the genes encoding the Paf1 complex share a similar H3 K36 tri-methylation defect as was seen previously in bur1, bur2, and rpb1 CTD truncation strains (Xiao et al, 2003; Chu et al, 2006), we next tested whether genetic interactions could be detected by double mutant analysis. First, double mutants were constructed from a series of rpb1 CTD truncation alleles and all Paf1 complex deletion mutations using a plasmid shuffle strategy. paf1Δ and ctr9Δ displayed strong synthetic lethality with CTD truncations, while leo1Δ and rtf1Δ exhibited no interactions with any CTD truncations, and cdc73Δ showed a synthetic growth defect only with the most severe CTD truncation, indicative of an intermediate functional interaction (Figure 2A). These double-mutant phenotypes strongly parallel the extent of the tri-methylation defect and the growth defect observed in individual Paf1 complex mutants (paf1Δ=ctr9Δ>cdc73Δ>leo1Δ=rtf1Δ). Second, a similar pattern of genetic interactions was observed between a subset of the Paf1 complex deletions and a deletion of SET2, with set2Δ causing strong synthetic growth defects with paf1Δ and ctr9Δ, synthetic lethality with cdc73Δ at 38.5°C and partial suppression at 30°C, and no interactions with leo1Δ and rtf1Δ (Figure 2B, and data not shown). These results differ from those obtained by a large-scale screen that described synthetic lethal interactions of all Paf1 complex deletion mutations, except leo1Δ, with set2Δ (Krogan et al, 2003b). The cause of this discrepancy is unknown but likely reflects limitations of the yeast deletion collection and the use of large-scale SGA screening methodology instead of creating fresh knockouts and tetrad analysis to follow double-mutant phenotypes. The consistent segregation of these interactions as detected by tetrad analysis, the overlap with the K36 tri-methylation defect, and concordance with the data of Biswas et al (2006) provide confidence in our results. Third, genetic interactions were also observed between Paf1 complex mutations and bur2-1, a mutation that causes a moderate growth defect by itself. Double-mutant combinations of bur2-1 with paf1Δ, ctr9Δ, cdc73Δ, and leo1Δ all revealed synthetic growth defects, whereas rtf1Δ partially suppressed the bur2-1 growth defect (Figure 2C and D). Although this pattern differed from that observed for the H3 K36 tri-methylation defect and for the genetic interactions with the CTD truncations and set2Δ, it is consistent with, and expands upon, prior results describing suppression of bur1Δ by rtf1Δ and synthetic growth defects between bur1-23 and leo1Δ, paf1Δ, and cdc73Δ (Keogh et al, 2005). In summary, a subset of Paf1 complex mutations displays extensive genetic interactions with rpb1 CTD truncations, set2Δ, and bur2-1 that overlap significantly, but not completely, with the K36 tri-methylation defect. These systematic tests of double-mutant interactions further demonstrate functional distinctions between the subunits of the Paf1 complex and functional overlap with the pol II CTD, Set2, and the Bur1–Bur2 kinase complex.
Figure 2.
Paf1 complex mutations interact genetically with CTD truncations, a bur2 mutation, and set2Δ. (A) The growth phenotypes of strains containing deletions of Paf1 complex subunits and truncations of the pol II CTD. The numbers in the diagram on the right indicate the number of CTD repeats remaining. Strains were constructed by a plasmid shuffle approach and grown on 5FOA medium for 4 days at 30°C. (B) set2Δ causes synthetic sickness selectively with paf1Δ and ctr9Δ. Five-fold serial dilutions of the strains listed were spotted onto YPD plates and photographed after 3–4 days of growth at 30°C. (C) Summary of the growth phenotypes of double mutants between bur2-1 and deletions of the Paf1 complex subunits. (D) Strains with the indicated genotypes were streaked onto a YPD plate and grown at 30°C for 3 days.
