Abstract
OPA1, a dynamin-related guanosine triphosphatase mutated in dominant optic atrophy, is required for the fusion of mitochondria. Proteolytic cleavage by the mitochondrial processing peptidase generates long isoforms from eight messenger RNA (mRNA) splice forms, whereas further cleavages at protease sites S1 and S2 generate short forms. Using OPA1-null cells, we developed a cellular system to study how individual OPA1 splice forms function in mitochondrial fusion. Only mRNA splice forms that generate a long isoform in addition to one or more short isoforms support substantial mitochondrial fusion activity. On their own, long and short OPA1 isoforms have little activity, but, when coexpressed, they functionally complement each other. Loss of mitochondrial membrane potential destabilizes the long isoforms and enhances the cleavage of OPA1 at S1 but not S2. Cleavage at S2 is regulated by the i-AAA protease Yme1L. Our results suggest that mammalian cells have multiple pathways to control mitochondrial fusion through regulation of the spectrum of OPA1 isoforms.
Introduction
Mitochondrial fusion plays an important role in controlling the shape and function of mitochondria (Okamoto and Shaw, 2005; Chan, 2006). In mammalian cells, the dynamin-related GTPase OPA1 is essential for mitochondrial fusion (Cipolat et al., 2004; Chen et al., 2005). The yeast OPA1 orthologue Mgm1 is also essential for fusion (Wong et al., 2000, 2003; Sesaki et al., 2003b) and has been shown to form oligomers important for tethering and fusion of the inner membranes (Meeusen et al., 2006). OPA1 is associated with the inner membrane and protects cells from apoptosis by regulating inner membrane dynamics (Olichon et al., 2003; Frezza et al., 2006). Mutation of OPA1 causes the disease dominant optic atrophy, a degeneration of the retinal ganglion cells (Alexander et al., 2000; Delettre et al., 2000).
Both OPA1 and Mgm1 undergo proteolytic processing, and, in the case of Mgm1, such processing has been shown to be essential for mitochondrial fusion activity (Herlan et al., 2003; McQuibban et al., 2003; Sesaki et al., 2003a; Ishihara et al., 2006). Yeast Mgm1 is produced as a precursor with a mitochondrial leader sequence that is cleaved by the mitochondrial processing peptidase (MPP). Mgm1 processed by only MPP leads to the long isoform, l-Mgm1. The mitochondrial rhomboid Pcp1/Rbd1 further cleaves a subset of Mgm1 to form the short isoform, s-Mgm1 (Herlan et al., 2003; McQuibban et al., 2003; Sesaki et al., 2003a). Loss of Pcp1 greatly reduces mitochondrial fusion because a mixture of both the long and short Mgm1 isoforms is essential for normal activity (Herlan et al., 2003).
Several issues concerning OPA1 processing remain enigmatic. In contrast to the two isoforms produced by Mgm1, OPA1 produces many more isoforms. OPA1 is encoded by a complicated set of at least eight mRNA splice forms that are produced by differential splicing (Delettre et al., 2001). In addition to the MPP processing site, the polypeptides encoded by each mRNA splice form contain an S1 cleavage site, and some also contain a more C-terminal S2 cleavage site (Fig. 1 A; Ishihara et al., 2006). In principle, therefore, each mRNA splice form can produce a long isoform (produced by cleavage with MPP alone) and one or more short isoforms (produced by cleavage at S1 or S2). The proteases acting at sites S1 and S2 are poorly understood. There is evidence for the involvement of both the rhomboid protease presenilin-associated rhomboid-like (PARL) and the m-AAA protease paraplegin in OPA1 processing (Cipolat et al., 2006; Ishihara et al., 2006). However, cells lacking PARL or paraplegin have normal OPA1 processing (Duvezin-Caubet et al., 2007), suggesting that other proteases remain to be identified. In addition, when reconstituted in yeast cells, OPA1 cleavage appears to depend on two other m-AAA proteases, Afg3L1 and Afg3L2 (Duvezin-Caubet et al., 2007). In contrast to Mgm1, it has been suggested that the long form of OPA1 isoform 1 is the fusion-active species (Ishihara et al., 2006). As a result, it is unclear whether proteolytic processing is simply a way to inactive OPA1 or whether the short isoforms have another function. To clarify these issues, we developed a cellular system to study the role of specific OPA1 isoforms in mitochondrial fusion. We find that a combination of long and short OPA1 isoforms is important for mitochondrial fusion activity. Moreover, we find that the i-AAA protease Yme1L regulates OPA1 processing.
