Abstract
EPI64 is a TBC domain–containing protein that binds the PDZ domains of EBP50, which binds ezrin, a major actin-binding protein of microvilli. High-resolution light microscopy revealed that ezrin and EBP50 localize exclusively to the membrane-surrounded region of microvilli, whereas EPI64 localizes to variable regions in the structures. Overexpressing EPI64 results in its and EBP50's relocalization to the base of microvilli, including to the actin rootlet devoid of ezrin or plasma membrane. Uncoupling EPI64's binding to EBP50, expression of any construct mislocalizing its TBC domain, or knock down of EBP50 results in loss of microvilli. The TBC domain of EPI64 binds directly to Arf6-GTP. Overexpressing the TBC domain increases Arf6-GTP levels, and expressing dominant-active Arf6 results in microvillar loss. These data reveal that microvilli have distinct cytoskeletal subdomains and that EPI64 regulates microvillar structure.
Introduction
Microvilli are slender F-actin–containing structures on the apical surface of many epithelial cells. Perhaps the best-studied examples are the densely packed microvilli of brush borders on intestinal and kidney proximal tubule epithelial cells, where microvilli are believed to increase the surface area for absorption. Less well ordered microvilli, having a somewhat different complement of actin binding proteins, are found on the apical aspect of other epithelial cells, such as the placental syncytiotrophoblast (Bretscher, 1991; Bartles, 2000). In neither case is the regulation of these structures understood.
One protein common to both types of microvilli is ezrin, a member of the ezrin/radixin/moesin (ERM) family of membrane-cytoskeletal linking proteins. Members of this family have an ∼300-residue N-terminal 4.1 ERM (FERM) domain, followed by a central ∼150-residue region predicted to be largely α-helical, and terminate in an ∼100-residue domain known as the C-ERMAD (C-terminal ERM association domain), as it has the ability to bind the FERM domain of all family members (Gary and Bretscher, 1993); an F-actin binding site lies in the last ∼30 residues of the C-ERMAD (Turunen et al., 1994; Pestonjamasp et al., 1995). ERM proteins are conformationally regulated, as the F-actin binding site in the C-ERMAD, and some sites for association with membrane proteins in the FERM domain, are masked in dormant molecules when these two domains are associated (Bretscher et al., 2002). Activation to release the association and expose these binding sites can occur through PIP2 binding and subsequent phosphorylation of a C-terminal threonine (567 in ezrin), found on the interface between the FERM domain and the C-ERMAD (Hirao et al., 1996; Matsui et al., 1998; Simons et al., 1998; Gautreau et al., 2000; Fievet et al., 2004).
The cytoskeletal-membrane linking properties of ERM proteins are attributed to their ability to bind F-actin through their C-terminal domain and membrane proteins, such as CD44, CD43, and ICAM-1-3, through their FERM domain (Tsukita et al., 1994; Helander et al., 1996; Hirao et al., 1996; Serrador et al., 1997, 1998; Heiska et al., 1998; Yonemura et al., 1998). In addition to this direct linkage with transmembrane proteins, the FERM domain binds the related scaffolding proteins EBP50 (ERM binding phosphoprotein of 50 kD)/NHERF1 and E3KARP/NHERF2, proteins enriched in epithelial microvilli (Reczek et al., 1997; Yun et al., 1997). These proteins consist of two N-terminal PDZ (postsynaptic density/95-discs large/zona occludens-1) domains and a C-terminal region that binds tightly to isolated FERM domains (Reczek et al., 1997). The EBP50 binding site on the FERM domain lies on the same surface occupied by the last helix of the C-ERMAD in the dormant protein, thereby providing a physical explanation for its masking in dormant ezrin (Reczek and Bretscher, 1998; Finnerty et al., 2004). EBP50 binds the C-terminal tails of many transmembrane proteins, including the CFTR, the β2-adrenergic receptor, and the PDGF receptor to regulate aspects of their function (Hall et al., 1998; Short et al., 1998; Cao et al., 1999; Moyer et al., 1999; Maudsley et al., 2000; James et al., 2004; Li et al., 2005). Previously, we identified EPI64 (EBP50-PDZ interactor of 64 kD) from extracts of placental microvilli, the richest known source of both ezrin and EBP50, as a protein that binds the PDZ domains of EBP50 (Reczek and Bretscher, 2001). EPI64 is a cytosolic protein of 508 residues containing an N-terminal Tre-2/Bub2/Cdc16 (TBC) domain and ending in the sequence DTYL, which binds preferentially to the first PDZ domain of EBP50 (Reczek and Bretscher, 2001).
The current model based on biochemical interactions (Fig. 1 A) suggests that microvilli should exhibit a uniform distribution of phosphorylated ezrin, EBP50, and EPI64 along their length. We used high-resolution fluorescence microscopy to assess this prediction and found that distinct subdomains exist in microvilli whose distribution can be regulated by EPI64. By exploring the effects of expressing different domains of EPI64, we found that when EPI64 is unable to be linked to ezrin, cells have a reduced number of microvilli. We have traced this phenomenon to the mislocalization of EPI64's TBC domain, which we find binds directly to Arf6-GTP.
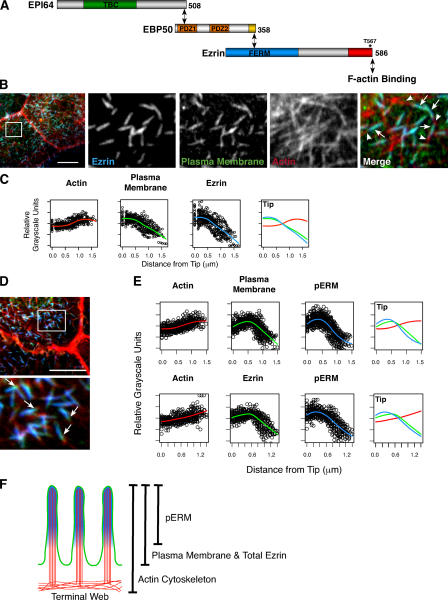
Figure 1.
Ezrin and phospho-ERM localization in microvilli. (A) Schematic representation of biochemical interactions between ezrin, EBP50, EPI64, and F-actin. The asterisk denotes the C-terminal T567 phosphorylation site in ezrin. (B) JEG-3 cells stained with antibodies to ezrin (blue), the plasma membrane with fluorescently conjugated WGA (green), and F-actin with fluorescently conjugated phalloidin (red). Individual channels of the boxed region are enlarged in the right panels. The arrows identify regions of ezrin localization with the plasma membrane, whereas the arrowheads identify the microvillar rootlet. (C) Fitted curves to the total dataset localizing ezrin, the plasma membrane, and F-actin regions in JEG-3 cell microvilli. Distance is measured from the microvillus tip to its base. (D) JEG-3 cells stained for pERM (blue), the plasma membrane (green), and F-actin (red); the boxed region is enlarged in the bottom panel. Arrows indicate membrane-bound regions weakly stained for pERM proteins. (E) Fitted curves to the total dataset localizing pERM, F-actin, and the plasma membrane regions in JEG-3 cell microvilli (top) or pERM, F-actin, and total ezrin (bottom). Unlike ezrin, pERM levels are more intense toward the microvillar tips and drop off before the plasma membrane (compare with C). (F) Schematic representation of microvilli depicting the localization of pERM (blue), F-actin (red), the plasma membrane, and total ezrin (green). Bars, 10 μm.
Results
Microvilli contain subdomains distinguished by the localization of ezrin and EPI64
To localize specific proteins in microvilli by immunofluorescence microscopy, 10–15 confocal sections 0.2 μm apart covering just the apical aspect of stained human JEG-3 syncytiotrophoblast cells were collected and merged. When the plasma membrane was stained with fluorescently tagged WGA and ezrin localized with specific antibodies, a high degree of colocalization was found. Comparing these with phalloidin staining for F-actin, the microvillar rootlet extending into the cell body was clearly visible, and this allowed us to determine the orientation of microvilli in triple-label studies (Fig. 1 B). To quantitate the degree of colocalization of three markers, the fluorescent intensity along the length of a minimum of 25 microvilli was determined, and the mean was assessed. It should be noted that because microvilli vary in length (from ∼0.75 to 1.25 μm), this mean gives a measure of colocalization but does not accurately reflect sharp transitions seen in individual microvilli (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200604046/DC1). As expected, the distribution of ezrin closely follows that of the plasma membrane (Fig. 1 C).