Histone H3 K36 tri-methylation patterns at specific genes
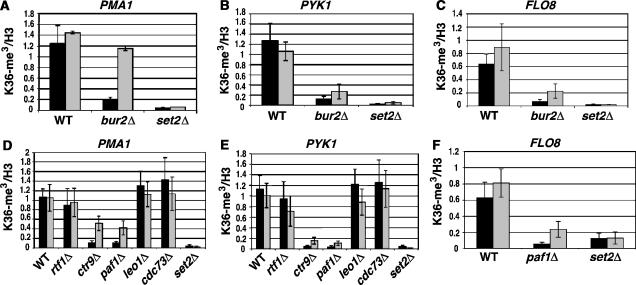
Because H3 K36 tri-methylation defects were detected in paf1Δ, ctr9Δ, and bur2Δ crude extracts, it was expected that tri-methylation would also be affected at individual genes. ChIP assays were performed to independently assess K36 tri-methylation levels, to determine whether all genes being tested were equally affected, and to examine whether any detectable effects were equally distributed throughout the ORFs. Previous studies revealed that tri-methylation occurs throughout the ORF of transcribed genes, with a genome-wide average peaking toward the 3′ end, although the 3′ end peak does not occur at every gene (Pokholok et al, 2005). Several genes were examined by ChIP, using primers specific for the 5′ and 3′ ends of their respective ORFs. In broad agreement with the results observed by western analysis, K36 tri-methylation at PMA1, PYK1, and FLO8 was severely reduced in a bur2Δ strain (Figure 3A–C). This defect likely operates through the Paf1 complex, since paf1Δ and ctr9Δ strains displayed the same reduction of tri-methylated H3 at PMA1, PYK1, FLO8, PDR5, and GLN4, whereas little if any effect was observed in cdc73Δ, leo1Δ, and rtf1Δ strains (Figure 3D–F; Supplementary Figure 1). Interestingly, a greater reduction was consistently observed at the 5′ end than at the 3′ end in bur2Δ, paf1Δ, and ctr9Δ strains. The reduced tri-methylation signal is not due to a loss of histone H3 occupancy, since H3 levels are unaffected at PMA1 and FLO8 in bur2Δ and paf1Δ strains (Supplementary Figure 2). Three conclusions emerge from these ChIP results: strong Paf1c effects on H3 K36 tri-methylation were limited to paf1Δ and ctr9Δ, the greatest reduction occurred at the 5′ ends of the ORFs, and the reduction of 5′ H3 K36 tri-methylation was shared with bur2Δ.
Figure 3.
Histone H3 K36 tri-methylation levels at specific genes in bur2Δ and Paf1 complex mutants. (A–C) Total histone H3 or histone H3 tri-methylated at K36 was immunoprecipitated from extracts of formaldehyde-treated wild type, bur2Δ, and set2Δ strains. Levels of tri-methylated K36 or total H3 were assessed at the 5′ (black bars) and 3′ ends (gray bars) of the PMA1 (A), PYK1 (B), and FLO8 (C) coding regions by ChIPs. (D–F) ChIPs of total histone H3 and H3 tri-methylated at K36 were performed in Paf1 complex deletion strains. Enrichment of tri-methyl K36 was measured at the 5′ (black bars) and 3′ (gray bars) ends of PMA1 (D), PYK1 (E), and FLO8 (F). Tri-methyl K36 levels were normalized to total H3 levels. Standard error is provided for values representing three experiments.
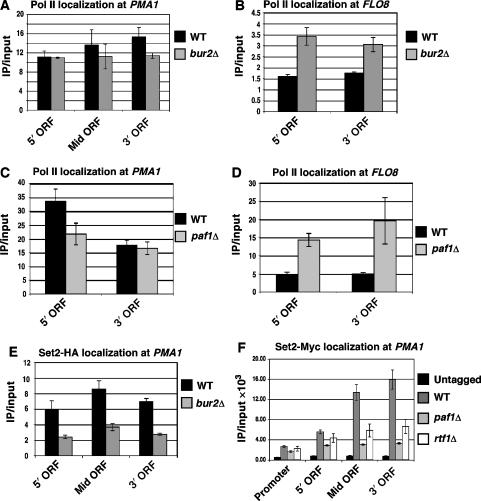
The reduction of H3 K36 tri-methylation in bur2Δ and paf1Δ mutants could conceivably result from decreased association of pol II or Set2 with transcribed genes. The occupancy of pol II as measured by ChIP assays with antibodies against unphosphorylated CTD (8WG16) or Ser5-phosphorylated CTD (H14), however, was only slightly reduced at the 5′ and 3′ ends of PMA1 in bur2Δ and paf1Δ strains (Figure 4A and C; Supplementary Figure 3). At FLO8, pol II levels actually increased in the mutant strains (Figure 4B and D). These data indicate that the dramatic reduction of H3 K36 tri-methylation in bur2Δ and paf1Δ strains is not caused by a pol II recruitment defect. In contrast to the pol II results, deletion of BUR2 or PAF1 caused a significant decrease in Set2 levels at PMA1 (Figure 4E and F), PYK1, and GLN4 (Supplementary Figure 4C and D), without affecting total cellular levels of Set2 (Supplementary Figure 4A and B). This result agrees with previous observations of decreased Set2 recruitment in rtf1Δ and cdc73Δ strains (Krogan et al, 2003b). Interestingly, in paf1Δ strains, association of Set2 was reduced more severely at the middle and 3′ ends of PMA1, PYK1, and GLN4 than at the 5′ ends (Figure 4F; Supplementary Figure 4), even though the reduction in H3 K36 tri-methylation was more pronounced at the 5′ ends (Figure 3). These results suggest that the H3 K36 tri-methylation defect does not correlate simply with a reduction in Set2 localization or levels. Consistent with this idea, deletion of RTF1, which did not affect H3 K36 methylation, and deletion of PAF1, which dramatically lowered H3 K36 tri-methylation, caused a similar decrease in Set2 recruitment (Figure 4F; Supplementary Figure 4C and D).
Figure 4.