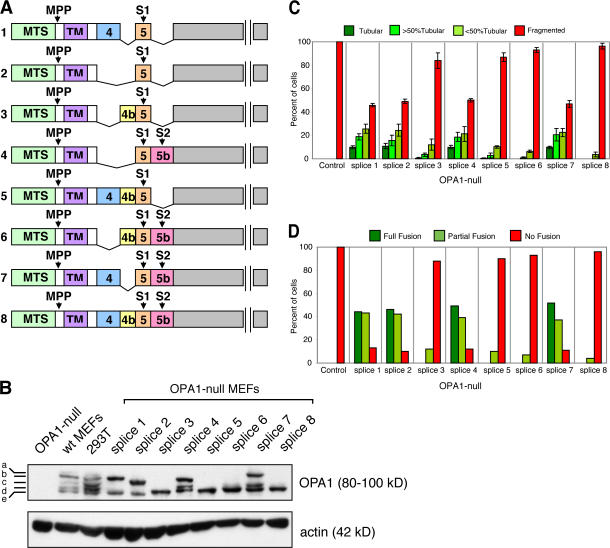
Figure 1.
OPA1 mRNA splice forms that are partially processed have substantial mitochondrial fusion activity in isolation. (A) Schematic of the eight OPA1 mRNA splice forms. The mRNA splice forms differ in the presence or absence of exons 4, 4b, and 5b. Cleavage of the mitochondrial targeting sequence (MTS) by MPP leads to the long isoforms. Additional cleavage at sites S1 (exon 5) or S2 (exon 5b) leads to the short isoforms. TM, transmembrane. (B) Processing of polypeptides encoded by individual OPA1 mRNA splice forms. The eight OPA1 mRNA splice forms were expressed in OPA1-null MEFs, which contain no OPA1 protein. OPA1 was detected by Western blot analysis with an anti-OPA1 antibody. The five bands detected in wild-type cells are indicated. Actin was used as a loading control. (C) Tubulation of mitochondria in OPA1-null cells by individual OPA1 mRNA splice forms. The eight OPA1 mRNA splice forms were expressed in OPA1-null cells containing mitochondrially targeted DsRed, and mitochondrial morphology was scored according to the criteria detailed in Materials and methods. 100 cells were scored in each experiment; error bars indicate SD in three independent experiments. (D) Mitochondrial fusion activity of individual OPA1 mRNA splice forms. The eight OPA1 splice forms were expressed in OPA1-null cells, and mitochondrial fusion was scored in the PEG cell hybrid assay at 7 h after cell fusion.
Results and discussion
Mitochondrial fusion activity of distinct OPA1 mRNA splice forms
As a result of eight mRNA splice forms (Delettre et al., 2001) and subsequent proteolytic processing, OPA1 isoforms migrate as a complex mixture of at least five bands (a–e) on gel electrophoresis (Fig. 1, A and B). The bands a and b are thought to be a mixture of long isoforms of OPA1, whereas the shorter bands (c–e) are thought to result from additional proteolytic processing (Ishihara et al., 2006). The complexity of this mixture prevents the definitive assignment of function to specific mRNA splice forms and their processed polypeptides. To circumvent these complications, we analyzed the mitochondrial fusion activity of the eight known human OPA1 mRNA splice forms upon expression in OPA1-null mouse embryonic fibroblasts (MEFs; Fig. 1). The parental OPA1-null cells contain no OPA1 protein and have completely fragmented mitochondria as a result of the lack of mitochondria fusion (unpublished data; Fig. 1, B and C). Of the eight OPA1 mRNA splice forms, the expression of isoforms 1, 2, 4, and 7 result in the robust tubulation of mitochondria in OPA1-null cells (Fig. 1 C). In contrast, splice forms 3, 5, 6, and 8 have modest or barely detectable tubulation activity. The ability of OPA1 splice forms to tubulate mitochondria in OPA1-null cells correlates well with their mitochondrial fusion activity in a polyethylene glycol (PEG)–induced cell hybrid assay (Fig. 1 D and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200704110/DC1).