When the distribution of T567 phosphorylated ezrin (pERM) was compared with the plasma membrane, it was found that they were largely overlapping but that the pERM localization was slightly more enriched in the distal portion of microvilli and reduced toward their base (Fig. 1 D, arrows; and Fig. S1). Although this is a subtle effect in the means (Fig. 1 E), it is also seen in other cell types, such as intestinal Caco-2 epithelial cells and porcine kidney proximal tubule LLC-PK1 cells (not depicted). In similar analyses, EBP50 was found to precisely colocalize with the plasma membrane–like ezrin (Fig. 2 A).
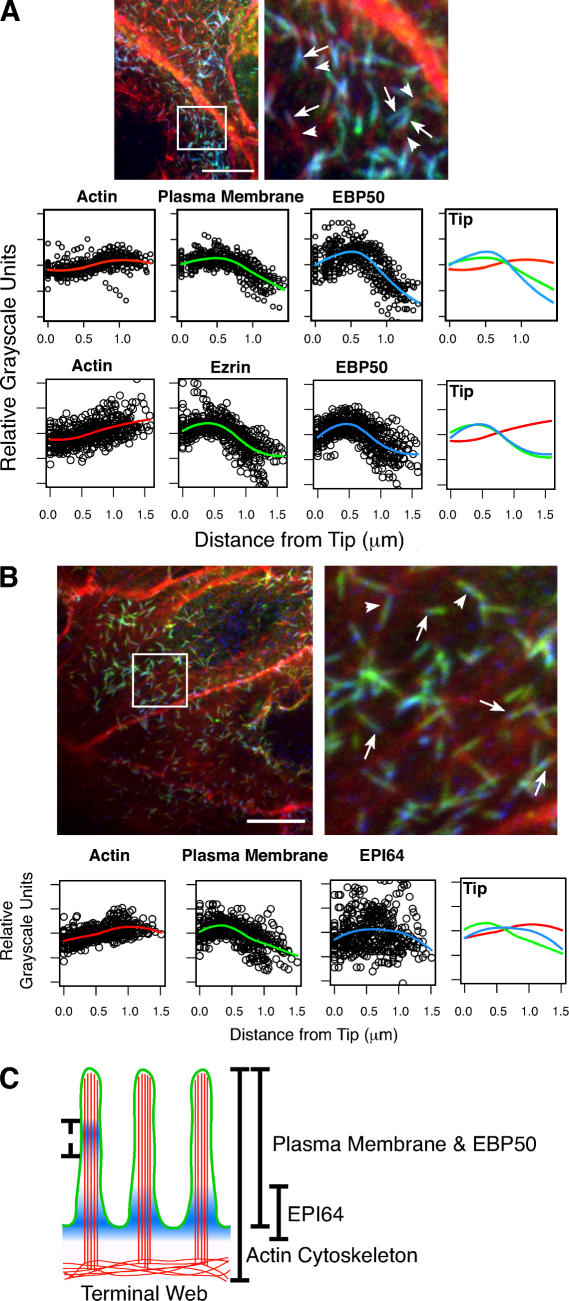
Figure 2.
EBP50 and EPI64 localization in microvilli. (A, top) JEG-3 cells stained for EBP50 (blue), F-actin (red), and the plasma membrane (green). The boxed region is enlarged in the right panel. Arrows identify regions of EBP50 localization with the plasma membrane, and arrowheads identify the microvillar rootlet. (bottom) Fitted curves to the total dataset localizing EBP50, F-actin, and the plasma membrane or ezrin regions in JEG-3 cell microvilli. (B, top) JEG-3 cells stained for EPI64 (blue), F-actin (red), and the plasma membrane (green). The boxed region is enlarged in the right panel. Arrows identify regions of EPI64 localization near microvilli bases; instances of more distal staining can also be seen (arrowheads). (bottom) Fitted curves to the total dataset localizing EPI64, F-actin, and the plasma membrane regions in JEG-3 cell microvilli. Distance is measured from a microvillus tip to its base. (C) Schematic representation of microvilli depicting the localization of EPI64 (blue), F-actin (red), and the plasma membrane and EBP50 (green). The dashed line indicates occasional additional localization for EPI64. Bars, 10 μm.
In contrast to the localizations of ezrin and EBP50 in JEG-3 cell microvilli, the localization of EPI64 was more variable. Traces of individual microvilli never showed a clear colocalization with other markers but, rather, were enriched in different regions of individual microvilli (Fig. 2 B and Fig. S1). As such, the mean distribution curve shows no specific localization, though the peak of the curve is near the microvilli bases. Remarkably, EPI64 was sometimes enriched in the region of the microvillus rootlet with no ezrin or plasma membrane colocalization.
Overexpression of EPI64 affects microvillar domains
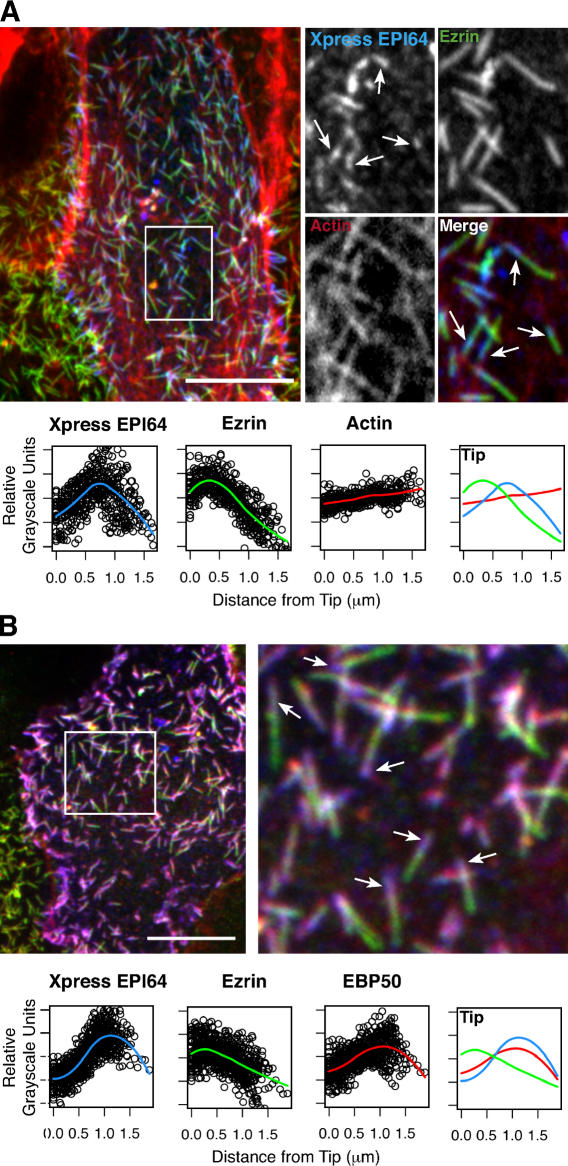
To begin examining how microvillar subdomains arise, we investigated the effects of overexpressing EPI64 and EBP50. Overexpressing EPI64 resulted in the majority of microvilli exhibiting enrichment of EPI64 in a region immediately below ezrin and associated with the microvillar rootlet (Fig. 3 A, arrows). Moreover, overexpressing EPI64 caused EBP50 to now be largely coincident with EPI64 and the distribution of both EBP50 and EPI64 to be distinct from that of ezrin (Fig. 3 B). Thus, overexpressing EPI64 results in redistribution of EBP50 and enhances the distinction between microvillar subdomains. In contrast to EPI64, overexpressing EBP50 had no discernable effect on the distribution of ezrin, pERM, EBP50, or EPI64 in JEG-3 cells (unpublished data). We therefore focused our studies on the role of EPI64.
Figure 3.
