Abstract
The human cytomegalovirus (HCMV) immediate-early 2 (IE2) transactivator has previously been shown to form intranuclear, dot-like accumulations in association with subnuclear structures known as promyelocytic leukemia protein (PML) nuclear bodies or ND10. We recently observed that IE2 can form dot-like structures even after infection of PML knockdown cells, which lack genuine ND10. To further analyze the determinants of IE2 subnuclear localization, a recombinant HCMV expressing IE2 fused to the enhanced green fluorescent protein was constructed. We infected primary human fibroblasts expressing Sp100 fused to the autofluorescent protein mCherry while performing live-cell imaging experiments. These experiments revealed a very dynamic association of IE2 dots with ND10 structures during the first hours postinfection: juxtaposed structures rapidly fused to precise colocalizations, followed by segregation, and finally, the dispersal of ND10 accumulations. Furthermore, by infecting PML knockdown cells we determined that the number of IE2 accumulations was dependent on the multiplicity of infection. Since time-lapse microscopy in live-infected cells revealed that IE2 foci developed into viral replication compartments, we hypothesized that viral DNA could act as a determinant of IE2 accumulations. Direct evidence that IE2 molecules are associated with viral DNA early after HCMV infection was obtained using fluorescence in situ hybridization. Finally, a DNA-binding-deficient IE2 mutant could no longer be recruited into viral replication centers, suggesting that the association of IE2 with viral DNA is mediated by a direct DNA contact. Thus, we identified viral DNA as an important determinant of IE2 subnuclear localization, which suggests that the formation of a virus-induced nucleoprotein complex and its spatial organization is likely to be critical at the early stages of a lytic infection.
Human cytomegalovirus (HCMV), a member of the beta subgroup of herpesviruses, generally causes asymptomatic infections in immunocompetent individuals. However, it is of considerable clinical importance in immunocompromised persons such as organ transplant recipients, tumor and AIDS patients, and prenatally infected newborns (9).
Similar to those of other herpesviruses, the HCMV open reading frames are expressed in a temporally regulated cascade consisting of three sequential phases, termed immediate-early (IE), early, and late (13, 35, 52). IE gene expression is driven mainly by a very strong and complex regulatory element known as the major IE enhancer promoter (MIEP) and results in synthesis of the two most abundant IE gene products, the important viral regulatory factors IE1p72 (IE1) and IE2p86 (IE2) (45). Both viral IE proteins are expressed via differential splicing from a single transcript consisting of five exons (47, 48). IE1 mRNA comprises exons 1 to 4, whereas IE2 transcripts contain exons 1 to 3 and 5. Consequently, the two major IE proteins share a common N-terminal domain of 85 amino acids.
IE1 and IE2 both cooperate in stimulating viral early and late gene expression and thus play a crucial role in the regulation of a productive HCMV infection (33, 46). While the generation of HCMV mutants has demonstrated that IE2 is indispensable for viral replication, since a deletion of exon 5 of the ie1/2 gene region results in a nonviable virus (34), viruses with IE1 deleted exhibit a growth defect only at a low multiplicity of infection (MOI) but replicate efficiently at a high MOI (19). Consequently, IE2 is thought to be the key viral transactivator, as it is absolutely required for lytic infection and progression of the replicative cycle from the IE phase to the early phase.
To fulfill their fundamental transactivator functions, IE1 and IE2 are transported to the nucleus of an infected cell during the initial stage of infection, where they accumulate in distinct foci that are directly associated with a cellular subnuclear structure known as promyelocytic leukemia protein (PML) nuclear bodies or nuclear domain 10 (ND10) (3, 26). ND10 domains are spherical nuclear substructures that represent accumulations of multiple cellular proteins such as Sp100, hDaxx, BLM, or SUMO-1, which require the PML protein for their assembly (36). For a long time, this subcellular compartment, which colocalizes with sites of input viral genome accumulation, was hypothesized to be essential for viral replication, since only viral DNA deposited at ND10 had been demonstrated to initiate transcription (4, 24, 25). However, recent studies using primary human foreskin fibroblasts (HFFs) with a small interfering RNA-mediated knockdown (kd) of PML revealed enhanced replication efficacy of either HCMV or an ICP0-deleted herpes simplex virus type 1 (HSV-1) in the absence of genuine ND10, thus strongly suggesting an antiviral function of this subnuclear structure (16, 50).
While IE1 demonstrates perfect colocalization with ND10 during the first hours after infection, IE2 can only be found juxtaposed to these domains in infected cells (3, 4, 25, 26). Hence, these viral regulatory proteins do not occupy exactly the same space but are positioned next to each other during infection. These findings imply that the determinants for the subnuclear distribution of these proteins differ. For the IE1 protein, a direct interaction with the ND10 constituent PML has been reported (2). Consistent with this finding, a diffuse staining pattern of IE1 is detectable in the apparent absence of PML immediately after infection of PML-deficient cells (50). The presence of PML-associated IE1 aggregations in normal HFFs, however, is only temporary, as IE1 is able to induce a dispersal of ND10 accumulations starting at about 3 h postinfection (hpi) (3, 26, 54). Consequently, both proteins, IE1 and PML, redistribute into a nuclearly diffuse form early after infection. For IE2, however, ND10 domains appear not to be the structures that decisively determine the subnuclear localization of this viral regulatory protein. This assumption is based on the following observations: (i) only transiently expressed IE2 perfectly colocalizes with ND10 (3); (ii) during infection, IE2 is not positioned precisely at these subnuclear structures but accumulates adjacent to them (3, 4, 25, 26); (iii) IE2 retains its dot-like distribution pattern although ND10 domains are disrupted by the IE1 protein during the initial hours after infection (3); (iv) IE2 accumulation in distinct foci can be observed during infection of PML-depleted cells devoid of genuine ND10 structures, as demonstrated recently (50). This finding strongly suggests that other viral factors in addition to ND10 domains determine the subnuclear localization of this viral regulatory protein.
Interestingly, previous research on the immediate-early regulatory proteins ICP0 and ICP4 of HSV-1 revealed a similar situation: while ICP0 was shown to colocalize precisely with PML, ICP4 formed foci that were only juxtaposed to ND10 (18). Due to the observation that ICP4 foci developed into replication compartments, Everett et al. (18) proposed that ND10-associated ICP4 foci represent ICP4 molecules being recruited onto parental viral genomes (18). This concept was confirmed in subsequent publications of this group, giving rise to the hypothesis that formation of a nucleoprotein complex between viral DNA and a DNA-binding transactivator protein may constitute a critical step early in lytic infection that is common to the alpha- and betaherpesviruses (15, 17, 18).
In order to further investigate the determinants of IE2 subnuclear localization, a recombinant HCMV expressing an enhanced green fluorescent protein (EGFP)-tagged version of the protein (termed AD169/IE2-EGFP) was generated using bacterial artificial chromosome (BAC) technology. Furthermore, via retroviral transduction of primary HFFs, cells stably expressing a chimeric protein consisting of the autofluorescent protein mCherry and the ND10 component Sp100 were generated (44), thus allowing us to analyze the in vivo dynamics of IE2 accumulation with regard to ND10 localization using live-cell microscopy. These experiments revealed a very dynamic spatial association of IE2 dots with ND10 domains during the first hours postinfection. Furthermore, we showed that dot-like IE2 accumulations colocalize with input viral genomes and develop into replication compartments during progression of the replicative cycle. Since a DNA-binding-deficient mutant of IE2 could no longer be recruited into viral replication compartments, we concluded that, similarly to the ICP4 of HSV-1, viral DNA acts as an important determinant of IE2 subnuclear localization.