Interestingly, all of the mRNA splice forms with high levels of fusion activity produce a long form of OPA1 in addition to one or more further processed short forms (Fig. 1 B). All OPA1 mRNA splice forms encode a protease processing site in exon 5 (S1 site), and some contain a second site (S2) in the alternative exon 5b (Ishihara et al., 2006). mRNA splice forms 1 and 2 encode only site S1 and yield a single long and a single short form. mRNA splice forms 4 and 7 encode sites S1 and S2 and therefore yield two short forms in addition to a single long form. These results illustrate the complexity of analyzing endogenous OPA1 bands from wild-type cells. The top two bands (a and b) are generally thought to be the two long forms of OPA1, but our analysis indicates that there are at least four species of long forms.
In contrast, all of the mRNA isoforms with little fusion activity are processed with greater efficiency to yield only short forms of OPA1 (Fig. 1 B). These results suggest that the short isoforms produced by cleavage at S1 or S2 have little activity on their own. Interestingly, all of the mRNA splice forms producing highly processed polypeptides (3, 5, 6, and 8) contain exon 4b (Fig. 1 A). Polypeptides produced from mRNA splice forms 1 and 2 are partially processed, whereas those from splice forms 5 and 3, which are identical, respectively, except for the 4b insertion, are fully processed. Similarly, polypeptides from splice forms 6 and 8 are identical to those from splice forms 4 and 7, respectively, except for the 4b insertion. These results indicate that peptide sequences encoded by exon 4b stimulate proteolytic processing. In future experiments, it will be interesting to determine whether exon 4b–containing transcripts are preferentially expressed under specific cellular conditions. In yeast, production of the short isoform of Mgm1 requires adequate ATP levels in the mitochondrial matrix, suggesting that Mgm1 processing is regulated by cellular metabolism (Herlan et al., 2004).
It should be noted that in our expression system, the total amount of exogenously expressed OPA1 is comparable with the total endogenous OPA1 in wild-type MEFs (Fig. 1 B). In contrast, gross overexpression of exogenous OPA1 is unable to rescue mitochondrial fusion activity in OPA1-null cells (unpublished data).
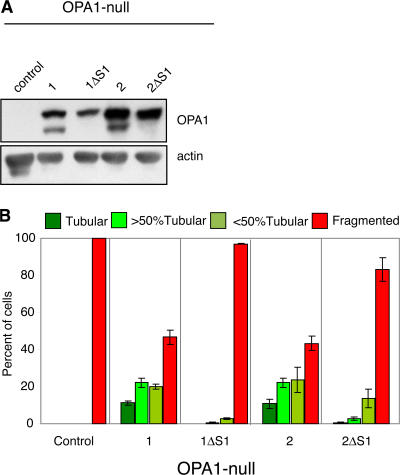
Long forms of OPA1 isoforms 1 and 2 in isolation have little or no fusion activity
Previous experiments suggested that the long form of isoform 1 is the fusion-active species (Ishihara et al., 2006). This conclusion was based on analysis of an isoform 1 construct in which the S1 cleavage site was removed by the deletion of 10 residues surrounding alanine 195 (ΔS1 mutation). To reexamine this issue, we analyzed the function of polypeptides containing the ΔS1 mutation. As expected, OPA1 1ΔS1 and 2ΔS1 produce only the long form (Fig. 2 A). Splice forms 1 and 2 are the only ones lacking both the S2 cleavage site and exon 4b, thereby simplifying the analysis of processing. Whereas wild-type splice forms 1 and 2 are able to induce tubulation in >50% of OPA1-null cells, the same splice forms lacking the S1 site, especially OPA1-1ΔS1, have barely detectable activity (Fig. 2 B). Because the long forms of isoforms 1 and 2 are ineffective for mitochondrial fusion, these results indicate that proteolytic processing is important for OPA1 function.
Figure 2.
The long forms of isoform 1 or isoform 2 lack substantial mitochondrial fusion activity. (A) Processing of mRNA splice forms 1 and 2 and their ΔS1 mutants. After expression in OPA1-null cells, Western blot analysis was used to detect OPA1. Actin was used as a loading control. (B) Tubulation of mitochondria in OPA1-null cells. After expression of the indicated splice forms in OPA1-null cells containing mitochondrially targeted DsRed, cells were scored as in Fig. 1 C; error bars indicate the SD from triplicate experiments.
