Abstract
Hypoxia-inducible factor 1 (HIF-1) is a ubiquitously expressed transcriptional regulator involved in induction of numerous genes associated with angiogenesis and tumor growth. Kaposi's sarcoma, associated with increased angiogenesis, is a highly vascularized, endothelial cell-derived tumor. Previously, we have shown that the latency-associated nuclear antigen (LANA) encoded by Kaposi's sarcoma-associated herpesvirus (KSHV) targets the HIF-1α suppressors von Hippel-Lindau protein and p53 for degradation via its suppressor of cytokine signaling-box motif, which recruits the EC5S ubiquitin complex. Here we further show that HIF-1α was aberrantly accumulated in KSHV latently infected primary effusion lymphoma (PEL) cells, as well as HEK293 cells infected with KSHV, and also show that a potential α-helical amino-terminal domain of LANA was important for HIF-1α nuclear accumulation in normoxic conditions. Moreover, we have now determined that this association was dependent on the residues 46 to 89 of LANA and the oxygen-dependent degradation domain of HIF-1α. Introduction of specific small interfering RNA against LANA into PEL cells also resulted in a diminished nuclear accumulation of HIF-1α. Therefore, these data show that LANA can function not only as an inhibitor of HIF-1α suppressor proteins but can also induce nuclear accumulation of HIF-1α during KSHV latent infection.
Kaposi's Sarcoma-associated herpesvirus (KSHV) is a human gammaherpesvirus and is the major etiological agent associated with Kaposi's sarcoma (KS) (6). KS is a highly vascularized, inflammatory angiogenic tumor of endothelial cells commonly found in untreated AIDS patients (41). Infection by KSHV is required for the development of KS tumor (8). KSHV is also associated with two lymphoproliferative disorders, primary effusion lymphoma (PEL), and multicentric Castleman's disease (5, 38). Like other herpesvirus, KSHV infection has two distinct modes of life cycle: a latent and a lytic replication cycle (25). In KS tumors, KSHV undergoes a predominantly latent form of infection that is critical for KSHV long-term infection and KSHV-mediated sarcomagenesis (40). with chemical induction or hypoxia treatment, KSHV was able to reactivate from latency to lytic replication (7, 49).
Although there have been over 90 open reading frames (ORFs) identified in the KSHV genome, only a small subset of these genes are typically expressed during latency (32). The latency-associated nuclear antigen (LANA), one of the major latent genes, is a multifunctional protein encoded by KSHV that is important for maintaining latent infection and plays a crucial role in persistence of the viral episome. In addition, LANA also has been shown to be involved in the regulation of a number of cellular genes by functioning as a coactivator or a corepressor (1, 3, 10, 15, 16, 19, 27-29, 43). More recently, our previous studies showed that LANA can also act as an adaptor for the ubiquitin complex that specifically promotes ubiquitylation of the tumor suppressors von Hippel-Lindau protein (VHL) and p53 leading to degradation (2).
In contrast to normoxia, hypoxia (low oxygen) is known to exist within many tumors, and the extent of tumor hypoxia correlates with prognosis in a number of tumor types (13, 39, 42). As a key hypoxia responder, hypoxia-inducible factor 1 (HIF-1) is a heterodimeric transcription factor composed of an oxygen-sensitive α protein, HIF-1α, and a constitutively expressed β subunit, HIF-1β, also known as the aryl hydrocarbon receptor nuclear translocator (44). HIF-1 regulates more than 60 specific cellular proteins involved in a number of core processes such as angiogenesis, erythropoiesis, and glycolysis (18, 34). In normoxic conditions, the tumor suppressor VHL or p53 can recruit HIF-1α for rapid ubiquitylation and proteasomal degradation (24, 30). However, in hypoxic response, HIF-1α hydroxylation is blocked, and it is translocated to the nucleus, where it heterodimerizes with HIF-1β to activate expression of downstream genes (i.e., vascular epidermal grow factor [VEGF] and Glut-1) by targeting hypoxia responsive elements (HREs) within the promoters (18).
The evidence of HIF-1α upregulation in a broad range of cancers and its striking correlation with tumor grade and vascularization suggest that HIF-1 is essential for tumor cell dissemination and proliferation (46, 48). Therefore, oncogene activation or tumor suppressor inactivation of HIF-1α can lead to deregulated cell proliferation (35, 36). In the KSHV-associated B lymphoma tumor cells, we previously showed that the viral latent antigen LANA can target both HIF-1α suppressors VHL and p53 for degradation (2). Here, we further demonstrate that HIF-1α is aberrantly accumulated in KSHV latently infected cells and also show that an amino-terminal domain of LANA is important for HIF-1α nuclear accumulation in normoxic conditions. Moreover, using green fluorescent protein (GFP) fusion protein constructs, and a series of truncated LANA polypeptides, we show that this accumulation is mediated by a potential α-helical structure located at the amino termini of LANA. The results at least in part demonstrate another mechanism by which LANA mediates HIF-1α accumulation in KSHV-associated cancer during latency infection.
MATERIALS AND METHODS
DNA constructs.
Plasmids pDSRed/LANA (LANA-RFP), pA3M/LANA1-329^925-1162 (LN-1), pA3M/LANA1-340 (LN-2), and pA3M were described previously (3, 37). Plasmids pA3M/LANA1-233 (LN-3), pA3M/LANA46-145 (LN-4), pA3M/LANA89-233 (LN-5), pA3M/LANA1-46<942>129-233 (LN-6), and pA3M/LANA1-233H47AC57A (LN-7) were generated by PCR amplicons subcloned into the pA3M vector by KpnI/EcoRV digestion. Construct pEGFP/HIF-1α was prepared by PCR amplification of the ORF from pCEP4/HIF-1α (26). The amplicon was digested with BamHI/KpnI and cloned into the BglII and KpnI sites of pEGFP-C1 (Clontech, Inc., Mountain View, CA). pEGFP/HIF-1α 1-330(N) was generated by pEGFP/HIF-1α digestion with EcoRI and the self-ligation of the large fragment. pEGFP/HIF-1α 1-3005̂30-826(NC) was obtained from the large fragment of the EcoRI-digested pEGFP/HIF-1α, and its ends were filled by Klenow enzyme followed by blunt-end ligation. pEGFP/HIF-1α 300-530(M) was generated by PCR from pCEP4/HIF-1α, and then the amplicon was digested with BamHI/EcoRI cloned into the BglII and EcoRI sites of pEGFP-C1. All clones were confirmed by DNA sequencing. pCEP4/HIF-1α was provided by Gregg L. Semenza of Johns Hopkins University (26). The wild-type hypoxia response element (wHRE) consisted of a trimerized 24-mer containing 18 bp of sequence from the PGK promoter, including the HRE (5-TGTCACGTCCTGCACGACTCTAGT [HRE is underlined]), and its mutant (mHRE) had the ACG of the HIF-1 binding site mutated to CAT, abolishing binding, in the pGL2-Basic vector provided by Craig B. Thompson (University of Pennsylvania School of Medicine). pGL3-1.5VEGF plasmid containing the 47-bp HRE and pGL3-1.2VEGF with HRE deletion were provided by Amit Maity (University of Pennsylvania School of Medicine, Philadelphia) (22).
Cell cultures and transfection.
KSHV-positive type cells (BCBL-1 and BC-3) and KSHV-negative type cells (BJAB and DG75) were maintained in RPMI 1640 medium with 7% fetal bovine serum (FBS), 4 μM l-glutamine, penicillin, and streptomycin. The renal carcinoma VHL-null cell line 786-O (provided by Volker H. Haase from the University of Pennsylvania School of Medicine), the human osteosarcoma p53-null cell line Saos-2 (provided by Jon Aster from Brigham and Women's Hospital, Boston, MA), and human embryonic kidney 293 (HEK293) and U2OS cells (from the American Type Culture Collection) were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 5% FBS, 4 μM l-glutamine, penicillin, and streptomycin. All cells were incubated at 37°C in a humidified environmental incubator supplemented with 5% CO2. Ten million cells with 400 μl of medium were transfected by electroporation with a Bio-Rad Gene Pulser in 0.4-cm-gap cuvettes at 220 V and 975 μF. To create a hypoxic environment, cells were allowed to grow for 24 h with 100 μM CoCl2 treatment before use (31).