MATERIALS AND METHODS
Oligonucleotides and plasmids.
Oligonucleotides used for cloning, site-directed mutagenesis, and generation of recombinant BACs were obtained from Biomers (Ulm, Germany); the sequences are listed in Table 1. Plasmid pHM990, coding for IE2 fused to EGFP, was constructed as described elsewhere (50). For generation of expression plasmid pHM2626, which encodes a DNA-binding-deficient IE2 mutant fused to EGFP, nucleotide exchanges were introduced via site-directed mutagenesis, using the primer pair delDNAbinIE2-5′ and delDNAbinIE2-3′, resulting in the substitution of two histidines (amino acids 446 and 452 of IE2) of a putative zinc finger motif within IE2 with leucines, thus abolishing the DNA binding of IE2 (32). Site-directed mutagenesis was performed as described previously (29). Plasmid pHM2397, expressing Sp100 isoform A fused with the monomeric red fluorescent protein mCherry, was generated by PCR amplification of the mCherry coding sequence, using primers 5′- and 3′mCherry together with plasmid pRSET-B-mCherry as the template (44). In parallel, the coding sequence of Sp100 was amplified with primers 5′Sp100-mCherry and 3′FL-Sp100. Finally, primers 5′mCherry/BstXI-Sp100 and 3′Sp100/ApaI-mCherry were used for a hybrid PCR to generate the mCherry-Sp100 fusion gene, which was then inserted via BstXI and ApaI into the pLenti6/V5-D-Topo vector (Invitrogen). The recombinant vector pHM1471 was constructed as follows for generation of a recombinant BAC expressing IE2 fused to EGFP. First, the 5′ end of the genomic sequence required for homologous recombination was amplified, using plasmid pRR47 as the template and oligonucleotides FragA-5 and FragA-3mul. The resulting PCR product was ligated with plasmid pHM134 via HindIII and MluI, resulting in plasmid pHM1438. Next, the 3′ homologous region was amplified by PCR, using cosmid pCM1058 as the template and oligonucleotides FragB-5 and FragB-3, followed by insertion into pHM1438, using StuI and SphI, giving rise to plasmid pHM1469. Finally, the recombination cassette was released from vector pHM1469 by restriction with Asp718 and SphI and inserted into pST76K-SR to yield recombination vector pHM1471.
TABLE 1.
Oligonucleotides used for construction of plasmids and recombinant BACs
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| 5′Red1-N1-IE2/Frag.B | CATAGCGGCCGCGAGAGACATGGACTCTTGTAC |
| 3′Red1-N1-IE2/Frag.B | CATAGCGGCCGCGCATGCGCCATCGTTACCACGTCCCATC |
| delDNAbinIE2-5′ | CCTCATGGAGCTTACCATGCCCGTGACACTTCCACCCGAAG |
| delDNAbinIE2-3′ | CTTCGGGTGGAAGTGTCACGGGCATGGTAAGCTCCATGAGG |
| 5′mCherry | CACCATGGTGAGCAAGGGCGAGGAG |
| 3′mCherry | CTTGTACAGCTCGTCCATGCCG |
| 5′Sp100-mCherry | GGCATGGACGAGCTGTACAAGATGGCAGGTGGGGGCGGC |
| 3′FL-Sp100 | AGTCGATATCCTAATCTTCTTTACCTGACCC |
| 5′mCherry/BstXI-Sp100 | CATACCAGTGTGGTGGATGGTGAGCAAGGGCGAGGAG |
| 3′Sp100/ApaI-mCherry | CATAGGGCCCCTAATCTTCTTTACCTGACCC |
BAC mutagenesis.
For construction of the IE2-EGFP BAC, a two-step replacement procedure was performed as described previously (8). Starting with a BACmid (ΔkΔex5) already lacking the exon 5 sequence of the ie1/2 gene locus (20), an exon 5-EGFP hybrid gene was reinserted, using a recombination strategy involving cointegrate formation. For this, an 800-nucleotide fragment comprising 3′ genomic flanking sequences of exon 5 required for homologous recombination was amplified, using oligonucleotides 5′ and 3′Red1-N1-IE2/Frag.B and plasmid pHM1471 as templates. The resulting PCR product was inserted into plasmid pHM990 via NotI, giving rise to plasmid pHM2290. Thereafter, the IE2-EGFP/Frag.B fusion sequence was recloned into pHM1471, using restriction enzymes SphI and MluI, finally resulting in recombination vector pHM2291. In the next step, pHM2291 was transformed into Escherichia coli (strain DH10B) cells already harboring BACmid ΔkΔex5. Transformants were selected at 30°C on Luria-Bertani (LB) agar plates containing chloramphenicol (15 mg/ml) and kanamycin (15 mg/ml) to allow for cointegrate formation. Clones with cointegrates were subsequently identified by incubating the bacteria on chloramphenicol- and kanamycin-positive agar plates at 43°C. To achieve a resolution of the cointegrates, clones were streaked on LB plates containing chloramphenicol only and incubated at 30°C. Finally, to select for clones with a loss of the cointegrate, the bacteria were subjected to chloramphenicol agar plates additionally supplemented with 5% sucrose.
Viral nucleic acid isolation and analysis.
BACmid DNA was isolated from bacteria by a standard alkaline lysis minipreparation procedure from 5-ml liquid cultures and characterized by restriction digestion using the enzyme BamHI. Thereafter, DNA fragments were separated via electrophoresis at 30 V overnight, using a 0.7% agarose gel. For Southern blot analysis, the DNA was transferred onto a nylon membrane (Biodyne B transfer membrane; PALL, Portsmouth, United Kingdom) and probed with biotinylated DNA spanning the IE2 cDNA sequence of the ie1/2 gene region of HCMV (exons 2, 3, and 5) according to the instructions of the manufacturer (NEBlot Phototope kit; Biolabs, Frankfurt, Germany).
Cells and viruses.
HFF cells were maintained in Eagle's minimal essential medium (GIBCO/BRL, Eggenstein, Germany) supplemented with 5% fetal calf serum. HFFs with a small interfering RNA-mediated kd of PML (PML-kd cells) were cultured in Dulbecco's minimal essential medium (GIBCO/BRL, Eggenstein, Germany) supplemented with 10% fetal calf serum and 5 μg/ml puromycin (50). HFFs stably expressing Sp100 fused to the monomeric red autofluorescent protein mCherry were generated via retrovirus transduction, using the ViraPower lentiviral system according to the manufacturer's protocol (Invitrogen, Karlsruhe, Germany), and maintained in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum plus 2 μg/ml blasticidin. For transfection experiments, HFFs or mCh-Sp100 cells were seeded either on coverslips or into 4-well, chambered coverglass units with coverslip quality glass bottoms (Lab-Tek; Nunc). Transfection was performed using a TransPEI transfection reagent (Eurogentec, Belgium) according to the manufacturer's instructions. Viral stocks of wild-type (wt) AD169 and the recombinant AD169/IE2-EGFP virus were prepared and titrated via IE1p72 fluorescence as described previously (31). Quantification of viral input DNA by real-time PCR was also performed exactly as described previously (31, 50).