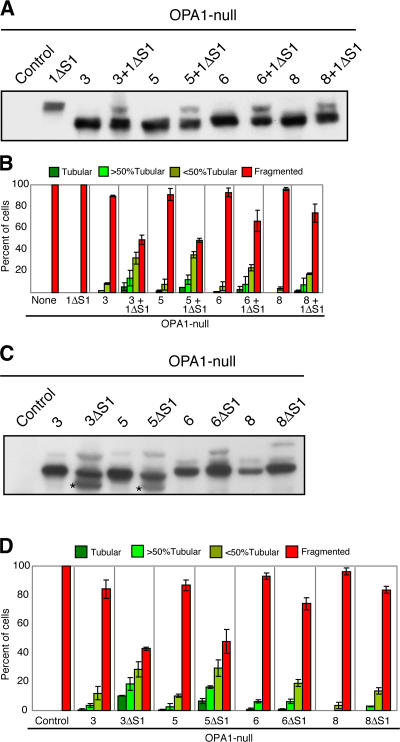
Long and short forms of OPA1 complement each other to reconstitute mitochondrial fusion activity
The aforementioned experiments indicate that neither the long forms nor the short forms in isolation are sufficient for mitochondrial fusion. Therefore, we tested whether a combination of long and short forms might be necessary. We analyzed whether 1ΔS1, which produces only a single long form with essentially no fusion activity (Fig. 2, A and B), could be complemented by the additional expression of splice forms 3, 5, 6, or 8, which produce fully processed isoforms that also have little fusion activity (Fig. 1, B–D). As expected, coexpression produces a mixture of long and short forms (Fig. 3 A). All four combinations of coexpression resulted in a synergistic increase in mitochondrial tubulation in OPA1-null cells (Fig. 3 B). These results demonstrate that 1ΔS1 and short forms work together to promote fusion.
Figure 3.
A combination of long and short OPA1 isoforms increases mitochondrial fusion. (A) Coexpression of 1ΔS1 with short isoforms. The indicated constructs were expressed in OPA1-null cells and analyzed by Western blotting. (B) Functional complementation of 1ΔS1. OPA1-null cells were infected with retrovirus expressing splice forms 3, 5, 6, or 8. Each infected line along with an uninfected control was transfected with a control GFP vector or the GFP vector and OPA1-1ΔS1. (C) Processing of ΔS1 mutants. The indicated splice forms and mutants were expressed in OPA1-null cells. OPA1 was detected by Western blot analysis. Asterisks indicate novel cleavage products for 3ΔS1 and 5ΔS1. (D) Enhanced tubulation of mitochondria by ΔS1 mutants. The indicated OPA1 splice forms and mutants were expressed in OPA1-null cells. (B and D) Mitochondrial morphology was scored as in Fig. 1 C; error bars indicate the SD from triplicate experiments.
To test this idea further, we analyzed ΔS1 versions of splice forms 3, 5, 6, and 8. Because of the 4b exon, the polypeptides produced from these splice forms are normally fully processed to yield only short forms that have little fusion activity. We hoped that by inhibiting cleavage at site S1, we could produce a mixture of long and short forms. In each case, the ΔS1 versions yield only a modest amount of the long form because OPA1 is still mostly processed (Fig. 3 C). In the case of 3ΔS1 and 5ΔS1, processing is caused by increased cleavage at sites other than S1. In the case of 6ΔS1 and 8ΔS1, cleavage still occurs efficiently at S2 based on the size of the processed bands. Remarkably, in each case, the ΔS1 mutants contain more mitochondrial fusion activity than the wild-type proteins (Fig. 3 D). These effects were most dramatic for 3ΔS1 and 5ΔS1. These results again indicate that a combination of long and short OPA1 isoforms is needed for efficient fusion activity.
Our results differ from a previous experiment showing that the expression of rat splice form 1ΔS1 alone can tubulate mitochondria in cells treated with siRNA against OPA1 (Ishihara et al., 2006). We cannot rule out that rat and human OPA1 isoforms behave differently. However, because knockdown experiments do not completely deplete protein, it is likely that some short forms of OPA1 are retained in the siRNA-treated cells, thereby accounting for the apparent activity of the long isoform.