Immunofluorescence assays.
Cells transfected on coverslips were fixed in 3% paraformaldehyde for 10 min at 4°C at 24 h posttransfection. Antibody working dilutions were as follows: mouse anti-HIF-1α (BD Transduction Laboratory, San Jose, CA) and anti-myc (9E10) or rabbit anti-LANA polyclonal antibodies (kindly provided by Bala Chandran of Rosalind Franklin University of Medicine and Science, North Chicago, IL) at 1:50. Cells were permeabilized in immunofluorescence buffer (0.2% fish skin gelatin and 0.2% Triton X-100 in phosphate-buffered saline [PBS]) for 5 min, followed by 1 h of incubation in immunofluorescence buffer with primary antibodies. Cells were washed three times in PBS. Fluorescence-labeled Alexa Fluor 488 or 594 anti-mouse or anti-rabbit antibodies (Molecular Probes, Inc., Eugene, OR) were used as secondary antibodies for 30 min. DAPI (4′,6′-diamidino-2-phenylindole at 0.5 μM was used for nuclear staining (Pierce, Inc., Rockford, IL), and coverslips were washed in PBS and mounted on glass slides using Prolong antifade mounting medium (Molecular Probes). All steps were performed at room temperature. Fluorescence confocal microscopy was performed with an Olympus microscope using FluoView FV300 software (Olympus, Inc., Melville, NY).
Fractionation of nuclear or cytoplasm proteins.
Twenty million transfected cells were harvested and washed twice with ice-cold PBS, followed by resuspension of the cell pellet in hypotonic buffer A (10 mM HEPES-K+ [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 0.5 dithiothreitol) in the presence of protease inhibitor cocktail (PIC; 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 10 μg of pepstatin A/ml, 10 μg of phenanthroline/ml, 16 μg of benzamidine/ml). Cells were pelleted by spinning them at 1,000 rpm 5 min. The cells were lysed in ice-cold 0.5% NP-40 containing buffer A with PIC on ice for 10 min. The nuclei were pelleted by centrifugation at 3,000 rpm for 2 min at 4°C. The supernatant (cytoplasm protein) was harvested and frozen at −80°C for use. The nuclear pellets were washed with buffer A (without NP-40), followed by resuspension in buffer C (20 mM HEPES-K+ [pH 7.9], 420 mM NaCl, 0.2 mM EDTA, 1.5 mM MgCl2, 0.5 dithiothreitol, 25% glycerol) with PIC. Nuclei were incubated on ice for 30 min and vortex mixed periodically. Supernatants containing nuclear protein were collected by spinning at 14,500 rpm for 10 min at 4°C and then snap-frozen for further use. Antibodies to nuclear protein Sp1 (1C6; Santa Cruz, Inc., Santa Cruz, CA) and cytoplasm protein Hsp70 (BD Transduction Laboratory) were used as markers.
Immunoprecipitation and immunoblotting.
For immunoprecipitation, B-cell lysates were extracted by using radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, aprotinin [1 μg/ml], pepstatin [1 μg/ml], 25 mM N-ethylmaleimide). The protein concentration was determined by using a Bio-Rad protein assay. After one-step preclearing (rotation 1 h, 4°C) with protein A/G (50/50) Sepharose Fast-Flow (Amersham Biosciences, Inc., Piscataway, NJ), the antibodies were added to the cell protein extract in a binding buffer adjusted to 20 mM Tris (pH 7.5)-200 mM NaCl-0.1% Nonidet P-40. After overnight incubation, immunocomplexes were recovered with protein A/G (50/50) Sepharose after 1 h of incubation. After three washes with binding buffer, the proteins were eluted in sample buffer.
For immunoblotting, proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membrane. After a blocking step with 5% nonfat dry milk in PBS (2 mM KCl, 120 mM NaCl, 1.5 mM KH2PO4, 8 mM Na2HPO4) containing 0.1% Tween 20, the membrane was incubated with primary antibodies overnight at 4°C. The next day, the appropriate infrared-conjugated secondary antibodies (Alexa Fluor 800 or 680) were incubated for 1 h at room temperature, followed by scanning with a Li-Cor Odyssey scanner (Li-Cor, Inc., Lincoln, NE). The blot was washed three times with TBST (25 mM Tris-HCl [pH 8.0], 125 mm NaCl, 0.1% Tween 20), and each wash was for an interval of 5 min.
RNA interference.
The small hairpin RNAs complementary to the C-terminal (GCTAGGCCACAACACATCT) fragment of LANA as described previously (11) were cloned into the pSIREN vector according to the instructions of manufacture (BD Clontech) to generate sh-LANA (2). pSIREN vector with luciferase target sequence (sh-Luc) was used as a control. Ten million BCBL-1 cells were transfected by electroporation with 5 μg of sh-LANA or sh-Luc. BCBL-1 stable cells knockdown for LANA were selected and maintained in 4 μg of puromycin/ml. Cells were fixed in 3% paraformaldehyde, and an immunofluorescence assay was performed as described previously (3).
Infection of HEK293 cells with KSHV virus and induction for virus production.
HEK293 cells grown at 60 to 80% confluence in 100-mm-diameter tissue culture dishes were infected with the same amount of concentrated virus from an identical number of BCBL-1 cells. Virus produced from approximately 20 million BCBL-1 cells was used for infecting a single 100-mm culture dish in the presence of Polybrene (Sigma, St. Louis, MO) at different time points.
For virus induction, five hundred million exponentially growing BCBL-1 cells were induced with 20 ng of tetradecanoyl phorbol acetate/ml and 1.5 mM sodium butyrate (Sigma-Aldrich, St. Louis, MO) for 5 days at 37°C with 5% CO2. The supernatant was collected and filtered through a 0.45-μm-pore-size filter, and viral particles were spun down at 25,000 rpm for 2 h at 4°C. The concentrated virus was collected and used for infection experiments.
Determination of HIF-1α stability.
HEK293 cells were cotransfected pCEP4/HIF-1α with or without pA3M/LANA as described above. After 24 h of transfection, the cells were incubated with 100 μM cycloheximide (C4859; Sigma) for 0 to 4 h and then harvested. Equal amounts of total proteins from each treatment were taken to perform Western blotting or immunoprecipitation.
VEGF expression.
VEGF expression in the cell supernatant was monitored by using a Quantikine human VEGF immunoassay kit (R&D Systems, Minneapolis, MN). Briefly, when cells were 70 to 80% confluent (2 × 106 cells/well), they were washed twice in DMEM and further incubated in phenol red-free DMEM supplemented with 5% FBS at 37°C. After 24 h of incubation, supernatants were collected in 1.5-ml vials, spun at 4°C to remove the particulates, and then stored at −80°C. The supernatant (200 μl) was thawed and used to test VEGF expression by performing an enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's recommendations.
Luciferase assay.
Ten million cells were transfected with luciferase reporter (5 μg) combined with specific expression constructs. The transfected cells were harvested at 48 h posttransfection and subsequently washed once with PBS (HyClone, Inc., Logan, UT), followed by lysis with 200 μl of reporter lysis buffer (Promega, Inc., Madison, WI). Then, 40 μg of total protein of the lysate was mixed with 100 μl of the luciferase assay reagent. Luminescence was measured for 10 s with an OptiComp luminometer (MGM Instruments, Inc., Hamden, CT). The luciferase activity was normalized with cotransfected β-galactosidase activity (i.e., the optical density at 450 nm), which was driven by the cytomegalovirus promoter. The relative luciferase activity was expressed as the fold activation relative to the reporter construct alone. Assays were performed in triplicate.