Pol II and Set2 occupancy in bur2Δ and paf1Δ mutant strains. (A–D) 8WG16 antibody was incubated with chromatin samples to test the total pol II level at PMA1 and FLO8 from wild-type and bur2Δ strains (A, B) and from wild-type and paf1Δ strains (C, D). (E, F) ChIP assays measuring the recruitment of tagged Set2 to the promoter, 5′ ORF, Mid ORF and 3′ ORF of PMA1 in bur2Δ, paf1Δ and rtf1Δ strains. Standard error is provided for values representing three experiments.
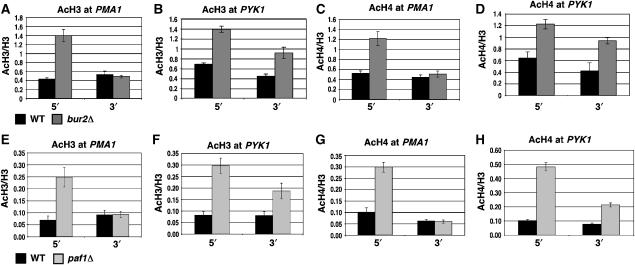
Histone acetylation increases at several active genes in bur2Δ and paf1Δ mutants
A functional role for histone H3 K36 methylation was described recently, in which methylated K36 recruits the Rpd3S HDAC complex to the 3′ ends of ORFs, triggering histone deacetylation and the repression of cryptic intragenic transcripts (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005). The reduction of K36 tri-methylation in bur2Δ and paf1Δ strains allowed a test of whether a similar coupling of acetylation and K36 methylation occurs at the 5′ ends of ORFs. Antibodies that recognize di-acetylated H3 and tetra-acetylated H4 were used in ChIP assays to assess H3 and H4 acetylation at the 5′ and 3′ ends of several genes in bur2Δ and paf1Δ strains. Mirroring the reduction of K36 tri-methylation, acetylation of H3 and H4 was increased preferentially near the 5′ ends of PMA1 and FLO8 in a bur2Δ strain (Figure 5A and C; Supplementary Figure 5A and B). In PYK1, H3 and H4 acetylation was increased at the 5′ end, but also at the 3′ end in the bur2Δ strain (Figure 5B and D). Similar histone acetylation changes were detected in a paf1Δ strain, where histone H3 and H4 acetylation was increased at the 5′ ends of PMA1 and FLO8 (Figure 5E and G; Supplementary Figure 5G and H), and at both the 5′ and 3′ ends of PYK1 (Figure 5F and H). The acetylation levels at two other genes were also tested in a paf1Δ strain; at PDR5, H3 and H4 acetylation increased at the 5′ end, while acetylation was unaffected at GLN4 (Supplementary Figure 5). These data indicate that the loss of BUR2 or PAF1 results in significantly increased histone H3 and H4 acetylation at the 5′ ends of PMA1, PYK1, FLO8, and PDR5, and additionally affects the 3′ end of PYK1, while some genes, exemplified by GLN4, remain unaffected.
Figure 5.
bur2Δ and paf1Δ affect histone acetylation at the 5′ regions of ORFs. (A–D) ChIP analysis of wild type and bur2Δ strains using antibodies against acetylated histones H3 and H4. Levels of H3 acetylation and H4 acetylation were assessed at the 5′ and 3′ ends of PMA1 (A, C) and PYK1 (B, D). (E–H) ChIP analysis of wild type and paf1Δ strains using antibodies against acetylated histones H3 and H4 at the 5′ and 3′ ends of PMA1 (E, G) and PYK1 (F, H). Standard error is provided for values representing three experiments.
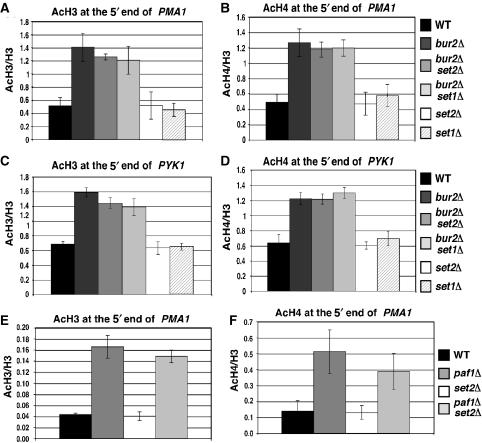
The increase in histone acetylation at the 5′ ends of genes is independent of H3 K36 methylation
Since we observed a parallel decrease in H3 K36 tri-methylation with an increase in histone H3 and H4 acetylation at the 5′ ends of several genes, we asked whether the changes in methylation and acetylation are functionally linked. The increased acetylation at the 5′ ends of PMA1 and PYK1 observed in bur2Δ and paf1Δ strains did not occur in set2Δ or set1Δ strains (Figure 6A–D), indicating that complete loss of K36 or K4 methylation does not cause the increased histone acetylation at the 5′ end. Moreover, the increased acetylation observed in a bur2Δ strain is not reduced in bur2Δ set2Δ or Δbur2Δ set1Δ strains (Figure 6A–D), indicating that the increased acetylation is not dependent on either K36 or K4 methylation. Similarly, the increase in H3 and H4 acetylation at PMA1 in a paf1Δ strain was SET2 independent (Figure 6E and F). Based on these results, we conclude that the increased H3 and H4 acetylation observed near the 5′ ends of PMA1 and PYK1 in bur2Δ and paf1Δ strains does not require K36 methylation and is not caused by partial methylation of K36. It remains possible that the increased histone acetylation causes the reduced H3 K36 tri-methylation at the 5′ ends of genes, but a test of this hypothesis awaits identification of the responsible histone acetyltransferase (HAT) and HDAC.