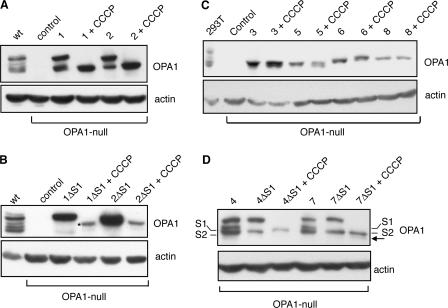
Isoform-specific control of OPA1 processing and stability by membrane potential
Disruption of mitochondrial membrane potential by carbonyl cyanide m-chlorophenyl hydrazone (CCCP) has been shown to enhance the processing of OPA1, leading to accumulation of the short isoforms (Duvezin-Caubet et al., 2006; Ishihara et al., 2006). Consistent with this idea, we found that CCCP treatment of OPA1-null cells expressing splice form 1 or 2 leads to accumulation of the S1-cleaved short isoform (Fig. 4 A). However, this effect appears to involve not only increased processing at S1 but also induced degradation of the long isoform. Upon treatment with CCCP, cells expressing 1ΔS1 show a complete loss of the long isoform (Fig. 4 B). Under these conditions, a minor fraction of OPA1 is cleaved at a site other than S1, and this minor species is the only OPA1 that remains. With 2ΔS1, most of the long isoform is also degraded. It should be noted that we observed this degradation only with a 4-h CCCP treatment, which is longer than in previous studies (Duvezin-Caubet et al., 2006; Ishihara et al., 2006). Because CCCP treatment leads to the accumulation of short isoforms, we tested directly whether short isoforms are stable in the presence of CCCP. Expression of the exon 4b–containing splice forms (3, 5, 6, and 8) leads to fully processed isoforms that are stable after CCCP treatment (Fig. 4 C).
Figure 4.
CCCP affects OPA1 processing and stability. (A) Effect of CCCP on isoforms 1 and 2. The splice forms 1 and 2 were expressed in OPA1-null cells and treated with CCCP as indicated. OPA1 was detected by Western blotting. Actin was used as a loading control. (B) Degradation of long forms of isoforms 1 and 2. Splice forms 1ΔS1 and 2ΔS1 were expressed in OPA1-null cells and treated with CCCP as indicated. The asterisk indicates a new cleavage product with CCCP treatment of 1ΔS1. (C) Resistance of short isoforms to degradation. OPA1 constructs were expressed in OPA1-null cells and treated with CCCP as indicated. (D) CCCP effect on site S1 versus S2. OPA1 splice forms and their mutants were expressed in OPA1-null cells and analyzed as in A. The S1- and S2-cleaved products for isoforms 4 and 7 are labeled. The arrow indicates a novel cleavage product with CCCP treatment.
Given the enhanced cleavage at site S1 in the presence of CCCP, we also examined its effect on cleavage at S2. Variants 4ΔS1 and 7ΔS1 produce only a single short isoform as a result of cleavage at site S2 (Fig. 4 D). Upon CCCP treatment, the long isoform disappears, but without an increased accumulation of the S2-cleaved isoform. In addition, there is a small accumulation of a novel cleavage product. Collectively, these results indicate that the loss of membrane potential enhances OPA1 processing at S1 but not S2, suggesting that these sites are under differential regulation. In addition, the loss of membrane potential leads to degradation of the long isoforms but not the short ones.
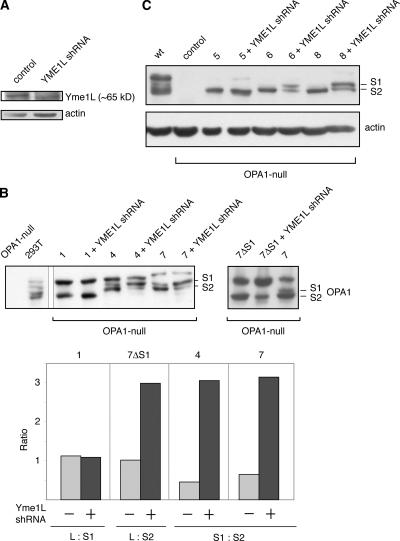
The intermembrane space protease Yme1L regulates OPA1 cleavage at site S2 but not S1
The different response of S1 versus S2 cleavage to the loss of membrane potential suggests that the two sites may be regulated by different proteases. Previous studies have implicated both m-AAA proteases and the rhomboid protease PARL in OPA1 processing (Cipolat et al., 2006; Ishihara et al., 2006; Duvezin-Caubet et al., 2007). However, the depletion of these proteases had only limited effects on OPA1 processing (Duvezin-Caubet et al., 2007), indicating that additional proteases might be involved. Yme1L is an i-AAA protease anchored in the inner membrane with its protease domain located within the intermembrane space (Shah et al., 2000; Arnold and Langer, 2002). Short hairpin RNA (shRNA) against Yme1L effectively reduces protein levels (Fig. 5 A), and we examined its effect on OPA1 processing. Knockdown of Yme1L has no effect on isoform 1 processing, which occurs exclusively at site S1 (Fig. 5 B). In contrast, for isoforms 4 and 7, which contain both sites S1 and S2, Yme1L shRNA reduces the level of the S2-cleaved product and increases the level of the S1-cleaved product. By densitometry of the Western blots, the ratio of the S1-cleaved band to the S2-cleaved band was 0.4 and 0.6 for isoforms 4 and 7, respectively, without treatment. With the Yme1L knockdown, this ratio rose to >3. Likewise, 7ΔS1, which produces only the S2-cleaved product, shows reduced processing after shRNA treatment. In this case, the ratio of the long isoform to the S2-cleaved isoform rose from 1 to 3. Similar effects on the S1 and S2 cleavage products were observed with the exon 4b–containing splice forms 6 and 8 (Fig. 5 C). These results indicate that Yme1L is important for proteolytic processing at site S2. Isoform 5 has no S2 cleavage site, but shRNA resulted in the slight accumulation of a long isoform. It may be that Yme1L is also involved in cleavage at sites other than S2.