Reconstitution of recombinant cytomegaloviruses.
One day before transfection, HFFs were seeded into 6-well dishes at a density of 3.5 × 105 cells/well. About 1 μg of BACmid DNA and 1 μg of the pp71 expression plasmid pCBpp71 were cotransfected, using SuperFECT transfection reagent (QIAGEN, Hilden, Germany) according to the instructions of the manufacturer. One week after transfection, the cells were transferred into 25-cm2 flasks and incubated until plaques appeared.
Antibodies, indirect immunofluorescence analysis, and Western blot analysis.
The polyclonal antisera raised against exon 5 of IE2p86 (referred to as anti-pHM178) or pUL84 of HCMV were generated by immunizing rabbits with the respective prokaryotically expressed proteins. MAb-UL44 BS 510, MAb-MCP 28-4, and MAb-pp65 65-33 were kindly provided by B. Plachter (University of Mainz, Germany) and W. Britt (University of Birmingham, AL). MAb p63-27 and MAb 41-18 directed against IE1p72 and pp28 (UL99) were described previously (5, 39). MAb-UL69 69-66 (55) was used for detection of pUL69. For detection of endogenous PML and hDaxx, respectively, the monoclonal antibodies PG-M3 (Santa Cruz Biotechnology, Santa Cruz, CA) and MCA2143 (Serotec, Oxford, United Kingdom) were used. The monoclonal antibody Ac-15, which recognizes β actin, was purchased from Sigma (Deisenhofen, Germany). Anti-mouse as well as anti-rabbit horseradish peroxidase-conjugated secondary antibodies were obtained from Dianova (Hamburg, Germany), while Alexa 488-, Alexa 555-, and Cy3-conjugated secondary antibodies were purchased from Molecular Probes and GE Healthcare.
For indirect immunofluorescence analysis, primary HFF cells (1 × 105) were grown on coverslips for transient transfection or HCMV infection. The conditions for transfection of the HFFs and the fixation and immunodetection of viral and cellular proteins were as previously described (22).
For Western blotting, extracts from infected cells were prepared in sodium dodecyl sulfate loading buffer, separated on 8 to 15% polyacrylamide gels containing sodium dodecyl sulfate, and transferred to nitrocellulose membranes. Western blotting and chemiluminescence detection were performed as described previously (30).
Time-lapse microscopy of live cells.
Live cells were observed using a Leica DMIRE2 inverted fluorescence microscope equipped with a Leica DFC300 FX digital camera. The excitation wavelength was controlled by mercury lamp illumination and a manual filter wheel equipped with filters suitable for EGFP, the monomeric red fluorescent protein mCherry, and DAPI (4′,6′-diamidino-2-phenylindole). Camera image acquisition was controlled by IM50 software (Leica). Single images or timed image sequences were exported as TIFF files from the IM50 software. Individual frames were prepared for presentation using Photoshop. An essential part of the system was the ability to maintain the cells for long periods in a controlled environment with constant temperature, CO2, and humidity. The conditions within the box were maintained at 37°C, 5% CO2, and 90% humidity to keep the cells healthy indefinitely. The cells were seeded into 4-well, chambered coverglass units with coverslip quality glass bottoms (Lab-Tek; Nunc) at a density of 1 × 105 cells per well the day before each experiment, and the samples were examined with a ×63 dry objective lens.
Fluorescence in situ hybridization.
Fluorescence in situ hybridization was performed according to a protocol described by Everett et al. (15). HFF cells were grown on 13-mm-diameter glass coverslips and then infected with HCMV AD169 at an MOI of 0.5 PFU per cell. At various times thereafter, the cells were washed twice with phosphate-buffered saline (PBS) containing 1% fetal calf serum (FCS) and then fixed by incubation for 5 min at −20°C with precooled 95% ethanol-5% glacial acetic acid. The cells were then washed three times with PBS-1% FCS and stored at 4°C until further use. The probe for in situ hybridization was a BAC DNA containing the entire HCMV genome (8) (a gift from Ulrich H. Koszinowski, Munich, Germany). After a DNaseI treatment, the probe was labeled by nick translation with Cy3-dCTP (Amersham) according to the manufacturer's protocol. The cells were prehybridized by incubation for 30 min at 37°C with 20 μl of hybridization buffer (50% formamide, 10% dextran sulfate, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) in a humidified microarray hybridization chamber (Camlab). The coverslips were then removed from the chamber and blotted dry. The probe was added to the hybridization buffer to a concentration of 1 ng/μl, after which each coverslip was incubated at 95°C for 2 min to denature the probe and the sample. The coverslips and hybridization mixture were then placed into the humidified chamber, and hybridization was continued overnight at 37°C. The cells were then washed for 5 min at 60°C with 2× SSC and once with 2× SSC at room temperature. After a further washing with PBS-1% FCS, the coverslips were incubated with the anti-IΕ2 polyclonal antibody anti-pHM178 for 1 h. After several washings, the cells were incubated with fluorescein isothiocyanate-conjugated sheep anti-mouse immunoglobulin G (Sigma) for another hour. The cells were washed several times, air dried, and then mounted on glass slides by using a glycerol-PBS mounting solution (CitiFluor). Images were analyzed using a Zeiss Axioplan-2 microscope and recorded with a cooled SPOT color digital camera (Diagnostic Instruments, Sterling Heights, MI) under image capture conditions that eliminated channel overlap. The images were exported as TIFF files and then processed with Photoshop.
RESULTS
Construction and verification of a recombinant HCMV expressing an IE2-EGFP fusion protein.
In order to investigate the in vivo dynamics of IE2 protein accumulation during infection, we generated a recombinant HCMV expressing EGFP fused to the C terminus of IE2. For construction of this recombinant virus, termed AD169/IE2-EGFP, BAC technology was employed (for a review, see reference 1). Starting with a BAC derived from the HCMV genome of strain AD169 carrying a deletion of the complete exon 5 sequence of the ie1/2 gene locus (designated ΔkΔex5) (20), we reintroduced the exon 5 sequence fused to EGFP via homologous recombination between up- and downstream-flanking regions of exon 5. For this purpose, a recombinant vector (pHM2291) based on plasmid pST76K-SR and containing the exon 5-EGFP fusion sequence enframed by the 5′- and 3′-ends of the homologous genomic sequences of ie2 exon 5 was used. The pST76K-SR-derived vector backbone also harbors a temperature-sensitive origin of replication, a kanamycin resistance gene, the recombinase gene recA, and the negative selection marker gene sacB, thus allowing a recombination strategy that involves the formation of a cointegrate (for a detailed description, see Materials and Methods).