Overexpressed EPI64 localizes to the bases of microvilli and causes a redistribution of EBP50. (A, top) JEG-3 cells transfected with EPI64 (containing an N-terminal Xpress tag) and stained for the tag (blue), F-actin (red), and ezrin (green). A transfected cell is shown in the middle, surrounded by nontransfected ones. Individual channels and the merge of the boxed region are enlarged to the right. Arrows identify regions of EPI64 localization near microvilli bases, flanked by red and green staining on either side. (bottom) Fitted curves to the total dataset. The distribution of ezrin does not appear affected by the overexpression of EPI64. (B, top) JEG-3 cells transfected with tagged EPI64 and stained for the tag (blue), anti-ezrin (green), and anti-EBP50 (red). The boxed region is enlarged to the right. The arrows identify regions of EPI64 localization near microvilli bases, which appear purple because of colocalization with EBP50. (bottom) Fitted curves to the total dataset. Bars, 10 μm.
The TBC domain of EPI64 is a regulator of microvilli
To further investigate the role of EPI64, we generated several N-terminally tagged mutants (Fig. 4 A). Because the C-terminal DTYL sequence of EPI64 binds the PDZ domains of EBP50, we made a mutant (EPI64-LA) with an additional C-terminal alanine known to abolish this interaction (Reczek and Bretscher, 2001). Some TBC domains are known to accelerate the catalytic activity of Rab GTPases through a catalytic arginine finger motif (Neuwald, 1997; Albert et al., 1999), so we mutated this residue, arginine 160 in EPI64, to generate EPI64-R160A. In addition, we made an N-terminal construct (EPI64-NT) containing the TBC domain and a C-terminal construct (EPI64-CT) consisting of the C-terminal 189 residues. The expression of these constructs modestly enhanced the level of EPI64 expression as revealed by Western blotting (Fig. 4 B).
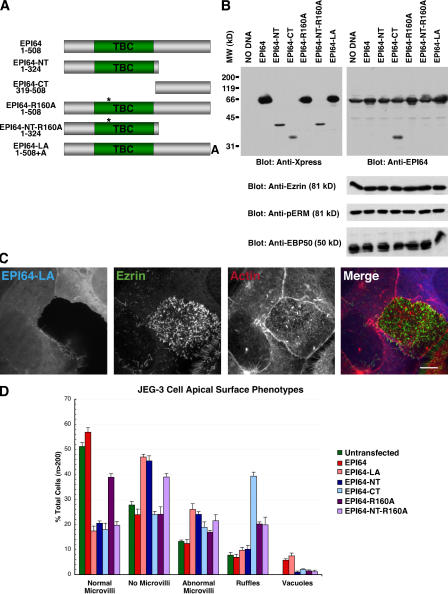
Figure 4.
Mislocalization of EPI64 causes a loss of microvilli in JEG-3 cells. (A) Schematic representation of the EPI64 constructs introduced into cells. The asterisk denotes the putative catalytic arginine, R160. (B) The constructs shown in A were transfected into JEG-3 cells and allowed to express for ∼24 h. 25 μg of total lysates were separated by 12% SDS-PAGE followed by a Western blot. (bottom) Levels of ezrin, pERM, and EBP50 in transfected cells. (C) JEG-3 cells were transfected with tagged EPI64-LA for 18 h and stained for the tag (blue), ezrin (green), and F-actin (red). Merged confocal sections of the apical regions are shown. Bar, 10 μm. (D) Quantification of cells transfected with tagged EPI64 constructs. Cells were stained for the tag, ezrin, and F-actin and scored as described in Materials and methods. Error bars indicate mean ± SD.
Overexpressing EPI64-LA resulted in the reduction or loss of microvilli from JEG-3 cells as seen by ezrin or F-actin staining, without noticeably affecting the levels of ezrin, pERM, or EBP50 in these cells (Fig. 4 B). Instead of being enriched in the apical aspect of cells, like overexpressed wild-type EPI64, EPI64-LA was found diffuse in the cytoplasm (Fig. 4 C). To quantify the microvillar loss phenotype, at least 200 transfected cells were assessed for the presence of microvilli, with 46% of the transfected cells lacking or exhibiting greatly reduced numbers of microvilli, compared with 25% of untransfected or wild-type EPI64 overexpressing cells that had reduced numbers of microvilli. In addition, a small percentage of EPI64 and EPI64-LA overexpressing cells contained vacuoles (Fig. 4 D), which are discussed in the next section. In contrast to EPI64-LA, EPI64-R160A overexpression was similar to overexpressing wild-type EPI64.
To investigate the mechanism of microvillar loss by EPI64-LA, we overexpressed domains of the protein. Overexpressing EPI64-CT resulted in a reduction of microvilli and an increase in the number of ruffling cells (Fig. 4 D), with the ruffles enriched in the transfected protein (not depicted). Similar to overexpressing EPI64-LA, overexpressing EPI64-NT, containing the TBC domain, resulted in microvillar loss. A similar effect was seen in cells expressing a mutant TBC domain, EPI64-NT-R160A (Fig. 4 D). To explore whether the loss of microvilli resulted from a defect in cell polarity, the localization of the tight junction marker ZO-1 and the adherens junction marker E-cadherin was examined in cells overexpressing EPI64 or EPI64-LA. In neither case did cell junctions appear significantly different between transfected and nontransfected cells (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200604046/DC1).
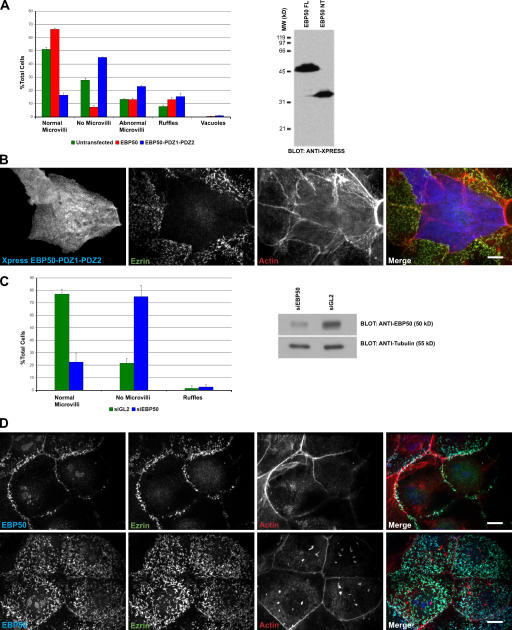
The data imply that mislocalizing the TBC domain from the apical region results in loss of microvilli. Because perturbing the binding of EBP50 to ezrin should reproduce this effect, we explored the consequences of overexpressing EBP50-PDZ1-PDZ2, which lacks the C-terminal ezrin binding site. Consistent with this notion, whereas overexpressing wild-type EBP50 had no effect on microvilli, overexpressing EBP50-PDZ1-PDZ2 resulted in the loss of microvilli (Fig. 5, A and B). Moreover, knock down of EBP50 by siRNA treatment for 48 h also resulted in the loss of surface microvilli, whereas treatment with control siRNA did not (Fig. 5, C and D). Interestingly, the microvilli located above the adherens junctions were the most resistant to EBP50 knockdown, perhaps suggesting that the lateral border provides a signal for microvillus formation that can normally be propagated across the apical surface.
Figure 5.
Uncoupling EBP50 from ezrin results in loss of microvilli. (A) Cells were transfected with either Xpress-tagged EBP50 or EBP50-PDZ1-PDZ2 and stained for the tag, ezrin, and F-actin, and scored as described in Materials and methods. (right) Western blot of total cell lysates probed with antibodies to the Xpress tag epitope. (B) Cells were transfected to Xpress-tagged EBP50-PDZ1-PDZ2 and stained for the tag, ezrin and F-actin. (C) Cells were transfected with siRNA to EBP50 (siEBP50) or luciferase (siGL2) and either stained for EBP50, ezrin, and actin or collected for Western blot analysis. (left) Results of scoring cells for the presence of microvilli; (right) Western blot of total cell lysates probed with antibodies to EBP50 and β-tubulin. Error bars indicate mean ± SD. (D) Localization of the indicated proteins in cells treated with siEBP50 (top) and siGL2 (bottom). Bars, 10 μm.