Figure 6.
Increased acetylation at the 5′ end is independent of histone methylation in bur2Δ and paf1Δ strains. (A–D) ChIP analysis with antibodies against acetylated histones H3 and H4 in wild type, bur2Δ, bur2Δ set2Δ, bur2Δ set1Δ, set1Δ, and set2Δ strains. Levels of H3 acetylation and H4 acetylation were assessed at the 5′ ends of PMA1 (A, B) and PYK1 (C, D). (E, F) ChIP analysis of acetylated histone H3 and H4 in wild type, paf1Δ, paf1Δ set2Δ, and set2Δ strains at the 5′ end of PMA1 (E, F). Standard error is provided for values representing three experiments.
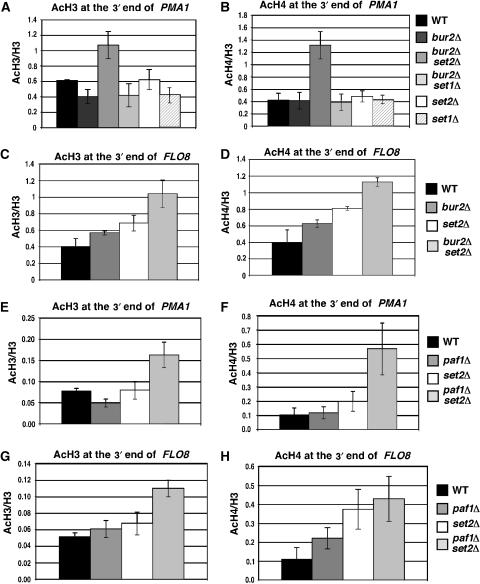
Bur2 and Paf1 function redundantly with Set2 in regulating histone acetylation
The major difference observed at PMA1 and PYK1 in bur2Δ and paf1Δ strains was that acetylation increased only near the 5′ end of PMA1 but at both the 5′ and 3′ ends of PYK1 (Figure 5). This could be because BUR2 and PAF1 do not control acetylation near the 3′ end of PMA1, or because they function redundantly with another pathway. An excellent candidate is the Set2–Rpd3S pathway that controls histone acetylation and the appearance of cryptic transcripts at the 3′ ends of some ORFs (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005). To investigate whether BUR2 and PAF1 are redundant with the Set2–Rpd3S pathway, acetylation at the 3′ end of PMA1 and FLO8 in bur2Δ and paf1Δ strains was compared to that of set2Δ, bur2Δ set2Δ, and paf1Δ set2Δ strains (Figure 7). set2Δ by itself did not increase histone H3 or H4 acetylation at the 3′ end of PMA1, consistent with previous observations that H3 and H4 acetylation did not increase at the 3′ end of PMA1 in a strain lacking the Eaf3 subunit of the Rpd3S complex (Reid et al, 2004; Joshi and Struhl, 2005; Pokholok et al, 2005). In bur2Δ set2Δ and paf1Δ set2Δ double mutants, acetylation was increased at PMA1 and FLO8 compared with the single mutants (Figure 7). These results indicate that Bur2 and Paf1 have redundant functions with K36 methylation that are required to inhibit acetylation at the 3′ ends of genes.
Figure 7.
Bur2 and Paf1 have redundant roles with Set2 in controlling histone acetylation at the 3′ end of ORFs. (A–D) ChIP analysis with antibodies against acetylated histones H3 and H4 in wild type, bur2Δ, bur2Δ set2Δ, bur2Δ set1Δ set1Δ, and set2Δ strains. Levels of H3 acetylation and H4 acetylation were assessed at the 3′ ends of PMA1 (A, B) and FLO8 (C, D). (E–H) ChIP analysis of acetylated histone H3 and H4 in wild type, paf1Δ, paf1Δ set2Δ, and set2Δ strains at the 3′ ends of PMA1 (E, F) and FLO8 (G, H). Standard error is provided for values representing three experiments.