Figure 5.
Yme1L knockdown reduces cleavage at site S2. (A) Knockdown of Yme1L. Lysates from control cells and cells expressing shRNA against Yme1L were analyzed by Western blotting against Yme1L. Actin was used as a loading control. (B) Effect of Yme1L knockdown on the proteolytic processing of isoforms 1, 4, and 7. OPA1 splice forms and mutants were expressed in OPA1-null cells and treated with shRNA against Yme1L as indicated. OPA1 was detected by Western blotting. For isoforms 4 and 7, the S2-cleaved short isoform is the lowest band, as indicated. These results are quantitated by densitometry in the bottom panel. For splice form 1 and 7ΔS1, the ratio of the long isoform to the short isoform is presented. For splice forms 4 and 7, which produce S1- and S2-cleaved products, the ratio of the S1-cleaved band to the S2-cleaved band is presented. (C) Effect of Yme1L knockdown on the proteolytic processing of isoforms 5, 6, and 8. Samples were analyzed as in B. The S1- and S2-cleaved products of isoforms 6 and 8 are labeled.
As with paraplegin and PARL, it is not clear whether Yme1L directly cleaves OPA1 or affects another protease. Thus far, we have not been able to demonstrate a direct physical interaction between Yme1L and OPA1 by coimmunoprecipitation. Cleavage of site S2 by YmeL1 is topologically sensible because the S2 region would be expected to reside in the intermembrane space, where the protease domain of Yme1L is located. In contrast, the protease activity of PARL resides within the inner membrane, and the protease activities of m-AAA proteases reside within the matrix. If Yme1L does directly cleave OPA1, it would be the first example of a protein-processing function through site-specific cleavage for Yme1L. Previously, the role of Yme1L was thought to be in degrading misfolded mitochondrial proteins in the intermembrane space (Arnold and Langer, 2002). Analogously, the m-AAA proteases have a role in protein quality control through protein degradation but have recently been found to also process proteins at specific sites (Nolden et al., 2005; Ishihara et al., 2006; Duvezin-Caubet et al., 2007).
Our results provide insight into the function of distinct OPA1 isoforms. Importantly, they indicate that OPA1, like Mgm1, requires both long and short isoforms for efficient mitochondrial fusion activity. With this unifying property, it is likely that OPA1 and Mgm1 have similar mechanisms of action. It is interesting to note that any combination of long and short OPA1 isoforms is able to reconstitute fusion activity. This observation suggests that the exact cleavage sites (S1 versus S2) may not be important in terms of protein function. Instead, the presence of multiple sites may enable increased flexibility in regulating OPA1 processing because the two sites appear to be cleaved by distinct proteases and respond differently to changes in membrane potential.
Finally, at least three types of mitochondrial proteases—m-AAA proteases, the rhomboid PARL, and now the i-AAA protease Yme1L—appear to regulate OPA1 processing. By regulating OPA1 isoforms through mRNA splicing, degradation of long isoforms, and multiple modes of proteolytic processing, mammalian cells may be able to rapidly control mitochondrial fusion in response to environmental stimuli.
Materials and methods
Cell culture, transfection, and PEG fusion assay
MEFs were maintained in DME containing 10% heat-inactivated FBS, 1 mM l-glutamine, and penicillin/streptomycin. For transfection of cells with expression constructs, LipofectAMINE 2000 (Invitrogen) was used according to the manufacturer's specifications. In the experiments in Fig. 4, CCCP was used at 20 μM, and cells were analyzed at 4 h.