Overexpressing EPI64 and EPI64-LA can cause the production of F-actin–coated vacuoles
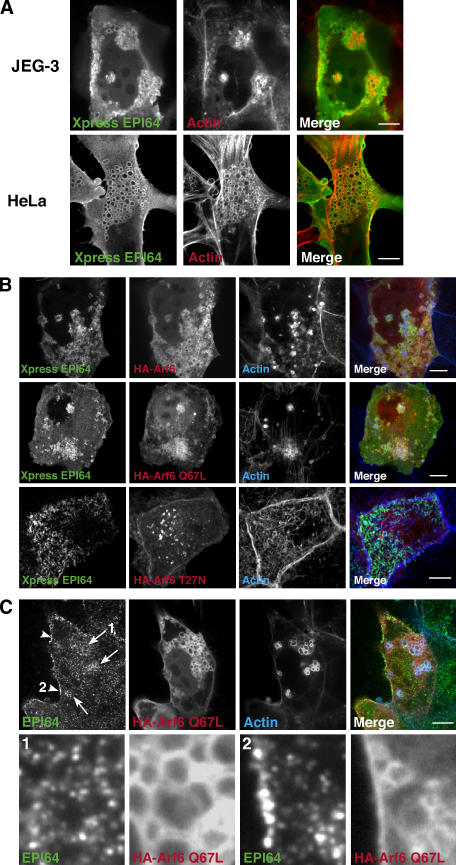
Overexpressing the full-length EPI64 constructs caused a small percentage of cells to contain intracellular vacuoles often decorated by EPI64 and F-actin both in JEG-3 cells but more dramatically in HeLa cells (Fig. 6 A). Because similar F-actin– decorated structures are seen in cells overexpressing dominant-active Arf6 (Brown et al., 2001), we cotransfected tagged EPI64 and Arf6 and found that the vacuoles were also enriched in Arf6 (Fig. 6 B), suggesting a relationship between Arf6 and EPI64. We thus examined cells expressing EPI64 and wild type or dominant-active (Q67L) or dominant-negative (T27N) mutants of Arf6. EPI64 colocalized on vacuoles with both Arf6 and Arf6 Q67L, but not with Arf6 T27N (Fig. 6 B). An enrichment of endogenous EPI64 is seen in the region of vacuoles produced by Arf6 Q67L overexpression and on areas of the plasma membrane to which it localizes, suggesting that Arf6 may have the ability to recruit EPI64 to these regions (Fig. 6 C).
Figure 6.
Overexpression of EPI64 or EPI64-LA: role of Arf6. (A) JEG-3 or HeLa cells were transfected to Xpress-tagged EPI64. After 24 h, cells were stained for the tag (green) and F-actin (red). The vacuoles also sometimes stain for ezrin and EBP50 (not depicted). (B) JEG-3 cells were cotransfected with Xpress-tagged EPI64 and HA-Arf6, HA-Arf6 Q67L, or HA-Arf6 T27N and stained for the Xpress-tagged EPI64 (green), F-actin (blue), and HA-tagged Arf6 (red). A projection through the cell is shown. EPI64 can still be seen in the cell microvilli in cells expressing Arf6 T27N. (C) Endogenous EPI64 is enriched in regions expressing HA-Arf6 Q67L. Enrichment can be seen on intracellular vacuoles (arrows) and on the plasma membrane (arrowheads). (bottom) Enlargement of the regions indicated by the arrow labeled 1 and the arrowhead labeled 2. Bars, 10 μm.
The TBC domain of EPI64 binds active Arf6
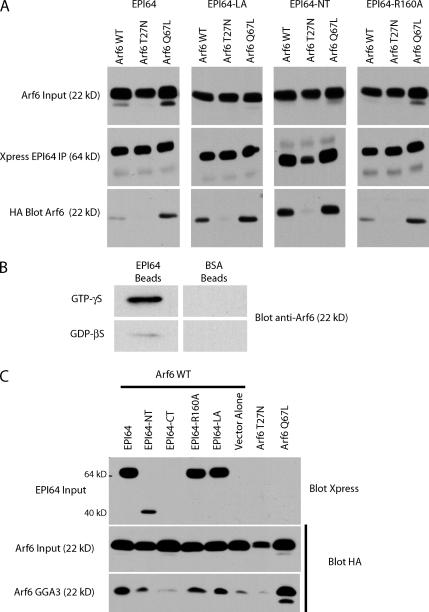
It has been found that the TBC domain of TRE17 binds Arf6-GDP (Martinu et al., 2004). Thus, we explored the possibility that Arf6 binds to the TBC domain of EPI64. Cells were transfected with Xpress-tagged EPI64 and HA-tagged Arf6, Arf6 T27N, or Arf6 Q67L. Immunoprecipitates of EPI64 contained a small amount of wild-type Arf6, were enriched in Arf6 Q67L, and lacked Arf6 T27N (Fig. 7 A). Immunoprecipitates from cells expressing EPI64-LA or -NT recovered both Arf6 and Arf6 Q67L efficiently, whereas Arf6 T27N was absent (Fig. 7 A). Notably, there was a consistent increase in the relative amount of wild-type Arf6 recovered by these constructs when compared with EPI64, indicating that the mutated proteins have increased access, or bind more efficiently, to Arf6-GTP. In addition, both constructs also decorated Arf6 Q67L–induced vacuoles, as is seen for EPI64 (unpublished data). In coimmunoprecipitation experiments, EPI64-R160A behaved indistinguishably from EPI64 (Fig. 7 A), suggesting that the R160A mutation has no effect on Arf6 binding.
Figure 7.
EPI64 interacts directly with Arf6. (A) EPI64 interacts through its TBC domain with Arf6-GTP. Xpress-tagged EPI64 constructs and HA-Arf6 wild type, HA-Arf6 T27N, or HA-Arf6 Q67L were cotransfected into JEG-3 cells for 6 h, after which Xpress-EPI64 was immunoprecipitated and immunoprecipitates were blotted for the tagged Arf6. Arf6 input is 4% of input to immunoprecipitates. A representative example of three independent experiments is shown. (B) Recombinant EPI64 immobilized on Sepharose beads binds preferentially to purified myristoylated Arf6-GTPγS over -GDPβS. No binding of Arf6 is seen on control BSA beads. (C) EPI64 is not a GAP for Arf6. Wild-type HA-tagged Arf6 was coexpressed in JEG-3 cells with Xpress-tagged EPI64 constructs or with an empty vector. Cells were lysed, and the amount of HA-Arf6 recovered on GST-GGA3 resin was determined by Western blotting. Arf6 input is 2% of lysate used for binding assay. A representative example of three independent experiments is shown.
An in vitro binding assay using purified recombinant proteins was used to determine whether the interaction between EPI64 and Arf6-GTP is direct (Fig. 7 B). Immobilized EPI64 bound Arf6-GTP with a greater affinity than Arf6-GDP, whereas no binding was observed to immobilized BSA. Collectively, these results indicate that, unlike the TBC domain of TRE17, which preferentially binds Arf6-GDP (Martinu et al., 2004), the TBC domain of EPI64 specifically binds Arf6-GTP.
To evaluate whether EPI64 might be a GTPase activating protein (GAP) for Arf6, we assessed the level of Arf6-GTP in cells by making use of the ability of GST-GGA3 to bind selectively to Arf6-GTP (Dell'Angelica et al., 2000). Cells were transfected with HA-tagged Arf6, together with full-length EPI64 or mutants, and lysed. Lysates were incubated with GST-GGA3 beads, and the amount of bound Arf6-GTP was determined. Expression of all constructs containing the TBC domain did not reduce the level of Arf6-GTP and, in fact, enhanced it, when compared with the vector alone control (Fig. 7 C), making it unlikely that the TBC domain of EPI64 is a GAP for Arf6.
Arf6 regulates the abundance of microvilli
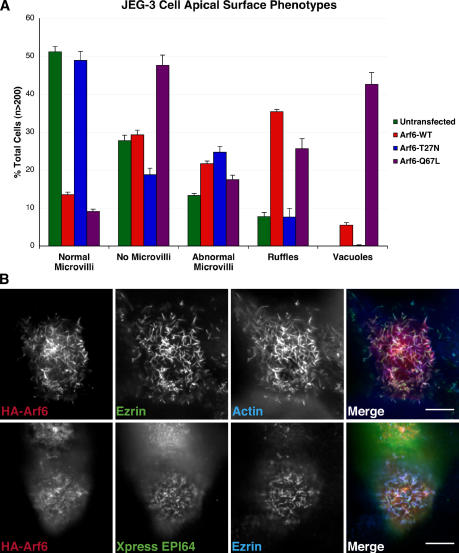
The connection between EPI64 and Arf6 prompted us to explore the effect of expressing wild-type, dominant-active, or dominant-negative Arf6 on microvilli of JEG-3 cells. Overexpressing wild-type Arf6 and Arf6 Q67L both led to microvillar loss, whereas the dominant-negative Arf6-T27N had no effect (Fig. 8 A). Dominant-active Arf6 also greatly enhanced the percentage of cells with vacuoles, as reported previously (Brown et al., 2001). In cells transfected to express HA-Arf6 yet still containing microvilli, the Arf6 colocalized with ezrin and EPI64 in the apical microvilli (Fig. 8 B), confirming observations of Arf6 at the apical surface of epithelial cells (Altschuler et al., 1999).