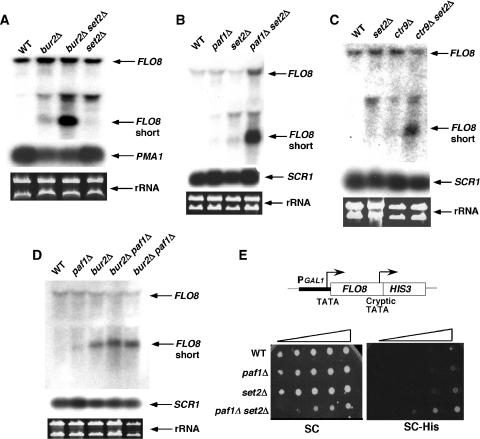
Bur2 and Paf1 function redundantly with Set2 in regulating cryptic transcription
To determine whether the increased acetylation at the 3′ end has any functional consequences, we tested for the appearance of cryptic internal transcripts in single and double mutants. The effect of bur2Δ, paf1Δ, and set2Δ was examined at FLO8, which has been characterized as a reporter for cryptic transcription initiation (Kaplan et al, 2003; Carrozza et al, 2005). Paralleling the effects observed on 3′ acetylation, the internal FLO8 cryptic transcript was detected at a low level in bur2Δ, paf1Δ, ctr9Δ, and set2Δ single mutants, but a strong combinatorial effect was observed in bur2Δ set2Δ, paf1Δ set2Δ, and ctr9Δ set2Δ double mutants (Figure 8A–C). As expected for components of the same linear pathway, combinatorial effects on cryptic transcription were not observed in bur2Δ paf1Δ double mutants (Figure 8D). These results were confirmed in two ways: by transforming BUR2 and SET2 plasmids into the bur2Δ set2Δ strain, and by using an in vivo assay in which the FLO8 cryptic TATA box drives expression of a HIS3 reporter (Prather et al, 2005). In the complementation assay, plasmids containing BUR2 or SET2 reduced internal FLO8 transcript levels in the bur2Δ set2Δ strain to the same level as observed in a set2Δ strain or a bur2Δ strain (data not shown), indicating that the increased cryptic transcription was due to bur2Δ and set2Δ, and not some unsuspected mutations in that strain. In the HIS3 reporter assay, paf1Δ and set2Δ individually caused an extremely weak His+ phenotype, whereas the paf1Δ set2Δ double mutant was stronger His+ (Figure 8E), confirming the northern results in vivo. Based on these combined results, the Bur1–Paf1 pathway and Set2 have redundant roles in regulating histone acetylation and cryptic transcription at the 3′ ends of some genes.
Figure 8.
The Bur1 and Paf1 complexes are redundant with Set2 for repressing cryptic transcription. (A–D) Northern blot analysis of total RNA prepared from the indicated strains. Filters were probed with sequences complementary to the 3′ region of FLO8 (+1672/+2399), PMA1 (+168/+2290), or SCR1. Ethidium bromide-stained gels showing the levels of ribosomal RNA serve as additional loading controls. The full-length and short cryptic transcript signals for FLO8 are indicated. (E) Strains with the indicated genotypes were tested for effects on an integrated pGAL1∷FLO8-HIS3 reporter gene (Prather et al, 2005). Ten-fold serial dilutions were spotted onto SC and SC-His plates and plates were incubated at 30°C for 3 days. His+ growth is indicative of transcription from the internal FLO8 promoter.
Discussion
Expanded functions of the Bur1–Paf1c pathway
The genetic screens that originally identified Bur1–Bur2 and the Paf1 complex subunits relied on very different mutant phenotypes (Reed et al, 1988; Prelich and Winston, 1993; Di Como et al, 1995; Stolinski et al, 1997; Costa and Arndt, 2000), providing no obvious indication that these proteins were functionally related. Subsequent studies, however, revealed extensive functional overlap between Bur1–Bur2 and the Paf1 complex, including shared sensitivity to base analogs, similar Spt− phenotypes (Murray et al, 2001; Krogan et al, 2002; Squazzo et al, 2002; Keogh et al, 2003), association with the coding regions of active genes (Krogan et al, 2002; Pokholok et al, 2002; Keogh et al, 2003), and regulation of H2B ubiquitylation and K4 methylation (Laribee et al, 2005; Wood et al, 2005). The results presented here identify new functions for the Bur1 and Paf1 complexes, revealing shared roles in promoting K36 tri-methylation, reducing H3 and H4 acetylation, and cooperating with Set2 to repress cryptic transcription. The number of shared characteristics strongly implies that the Bur1–Bur2 and Paf1 complexes are components of the same pathway for the regulation of histone methylation and acetylation. Indeed, Bur1–Bur2 is required for Paf1c occupancy at ORFs (Laribee et al, 2005; Wood et al, 2005; Qiu et al, 2006), although Paf1 is not required for Bur2 occupancy (Y Chu and G Prelich, data not shown). This ordering of the pathway suggests that the Paf1 complex might be a direct substrate for the Bur1 kinase. We have not been able to detect Bur1-dependent phosphorylation of the Paf1 complex in vitro, suggesting that Bur1 targets an additional component of the pathway. It is important to note that Bur1–Bur2 and the Paf1 complex also have roles that are independent of each other, exemplified by the near inviability of bur1Δ strains compared with the viable Paf1 complex deletion mutants, and by the observation that set2Δ partially suppresses bur1Δ and bur2Δ, yet causes synthetic growth defects with paf1Δ and ctr9Δ. The molecular basis for these differences is not understood, but must be due to some unshared function.