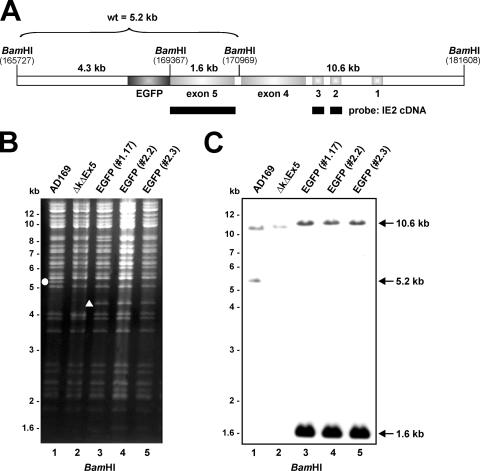
The successful and correct integration of the exon 5-EGFP fusion sequence into the BAC ΔkΔex5 was subsequently determined by PCR and sequencing reactions with primer pairs binding either within the exon 5 sequence or within exon 5 and the upstream- and downstream-flanking regions (data not shown). Furthermore, the structural integrity of the recombinant BACs was verified by restriction enzyme digestion and Southern blot analysis (Fig. 1). The cleavage of three independent IE2-EGFP BACs (Fig. 1B, lanes 3 to 5) with BamHI yielded complex restriction patterns identical to those of HCMV AD169 wt DNA and the parental ΔkΔex5 BAC, with the following predicted exceptions. The insertion of an additional BamHI restriction site together with the EGFP sequence in the recombinant BACs resulted in the replacement of a 5.2-kb fragment of the wt BAC (Fig. 1B, lane 1) with a 4.3-kb fragment in a positive IE2-EGFP BAC (Fig. 1B, lanes 3 to 5). The diagram in Fig. 1A indicates all existing BamHI restriction sites of the relevant ie2 gene region plus the additional BamHI cleavage site of the inserted EGFP sequence. Due to deletion of exon 5, the 5.2-kb wt fragment is also absent in the parental ΔkΔex5 BAC (Fig. 1B, lane 2). The correct integration of the IE2-EGFP recombination cassette was finally confirmed by Southern blot hybridization experiments using the BamHI-digested BAC DNAs together with a biotinylated probe representing the IE2 cDNA sequence, thus comprising exons 2, 3, and 5 of the MIE gene locus, as indicated in Fig. 1A. Consistent with the integration of an additional BamHI site into the IE2-EGFP BAC, successful recombination (Fig. 1C, lanes 3 to 5) resulted in a 1.6-kb fragment that migrates faster than the 5.2-kb fragment of the wt BAC (Fig. 1C, lane 1). The latter fragment was missing in the ΔkΔex5 BAC (Fig. 1C, lane 2). Taken together, the results of Southern blotting, PCR, and nucleotide sequence analysis showed that recombination events took place at the correct sites of the HCMV genome. Thus, we successfully generated a recombinant HCMV BAC with the insertion of an EGFP-tagged IE2 sequence.
FIG. 1.
Generation of a recombinant HCMV expressing an EGFP-tagged version of the viral transactivator protein IE2. The EGFP sequence was fused to the 3′ end of the open reading frame UL122 (exon 5 of the IE1/2 gene region), and an exon 5-EGFP hybrid gene was integrated into the HCMV genome via homologous recombination in E. coli, using an HCMV BAC lacking exon 5 of the MIE gene locus. (A) Schematic representation of the HCMV genomic region where the exon 5-EGFP fusion gene was inserted. To check for the integrity of the obtained recombinant BACs, BamHI digestion and subsequent Southern blot analysis were performed. The BamHI restriction sites and the probe used for Southern blotting are indicated in the diagram. (B) Wild-type AD169 and recombinant AD169/IE2-EGFP BACs were digested with BamHI, and the fragments were separated by agarose gel electrophoresis. Due to an additional BamHI restriction site inserted together with the EGFP sequence, the wt 5.2-kb fragment (indicated by a circle in lane 1) was digested into a 4.3-kb fragment for successful insertion of the IE2-EGFP fusion gene (indicated by a triangle in lane 3). (C) Southern blot analysis of BamHI digests of BAC DNAs using an IE2 cDNA probe.
Reconstitution and growth analysis of the AD169/IE2-EGFP virus.
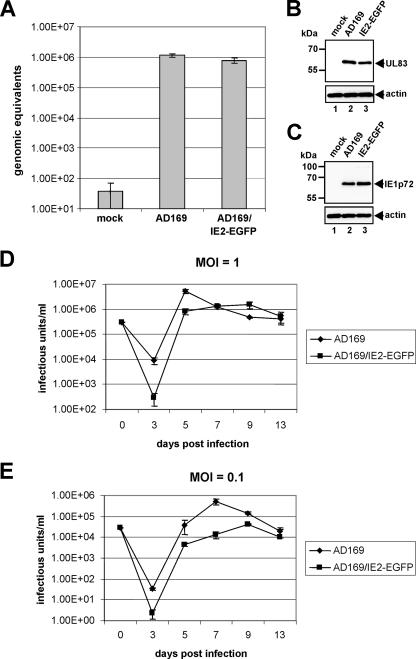
Next, infectious viruses were reconstituted by transfection of HFF cells with purified AD169/IE2-EGFP BAC DNA. One week after being transfected, the cells were transferred to 25-cm2 flasks and cultured for about 2 weeks, until plaques appeared. Thereafter, viral progeny particles were recovered and used to infect fresh HFF cultures for the preparation and titration of virus stocks via IE1p72 fluorescence (see Materials and Methods). In order to make sure that the particle-to-PFU ratios of the recombinant AD169/IE2-EGFP virus and the wild-type virus were comparable, we either quantified viral input DNA by real-time PCR (Fig. 2A) or investigated the uptake of viral particles via Western blot analysis using antibodies against the tegument protein pp65 (serving as the indicator protein for viral particle uptake; Fig. 2B) and the immediate-early protein IE1p72 (serving as the indicator protein for active viral transcription; Fig. 2C). As shown in Fig. 2A to C, no significant differences between the recombinant and wt viruses were observed in these experiments.
FIG. 2.
Determination of the particle-to-PFU ratio followed by multistep growth curve analysis of wt HCMV AD169 and the recombinant AD169/IE2-EGFP virus. (A) Results of quantitative real-time PCR for evaluation of the input viral DNA load after infection with wt AD169 or AD169/IE2-EGFP at an MOI of 1. DNA, extracted at 14 hpi from infected cells, was used for real-time PCR in order to determine the number of input HCMV genome copies. (B) Western blot for detection of pp65 (UL83) protein levels in cells harvested at 2 hpi. (C) Western blot for detection of IE1p72 protein levels in cells harvested at 48 hpi. (B and C) Lanes: 1, mock infection; 2, infection with wt AD169; 3, infection with AD169/IE2-EGFP. Infection was performed at an MOI of 1; detection of actin by Western blot analysis (see lower panels of B and C) served as a loading control. (D and E) HFF cells were infected in parallel at a high (panel D; MOI = 1) or low (panel E; MOI = 0.1) MOI with viral inocula of wt AD169 (filled diamonds) and AD169/IE2-EGFP (filled rectangles) which were standardized for equal IE1p72 expression at 24 hpi. The virus progeny in the supernatants of infected cell cultures were harvested at different time points postinfection as indicated, followed by quantification of the viral load via IE1p72 fluorescence. Error bars indicate the standard deviation derived from three independent experiments.