Figure 8.
Arf6 localizes to microvilli, and its overexpression results in loss of microvilli. (A) JEG-3 cells transfected to express HA-Arf6 constructs were stained for the tag, ezrin, and F-actin. Cells were scored as described in Materials and methods. Error bars demonstrate mean ± SD. (B) JEG-3 cells transfected with HA-tagged wild-type Arf6 for 6 h and stained for the HA tag (red), ezrin (green), and F-actin (blue). (bottom) JEG-3 cells were transfected with HA-Arf6 and Xpress-EPI64 for 6 h and stained for the HA-tag (red), Xpress tag (green), and ezrin (blue). Bars, 10 μm.
Discussion
We have uncovered two unexpected findings relating to the organization of the apical domain of epithelial cells. First, using high-resolution light microscopy, we found that the microvilli of epithelial cells do not have a uniform composition but, rather, have distinguishable subdomains that are influenced by mild overexpression of EPI64. Second, we have found that expressing an EPI64 construct unable to bind the PDZ domains of EBP50 results in a dramatic loss of microvilli. Moreover, knock down of EBP50, or expression of an EBP50 construct that binds EPI64 but not ezrin, also results in the loss of microvilli. Thus, our studies suggest that mislocalization of EPI64 results in loss of microvilli. Because a major recognizable feature in EPI64 is the presence of a TBC domain that we show binds directly to Arf6-GTP, it is likely that Arf6 is also involved, directly or indirectly, in regulating microvilli on cells.
As far as we are aware, all previous studies have suggested that microvilli are of uniform cytoskeletal composition along their length. Here, we demonstrate that phospho-T567 ezrin is enriched toward the distal ends of microvilli, whereas total ezrin seems to be uniformly distributed, colocalizing with the plasma membrane marker WGA. Additionally, EPI64 is not evenly distributed in microvilli but appears to be enriched toward the middle of some and at the base of others. What could account for this uneven distribution? It has been shown that the microvilli of cultured cells are not static structures but, rather, undergo growth and retraction over a period of minutes (Gorelik et al., 2003). The actin filaments in microvilli, in which the barbed ends are associated with the tip, are presumably treadmilling, as has been documented in other systems (Tyska and Mooseker, 2002; Loomis et al., 2003). For example, ezrin might become activated at the microvillus tip, associate with the newly assembled filaments there, and ride down the microvillus with treadmilling actin and become dephosphorylated as it nears the base. In this way, its membrane-cytoskeletal linking role might be inactivated. The finding that EBP50 colocalizes with ezrin along the membrane-associated part of the microvillus, extending slightly beyond the region of pERM, was also initially surprising because EBP50 is proposed to only bind activated ezrin molecules (Reczek and Bretscher, 1998). Perhaps as ezrin becomes dephosphorylated it takes time to return to its closed form (depending on the rates of dissociation from actin and the membrane) and so retains some ability to bind EBP50 after dephosphorylation.
Analysis of cells overexpressing EPI64 reinforces the idea that microvilli have different domains: ezrin is found colocalizing with the plasma membrane of microvilli, whereas EBP50 and EPI64 are now enriched on the rootlet ∼0.25 μm below the membrane; how this enrichment might occur remains unknown. To investigate how EPI64 alters the distribution of proteins in microvilli, we began to study its functional domains. To evaluate the importance of the C-terminal linkage of EPI64 to the PDZ domains of EBP50, we examined cells expressing EPI64 with an additional single alanine residue to abolish binding to EBP50. Surprisingly, the transfected cells lost microvilli, indicating that the proper localization of EPI64 is necessary for microvillar maintenance. By expressing domains of EPI64, we found that expressing any construct mislocalizing the TBC domain resulted in microvillar loss.
Sequence comparisons suggest that the TBC domain of EPI64 might have RabGAP activity (Reczek and Bretscher, 2001). However, biochemical studies did not identify any GAP activity on several purified Rab proteins using full-length recombinant EPI64 (unpublished data; see the following paragraph). A clue to the possible role of the TBC domain came from two sources. First, we noticed that in 5–10% of cells overexpressing EPI64 vacuoles formed, often associated with F-actin, highly reminiscent of the vacuoles seen in dominant-active Arf6-expressing cells (Brown et al., 2001). Second, it was recently reported that the TBC domain of TRE17 binds to Arf6-GDP (Martinu et al., 2004). Immunoprecipitation experiments and in vitro studies using recombinant proteins revealed that the TBC domain of EPI64 also binds Arf6, but with a preference for Arf6-GTP over Arf6-GDP. Initially, we wondered if EPI64 was a GAP for Arf6. RabGAPs have a conserved arginine residue necessary for enhancing the hydrolysis of the γ-phosphate of GTP (Albert et al., 1999). Cells expressing EPI64-R160A, a construct in which this catalytic arginine was mutated to alanine, showed no reduction in microvilli, and the level of Arf6 associated with EPI64 was not diminished. Indeed, overexpression of either EPI64 or EPI64-R160A resulted in an enhanced level of Arf6- GTP. Therefore, EPI64 is not a GAP for Arf6 and, because EPI64 has a preference for Arf6-GTP, it is not a guanine nucleotide exchange factor for Arf6 either. At present, we believe that EPI64 is an effector of Arf6-GTP and that, by binding Arf6-GTP, it protects it from inactivation by a GAP.
While this manuscript was in the final stages of review, Itoh and Fukuda (2006) reported the important finding that EPI64 is a GAP for Rab27A and that this activity is abolished by a R160K mutation. Combining their results with ours implies that the TBC domain of EPI64 has two distinct functions: as a RabGAP for Rab27A and as a binding domain for Arf6-GTP that is unaffected by the R160A mutation.
Why does expression of EPI64 defective in binding EBP50 result in microvillar loss? Loss of microvilli is also seen in cells transfected to express just the PDZ domains of EBP50 or in cells with EBP50 levels reduced by siRNA treatment, thereby breaking the linkage to ezrin. Furthermore, overexpressing wild-type Arf6 or its mutant that binds GTP constitutively also results in microvillar loss. Because overexpression of the dominant-active Arf6 results in the formation of actin-containing vacuoles in the majority of cells, it is possible that the loss of microvilli is simply due to sequestration of F-actin internally. However, we do not think this is very likely, as overexpression of wild-type Arf6, which induces a very much lower level of vacuoles, also results in microvillar loss, including cells where no vacuoles are evident. Although these data suggest a correlation between elevated levels of mislocalized Arf6-GTP and the loss of microvilli, we also attempted to explore the consequences of Arf6 depletion by siRNA. Although we were able to knock down expression of transfected HA-Arf6 about fivefold, no effect on microvilli was seen in siRNA-treated HA-Arf6 expressing or otherwise wild-type cells (unpublished data). Whether these data suggest that microvilli can exist in the absence of Arf6 function, which is consistent with the lack of interference by expression of the dominant-negative Arf6-T27N mutant, or that Arf6 was not sufficiently reduced, remains an open question. If Arf6 is directly relevant to the regulation of microvilli, an attractive scenario is that Arf6-GDP is returned to the plasma membrane through a membrane-recycling pathway, where it is acted upon at the plasma membrane by Arf6 guanine nucleotide exchange factors such as ARNO or EFA6 (Franco et al., 1998, 1999) and then captured by EPI64. Microvillar formation or maintenance might first require capture of Arf6-GTP by EPI64, followed by Arf6 inactivation, when it is brought in close proximity to a microvillar ARF-GAP. Consistent with this model, we found that the association of Arf6-GTP with EPI64 was elevated in mutants defective in microvillar localization. Moreover, in a preliminary report, a novel Arf6 GAP (ARF-GAP) was shown to bind the activated form of ezrin (Wu, F., H. Deng, R. Zhou, Z. Guo, Z. Fan, K. Yuan, X. Cao, J. Forte, and X. Yao. 2005. American Society for Cell Biology Annual Meeting. Abstr. 1804).