Paf1 and Ctr9 are required for proper histone H3 K36 tri-methylation
An ongoing question in the chromatin field is whether the mono-, di-, and tri-methylation marks on histones confer different functions. Our study was motivated by the idea that identifying factors that are specifically required for K36 tri-methylation would be useful for investigating its potential roles. Indeed, we found that, like mutations in BUR1, BUR2, and some pol II CTD truncations, PAF1 and CTR9 are required for K36 tri-methylation, with little, if any effects on total mono- or di-methyl K36 level. Strikingly, the K36 tri-methylation function of the Paf1 complex is strongly dependent on Paf1 and Ctr9, with Cdc73 having a moderate role and Rtf1 and Leo1 having no detectable effect on K36 tri-methylation levels in crude extracts or at specific genes. This pattern contrasts with the essential role that Rtf1 plays in the ubiquitylation of H2B and methylation of H3 K4 and K79 (Wood et al, 2003; Ng et al, 2003a) and further supports the idea that the Paf1 complex carries out multiple functions during transcription elongation.
For any histone modification, functional significance is best inferred from its differential localization or by the identification of factors that recognize specific modified forms. The spatial distribution of tri-methyl K36, however, is similar to that observed for di-methyl K36 (Bannister et al, 2005; Pokholok et al, 2005; Rao et al, 2005), although the tri-methyl K36 pattern correlates more closely with transcriptional frequency. Moreover, the chromodomain of Eaf3, which is the only factor currently known to bind K36, binds equally well to mono-, di-, and tri-methylated K36 peptides (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005). Thus, by these criteria, the functional significance of K36 tri-methylation remains unknown. In genome-wide analyses, the level of K36 tri-methylation in wild-type strains correlated with transcriptional activity (Pokholok et al, 2005), but we have not observed a strict correlation between changes in K36 tri-methylation level and transcription in bur2Δ or paf1Δ mutant strains. For example, K36 tri-methylation levels are reduced at both the 5′ and 3′ ends of FLO8 even though pol II occupancy increases. This suggests that the changes in K36 tri-methylation are not simply a consequence of altering transcription, but instead indicate that the Bur1–Paf1c pathway can affect the activity of Set2 or a K36 demethylase independent of transcription.
Potential crosstalk between histone acetylation and H3K36 methylation
Several precedents establish that one histone modification can affect the appearance of a second histone modification (Fischle et al, 2003). Particularly relevant to this study is the established role of H3 K36 methylation in recruiting the Rpd3S complex and consequently causing histone deacetylation at the 3′ ends of genes (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005). The more severe reduction of K36 tri-methylation in the 5′ region of four different ORFs in bur2Δ and paf1Δ strains and the correlation of reduced tri-methylation with increased acetylation in these strains suggested that a similar process might operate at the 5′ ends of genes. The links between acetylation and K36 methylation at the 5′ and 3′ ends of genes are mechanistically distinct, however, since acetylation at the 5′ end did not increase in a set2Δ strain, and the increased acetylation observed in bur2Δ and paf1Δ strains was unaffected by loss of SET2. The increased 5′ acetylation therefore is not caused by the loss of K36 methylation and does not require K36 methylation. Two other possibilities remain; the first is that the increased acetylation causes the decreased K36 tri-methylation, possibly by recruiting a K36 demethylase. Testing this model will require identification of the HAT, HDAC, and the K36 demethylase that act at the 5′ end. The second possibility is that the increased acetylation and decreased K36 tri-methylation are coincidental. This seems unlikely, however, since four out of five genes that have increased 5′ acetylation in bur2Δ or paf1Δ strains also have decreased K36 tri-methylation. The results presented here constitute another possible link between different histone modifications, indicate that the crosstalk between histone acetylation and H3 K36 methylation at the 5′ region is different from the one existing at the 3′ ends of ORFs, and raise the intriguing idea of a regulatory loop whereby acetylation reduces methylation at the 5′ end, and methylation reduces acetylation at the 3′ end via the Rpd3S pathway.
The Bur1–Paf1 pathway acts redundantly with Set2 for repressing cryptic transcription
Bur1–Bur2 and the Paf1 complex do not only function at the 5′ ends of genes. Both complexes localize throughout ORFs (Keogh et al, 2003; Mueller et al, 2004) and individually contribute to repression of internal cryptic transcription (Kaplan et al, 2003; Nourani et al, 2006). Because bur2Δ, paf1Δ, and set2Δ each increase cryptic transcription and reduce K36 methylation, it was possible that they all increased cryptic transcription through the Set2–Rpd3S pathway. Our results, however, indicate that this is not the case, since a significant increase in cryptic transcription occurs in both bur2Δ set2Δ and paf1Δ set2Δ double mutants relative to the individual mutants. We infer that the Bur1–Paf1c pathway represses cryptic transcription by a mechanism that is redundant with the Set2–Rpd3S pathway. This result raises the questions of how many pathways control cryptic transcription and what their components are. Other factors known to affect cryptic transcription include histones, Spt6, Spt16, Spt10, Spt2, Hir1, and Elf1 (Kaplan et al, 2003; Prather et al, 2005; Nourani et al, 2006). Double-mutant analyses such as those reported here will help to elaborate the organization of the pathways and to identify additional components that repress spurious transcription of the genome. Because increased cryptic transcription at the 3′ end of FLO8 correlates with increased acetylation in bur2Δ and paf1Δ strains, we infer that the redundant pathway impacts directly or indirectly upon a HAT or HDAC that functions within transcribed ORFs. Efforts are underway to identify the responsible factor(s).