RESULTS
HIF-1α is accumulated in KSHV-infected cells.
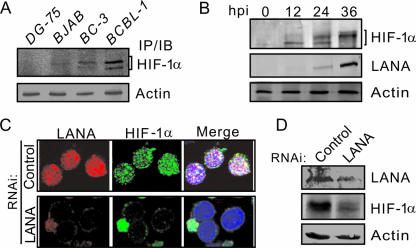
Previously, we observed an interaction between the key KSHV latent antigen LANA and HIF-1α, which led to virus lytic replication under hypoxic conditions (3). More recently, we saw that both the HIF-1α suppressors VHL and p53 were downregulated by LANA under normoxic conditions (2). To explore this potential effect of LANA on HIF-1α accumulation in KSHV latent infection, we compared the expression levels of HIF-1α in two KSHV-positive (BCBL-1 and BC-3) PEL cells and two KSHV-negative (BJAB and DG75) B-lymphocyte cell lines by immunoprecipitation and immunoblot assays targeting HIF-1α. The results clearly showed that HIF-1α levels were dramatically elevated in the KSHV positive BCBL-1 cells and slightly increased in the BC-3 cells (Fig. 1A). To demonstrate the role of LANA as an inducer of HIF-1α expression during the KSHV latent infection, we performed KSHV latent infection in vitro using the susceptible cell line HEK293. The data from Western blot analysis of infected 293 cells at 12, 24, and 36 h postinfection by KSHV showed that increased HIF-1α expression was consistent with increased expression of LANA compared to cells uninfected at the zero time point (Fig. 1B). Unexpectedly, we also observed an increase in modification of HIF-1α at the 24- and 36-h time points when LANA was expressed at relatively higher levels (Fig. 1B). Previous studies have shown that except for latent genes there are many other KSHV immediate-early- and early-stage genes expressed (17). To verify the role of LANA on HIF-1α accumulation, we tested the HIF-1α levels in the BCBL-1 cells with specific small interfering RNA targeting LANA mRNA. The data from both immunofluorescence and immunoprecipitation assays showed that the increased expression of HIF-1α in the PEL cell lines was significantly diminished once the endogenous viral LANA was knockdown (Fig. 1C and D). This indicates that LANA contributes to HIF-1α accumulation in KSHV latent infections.
FIG. 1.
HIF-1α accumulates in PEL cells. (A) Western blot analysis of KSHV-positive cell lines (BCBL-1 and BC-3) and KSHV-negative cell lines (BJAB and DG75) with anti-HIF-1α antibody. Twenty million cells were lysed, and coimmunoprecipitation (IP) with anti-HIF-1α antibody, followed by immunoblot (IB) detection with anti-HIF-1α, was performed. (B) Western blot analysis of HEK293 cells infected by KSHV virus. Ten million 293 cells infected with BCBL-1 KSHV virion were harvested at the indicated times (0, 12, 24, and 36 h). Cell lysates were analyzed by immunoblotting (IB) with rabbit anti-LANA or mouse anti-HIF-1α antibodies. The data showed that the levels of HIF-1α are increased, along with LANA expression, and HIF-1α was modified at 36 h postinfection. (C) HIF-1α accumulation in PEL cells is dependent on LANA expression. Immunofluorescence assays performed on BCBL-1 PEL cells transfected with sh-LANA or control sh-Luc. Cells were stained with antibodies to LANA (red) or HIF-1α (green). Nuclei were stained with DAPI (blue). (D) Western blotting of LANA knocked-down BCBL-1 cells to detect LANA, HIF-1α, and β-actin.
LANA enhances the stability of HIF-1α.
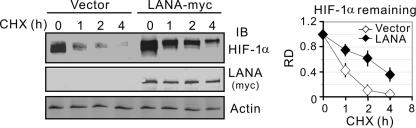
To determine whether the effect of LANA on HIF-1α results in an increase in HIF-1α stability, HEK293 cells expressing HIF-1α alone or both HIF-1α and LANA were treated with cycloheximide for 1 to 4 h after 24-h posttransfection. Immunoblotting against total HIF-1α showed that the stability of HIF-1α protein was significantly enhanced by LANA coexpressed compared to HIF-1α alone (Fig. 2, left panel). Determination of the relative percentage of HIF-1α remaining showed that the half-life HIF-1α was increased ∼2-fold with LANA coexpression (Fig. 2, right panel). These data suggest that LANA also contributes to stabilization of HIF-1α.
FIG. 2.
LANA increases the stability of HIF-1α. HEK293 cells were cotransfected HIF-1α with or without LANA-myc expression plasmids. At 24 h posttransfection, cell lysates (40 μg) treated with cycloheximide (CHX) for 0, 1, 2, and 4 h were subjected to immunoblotting as indicated. The relative quantitation of HIF-1α is shown in the right panel.
LANA promotes HIF-1α nuclear accumulation.
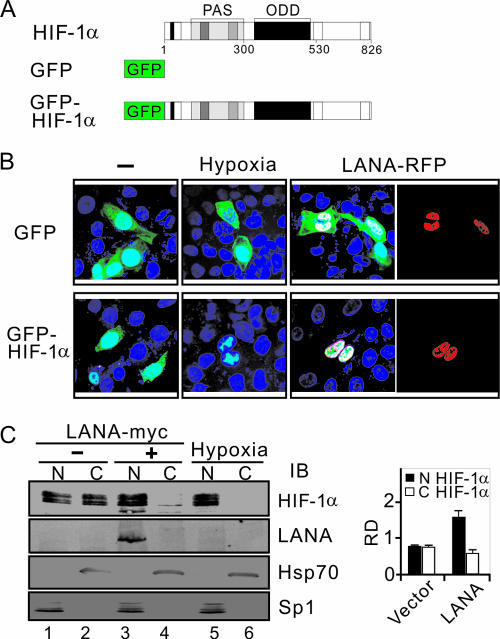
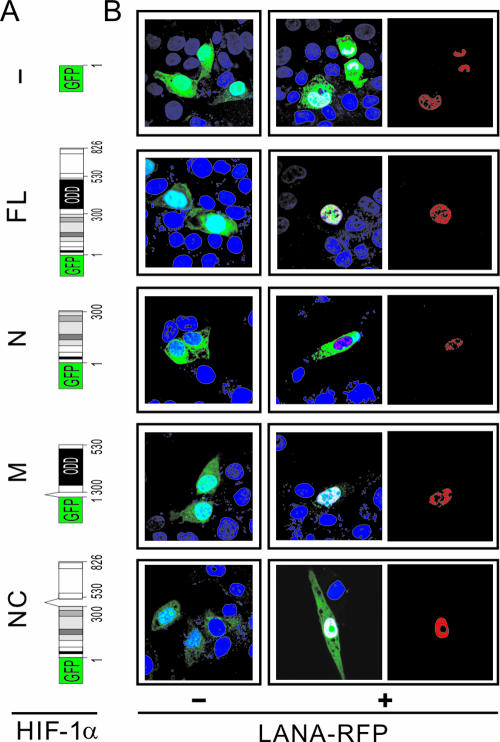
To further investigate the role of LANA in HIF-1α nuclear accumulation in KSHV latently infected cells, we wanted to look specifically at changes in HIF-1α localization in LANA-expressing cells. First, we constructed a vector carrying an in-frame GFP fusion of full-length HIF-1α (schematically shown in Fig. 3A). U2OS cells with clear cytoplasmic and nuclear compartments were then transiently transfected with either the parental GFP construct or the chimeric GFP-HIF-1α expression construct in the presence or absence of LANA-red fluorescence protein (RFP). Fluorescence was observed in ca. 20% of the cells, which reflected the transfection efficiency. As in previous studies (14, 20), the fluorescence of GFP or GFP-HIF-1α alone was uniformly distributed throughout the cell (Fig. 3B, left panels), and hypoxia dramatically increased the nuclear intensity of GFP-HIF-1α signals but had no effect on GFP alone (Fig. 3B, middle panels). However, the transient expression of GFP-HIF-1α with LANA-RFP constructs in normoxia resulted in clearly nuclear localization of HIF-1α, which also occurs by hypoxic treatment, even though the nuclear accumulation patterns of HIF-1α induced by LANA and hypoxia were distinctly different (Fig. 3B, right panels). To quantitatively compare the signals of HIF-1α in the nuclear and cytoplasmic compartments, the nuclear and cytoplasmic fractions from U2OS cells cotransfected with GFP-HIF-1α in the presence or absence of LANA-RFP were subjected to immunoblotting analysis to detect HIF-1α (GFP) and LANA. The nuclear protein Sp1 and cytoplasm protein Hsp70 immunoblot analysis were performed to determine the efficiency of nuclear and cytoplasm fractionation. The results showed that the signal for HIF-1α in the nucleus was significantly higher than in the cytoplasm when LANA was coexpressed, but similar signals were seen in both the cytoplasm and nucleus when GFP-HIF-1α was expressed alone (Fig. 3C). Therefore, this suggests a new role for LANA in inducing HIF-1α nuclear accumulation under normoxic conditions.