To test the fusion activity of OPA1 mRNA splice forms, OPA1- null cells stably expressing mitochondrially targeted GFP or DsRed were infected with retrovirus encoding each OPA1 mRNA splice form. Pairwise PEG fusion assays were performed as described previously (Chen et al., 2003).
Plasmid construction and retroviral transduction
The eight isoforms of human OPA1 cDNA were amplified by PCR from first-strand cDNA and subcloned into the retroviral vector pMSCV-puro. Retrovirus production and infection were performed as described previously (Chen et al., 2003). Western blots of cell lysates were probed with an affinity-purified anti-OPA1 antibody raised against the C-terminal region of human OPA1 (gift of L. Griparic and A. van der Bliek, University of California, Los Angeles, Los Angeles, CA; Griparic et al., 2004).
shRNA construction
shRNAi against Yme1L was performed using a modified retroviral vector with the H1 promoter to drive the expression of shRNAs (Chen et al., 2005). Three shRNAi constructs were tested, with the best results obtained with the vector constructed with the oligonucleotides 5′-GATCCCCGTGGCAGAGGAATTCATATTTCAAGAGAATATGAGTTCCTCTGCCACTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGTGGCAGAGGAACTCATATTCTCTTGAAATATGAATTCCTCTGCCACGGG-3′.
The annealed oligonucleotides were cloned into the retroviral vector linearized with BglII–HindIII. The targeted sequence of Yme1L was GAGTGGCAGAGGAACTCATAT. Western blot analysis was performed with a polyclonal antibody against Yme1L (gift of C. Koehler, University of California, Los Angeles, Los Angeles, CA).
Fluorescence microscopy
MEFs were grown on poly-l-lysine–treated coverslips and fixed in 10% formalin. Coverslips were mounted with GelMount (Biomeda). Images were acquired at room temperature with a camera (ORCA-ER; Hamamatsu) attached to a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) controlled by Axiovision version 4.5 software (Carl Zeiss MicroImaging, Inc.). A 100× plan Apochromat NA 1.4 objective was used. Mitochondria were identified by mitochondrially targeted GFP or DsRed.
To determine mitochondrial fusion activity, cells were visually scored under a fluorescent microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) into four morphological classifications. Tubular refers to cells with only tubular mitochondria. >50% tubular refers to cells in which greater than half of the mitochondrial mass existed as tubules as opposed to spherical fragments. <50% tubular refers to cells in which less than half of the mitochondrial mass existed as tubules. In addition, all cells in this class contained at least three clearly tubular mitochondria. Finally, fragmented refers to cells that contain spherical mitochondrial fragments with no more than two short tubules found. In the parental OPA1-null cell line, all cells lacked any tubular mitochondria and, thus, belonged to the last class.
Online supplemental material
Fig. S1 shows that OPA1-null cells expressing a single OPA1 RNA splice form have extensive fusion activity in the PEG cell hybrid assay. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200704110/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Lorena Griparic and Alex van der Bliek for communicating results before publication and for providing the anti-OPA1 antibody.
This work was supported by National Institutes of Health grant GM062967 to D.C. Chan. Z. Song is supported by an Elizabeth Ross postdoctoral fellowship.
Abbreviations used in this paper: CCCP, carbonyl cyanide m-chlorophenyl hydrazone; MEF, mouse embryonic fibroblast; MPP, mitochondrial processing peptidase; PARL, presenilin-associated rhomboid-like; PEG, polyethylene glycol; shRNA, short hairpin RNA.