Among the known effectors of Arf6 is phosphatidylinositol-4 phosphate 5 kinase-α (PI[4]P5Kα), which generates PIP2 at the plasma membrane (Honda et al., 1999). Excess PI(4)P5Kα, by either overexpression of the protein or overexpression of dominant-active Arf6, induces the formation of vacuoles rich in PIP2 and F-actin (Brown et al., 2001). This appears to be a consequence of greatly enhanced endocytosis and decreased membrane recycling through an actin-dependent mechanism, thereby depleting PIP2 from the plasma membrane, ultimately resulting in multiple phenotypic consequences (Brown et al., 2001; Aikawa and Martin, 2003). The similar phenotype conferred by EPI64 overexpression suggests that it drives this system by enhancing the level of Arf6-GTP, to hyperactivate PI(4)P5Kα to generate excess PIP2. Our results suggest that localized capturing or cycling of EPI64-Arf6-GTP is necessary for the maintenance of microvilli. If this is the case, one possibility is that PI(4)P5K has to be locally activated to maintain microvilli. As it has been reported that PIP2 binding to ezrin is the first step in its activation (Fievet et al., 2004), one attractive model is that transient locally elevated levels of PIP2 might be responsible for initiating or maintaining active ezrin necessary for microvilli.
In summary, we have shown that microvilli have subdomains and uncovered an involvement of EPI64 in regulating the presence of microvilli on the cell surface. We have also shown that the EPI64's TBC domain binds Arf6-GTP, which is the second example of a TBC domain interacting with Arf6, indicating that the function of TBC domains may extend beyond their RabGAP activities to encompass other small GTPases involved in membrane trafficking. A recent report suggested that actin organization and clathrin-mediated endocytosis at the apical surface of polarized epithelial cells are regulated by Arf6 (Hyman et al., 2006). The collected data suggest that EPI64 integrates actin cytoskeletal organization and membrane trafficking events mediated by Arf6 and Rab27A, thereby placing it at a pivotal point in balancing these processes. It will be fascinating to further unravel the connection between microvillar structure, endocytosis, and these proteins; such studies are under way.
Materials and methods
Cell culture
JEG-3 and HeLa cells (American Type Culture Collection) were maintained in a 5% CO2 humidified atmosphere at 37°C in MEM (Invitrogen) with 10% FBS (Invitrogen).
Antibodies and other reagents
Antisera and affinity-purified antibodies against human ezrin and EBP50 were described previously (Bretscher, 1989; Reczek et al., 1997). Hexahistidine-tagged EPI64 was expressed in insect cells, purified, and used to elicit antibody production in rabbits. The affinity-purified antibody recognized a single band at 64 kD in total extracts of several cultured cells (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200604046/DC1). Specific antibodies for phospho-T567 ezrin/T564 radixin/T558 moesin were obtained from Cell Signaling Technology or generated against the peptide CRDKYK(Tp)LRQIR (Matsui et al., 1998). Antibodies against the Xpress epitope were purchased from Invitrogen (anti-Xpress) or Santa Cruz Biotechnology, Inc. (OmniProbe M-21). Anti-HA (HA.11) was obtained from Covance Research Products. Mouse anti-Arf6 was provided by C. D'Souza-Schorey (University of Notre Dame, Notre Dame, Indiana). Mouse anti–ZO-1 and anti–E-cadherin were obtained from BD Biosciences. The antibody against tubulin (N356) was purchased from GE Healthcare. Donkey anti–rabbit Alexa Fluor 488, goat anti–mouse Alexa Fluor 568, goat anti–rabbit Rhodamine Red-X, fluorescently conjugated phalloidin, and WGA were obtained from Invitrogen. Donkey anti–rabbit and anti–mouse Cy5 were obtained from Jackson ImmunoResearch Laboratories. Goat anti–rabbit HRP was obtained from MP Biomedicals. GTPγS and GDPβS were obtained from Sigma-Aldrich. The siRNAs targeting human EBP50 (5′-CGGCGAAAACGTGGAGAAG-3′) and Luciferase GL2 (5′-CGUACGCGGAAUACUUCGA-3′) were obtained from Dharmacon.
Western blotting and immunofluorescence
Protein samples were separated by SDS-PAGE and transferred to Immobilon-P (Millipore). Western blotting was done as described previously (Reczek and Bretscher, 2001). For immunofluorescence, cells grown on glass coverslips were fixed in 3.7% formaldehyde/PBS for 10 min at room temperature. Cells were permeabilized in 0.2% Triton X-100/PBS for 5 min at room temperature, rinsed in PBS, and incubated with primary antibodies in PBS/2% FBS. After washing in PBS, secondary antibodies and additional markers (phalloidin and/or WGA) were added in PBS/2% FBS (unless WGA was included, in which case, FBS was omitted). Cells were mounted on glass slides in Vectashield (Vector Laboratories) and observed on a microscope (Eclipse TE-2000U; Nikon) using a 100× 1.4 NA lens (Nikon) on a confocal imaging system (UltraView LCI; PerkinElmer). Z-series images of single focal planes were obtained at 0.2-μm steps through the cells on a 12-bit digital output charge-coupled device camera (C4742-95-12ERG; Hamamatsu). Maximum projections of apical sections and identical contrast enhancements to all images were performed using ImageJ (NIH).
Measurements of microvillar protein distribution
Fluorescence intensity grayscale levels along the length of at least five random microvilli on each of at least five different cells were measured by selecting a linear region of interest along the length of each microvillus from its tip to actin rootlet (or the opposite end of staining if not stained for actin) using the UltraView software and recorded in Excel (Microsoft). R (http://www.R-project.org) was used to analyze protein distribution trends in the microvilli of each dataset. In brief, the mean intensity of the dataset was subtracted from each recorded point to center the numbers on a common point on the y axis (y = 0). These new numbers were used to generate a scatter plot of all data points followed by the application of a smooth curve fitted by LOESS linear regression.
DNA constructs
Xpress-tagged EPI64 and EPI64-LA constructs in pcDNA 3.1 His/Xpress were described previously (Reczek and Bretscher, 2001). Xpress-tagged EPI64 constructs expressing the N-terminal (residues 1–324, including the TBC domain) or C-terminal halves (residues 319–508) of EPI64 were generated by PCR. Xpress-tagged EPI64-R160A and EPI64-NT-R160A were made by PCR mutagenesis. Xpress-tagged EBP50 constructs were derived by subcloning from previously described GST constructs (Reczek and Bretscher, 1998) into pcDNA3.1 His/Xpress. HA-tagged Arf6 constructs were a gift from J. Donaldson (National Institutes of Health, Bethesda, MD; Peters et al., 1995). The construct expressing GST-GGA3VHS-GAT was a gift from J. Bonifacino (National Institutes of Health, Bethesda, MD; Dell'Angelica et al., 2000).
Cell transfection
Polyethylenimine (PEI) has been shown to be a nontoxic transfection reagent (Durocher et al., 2002). JEG-3 cells were seeded in 60-mm dishes to be 60–70% confluent at the time of transfection. 1 μg of DNA was mixed with 50 μl of serum and antibiotic-free MEM and 5 μl of 1 mg/ml PEI Linear (mol wt 25,000; Polysciences, Inc.), previously dissolved in water at room temperature for 10 min. Cells were washed with serum and antibiotic-free MEM and overlaid with the DNA/MEM/PEI mixture in 2 ml of serum and antibiotic-free MEM and incubated under normal growing conditions. After 4 h, 1.6 ml of serum and antibiotic-free medium and 400 μl of FBS was added to the cells. Cells were allowed to recover for 24–48 h. For 6-h transfections, cells 80–90% confluent were washed and incubated for 2 h with the DNA/MEM/PEI mixture before the supplementation of additional MEM and FBS. Under the conditions used, ∼50% of the cells were transfected.
siRNA transfection
JEG-3 cells were seeded in 60-mm dishes to be 30% confluent at the time of transfection. Cells were washed twice with PBS before adding 4 ml of OptiMEM medium (Invitrogen). 2.5 μl of 20 μM Block-iT Alexa Fluor Red Fluorescent Oligo (Invitrogen) and 2.5 μl of 20 μM duplex siRNA targeting either EBP50 or Luciferase (GL2) for control were diluted into 500 μl OptiMEM. 5 μl of Lipofectamine RNAiMAX (Invitrogen) was diluted into 500 μl OptiMEM per reaction, mixed with diluted siRNA, and incubated at room temperature for 15 min. The resulting 1-ml volume of Lipofectamine complexed with siRNA was added to cells and incubated at 37°C. After 4 h, the medium was replaced with 4 ml of MEM with 10% FBS and l-glutamine. 48 h after transfection, cells were either fixed and stained for immunofluorescence or lysed for Western blot analysis.