FIG. 3.
LANA stimulates HIF-1α nuclear accumulation in U2OS cells. (A) Schematic representation showing the GFP and GFP-HIF-1α protein constructs. For HIF-1α, the location of the PAS (Per/Arnt/Sim) and ODD domains are indicated. (B) LANA induces nuclear translocation of GFP-HIF-1α. U2OS cells were transiently transfected with either GFP or GFP-HIF-1α (green) expression vectors in the presence or absence of the LANA-RFP (red) construct. At 24 h posttransfection, cells were fixed with 3% paraformaldehyde and subjected to nuclear staining (blue) with DAPI. Cells expressing GFP or GFP-HIF-1α alone were exposed to hypoxia for 12 h with CoCl2 treatment as the nuclear localization control. (C) Immunoblot analysis. U2OS cells expressing HIF-1α in the presence or absence of LANA-myc or hypoxia treatment were subjected to nuclear (N) and cytoplasmic (C) protein extract, followed by immunoblotting against HIF-1α and myc antibodies. Nuclear protein Sp1 and cytoplasmic protein Hsp70 were blotted as the fraction positive control. The relative densities of HIF-1α are shown in the right panel.
LANA enhances the activities of a HIF-1α responsive promoter.
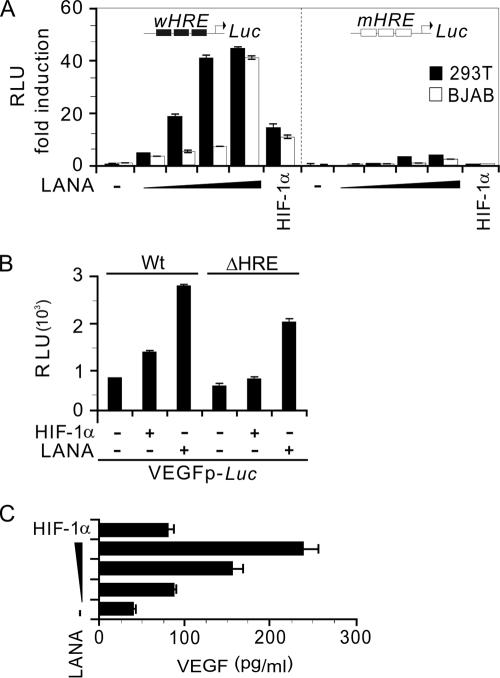
To investigate whether the effect of LANA enhancing HIF-1α accumulation would lead to an increase in transcriptional activities of HIF-1α under normoxic conditions, we analyzed the specific effects of LANA on the transcriptional activities of native HIF-1α targeting its HRE and compared this to HIF-1α expression alone. The promoter elements of a trimerized wild-type HRE (wHRE) and its corresponding mutant (mHRE) responsive elements fused upstream to the luciferase reporter individually were tested in these reporter assays using BJAB and HEK293T cell lines. The data showed that the wHRE promoter was highly responsive to LANA expression in both HEK293T and BJAB cell lines (Fig. 4A). However, little or no response was seen in the mutant HRE reporter (Fig. 4A). Specifically, using the luciferase reporters individually containing the well-known HIF-1α downstream genes VEGF promoter and its relative HRE deleted mutant, the reporter assays data showed that LANA dramatically enhance VEGF promoter activity and results in a drop in activity of ca. 30% once the HREs in the VEGF promoter were deleted (Fig. 4B). This suggests that the induction of VEGF by LANA is at least in part as a result of HIF-1α accumulation and its responsive element within the VEGF promoter.
FIG. 4.
LANA specifically increases the transcriptional activities of HIF-1α targeting downstream genes. (A) A trimerized HRE (wHRE) or its mutant (mHRE) fusion with luciferase was used for a reporter assay. BJAB and 293T cells were transfected with either pCEP4/HIF-1α (5 μg) or pA3M-LANA (0, 1, 2.5, 10, or 20 μg) in the presence of 5 μg of pGL/wHRE or pGL/mHRE reporter. The total amount was normalized by vector alone. Whole-cell lysates (40 μl) were subjected to luciferase reporter assay. The transcriptional activity of HIF-1α on HRE binding sites but not its mutant was enhanced by LANA in a dose-dependent manner. The transcriptional activity of HIF-1α mediated by LANA was expressed as the fold activation relative to the reporter construct alone. Means and standard deviations from three independent experiments are shown. RLU, relative luciferase unit. (B) Reporter assays of VEGF promoters and its HRE deletion mutant (ΔHRE). 293T cells were transfected with either pCEP4/HIF-1α (5 μg) or pA3M-LANA (10 μg) in the presence of the 5 μg of pGL3-1.5VEGF or pGL3-1.2VEGF reporter. (C) VEGF expression was increased in the culture medium of LANA-transfected 293T cells. We monitored the VEGF protein level in the cell culture supernatant by ELISA. The level of VEGF expression in the supernatant was increased by LANA in a dose-dependent manner. Values represent the means and standard deviations of the VEGF concentrations in the medium in three experiments.
To support these transcriptional studies, ELISAs were performed to determine the levels of VEGF, a downstream target of HIF-1α, which was previously demonstrated to be induced by KSHV infection (21, 47). The data obtained from cells transiently expressing LANA showed that the levels of VEGF produced were also significantly increased in HEK293T cells transfected with LANA compared to only HIF-1α expression (Fig. 4C). More importantly, increasing amounts of LANA showed a dramatic increase in VEGF levels as seen by ELISA (Fig. 4C).
HIF-1α nuclear accumulation mediated by LANA is dependent on its ODD domain.