Two of the key results presented here are that mutations in the Bur1–Paf1 pathway reduce Set2-dependent tri-methylation of K36, yet act independent of the Set2 pathway to block cryptic transcription. At first glance, these results might appear to be contradictory. Our interpretation is that the Bur1–Paf1 pathway regulates K36 tri-methylation, perhaps by directly stimulating Set2 or inhibiting a K36-specific demethylase, or indirectly by stimulating histone acetylation. Bur1–Bur2 and the Paf1 complex affect cryptic transcription through another mechanism that is redundant with the residual Set2-dependent mono-and di-methylation that occurs in bur2Δ and paf1Δ strains. The Bur1 and Paf1 complexes are clearly multifunctional and can impact different histone modifications. Although the redundant function that cooperates with Set2 to repress cryptic transcription remains unknown, these results provide a strong foundation for future investigation of the phenomenon.
Materials and methods
Strains and media
The S. cerevisiae strains used in this study are listed in Supplementary Table 1. All media, including yeast–peptone–dextrose (YPD), synthetic complete drop-out (e.g., SC-Ura), 5-fluoroorotic acid (5FOA), synthetic dextrose minimal (SD), and sporulation media were prepared as described previously (Rose et al, 1990). Standard genetic methods for mating, sporulation, transformation, and tetrad analysis were used throughout this study (Rose et al, 1990). Gene disruptions were constructed using a PCR-based method (Ausubel et al, 1997) and confirmed by PCR analysis. The C-terminus of Set2 was tagged with six copies of the HA epitope by PCR amplification from pYM3-6HA-klTRP1 (Knop et al, 1999), or with 13 copies of the c-Myc epitope by PCR amplification of pFA6a-13Myc-kanMX6 (Longtine et al, 1998), and transformation of the PCR product into a diploid strain. To test for genetic interactions between truncations of the pol II CTD and mutations in genes for members of the Paf1 complex, plasmid shuffle experiments were performed with LEU2-marked plasmids encoding wild-type Rpb1 (pRP114) or Rpb1 truncations containing 17 4/7 CTD repeats (pV7), 13 3/7 repeats (pV5), 11 2/7 repeats (pC1), 10 5/7 repeats (pC3), 9 3/7 repeats (pN15), or 6 3/7 repeats (pN56) (Nonet et al, 1987).
Western analysis
Preparation of yeast whole-cell lysates and western blotting were as described previously (Chu et al, 2006). Antibodies that recognize total histone H3 (ab1791), mono-methylated histone H3 K36 (ab9048), and tri-methylated histone H3 K36 (ab9050) were obtained from Abcam, and the antibody that recognizes di-methylated histone H3 K36 (07-369) was obtained from Upstate. Preparation of yeast extracts and western analysis of Set2-HA and Set2-Myc expression were performed as described previously (Shirra et al, 2005), using anti-HA antibody or anti-c-Myc antibody (Santa Cruz).
Chromatin immunoprecipitation
Yeast strains were grown at 30°C in YPD to a concentration of approximately 1–3 × 107 cells per ml. Chromatin was isolated and sonicated as described previously (Shirra et al, 2005). Anti-HA beads (Santa Cruz) or antibodies specific for polymerase II (8WG16 and H14) (Covance), c-Myc (Covance), histone H3 tri-methylated at K36, total H3, acetylated H3 (K9, K14) (Upstate, 06-599), or acetylated H4 (K5, K8, K12, K16) (Upstate, 06-866) were added to sonicated chromatin. Immunoprecipitation was performed for 4 h or overnight at 4°C. Immune complexes were captured by incubating with Protein A-coupled sepharose (Amersham Biosciences) for histone antibodies or Protein G-coupled sepharose (Amersham Biosciences) for c-Myc antibody and 8WG16 at 4°C for 2 h. Immune complexes with the H14 antibody were captured by incubating with Protein G-coupled sepharose beads (Sigma) at room temperature for 90 min. Precipitated DNA was purified using Qiagen PCR purification columns or phenol extraction and analyzed by PCR essentially as described (Komarnitsky et al, 2000; Keogh and Buratowski, 2004). Primers used for ChIP assays are listed in Supplementary Table 2. PCR signals were quantitated using a phosphorimager. For Set2-Myc ChIP assays, the average immunoprecipitated signal (IP) was divided by the average input signal for three independent chromatin preparations assayed in duplicate. For the recruitment of Set2-HA, pol II, and H3 occupancy, ChIP was quantified by normalizing band intensities for each sample using the following calculation: (specific gene IP/TEL IP)/(specific gene input/TEL input) as described (Komarnitsky et al, 2000; Carrozza et al, 2005). The background amplification of the chromosome V telomere region was performed as described (Komarnitsky et al, 2000). The normalized IP value calculated for H3 K36 tri-methyl, acetylated H3, or acetylated H4 was divided by the normalized IP value calculated for total H3.