Scoring the apical surface phenotype of transfected cells
Transfected JEG-3 cells were stained for ezrin, actin, and the expressed protein (for identifying transfected cells) to indicate the overall apical membrane phenotype. At least 200 cells were counted for each experiment and scored as normal, lacking/few microvilli, or ruffling. In instances where cells were ruffling but still had normal-looking microvilli, cells were scored as ruffling. Where surface structures did not clearly fall into the above categories, cells were scored as having abnormal microvilli. The siRNA-treated JEG-3 cells containing Alexa Fluor red fluorescent oligo (to identify transfected cells) and siRNA targeting either EBP50 or GL2 were stained for ezrin. 150 cells were counted for each experiment and scored as described.
Coimmunoprecipitation of EPI64 and Arf6
After a 6-h transfection in 100-mm dishes, JEG-3 cells were washed in prewarmed PBS at 37°C and treated for 2 min at 37°C with 5 ml of freshly made 1.25 mM DSP in PBS (Pierce Chemical Co.) to cross-link proteins. Cells were then washed in prewarmed TBS (50 mM Tris, pH 7.4, and 150 mM NaCl) at 37°C and incubated in 10 ml of fresh TBS for an additional 15 min at 37°C to quench the cross-linking reaction. Each dish was lysed in 1 ml of ice-cold RIPA buffer (0.1% SDS, 1% Triton X-100, 1% deoxycholate, 150 mM NaCl, 1 mM EDTA, and 25 mM Tris, pH 7.4) with protease inhibitors and clarified by centrifugation at 100,000 g in a TLA 100.3 rotor for 10 min. After removal of 100 μl of clarified supernatant, as a total lysate fraction, the remainder was added to a 25 μl volume of protein A Sepharose (Sigma-Aldrich) and 5 μg of anti-Xpress OmniProbe M-21 and incubated at 4°C for 2 h with gentle inversion. Bead-bound proteins were washed with ice-cold RIPA buffer and eluted by boiling in 50 μl of 2× SDS sample buffer.
In vitro binding assay
Hexahistidine-tagged EPI64 was purified from insect cells using standard methods and coupled to CNBr-activated Sepharose beads at ∼1 mg/ml as described previously (Bretscher, 1983). Purified myristoylated Arf6 was provided by P. Randazzo (National Cancer Institute, Bethesda, MD; Ha et al., 2005). Purified Arf6 was subjected to nucleotide exchange in the presence of GTPγS or GDPβS as described previously (D'Souza-Schorey et al., 1997). A molar excess of Arf6-GTP or -GDP was incubated with 25 μl of EPI64 or BSA beads for 2 h at 4°C with gentle inversion in binding buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 2 mM MgCl2, and 1 mM DTT). The bound protein was then washed extensively with ice-cold binding buffer and eluted by boiling in 50 μl of 2× SDS sample buffer. The eluted protein was separated by 12% SDS-PAGE and analyzed by Western blotting using anti-Arf6 antibodies.
Detection of relative levels of Arf6-GTP
A GST-GGA3 binding assay was used to determine the relative levels of Arf6-GTP (Santy and Casanova, 2001). In brief, the different EPI64 constructs were cotransfected with HA-tagged Arf6 into JEG-3 cells grown on 100-mm dishes. After 18–24 h, cells were lysed in 1 ml of ice-cold GGA3 buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 1% wt/vol Triton X-100, and protease inhibitors). Lysates were cleared by centrifugation at 16,000 g for 5 min at 4°C, and the resulting supernatant was applied to 50 μg of GST-GGA3VHS-GAT previously immobilized on glutathione Sepharose for 1 h at 4°C with continuous gentle inversion to bind Arf6-GTP to GGA3. The beads were washed in ice-cold GGA3 buffer, and proteins were eluted by boiling in sample buffer, followed by SDS-PAGE and Western blotting.
Online supplemental material
Fig. S1 shows sample plots of individual microvillar protein distributions for ezrin, pERM, and EPI64 with respect to the actin cytoskeleton and plasma membrane. Fig. S2 shows a Western blot of cell lysates demonstrating the specificity of the EPI64 antiserum compared with those detecting ezrin and EBP50. Fig. S3 shows immunofluorescent images of JEG-3 cells overexpressing EPI64 or EPI64-LA counterstained for the cell junction markers ZO-1 and E-cadherin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200604046/DC1.
Supplementary Material
Acknowledgments
We thank Julie Donaldson for supplying the Arf6 constructs, Juan Bonifacino for the GST-GGA3 construct, Paul Randazzo for providing purified Arf6, Crislyn D'Souza-Schorey for anti-Arf6 antibodies, James Booth (Cornell University) for help with statistical analysis, and Tal Ilani and Raghuvir Viswanatha for comments on the manuscript.
This work was supported by the National Institutes of Health grant GM36652.
D. Reczek's present address is Genzyme Corporation, Framingham, MA 01701.
D.N. Chamber's present address is Department of Biology, Concord University, Athens, WV 24712.
Abbreviations used in this paper: C-ERMAD, C-terminal ERM association domain; EBP50, ERM binding phosphoprotein of 50 kD; EPI64, EBP50 PDZ interactor of 64 kD; ERM, ezrin/radixin/moesin; FERM, 4.1 ERM; PDZ, postsynaptic density/95-discs large/zona occludens-1; GAP, GTPase activating protein; TBC, Tre-2/Bub2/Cdc16.