In order to characterize the growth properties of the AD169/IE2-EGFP virus, multistep growth curve analyses of AD169/IE2-EGFP and wt HCMV AD169 were then performed in parallel. For these analyses, virus replication was assayed after both high (MOI = 1) and low (MOI = 0.1) MOIs of HFF cells. The viral inocula used in these experiments were standardized for comparable expression levels of IE1p72 (5). At different time points postinoculation (3, 5, 7, 9, and 13 days), aliquots of supernatants of the infected cells were sampled for titration via IE1p72 staining. As Fig. 2E indicates, AD169/IE2-EGFP exhibited a slightly delayed kinetics of virus accumulation under conditions of low MOI compared to that for wt AD169, since a major release of infectious recombinant particles occurred after a retardation of about 3 days and the peak titers of AD169/IE2-EGFP remained 1 log below that of the wt virus. Under conditions of high MOI, however, no significant growth defect of AD169/IE2-EGFP could be detected (Fig. 2D), indicating that the production of infectious recombinant virus is not markedly impaired by the genomic insertion of the IE2-EGFP fusion sequence under the latter infection conditions.
Protein expression kinetics during AD169/IE2-EGFP lytic infection.
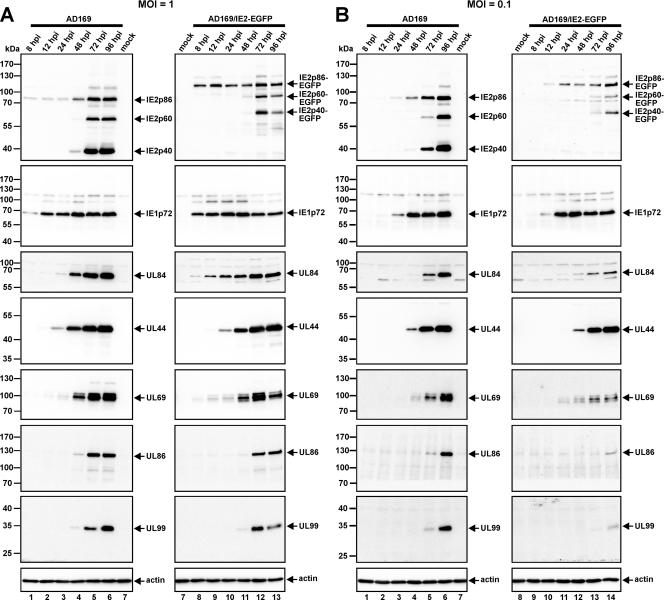
During lytic replication, the IE2 gene region gives rise to three major protein isoforms, termed IE2p86, IE2p60, and IE2p40. Since these proteins share identical C-terminal sequences, fusion to EGFP was predicted to affect all IE2 protein isoforms. In order to confirm this and to compare the kinetics of viral protein accumulation of wt AD169 and AD169/IE2-EGFP, HFF cells were infected in parallel with wt and recombinant viruses (MOIs of 1 and 0.1), respectively. At various times postinfection (8, 12, 24, 48, 72, and 96 hpi), cells were harvested and viral proteins were examined by immunoblotting with antibodies directed against viral immediate-early (IE1, IE2), early-late (UL44, UL69, UL84), and true late (UL86, UL99) proteins (Fig. 3). As expected, all IE2 isoforms of the AD169/IE2-EGFP virus exhibited an increased molecular mass, indicating correct expression of the respective IE2 proteins in fusion with EGFP (Fig. 3A and B, upper panels, lanes 10 to 14). Interestingly, however, we observed a slight difference in the expression kinetics of the IE proteins of wt AD169- and AD169/IE2-EGFP-infected cells under low MOI conditions: both IE1p72 and IE2p86-EGFP could be detected in higher abundances at early times after infection with the AD169/IE2-EGFP virus (Fig. 3B, upper panels, compare lanes 2 and 3 and 10 and 11). In contrast, the increase in abundance observed for IE2 proteins during the late phase of the replication cycle with wt virus was less pronounced with the AD169/IE2-EGFP virus (Fig. 3B, upper panel, compare lanes 5 and 6 and 13 and 14). Similarly, late expression of UL84, UL69, UL86, and UL99 was diminished after infection with AD169/IE2-EGFP, whereas other early-late genes (e.g., UL44) were apparently not affected. This suggests that the C-terminal fusion of IE2 to EGFP may negatively affect specific functions of IE2, which correlates with the observation that the release of infectious viruses is somewhat diminished under conditions of low MOI with AD169/IE2-EGFP (Fig. 2B). However, since these effects were less pronounced under conditions of high MOI (Fig. 2D and 3A), we assumed that all major functions of IE2 are preserved.
FIG. 3.
Comparison of the protein expression kinetics of wt HCMV AD169 and the recombinant virus AD169/IE2-EGFP. HFF cells were infected in parallel with AD169 and AD169/IE2-EGFP at an MOI of 1 (A) or 0.1 (B). At the indicated times postinfection, total cell lysates were prepared, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and subsequently transferred to a nitrocellulose membrane. Immediate-early (IE2p86, IE1p72), early-late (UL84, UL44, UL69), and true late (UL86, UL99) protein levels were analyzed by Western blotting as described in Materials and Methods. Cellular actin levels served as controls for equal protein loading.
Differential localization of IE2 and ND10 in live cells.
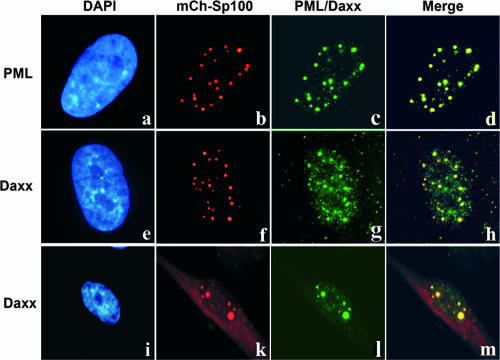
Previous indirect immunofluorescence studies with fixed cells have shown an intimate spatial association between the major regulatory protein IE2 and ND10 domains that is dependent on the mode of IE2 expression: while dot-like accumulations of transiently expressed IE2 display perfect ND10 colocalization (3, 25, 50), the viral IE protein can be found both colocalizing with and adjacent to this subnuclear structure during HCMV infection (3, 25, 26). Taking advantage of live-cell microscopy providing increased sensitivity, lower background signals, and the absence of potential artifacts caused by fixation and sample preparation, we investigated the subnuclear localization of IE2 in live cells. In order to visualize ND10 in live, HCMV-infected cells, primary human fibroblasts stably expressing Sp100 isoform A in fusion with the red autofluorescent protein mCherry (mCh-Sp100) were generated via retroviral transduction (see Materials and Methods). As shown in Fig. 4, the autofluorescent mCh-Sp100 fusion protein colocalized with both endogenous PML (Fig. 4, subpanels a to d) and hDaxx (Fig. 4, subpanels e to h) and could thus be used as a faithful marker for genuine ND10. Note that while most cells stably expressing mCh-Sp100 showed exclusive nuclear localization of the autofluorescent protein, we observed an additional cytoplasmic fluorescence (Fig. 4, subpanels i to m) in some cells (in particular those cells expressing high amounts of Sp100). This is not a novel observation, since cytoplasmic localization of Sp100 has been described previously and was correlated with a lack of SUMO modification of the respective Sp100 isoform (49).
FIG. 4.