In order to identify which structural motif involved in nuclear import of HIF-1α mediated by LANA, we generated four expression vectors containing GFP fused to wild type and three different domains of HIF-1α (schematically represented in Fig. 5A). After transient expression of these constructs with or without LANA-RFP in U2OS cells, the cells were analyzed for localization of the fusion proteins. As shown in Fig. 5B, the fusion protein containing the most central region of HIF-1α which spans the oxygen-dependent degradation (ODD) domain resulted in a predominantly nuclear accumulation when coexpressed with LANA-RFP (Fig. 5B). Moreover, equal cytoplasm and nuclear distribution of HIF-1α was seen when the ODD domain was deleted, indicating that its localization was not altered when LANA-RFP was coexpressed (Fig. 5B). This strongly supports the conclusion that the ODD domain of HIF-1α is critical for LANA-mediated HIF-1α nuclear translocation. Interestingly, unlike other truncated and full-length HIF-1α, the GFP fusion with HIF-1α amino acids 1 to 300 showed significant cytoplasmic fluorescence activity whether or not LANA-RFP was coexpressed in normoxic condition (Fig. 5B). To establish a quantitative set of analyses for our data, 50 fluorescent cells were routinely analyzed for compartmentalization of HIF-1α and subdivided into four categories according to previous studies (14, 45): N, exclusively nuclear fluorescence; N>C, nuclear fluorescence more than cytoplasmic fluorescence; N=C, equal distribution of nuclear and cytoplasmic fluorescence; and N<C, exclusively cytoplasmic fluorescence. The percentage of cells identified in each category is shown in Table 1. With LANA-RFP coexpression, 72% of cells transiently expressing the wild-type GFP-HIF-1α construct showed exclusively nuclear compartmentalization of HIF-1α (category N), whereas only a limited number of cells (6%) had similar signal with GFP-HIF-1α expression alone (Table 1). The GFP fused with truncated HIF-1α polypeptides showed that only the construct transiently expressing GFP-HIF-1α/330-530 had 10% of cells belonging to category N when LANA-RFP was coexpressed. No other truncated GFP-HIF-1α-expressing cells were seen in this category. This indicated that the ODD domain of HIF-1α is critical for LANA-mediated HIF-1α nuclear accumulation.
FIG. 5.
LANA facilitates HIF-1α nuclear accumulation by targeting the ODD domain of HIF-1α. (A) The schematic presents the truncated HIF-1α fusions with GFP tag. The full-length (1 to 826 amino acids [1-826 aa]) (FL), amino-terminal domain 1-300 aa (N), central domain with ODD domain 300-530 aa (M), and deleted central domain 1-3005̂30-826 aa (NC) are indicated. (B) LANA targets the ODD domain of HIF-1α to stimulate HIF-1α nuclear accumulation. U2OS cells were transfected with GFP or GFP with truncated HIF-1α (green) in the presence or absence of LANA-RFP (red) expression vectors. At 24 h posttransfection, cells were fixed with 3% paraformaldehyde and subjected to nuclear staining (blue) with DAPI.
TABLE 1.
Subcellular distribution of GFP-HIF-1α coexpressed with or without LANA in U2OS cellsa
| Construct | % Cells localized
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Vector
|
LANA
|
|||||||
| N | N>C | N=C | N<C | N | N>C | N=C | N<C | |
| GFP | 0 | 0 | 100 | 0 | 0 | 0 | 100 | 0 |
| GFP-HIF-1α | 6 | 20 | 74 | 0 | 72 | 26 | 2 | 0 |
| GFP-HIF-1α/1-330 | 0 | 0 | 16 | 84 | 0 | 0 | 18 | 82 |
| GFP-HIF-1α/330-530 | 0 | 36 | 64 | 0 | 10 | 76 | 14 | 0 |
| GFP-HIF-1α/Δ330-530 | 0 | 10 | 76 | 14 | 0 | 16 | 74 | 10 |
Cells were transfected with expression vectors as indicated and 24 h after transfection subjected to fixation and nuclear staining before microscopic observation and cell counting. N, exclusively nuclear fluorescence; N>C, nuclear fluorescence more than cytoplasmic fluorescence; N=C, equal distribution of nuclear and cytoplasmic fluorescence; and N<C, exclusively cytoplasmic fluorescence. At least 50 cells were analyzed for each sample. The numbers show the percentage of cells localized in the nucleus and cytoplasm of the transfected cells.
The amino-terminal residues 46 to 89 are required for LANA to mediate HIF-1α nuclear accumulation.
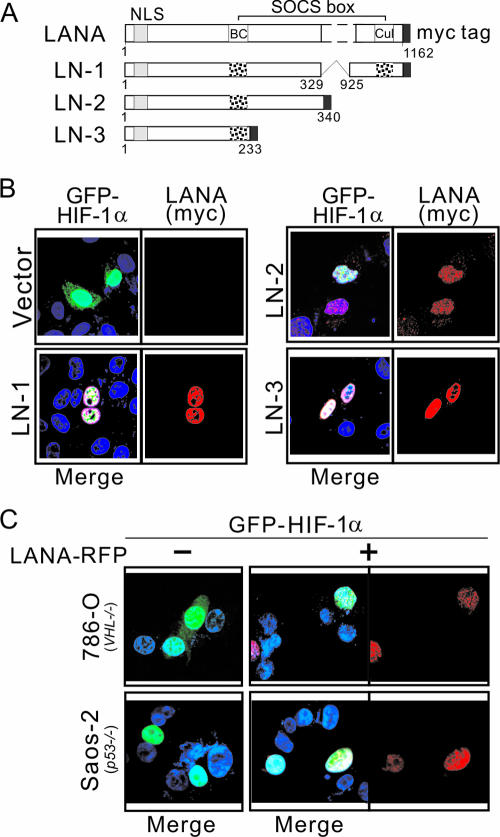
Previously, we demonstrated that LANA acts as an adapter of the EC5S ubiquitin complex blocking the HIF-1α suppressors VHL and p53 through its unconventional suppressors of cytokine signaling (SOCS)-box motif (Fig. 6A) (2). To test whether LANA-mediated HIF-1α nuclear accumulation is dependent on this motif, we coexpressed the LANA truncated mutants containing the entire (LN-1) or partly (LN-2 and LN-3) SOCS-box motif with GFP-HIF-1α in U2OS cells (Fig. 6A). Immunofluorescence assay data showed that the LANA mutant truncated from 1 to 233 residues containing only the BC-box component of the SOCS-box motif is sufficient to mediate HIF-1α nuclear accumulation (Fig. 6B). This suggests that the amino-terminal domain of LANA can mediate HIF-1α accumulation that is independent of its SOCS-box motif. Importantly, in the VHL-deficient 786-O cells, HIF-1α was also induced to nuclear accumulation by LANA coexpression but not HIF-1α alone, indicating that this accumulation is not dependent on VHL (Fig. 6C, top panel). Interestingly, in the p53-deficient Saos-2 cells, whether HIF-1α was expressed with LANA or not, the majority of HIF-1α was localized in nucleus (Fig. 5C, bottom panel).
FIG. 6.
The effect of LANA on HIF-1α nuclear accumulation is independent of its SOCS-box motif. (A) The schematic shows a truncated LANA fusion with a myc tag. LN-1 (1-329^925-1162 amino acids), which contains the entire SOCS-box (BC box and Cul box) motif, and LN-2 (1 to 340 amino acids) and LN-3 (1 to 233 amino acids), which contains part of the SOCS-box (only the BC box) motif, are indicated. (B) Immunofluorescence assays of U2OS cells cotransfected with GFP-HIF-1α and different truncated LANA polypeptides or vector alone. At 24 h posttransfection, cells were fixed with 3% paraformaldehyde and subjected to immunofluorescence assay, followed by the addition of mouse anti-myc antibody (red) and nuclear staining (blue) with DAPI. (C) LANA promotes HIF-1α nuclear accumulation in VHL-null 786-O and p53-null Saos-2 cell lines. The 786-O (top panel) and Saos-2 (bottom panel) cell lines were transfected with GFP-HIF-1α in the presence or absence of LANA-RFP. At 24 h posttransfection, the cells were fixed with 3% paraformaldehyde and subjected to immunofluorescence assay, followed by nuclear staining (blue) with DAPI.