References
- Aikawa, Y., and T.F. Martin. 2003. ARF6 regulates a plasma membrane pool of phosphatidylinositol(4,5)bisphosphate required for regulated exocytosis. J. Cell Biol. 162:647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, S., E. Will, and D. Gallwitz. 1999. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 18:5216–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler, Y., S. Liu, L. Katz, K. Tang, S. Hardy, F. Brodsky, G. Apodaca, and K. Mostov. 1999. ADP-ribosylation factor 6 and endocytosis at the apical surface of Madin-Darby canine kidney cells. J. Cell Biol. 147:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartles, J.R. 2000. Parallel actin bundles and their multiple actin-bundling proteins. Curr. Opin. Cell Biol. 12:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher, A. 1983. Purification of an 80,000-dalton protein that is a component of the isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J. Cell Biol. 97:425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher, A. 1989. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J. Cell Biol. 108:921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher, A. 1991. Microfilament structure and function in the cortical cytoskeleton. Annu. Rev. Cell Biol. 7:337–374. [DOI] [PubMed] [Google Scholar]
- Bretscher, A., K. Edwards, and R.G. Fehon. 2002. ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 3:586–599. [DOI] [PubMed] [Google Scholar]
- Brown, F.D., A.L. Rozelle, H.L. Yin, T. Balla, and J.G. Donaldson. 2001. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154:1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, T.T., H.W. Deacon, D. Reczek, A. Bretscher, and M. von Zastrow. 1999. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature. 401:286–290. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E.C., R. Puertollano, C. Mullins, R.C. Aguilar, J.D. Vargas, L.M. Hartnell, and J.S. Bonifacino. 2000. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., R.L. Boshans, M. McDonough, P.D. Stahl, and L. Van Aelst. 1997. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 16:5445–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30:E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fievet, B.T., A. Gautreau, C. Roy, L. Del Maestro, P. Mangeat, D. Louvard, and M. Arpin. 2004. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J. Cell Biol. 164:653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty, C.M., D. Chambers, J. Ingraffea, H.R. Faber, P.A. Karplus, and A. Bretscher. 2004. The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J. Cell Sci. 117:1547–1552. [DOI] [PubMed] [Google Scholar]
- Franco, M., P.J. Peters, J. Boretto, E. van Donselaar, A. Neri, C. D'Souza-Schorey, and P. Chavrier. 1999. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 18:1480–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, S., S. Upender, S.H. Hansen, and J.E. Casanova. 1998. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J. Biol. Chem. 273:23–27. [DOI] [PubMed] [Google Scholar]
- Gary, R., and A. Bretscher. 1993. Heterotypic and homotypic associations between ezrin and moesin, two putative membrane-cytoskeletal linking proteins. Proc. Natl. Acad. Sci. USA. 90:10846–10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau, A., D. Louvard, and M. Arpin. 2000. Morphogenic effects of ezrin require a phosphorylation-induced transition from oligomers to monomers at the plasma membrane. J. Cell Biol. 150:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik, J., A.I. Shevchuk, G.I. Frolenkov, I.A. Diakonov, M.J. Lab, C.J. Kros, G.P. Richardson, I. Vodyanoy, C.R. Edwards, D. Klenerman, and Y.E. Korchev. 2003. Dynamic assembly of surface structures in living cells. Proc. Natl. Acad. Sci. USA. 100:5819–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, V.L., G.M. Thomas, S. Stauffer, and P.A. Randazzo. 2005. Preparation of myristoylated Arf1 and Arf6. Methods Enzymol. 404:164–174. [DOI] [PubMed] [Google Scholar]
- Hall, R.A., R.T. Premont, C.W. Chow, J.T. Blitzer, J.A. Pitcher, A. Claing, R.H. Stoffel, L.S. Barak, S. Shenolikar, E.J. Weinman, et al. 1998. The beta2-adrenergic receptor interacts with the Na+/H+-exchanger regulatory factor to control Na+/H+ exchange. Nature. 392:626–630. [DOI] [PubMed] [Google Scholar]
- Heiska, L., K. Alfthan, M. Gronholm, P. Vilja, A. Vaheri, and O. Carpen. 1998. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J. Biol. Chem. 273:21893–21900. [DOI] [PubMed] [Google Scholar]
- Helander, T.S., O. Carpen, O. Turunen, P.E. Kovanen, A. Vaheri, and T. Timonen. 1996. ICAM-2 redistributed by ezrin as a target for killer cells. Nature. 382:265–268. [DOI] [PubMed] [Google Scholar]
- Hirao, M., N. Sato, T. Kondo, S. Yonemura, M. Monden, T. Sasaki, Y. Takai, and S. Tsukita. 1996. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J. Cell Biol. 135:37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, A., M. Nogami, T. Yokozeki, M. Yamazaki, H. Nakamura, H. Watanabe, K. Kawamoto, K. Nakayama, A.J. Morris, M.A. Frohman, and Y. Kanaho. 1999. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 99:521–532. [DOI] [PubMed] [Google Scholar]
- Hyman, T., M. Shmuel, and Y. Altschuler. 2006. Actin is required for endocytosis at the apical surface of Madin-Darby canine kidney cells where ARF6 and clathrin regulate the actin cytoskeleton. Mol. Biol. Cell. 17:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, T., and M. Fukuda. 2006. Identification of EPI64 as a GTPase-activating protein specific for Rab27A. J. Biol. Chem. 281:31823–31831. [DOI] [PubMed] [Google Scholar]
- James, M.F., R.L. Beauchamp, N. Manchanda, A. Kazlauskas, and V. Ramesh. 2004. A NHERF binding site links the betaPDGFR to the cytoskeleton and regulates cell spreading and migration. J. Cell Sci. 117:2951–2961. [DOI] [PubMed] [Google Scholar]
- Li, J., Z. Dai, D. Jana, D.J. Callaway, and Z. Bu. 2005. Ezrin controls the macromolecular complexes formed between an adapter protein Na+/H+ exchanger regulatory factor and the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 280:37634–37643. [DOI] [PubMed] [Google Scholar]
- Loomis, P.A., L. Zheng, G. Sekerkova, B. Changyaleket, E. Mugnaini, and J.R. Bartles. 2003. Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J. Cell Biol. 163:1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinu, L., J.M. Masuda-Robens, S.E. Robertson, L.C. Santy, J.E. Casanova, and M.M. Chou. 2004. The TBC (Tre-2/Bub2/Cdc16) domain protein TRE17 regulates plasma membrane-endosomal trafficking through activation of Arf6. Mol. Cell. Biol. 24:9752–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, T., M. Maeda, Y. Doi, S. Yonemura, M. Amano, K. Kaibuchi, and S. Tsukita. 1998. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J. Cell Biol. 140:647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudsley, S., A.M. Zamah, N. Rahman, J.T. Blitzer, L.M. Luttrell, R.J. Lefkowitz, and R.A. Hall. 2000. Platelet-derived growth factor receptor association with Na(+)/H(+) exchanger regulatory factor potentiates receptor activity. Mol. Cell. Biol. 20:8352–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, B.D., J. Denton, K.H. Karlson, D. Reynolds, S. Wang, J.E. Mickle, M. Milewski, G.R. Cutting, W.B. Guggino, M. Li, and B.A. Stanton. 1999. A PDZ-interacting domain in CFTR is an apical membrane polarization signal. J. Clin. Invest. 104:1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald, A.F. 1997. A shared domain between a spindle assembly checkpoint protein and Ypt/Rab-specific GTPase-activators. Trends Biochem. Sci. 22:243–244. [DOI] [PubMed] [Google Scholar]
- Pestonjamasp, K., M.R. Amieva, C.P. Strassel, W.M. Nauseef, H. Furthmayr, and E.J. Luna. 1995. Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol. Biol. Cell. 6:247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, P.J., V.W. Hsu, C.E. Ooi, D. Finazzi, S.B. Teal, V. Oorschot, J.G. Donaldson, and R.D. Klausner. 1995. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J. Cell Biol. 128:1003–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek, D., and A. Bretscher. 1998. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J. Biol. Chem. 273:18452–18458. [DOI] [PubMed] [Google Scholar]
- Reczek, D., and A. Bretscher. 2001. Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP. J. Cell Biol. 153:191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek, D., M. Berryman, and A. Bretscher. 1997. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J. Cell Biol. 139:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy, L.C., and J.E. Casanova. 2001. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrador, J.M., J.L. Alonso-Lebrero, M.A. del Pozo, H. Furthmayr, R. Schwartz-Albiez, J. Calvo, F. Lozano, and F. Sanchez-Madrid. 1997. Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J. Cell Biol. 138:1409–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrador, J.M., M. Nieto, J.L. Alonso-Lebrero, M.A. del Pozo, J. Calvo, H. Furthmayr, R. Schwartz-Albiez, F. Lozano, R. Gonzalez-Amaro, P. Sanchez-Mateos, and F. Sanchez-Madrid. 1998. CD43 interacts with moesin and ezrin and regulates its redistribution to the uropods of T lymphocytes at the cell-cell contacts. Blood. 91:4632–4644. [PubMed] [Google Scholar]
- Short, D.B., K.W. Trotter, D. Reczek, S.M. Kreda, A. Bretscher, R.C. Boucher, M.J. Stutts, and S.L. Milgram. 1998. An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J. Biol. Chem. 273:19797–19801. [DOI] [PubMed] [Google Scholar]
- Simons, P.C., S.F. Pietromonaco, D. Reczek, A. Bretscher, and L. Elias. 1998. C-terminal threonine phosphorylation activates ERM proteins to link the cell's cortical lipid bilayer to the cytoskeleton. Biochem. Biophys. Res. Commun. 253:561–565. [DOI] [PubMed] [Google Scholar]
- Tsukita, S., K. Oishi, N. Sato, J. Sagara, and A. Kawai. 1994. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J. Cell Biol. 126:391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen, O., T. Wahlstrom, and A. Vaheri. 1994. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J. Cell Biol. 126:1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska, M.J., and M.S. Mooseker. 2002. MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys. J. 82:1869–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura, S., M. Hirao, Y. Doi, N. Takahashi, T. Kondo, and S. Tsukita. 1998. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 140:885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, C.H., S. Oh, M. Zizak, D. Steplock, S. Tsao, C.M. Tse, E.J. Weinman, and M. Donowitz. 1997. cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc. Natl. Acad. Sci. USA. 94:3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.