Generation of primary human fibroblast cells stably expressing an mCherry-Sp100 (mCh-Sp100) autofluorescent fusion protein. Primary human fibroblast cells stably expressing mCherry-Sp100 were generated via retroviral transduction using plasmid pHM2397, followed by blasticidin selection. Indirect immunofluorescence analysis revealed colocalization of stably expressed mCh-Sp100 with both endogenously expressed PML (subpanels a to d) and hDaxx (subpanels e to m).
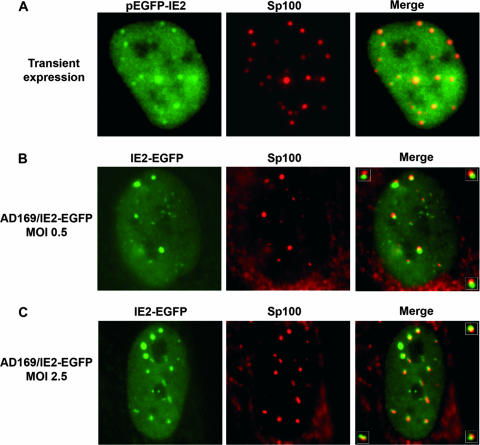
Having generated the mCh-Sp100 cells, we first investigated the subcellular localization of transiently expressed IE2 in live cells. For this experiment, mCh-Sp100 cells were transfected with plasmid pHM990, coding for IE2p86 fused to EGFP. As shown in Fig. 5A, precise colocalization of transiently expressed IE2 dots and mCh-Sp100 was detected by live-cell imaging. In contrast, the subnuclear localization of IE2 was different in the context of viral infection. The IE2-EGFP protein expressed from the recombinant AD169/IE2-EGFP virus was predominantly distributed in dot-like structures which were partially colocalized with or juxtaposed to Sp100 in live-infected cells as early as 2.5 hpi (Fig. 5B and C). Even in cases where the IE2 and Sp100 signals appeared to be colocalizing, detailed analyses at high magnification revealed that the signals from the two proteins were rather distinct (Fig. 5B and C, insets). These images were remarkably similar to those showing the juxtaposition of the HSV-1 ICP4 transcriptional regulatory protein in foci associated with ND10 (15, 17, 18). In summary, these observations not only confirm previous results obtained with fixed cells but also prove that the AD169/IE2-EGFP virus can serve as a valuable tool to visualize IE2 in live cells.
FIG. 5.
Live-cell imaging of the association of IE2p86 and ND10 in transfected or infected cells during the immediate-early phase of the replication cycle. Although IE2p86 precisely colocalizes with ND10 after transient expression of the IE2-EGFP protein in mCh-Sp100 fibroblasts (A), it is either associated with or juxtaposed to Sp100 during the course of an HCMV infection (panel B, MOI = 0.5; panel C, MOI = 2.5). Images were obtained from live, transfected or infected (4 hpi) mCh-Sp100 fibroblasts. The inset images show examples of association but not precise colocalization of IE2-EGFP and Sp100.
The number of IE2 foci increases at higher-input MOIs and is independent of the presence of genuine ND10 domains.
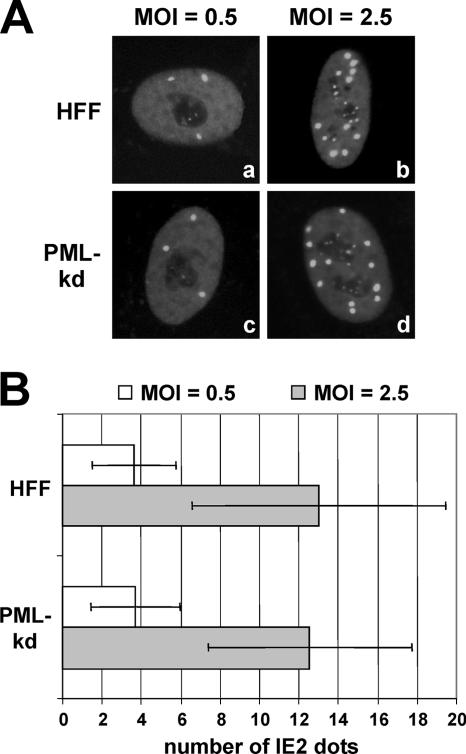
An interesting observation after infection with the AD169/IE2-EGFP virus was that the number of IE2 foci in the nucleus of the infected cells was dependent on the input MOI. HFF cells were infected with the recombinant HCMV virus at either a low or high viral input, and images were captured 4 hpi. The number of IE2 foci at an MOI of 0.5 was significantly lower (Fig. 6A, subpanel a, and B) than the corresponding number of IE2 dots when the MOI was 2.5 (Fig. 6A, subpanels b, and B). Similar observations of the dependence of the number of IE2 foci on the input MOI were made when PML-kd cells lacking intrinsic ND10 were infected with the virus (Fig. 6A, subpanels c and d, B). This observation indicates that, in contrast to the dependence of the dot-like localization of transiently expressed IE2 on the presence of PML-associated nuclear bodies (50), the major regulatory IE2 protein can be recruited onto foci independently of the presence of ND10 during the course of a productive infection. Furthermore, since we did not observe an MOI-dependent increase in the size of IE2 dots, it seems unlikely that another preexisting cellular structure serves as the nucleation site for IE2 accumulations. Rather, the finding that the amount of formation of IE2 foci is proportional to the MOI suggests that viral factors might contribute. In fact, we could argue that a viral component that is imported during infection may be responsible for the formation of IE2 dot-like structures.
FIG. 6.
The number of IE2 foci increases with the multiplicity of infection. The number of IE2-EGFP foci is dependent on the MOI of the infection but it is independent of the ND10 status of the cells. Both HFFs and PML-kd cells were infected with AD169/IE2-EGFP either at an MOI of 0.5 (panel A, subpanels a and c; panel B), or at an MOI of 2.5 (panel A, subpanels b and d; panel B). The graph in panel B shows mean values for 50 analyzed cells; standard deviations are indicated.
Dynamics of fluorescently labeled Sp100 and IE2 in live-infected cells.
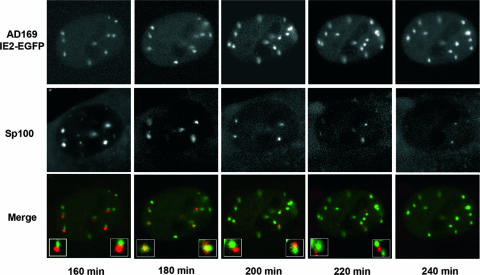
Construction of the recombinant AD169/IE2-EGFP virus allowed us to visualize the viral IE2 protein even at very early times of infection and thus to monitor its accumulation in relation to ND10. Time-lapse microscopy of mCh-Sp100 cells infected with AD169/IE2-EGFP at an MOI of 0.5 revealed a dynamic association of IE2 dots with ND10 structures during the first hours postinfection. Figure 7 shows a sequence of images of a single cell starting 160 min after addition of the virus. Several of the IE2 foci were strikingly juxtaposed next to Sp100 (Fig. 7), but some of them rapidly fused to precise colocalizations (Fig. 7), followed by segregation of IE2 and Sp100 (Fig. 7), and finally, the dispersal of ND10 accumulations (Fig. 7). Once IE2 and Sp100 were associated, there was a limited amount of movement between them, but those present in close association remained so throughout the sequence, until the disruption of ND10 occurred. We concluded that the IE2 foci and ND10 are separate structures that associate rapidly at early stages of HCMV infection and that this association is very dynamic and exceptionally reminiscent of the dynamics of its HSV-1 functional homolog, the ICP4 protein (17).