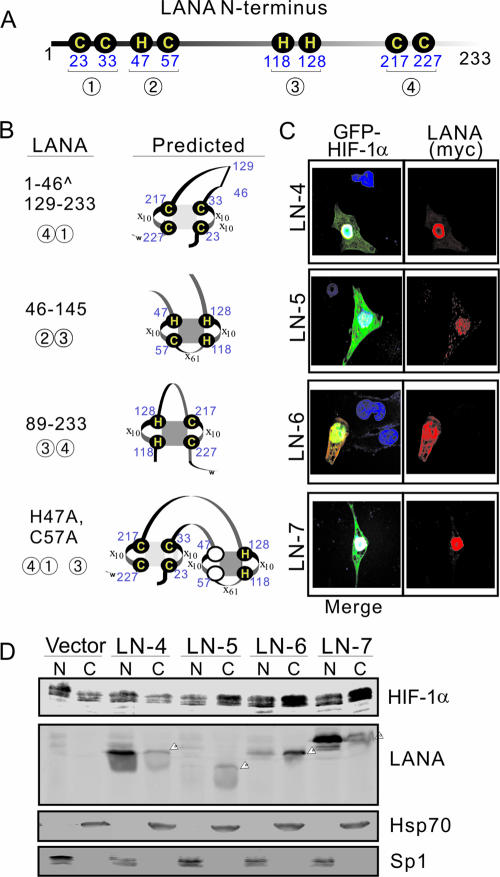
To further define which specific domain of LANA is necessary to mediate HIF-1α nuclear accumulation, we sought to identify the potential secondary structures that are located at the amino termini of LANA to associate with HIF-1α. The cysteine and histidine residues, within the region of LANA from amino acids 1 to 233 were then investigated. Figure 7A shows the positions of these residues. Based on the predicted potential functional RING domain formation via different disulfide bonds, three LANA mutants tagged with the myc epitope and mutated at these specific positions were generated (Fig. 7B). Immunofluorescence assays with these truncated LANA polypeptides coexpressed with GFP-HIF-1α constructs in U2OS cells were performed. The data showed that the LANA mutant with residues 47 to 129 deleted did diminish the level of HIF-1α nuclear accumulation (Fig. 7C, top panel). However, the LANA truncated mutant (LN5) containing only residues 46 to 145 was also not efficient in inducing HIF-1α nuclear localization (Fig. 7C). Interestingly, the LANA mutant with 1 to 89 residues deleted (LN6) not only lost the capability of inducing HIF-1α nuclear localization but was also dramatically reduced in its own ability to localize to the nucleus (Fig. 7C). To verify that the secondary structure containing histidine 47 and cysteine 57 is necessary for LANA to mediate HIF-1α nuclear accumulation, we generated double mutations of these two residues, which were converted to alanine and then performed similar immunofluorescence assays. Surprisingly, the data showed that mutation of these two residues is sufficient to destroy the effect of LANA on HIF-1α nuclear accumulation (Fig. 7C). In the cytoplasm and nuclear fractionation assays, the results also showed that the LANA mutants LN-5 with 46 to 129 residues deleted or LN-7 with only the H47 and C57 double mutations, the effect of LANA on HIF-1α nuclear accumulation was abolished (Fig. 7D). Expectedly, deletion of the nuclear localization sequence that existed in residues 1 to 23 of LANA resulted in LANA being mostly accumulated in the cytoplasmic compartment (Fig. 7D).
FIG. 7.
The secondary structure containing histidine 47 and cysteine 57 is required for LANA to mediate HIF-1α nuclear accumulation. (A) Analysis of histidine and cysteine positions in the amino termini of LANA residues 1 to 233. The residues (cysteines 23, 33, 57, 217, and 227 and histidines 47, 118, and 128), which potentially were involved in a RING finger domain, are classified into four groups (indicated in the figure by circled numbers 1 to 4) according to disulfide bond formation. (B) Amino-truncated mutants of LANA with potential secondary structures. LN-4 contains 1 to 46 and 129 to 233 residues with groups 1 and 4; LN-5 contains 46 to 145 residues with groups 2 and 3; LN-6 contains 89 to 233 residue with groups 3 and 4; LN-7 contains 1 to 233 residues including histidine 47 and cysteine 57 mutation with groups 1, 3, and 4. (C) Immunofluorescence assays of U2OS cells cotransfected with GFP-HIF-1α and different truncated LANA polypeptides. At 24 h posttransfection, the cells were fixed with 3% paraformaldehyde and subjected to immunofluorescence assay, followed by the addition of mouse anti-myc antibody (red) and nuclear staining (blue) with DAPI. (D) Immunoblotting analysis. U2OS cells expressing HIF-1α in the presence or absence of the truncated LANA mutants (LN-4, -5, -6, and -7) with a myc tag were subjected to nuclear (N) and cytoplasmic (C) protein extract, followed by immunoblotting against HIF-1α and myc antibodies. Nuclear protein Sp1 and cytoplasm protein Hsp70 were blotted as fraction positive controls.
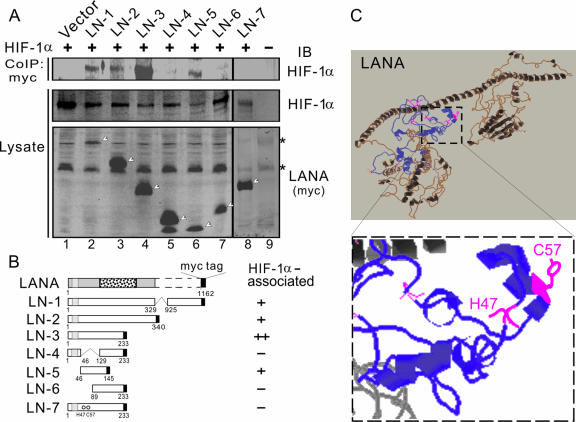
To determine whether the protein-protein interaction between LANA and HIF-1α is required for HIF-1α nuclear accumulation, we also performed immunoprecipitation assays in HEK293 cells using the LANA truncated mutants described above. As expected, the mutants with residues 46 to 129 deleted or with residues 1 to 89 deleted failed to interact with HIF-1α (Fig. 8A and B). However, compared to the truncated mutant of LANA 1 to 233, the mutant containing residues 46 to 145 associated with HIF-1α, even though it was not able to efficiently induce HIF-1α nuclear accumulation (Fig. 8A and B). This suggests that the secondary structure located in the region from residues 46 to 89 is necessary for LANA to interact with HIF-1α but not sufficient to induce HIF-1α nuclear accumulation. The construct with the histidine 47 and cysteine 57 both mutated also lost this association with HIF-1α, supporting this conclusion (Fig. 8A and B). Surprisingly, the secondary structure analysis of LANA reveals that residues H47 and C57 may be potentially located at separate α-helix motifs important for contact with HIF-1α (Fig. 8C).
FIG. 8.
The secondary domain with histidine 47 and cysteine 57 of LANA is associated with HIF-1α. (A) HEK293 cells were cotransfected HIF-1α with different LANA truncated expression plasmids (LN-1 to LN-7), respectively, as indicated. At 48 h posttransfection, cell lysates were subjected to immunoprecipitation and immunoblotting against myc or HIF-1α antibody. The arrow indicates the expressed LANA truncated proteins; the asterisks indicate nonspecific bands. (B) The relative binding ability of LANA truncated mutants to HIF-1α is indicated in the schematic in the right panel. (C) Proposed secondary domain of LANA associated with HIF-1α at its amino terminus. A three-dimensional structure of LANA, predicted by Robetta (http://robetta.bakerlab.org/), shows distinct amino- and carboxyl-terminal domains. The region from residues 1 to 233 of LANA that is involved in HIF-1α regulation is presented in blue. The HIF-1α associated residues histidine (H) 47 and cysteine (C) 57 are enlarged and marked in magenta in the lower panel.