References
- Alexander, C., M. Votruba, U.E. Pesch, D.L. Thiselton, S. Mayer, A. Moore, M. Rodriguez, U. Kellner, B. Leo-Kottler, G. Auburger, et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211–215. [DOI] [PubMed] [Google Scholar]
- Arnold, I., and T. Langer. 2002. Membrane protein degradation by AAA proteases in mitochondria. Biochim. Biophys. Acta. 1592:89–96. [DOI] [PubMed] [Google Scholar]
- Chan, D.C. 2006. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 22:79–99. [DOI] [PubMed] [Google Scholar]
- Chen, H., S.A. Detmer, A.J. Ewald, E.E. Griffin, S.E. Fraser, and D.C. Chan. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., A. Chomyn, and D.C. Chan. 2005. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280:26185–26192. [DOI] [PubMed] [Google Scholar]
- Cipolat, S., O. Martins de Brito, B. Dal Zilio, and L. Scorrano. 2004. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA. 101:15927–15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat, S., T. Rudka, D. Hartmann, V. Costa, L. Serneels, K. Craessaerts, K. Metzger, C. Frezza, W. Annaert, L. D'Adamio, et al. 2006. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 126:163–175. [DOI] [PubMed] [Google Scholar]
- Delettre, C., G. Lenaers, J.M. Griffoin, N. Gigarel, C. Lorenzo, P. Belenguer, L. Pelloquin, J. Grosgeorge, C. Turc-Carel, E. Perret, et al. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26:207–210. [DOI] [PubMed] [Google Scholar]
- Delettre, C., J.M. Griffoin, J. Kaplan, H. Dollfus, B. Lorenz, L. Faivre, G. Lenaers, P. Belenguer, and C.P. Hamel. 2001. Mutation spectrum and splicing variants in the OPA1 gene. Hum. Genet. 109:584–591. [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet, S., R. Jagasia, J. Wagener, S. Hofmann, A. Trifunovic, A. Hansson, A. Chomyn, M.F. Bauer, G. Attardi, N.G. Larsson, et al. 2006. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 281:37972–37979. [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet, S., M. Koppen, J. Wagener, M. Zick, L. Israel, A. Bernacchia, R. Jagasia, E.I. Rugarli, A. Imhof, W. Neupert, T. Langer, and A.S. Reichert. 2007. OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria. Mol. Biol. Cell. 10.1091/mbc.E07-02-0164. [DOI] [PMC free article] [PubMed]
- Frezza, C., S. Cipolat, O. Martins de Brito, M. Micaroni, G.V. Beznoussenko, T. Rudka, D. Bartoli, R.S. Polishuck, N.N. Danial, B. De Strooper, and L. Scorrano. 2006. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 126:177–189. [DOI] [PubMed] [Google Scholar]
- Griparic, L., N.N. van der Wel, I.J. Orozco, P.J. Peters, and A.M. van der Bliek. 2004. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J. Biol. Chem. 279:18792–18798. [DOI] [PubMed] [Google Scholar]
- Herlan, M., F. Vogel, C. Bornhovd, W. Neupert, and A.S. Reichert. 2003. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278:27781–27788. [DOI] [PubMed] [Google Scholar]
- Herlan, M., C. Bornhovd, K. Hell, W. Neupert, and A.S. Reichert. 2004. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 165:167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara, N., Y. Fujita, T. Oka, and K. Mihara. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25:2966–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuibban, G.A., S. Saurya, and M. Freeman. 2003. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 423:537–541. [DOI] [PubMed] [Google Scholar]
- Meeusen, S., R. DeVay, J. Block, A. Cassidy-Stone, S. Wayson, J.M. McCaffery, and J. Nunnari. 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 127:383–395. [DOI] [PubMed] [Google Scholar]
- Nolden, M., S. Ehses, M. Koppen, A. Bernacchia, E.I. Rugarli, and T. Langer. 2005. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 123:277–289. [DOI] [PubMed] [Google Scholar]
- Okamoto, K., and J.M. Shaw. 2005. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39:503–536. [DOI] [PubMed] [Google Scholar]
- Olichon, A., L. Baricault, N. Gas, E. Guillou, A. Valette, P. Belenguer, and G. Lenaers. 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 278:7743–7746. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., S.M. Southard, A.E. Hobbs, and R.E. Jensen. 2003. a. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem. Biophys. Res. Commun. 308:276–283. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., S.M. Southard, M.P. Yaffe, and R.E. Jensen. 2003. b. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell. 14:2342–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, Z.H., G.A. Hakkaart, B. Arku, L. de Jong, H. van der Spek, L.A. Grivell, and H.T. Jacobs. 2000. The human homologue of the yeast mitochondrial AAA metalloprotease Yme1p complements a yeast yme1 disruptant. FEBS Lett. 478:267–270. [DOI] [PubMed] [Google Scholar]
- Wong, E.D., J.A. Wagner, S.W. Gorsich, J.M. McCaffery, J.M. Shaw, and J. Nunnari. 2000. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, E.D., J.A. Wagner, S.V. Scott, V. Okreglak, T.J. Holewinske, A. Cassidy-Stone, and J. Nunnari. 2003. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 160:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.