FIG. 7.
In vivo dynamics of IE2 in relation to that of ND10. mCh-Sp100 cells live-infected with AD169/IE2-EGFP at an MOI of 0.5 were monitored at immediate-early times after infection. Presented are selected images that illustrate a dynamic pattern of association where, within minutes, Sp100 and IE2 dots, which are in the first instance next to each other, show perfect colocalization and then become juxtaposed until Sp100 is dispersed. The inset images show two regions of the cell at a higher magnification.
Development of IE2 foci into viral replication compartments.
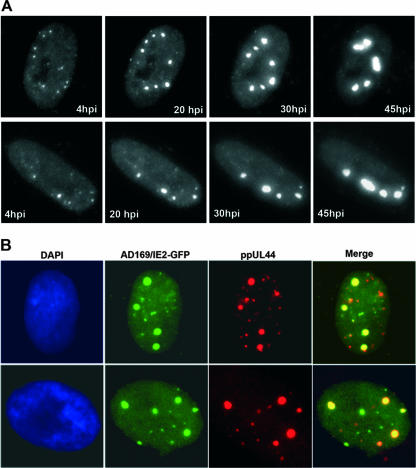
The IE2 protein is required for maximal efficiency of viral DNA replication in transient DNA cotransfection assays, while studies with fixed cells have also shown that it is incorporated into large nuclear structures corresponding to viral DNA replication compartments (4, 40). To investigate the fate of the initially formed IE2 foci during the entire course of a lytic HCMV infection, HFF cells were infected with the AD169/IE2-EGFP virus and examined thereafter at intervals comprising immediate-early, early, and late time points of the replication cycle. Time-lapse microscopy of individual live cells indicated that a proportion of the IE2 foci went on to expand and develop into larger nuclear structures (Fig. 8A). Most of the cells showed multiple separate structures within the nucleus, some of which became enlarged and some of which combined to form the fully developed oval or kidney-shaped viral DNA replication compartments at late times of infection. Additional evidence that the large nuclear structures represent viral replication compartments was obtained when 200 μg/ml phosphonoacetic acid was included in the medium: formation of viral DNA replication compartments but not of the initial IE2 foci was abrogated by this DNA replication inhibitor (data not shown). To further confirm the specificity of incorporation of IE2 into viral DNA replication compartments, we investigated the localization pattern of the protein along with the DNA polymerase processivity factor ppUL44, an essential HCMV replication protein known to be recruited into these compartments at late stages of the replication cycle (4). Infection of HFFs with AD169/IE2-EGFP and staining for ppUL44 demonstrated an accumulation of both proteins into large globular nuclear structures 30 hpi (Fig. 8B). These observations indicate that the IE2 protein can serve as a marker for monitoring the formation of viral replication compartments and suggest that the tiny IE2 accumulations at immediate-early times of infection may represent viral nucleoprotein complexes that develop into replication compartments as infection progresses.
FIG. 8.
Recruitment of IE2 into foci, some of which later develop into replication compartments. (A) HFF cells were infected with AD169/IE2-EGFP at an MOI of 0.5 and analyzed by live-cell microscopy. The localization of IE2p86 was detected via EGFP fluorescence. Times after addition of the virus are indicated in each panel. (B) Subnuclear colocalization of IE2-EGFP and ppUL44 was revealed after indirect immunofluorescence analysis with the antibody BS510 against ppUL44, confirming the development of the IE2p86 foci into viral replication compartments 30 hpi.
IE2 foci are associated with viral genomes at early stages of HCMV infection.
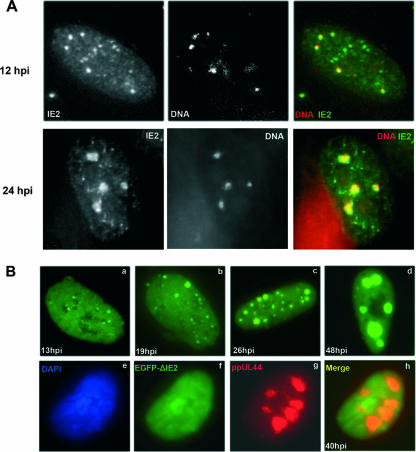
The ability of IE2 to directly bind to viral DNA (12, 27) along with our observations that the number of IE2 foci increased with increasing viral input and that many of these initially formed IE2 dots later developed into replication compartments gave rise to the idea that at least a proportion of the IE2 foci must represent an association of the IE2 protein with parental viral genomes. To address this hypothesis directly, in situ hybridization for the detection of HCMV DNA in combination with antibody detection of IE2 was applied in fixed HCMV-infected cells. As shown in Fig. 9A, the majority of IE2 foci formed early during infection were strongly associated with foci of viral DNA that were detected by in situ hybridization. A large number of cells were examined, showing that all distinct nuclear DNA foci exhibited an IE2 signal, while a subset of the IE2 foci, in particular the smaller ones, were not associated with a DNA signal. The formation of larger complexes between viral DNA and IE2 was obvious at 24 hpi, when viral DNA started to replicate, giving rise to the development of replication compartments (Fig. 9A, lower subpanels). Thus, these experiments provide direct evidence for an association of IE2 foci with parental viral genomes at early times after infection.
FIG. 9.
HCMV genomes as determinants of IE2p86 subnuclear localization. (A) Combined in situ hybridization of HCMV DNA and immunostaining of IE2 in HCMV-infected HFFs at an MOI of 0.5 shows an association between them 12 hpi or at later times during infection. Note the larger structures of HCMV DNA 24 hpi, indicating active replication of the viral genomes. (B) HFFs were transfected with plasmid pEGFP-IE2 and superinfected with HCMV AD169 (subpanels a to d). Images were obtained at different time points postinfection, demonstrating the progressive recruitment of IE2 into developing viral replication compartments. This was not the case when plasmid pEGFP-ΔIE2, encoding a DNA-binding-deficient mutant of IE2 fused to EGFP, was transfected instead, when IE2 failed to form foci even as late as 40 hpi (subpanel f) although viral replication had occurred, as indicated by the recruitment of ppUL44 (subpanel g). Subpanel h shows the two channels merged (ppUL44, red; IE2, green). Subpanel e shows DAPI staining.
DNA binding of IE2p86 is required for recruitment into viral replication compartments.