DISCUSSION
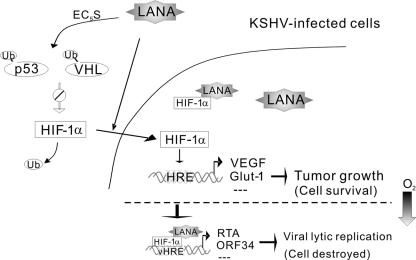
The data presented here demonstrate that the angiogenesis and tumor growth regulator HIF-1 is aberrantly accumulated in KSHV-infected PEL cell lines and that this accumulation is linked to the expression of the key latent viral protein LANA. LANA not only contains the SOCS motif, which can block the activities of the HIF-1α suppressors VHL and p53 as an adaptor of the ubiquitin complex (2), but also contains a specific 43-amino-acid domain that can mediate HIF-1α nuclear accumulation under normoxic conditions (Fig. 9). This highlights the role of LANA in regulating HIF-1α levels and nuclear translocation and suggests that HIF-1α is critical for KSHV-associated tumor growth during viral latent infection.
FIG. 9.
Schematic showing the LANA regulation of HIF-1α in KSHV latent infection. In the KSHV-infected B-lymphoma cells, the KSHV-encoded LANA acts as an adaptor of EC5S ubiquitin complex to induce HIF-1α suppressor (VHL and/or p53) for ubiquitylation and proteasomal degradation and, in turn, rescue HIF-1α (2). The rescued HIF-1α by LANA is associated with translocation to the nucleus, where HIF-1α activates the expression of downstream genes, including VEGF and Glut-1, to promote tumor growth and host cell survival. In hypoxic conditions, unknown factors (i.e., signaling events, including LANA modification) drive LANA to cooperate with HIF-1α to target viral lytic gene (e.g., RTA and ORF34) expression. This leads to the activation of virus lytic replication to destroy host cells.
VEGF, associated with angiogenesis, is an HIF-1α downstream target aberrantly activated in KS disease (23). The detection of abundant HIF-1α in PEL cell lines, along with enhanced expression of the HIF-1α target gene VEGF by LANA, confirmed that constitutive activation of genes regulated by LANA is an important contributing factor to the development of PEL. HIF-1α dysregulation is a consistent feature of KSHV-associated malignancies, which was reinforced by our observations that HIF-1α was overexpressed in the lesions of KS patient tissues (data not shown). Additional studies have shown that latent KSHV infection of endothelial cells can activate hypoxia-inducible factors, further supporting this conclusion (4).
Previously, we found that the nuclear extract from HEK293 cells transiently coexpressed with LANA and HIF-1α contains a complex of LANA and HIF-1α (3). To determine whether this nuclear accumulation of HIF-1α mediated by LANA is associated with the ability of LANA to functionally block the activities of the HIF-1α suppressors VHL and p53, we used the construct expressing LANA or HIF-1α with different fluorescence tags to transfect the VHL-deficient 786-O or p53-deficient Saos-2 cell lines, followed by immunofluorescence assays. The data showed that p53 deficiency alone was sufficient to induce HIF-1α nuclear accumulation, and LANA efficiently induced HIF-1α nuclear accumulation in the absence of VHL. This suggested that we could not exclude the possibility of LANA to promote HIF-1α nuclear accumulation via its SOCS-box motif through inhibition of p53 expression. However, immunofluorescence assays using a series of LANA truncated mutants identified a specific domain located within the amino-terminal residues 46 to 89 of LANA as important for HIF-1α nuclear accumulation, suggesting that this association mediated by LANA is independent on the inhibition of p53 at least through the SOCS motif (2).
To determine the likely secondary structure involved, we scanned the sequence for all of the histidine and cysteine residues with the potential of forming a disulfide bond within the region of 1 to 233 residues using the rule for constructing a RING finger domain (33). We assumed that this region can potentially form RING finger structures and generated different truncated mutants which were distinct in their potential ability to form a finger domain. Unexpectedly, the mutation of residues H47 and C57 destroyed the function of LANA to induce HIF-1α nuclear accumulation. This strongly showed that LANA-mediated HIF-1α nuclear accumulation was dependent on a specific motif containing H47 and C57 residues. The analysis of this region in a LANA three-dimensional structure model (http://www.robetta.com) reveals two relatively separate α helices containing the H47 and C57 residues strongly supporting this hypothesis. HIF-1α associated with residues 46 to 145 of LANA was not sufficient to induce HIF-1α nuclear accumulation, although the possibility existed that the secondary structure may be critical. The truncated mutants with only residues 46 to 145 deleted (LN-4) indicated that efficient nuclear import of LANA itself was also required for HIF-1α nuclear accumulation.
Only the wild-type HIF-1α and mutants that contained the ODD domain were capable of translocating to the nucleus in the presence of LANA. This suggested that in order to exert a dominant control of HIF-1α in KSHV-infected cells, LANA involvement with HIF-1α nuclear import through its ODD domain via another signaling pathway is quite possible. Further studies are required to determine the potential role of other signaling events and other cellular processes involved in the regulation of HIF-1α nuclear accumulation.
Here, our studies show that LANA contributes to HIF-1α nuclear accumulation, which therefore leads to the activation of downstream HIF-1-mediated transcriptional response. We have demonstrated that this function requires the potential α-helical structures of LANA which contains the H47 and C57 residues within its amino terminus. The specific domain of LANA interacting with HIF-1α was also shown to be distinct from that of other proteins (e.g., tumor suppressors p53 and Rb), which occur through its carboxyl-terminal domain (8, 10). This strengthens the case for a multifunctional role of LANA critical for KSHV-infected cell growth and proliferation. Thus, by combining the function of LANA which leads to an increase in HIF-1α accumulation and actively blocking the function of its suppressors, the resulting data provide compelling evidence that LANA contributes to the growth-stimulatory property of KSHV and that this activity is now linked to manipulation of the HIF-1 complex important for regulation of angiogenesis and tumor growth (Fig. 9).
LANA cooperates with HIF-1α to activate viral lytic gene expression in hypoxic conditions (3). However, LANA also dysregulates HIF-1α expression in latency without inducing lytic replication (2). Clearly, KSHV is capable of regulating cell survival and destruction (7, 40, 49). The regulation of HIF-1α levels and nuclear accumulation is likely to contribute at least in part to this regulation. The higher levels of HIF-1α accumulated during hypoxia, as distinct from latency, the different posttranslational modifications of LANA or HIF-1α in hypoxia and normoxic conditions, or other molecules associated with the LANA-HIF-1α complex in hypoxia, would further elucidate this complex regulatory network.
The evidence shows that intratumoral hypoxia usually occurs early during angiogenesis and tumor growth (9) and that elevated levels of HIF-1α during KSHV latent infection can induce the expression of a number of downstream genes essential for oxygen homeostasis and angiogenesis, including VEGF (12, 36). Given the role of HIF-1α in regulation of genes associated with cancer cell survival and tumor angiogenesis, the effect of LANA on HIF-1α nuclear accumulation raises the possibility that KSHV likely triggers the expression of hypoxia response factors, as well as survival factors, in KSHV-associated human malignancies early during infection and then later responds to hypoxia by activating viral lytic replication. This provides a fine balance between production of viral progeny and long-term survival in the intratumoral environment.
Acknowledgments
We are grateful to Amit Maity, Bala Chandran, Craig B. Thompson, Jon Aster, Gregg L. Semenza, and Volker H. Haase for generously providing reagents.