RNA isolation and northern analysis
Cells were grown in YPD medium to a concentration of 1–2 × 107 cells per ml. Total RNA was isolated and analyzed by northern analysis as described previously (Swanson et al, 1991). DNA probes were prepared by random prime labeling of PCR products for PMA1 (+168/+2290), FLO8 (+1672/+2399), and SCR1.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Information
Acknowledgments
We thank Rick Young for providing CTD truncation plasmids and Jeff Brodsky for providing anti-Sse1 antibody. This work was supported by National Institutes of Health grants GM52486 to GP and GM52593 to KMA.
References
- Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1997) Current Protocols in Molecular Biology. New York: John Wiley & Sons [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2005) Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736 [DOI] [PubMed] [Google Scholar]
- Biswas D, Dutta-Biswas R, Mitra D, Shibata Y, Strahl BD, Formosa T, Stillman DJ (2006) Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J 25: 4479–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Chu Y, Sutton A, Sternglanz R, Prelich G (2006) The Bur1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by Set2. Mol Cell Biol 26: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PJ, Arndt KM (2000) Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156: 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Chang H, Arndt KT (1995) Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol 15: 1835–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD (2003) Histone and chromatin cross-talk. Curr Opin Cell Biol 15: 172–183 [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Speer JL, Lindstrom DL (2002) Transcript elongation on a nucleoprotein template. Biochim Biophys Acta 1577: 276–286 [DOI] [PubMed] [Google Scholar]
- Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM (1993) Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 ‘tail' to DNA. J Biol Chem 268: 305–314 [PubMed] [Google Scholar]
- Joshi AA, Struhl K (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F (2003) Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Buratowski S (2004) Using chromatin immunoprecipitation to map cotranscriptional mRNA processing in Saccharomyces cerevisiae. Methods Mol Biol 257: 1–16 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (2005) Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Podolny V, Buratowski S (2003) Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol 23: 7005–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD (2005) A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 25: 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A (2003a) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J (2003b) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sutton A, Hesman T, Min J, Xu RM, Johnston M, Sternglanz R (2003) Set2-catalyzed methylation of histone H3 represses basal expression of GAL4 in Saccharomyces cerevisiae. Mol Cell Biol 23: 5972–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD (2005) BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol 15: 1487–1493 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR III, Workman JL (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278: 8897–8903 [DOI] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP (2002) Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 277: 49383–49388 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J (2005) Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell 18: 723–734 [DOI] [PubMed] [Google Scholar]
- Mueller CL, Jaehning JA (2002) Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol 22: 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA (2004) The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 14: 447–456 [DOI] [PubMed] [Google Scholar]
- Murray S, Udupa R, Yao S, Hartzog G, Prelich G (2001) Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol Cell Biol 21: 4089–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K (2003a) The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem 278: 33625–33628 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003b) Targeted recruitment of Set1 histone methylase by elongating pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Nonet M, Sweetser D, Young RA (1987) Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 50: 909–915 [DOI] [PubMed] [Google Scholar]
- Nourani A, Robert F, Winston F (2006) Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol Cell Biol 26: 1496–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 9: 799–809 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Prather D, Krogan NJ, Emili A, Greenblatt JF, Winston F (2005) Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae. Mol Cell Biol 25: 10122–10135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G, Winston F (1993) Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics 135: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Wong CM, Hinnebusch AG (2006) The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol Cell Biol 26: 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao B, Shibata Y, Strahl BD, Lieb JD (2005) Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol 25: 9447–9459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI, Ferguson J, Jahng KY (1988) Isolation and characterization of two genes encoding yeast mating pheromone signaling elements: CDC72 and CDC73. Cold Spring Harb Symp Quant Biol 53 (Part 2): 621–627 [DOI] [PubMed] [Google Scholar]
- Reid JL, Moqtaderi Z, Struhl K (2004) Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol Cell Biol 24: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1990) Methods in Yeast Genetics: a Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Shirra MK, Rogers SE, Alexander DE, Arndt KM (2005) The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics 169: 1957–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA (2002) The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J 21: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolinski LA, Eisenmann DM, Arndt KM (1997) Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol 17: 4490–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F (1991) SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol 11: 4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A (2003) The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem 278: 34739–34742 [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A (2005) The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell 20: 589–599 [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67: 545–579 [DOI] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 17: 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol 25: 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Neiman A, Prelich G (2000) BUR1 and BUR2 encode a divergent cyclin-dependent kinase–cyclin complex important for transcription in vivo. Mol Cell Biol 20: 7080–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Kassabov SR, Bartholomew B (2006) Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol 13: 339–346 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Information