This study was supported by Public Health Service grants NCI CA072510 and CA091792, NIDCR DE014136, NIAID AI067037 and DE17338 (to E.S.R.). E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.An, J., Y. Sun, and M. B. Rettig. 2004. Transcriptional coactivation of c-Jun by the KSHV-encoded LANA. Blood 103:222-228. [DOI] [PubMed] [Google Scholar]
- 2.Cai, Q., J. S. Knight, S. C. Verma, P. Zald, and E. S. Robertson. 2006. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLOS Pathog. 2:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, Q., K. Lan, S. C. Verma, H. Si, D. Lin, and E. S. Robertson. 2006. Kaposi's sarcoma-associated herpesvirus latent protein LANA interacts with HIF-1α to upregulate RTA expression during hypoxia: latency control under low oxygen conditions. J. Virol. 80:7965-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll, P. A., H. L. Kenerson, R. S. Yeung, and M. Lagunoff. 2006. Latent Kaposi's sarcoma-associated herpesvirus infection of endothelial cells activates hypoxia-induced factors. J. Virol. 80:10802-10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Davis, D. A., A. S. Rinderknecht, J. P. Zoeteweij, Y. Aoki, E. L. Read-Connole, G. Tosato, A. Blauvelt, and R. Yarchoan. 2001. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97:3244-3250. [DOI] [PubMed] [Google Scholar]
- 8.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folkman, J. 1992. The role of angiogenesis in tumor growth. Semin. Cancer Biol. 3:65-71. [PubMed] [Google Scholar]
- 10.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 11.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 12.Harris, A. L. 2002. Hypoxia-a key regulatory factor in tumour growth. Nat. Rev. Cancer 2:38-47. [DOI] [PubMed] [Google Scholar]
- 13.Hockel, M., K. Schlenger, S. Hockel, B. Aral, U. Schaffer, and P. Vaupel. 1998. Tumor hypoxia in pelvic recurrences of cervical cancer. Int. J. Cancer 79:365-369. [DOI] [PubMed] [Google Scholar]
- 14.Kallio, P. J., K. Okamoto, S. O'Brien, P. Carrero, Y. Makino, H. Tanaka, and L. Poellinger. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1α. EMBO J. 17:6573-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 17.Krishnan, H. H., P. P. Naranatt, M. S. Smith, L. Zeng, C. Bloomer, and B. Chandran. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, J. W., S. H. Bae, J. W. Jeong, S. H. Kim, and K. W. Kim. 2004. Hypoxia-inducible factor (HIF-1)α: its protein stability and biological functions. Exp. Mol. Med. 36:1-12. [DOI] [PubMed] [Google Scholar]
- 19.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 20.Liu, X. H., A. Kirschenbaum, M. Lu, S. Yao, A. Dosoretz, J. F. Holland, and A. C. Levine. 2002. Prostaglandin E2 induces hypoxia-inducible factor-1α stabilization and nuclear localization in a human prostate cancer cell line. J. Biol. Chem. 277:50081-50086. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Z. Y., R. K. Ganju, J. F. Wang, M. A. Ona, W. C. Hatch, T. Zheng, S. Avraham, P. Gill, and J. E. Groopman. 1997. Cytokine signaling through the novel tyrosine kinase RAFTK in Kaposi's sarcoma cells. J. Clin. Investig. 99:1798-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maity, A., N. Pore, J. Lee, D. Solomon, and D. M. O'Rourke. 2000. Epidermal growth factor receptor transcriptionally up-regulates vascular endothelial growth factor expression in human glioblastoma cells via a pathway involving phosphatidylinositol 3′-kinase and distinct from that induced by hypoxia. Cancer Res. 60:5879-5886. [PubMed] [Google Scholar]
- 23.Masood, R., J. Cai, T. Zheng, D. L. Smith, Y. Naidu, and P. S. Gill. 1997. Vascular endothelial growth factor/vascular permeability factor is an autocrine growth factor for AIDS-Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 94:979-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 25.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minet, E., I. Ernest, G. Michel, I. Roland, J. Remacle, M. Raes, and C. Michiels. 1999. HIF1A gene transcription is dependent on a core promoter sequence encompassing activating and inhibiting sequences located upstream from the transcription initiation site and cis elements located within the 5′UTR. Biochem. Biophys. Res. Commun. 261:534-540. [DOI] [PubMed] [Google Scholar]
- 27.Muromoto, R., K. Okabe, M. Fujimuro, K. Sugiyama, H. Yokosawa, T. Seya, and T. Matsuda. 2006. Physical and functional interactions between STAT3 and Kaposi's sarcoma-associated herpesvirus-encoded LANA. FEBS Lett. 580:93-98. [DOI] [PubMed] [Google Scholar]
- 28.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 29.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear. antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravi, R., B. Mookerjee, Z. M. Bhujwalla, C. H. Sutter, D. Artemov, Q. Zeng, L. E. Dillehay, A. Madan, G. L. Semenza, and A. Bedi. 2000. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1α. Genes Dev. 14:34-44. [PMC free article] [PubMed] [Google Scholar]
- 31.Ruas, J. L., L. Poellinger, and T. Pereira. 2005. Role of CBP in regulating HIF-1-mediated activation of transcription. J. Cell Sci. 118:301-311. [DOI] [PubMed] [Google Scholar]
- 32.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saurin, A. J., K. L. Borden, M. N. Boddy, and P. S. Freemont. 1996. Does this have a familiar RING? Trends Biochem. Sci. 21:208-214. [PubMed] [Google Scholar]
- 34.Seagroves, T. N., H. E. Ryan, H. Lu, B. G. Wouters, M. Knapp, P. Thibault, K. Laderoute, and R. S. Johnson. 2001. Transcription factor HIF-1 is a necessary mediator of the Pasteur effect in mammalian cells. Mol. Cell. Biol. 21:3436-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semenza, G. L. 2000. HIF-1 and human disease: one highly involved factor. Genes Dev. 14:1983-1991. [PubMed] [Google Scholar]
- 36.Semenza, G. L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3:721-732. [DOI] [PubMed] [Google Scholar]
- 37.Si, H., and E. S. Robertson. 2006. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 80:697-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric. Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 39.Stadler, P., A. Becker, H. J. Feldmann, G. Hansgen, J. Dunst, F. Wurschmidt, and M. Molls. 1999. Influence of the hypoxic subvolume on the survival of patients with head and neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 44:749-754. [DOI] [PubMed] [Google Scholar]
- 40.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturzl, M., C. Zietz, P. Monini, and B. Ensoli. 2001. Human herpesvirus-8 and Kaposi's sarcoma: relationship with the multistep concept of tumorigenesis. Adv. Cancer Res. 81:125-159. [DOI] [PubMed] [Google Scholar]
- 42.Talks, K. L., H. Turley, K. C. Gatter, P. H. Maxwell, C. W. Pugh, P. J. Ratcliffe, and A. L. Harris. 2000. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 157:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma, S. C., S. Borah, and E. S. Robertson. 2004. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 78:10348-10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ylikomi, T., M. T. Bocquel, M. Berry, H. Gronemeyer, and P. Chambon. 1992. Cooperation of proto-signals for nuclear accumulation of estrogen and progesterone receptors. EMBO J. 11:3681-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zagzag, D., H. Zhong, J. M. Scalzitti, E. Laughner, J. W. Simons, and G. L. Semenza. 2000. Expression of hypoxia-inducible factor 1α in brain tumors: association with angiogenesis, invasion, and progression. Cancer 88:2606-2618. [PubMed] [Google Scholar]
- 47.Zhang, X., J. F. Wang, B. Chandran, K. Persaud, B. Pytowski, J. Fingeroth, and J. E. Groopman. 2005. Kaposi's sarcoma-associated herpesvirus activation of vascular endothelial growth factor receptor 3 alters endothelial function and enhances infection. J. Biol. Chem. 280:26216-26224. [DOI] [PubMed] [Google Scholar]
- 48.Zhong, H., A. M. De Marzo, E. Laughner, M. Lim, D. A. Hilton, D. Zagzag, P. Buechler, W. B. Isaacs, G. L. Semenza, and J. W. Simons. 1999. Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 59:5830-5835. [PubMed] [Google Scholar]
- 49.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]