We next investigated whether the direct DNA-binding activity of IE2 is relevant for its association with viral DNA during the course of HCMV infection. Since previous studies demonstrated that recombinant cytomegaloviruses with mutations within the IE2 DNA binding domain are not viable (53), we chose a transient expression strategy in order to address this question. For this experiment, we first constructed a plasmid encoding a DNA-binding-deficient version of IE2 fused to EGFP. This was done via the substitution of two histidines (amino acids 446 and 452 of IE2) of a putative zinc finger motif within IE2 with leucines; this mutation has previously been shown to abrogate the DNA-binding activity of IE2 (32). Expression of the mutant pEGFP-ΔIE2 plasmid was tested by both Western blotting and immunofluorescence analysis, and no differences from wt protein could be detected in terms of expression levels or its subnuclear localization (data not shown). Next we investigated the capabilities of both wt and mutant IE2 proteins to be recruited into viral replication compartments in a transfection/superinfection approach. HFFs were initially infected with HCMV strain AD169 and subsequently transfected (2 hpi) with the plasmids expressing either wt EGFP-IE2 or the mutant EGFP-ΔIE2. Images were captured at different time points thereafter, starting at 13 hpi, when the EGFP signal was mature enough for visualization. As was expected, EGFP-IE2 foci appeared at early times of the replication cycle, and a proportion of them developed progressively into viral replication compartments (Fig. 9B, subpanels a to d). On the other hand, the mutant EGFP-ΔIE2 protein failed to form normal IE2 dot-like structures but was instead evenly distributed in the nuclei of infected cells. The mutant ΔIE2 protein remained in a diffuse form even at late times of the replicative cycle, when viral DNA replication compartments were formed, as indicated by the recruitment of ppUL44 into them (Fig. 9B, panels e to h). This finding suggests that the IE2 DNA-binding activity is a prerequisite for the formation of IE2 foci and recruitment into replication compartments. Based on the above observations and considering that both the wt EGFP-IE2 and its mutant version are able to colocalize with ND10 after transient expression, viral DNA appears to be an important determinant of IE2 subnuclear localization, suggesting that the formation of a virus-induced nucleoprotein complex and its spatial organization is likely to be critical at the early stages of a lytic infection.
DISCUSSION
In this paper, we present evidence that the previously described intranuclear, dot-like foci of IE2p86 that were observed in fixed cells to be juxtaposed to ND10 domains during HCMV infection (3, 25, 26) represent accumulations of IE2 protein on viral DNA. Therefore, we assume that viral DNA acts as the major determinant for the formation of IE2 dot-like accumulations, thus representing viral nucleoprotein complexes. This assumption is based on the following major observations. (i) IE2 dots can be detected even after infection of PML-depleted HFFs lacking genuine ND10 structures, indicating that ND10 structures do not serve as nucleation sites for these accumulations (50). (ii) Infection of both genuine and PML-depleted HFFs revealed that the number of IE2 dots is dependent on the MOI, strongly suggesting that a component of the virus particle contributes to their formation. (ii) By live cell imaging, we showed that IE2 foci, which can be observed as early as 2 h after infection, directly develop into viral replication compartments. (iii) In situ hybridization experiments demonstrated colocalization of IE2 accumulations with parental viral DNA. (vi) A DNA-binding-deficient mutant of IE2 could no longer be recruited into viral replication compartments, suggesting that direct binding of IE2 to DNA is required for the formation of this nucleoprotein complex. In this regard, note that previous studies had identified IE2 as a member of a small group of DNA-binding proteins that interact with AT-rich sequences within the minor groove of the DNA helix (28, 51). One major characteristic of minor-groove DNA-binding proteins is their ability to interact in a relatively nonspecific manner with various DNA recognition sequences and to assemble higher-order protein complexes (7). Indeed, both mutagenesis studies of known DNA recognition motifs (51) and results from a reiterative DNA-binding-site selection (S. Floess and T. Stamminger, unpublished results) confirmed that IE2 has the potential to recognize an extended spectrum of DNA sequences. Consequently, one would predict that in addition to the already identified recognition motifs (6, 23, 27, 42, 43, 57), several more IE2 binding sites exist on the HCMV genome. Together with the known propensity of IE2 to multimerize on DNA (51), this may contribute to the efficient formation of an IE2/DNA nucleoprotein complex immediately after the start of viral infection. Thus, from its earliest stages, virus infection must involve a structured assembly of viral proteins and DNA, implying that intracellularly replicating virus has a more discrete structural aspect than might be inferred from the apparently rather general localization of many of its regulatory proteins throughout the nucleus. This proposal would partly explain the discrepancy between the precise ND10 colocalization of the HCMV IE2 protein in transfected cells and its juxtaposition in infected cells: in transfected cells, IE2 may be aberrantly located because viral genomes are absent, so the protein is unable to assemble into the macromolecular complexes characteristic of a viral infection.
In order to analyze the formation of IE2 dots by live-cell imaging, we successfully generated a recombinant virus with a C-terminally EGFP-tagged IE2 as well as HFF cells expressing Sp100 fused to the red autofluorescent protein mCherry. This enabled us to monitor IE2 protein accumulation in relation to the localization of ND10, which revealed an unexpectedly high dynamic of the association between IE2 and ND10 dots. While previous experiments with fixed cells suggested that some IE2 foci may colocalize, others may be juxtaposed to ND10 (3, 25). We now report that the relative locations of IE2 and ND10 change within minutes: initially juxtaposed structures rapidly fuse, but fusion is followed by the segregation of IE2 and ND10 dots. This takes place within a narrow time window, since due to the action of IE1, ND10 domains are rapidly dispersed during HCMV infection (2, 26, 54). One important implication of these findings is that IE2 accumulations form independently of ND10; thus, ND10 and IE2 foci are distinct structures. This finding is supported by our previous observation that genuine ND10 are dispensable for the formation of IE2 foci (50). Both our own studies and studies from other groups recently provided good evidence for the assumption that ND10 may act to repress viral gene expression (11, 16, 37, 50, 56). In particular, it was shown that the ND10 component hDaxx has the propensity to mediate the formation of a repressive chromatin structure around the HCMV MIEP (56). Given our finding of a highly dynamic association of ND10 with viral nucleoprotein complexes on the supramolecular level, one may speculate that the induction of repressive chromatin modifications may not require prolonged or stable colocalization between these structures but occurs in a very rapid manner.
Determining that the number of IE2 dots within an infected cell is dependent on the MOI gave rise to speculation that a viral component rather than a cellular structure serves as a cofactor for the formation of IE2 accumulations. With a preexisting number of cellular structures serving as attachment sites for IE2, we expected to find that the number of dots is fixed but that the size increases depending on the viral input. An alternative possibility might be that cellular structures with a variable affinity exist: a higher amount of IE2 could then result in association with novel, low-affinity cellular sites, thus increasing the overall number of dot-like IE2 accumulations. However, the observation that after transient expression, the number of IE2 dots stays constant but the level of diffuse IE2 staining increases with higher expression levels argues against such a scenario. Furthermore, our live-cell imaging results showing that dot-like accumulations of IE2 developed directly into viral replication compartments strongly suggest that viral genomes are part of these structures. This was further corroborated by the fact that viral DNA colocalizing with IE2 could be detected by in situ hybridization, finally proving that IE2 foci represent viral nucleoprotein complexes.
Another interesting observation of our live-cell imaging experiments was that only a subset of IE2-DNA complexes enlarged and developed into viral replication centers, whereas others preserved their original sizes as tiny IE2 dots, even at late times of the replication cycle. It is tempting to speculate that these dots may represent silenced viral genomes that were inactivated due to the induction of a repressive chromatin structure by ND10 components. This may correlate with a recent observation made by Reeves and colleagues by chromatin immunoprecipitation (38) that already at 24 h postinfection, a proportion of viral MIEPs is associated with HP1, which is a defined marker of repressed chromatin. Thus, it might be interesting to analyze whether such IE2 dots, which do not develop into viral replication compartments, preferentially colocalize with specific HP1 isoforms. This should be feasible, since a previous study using YFP-tagged HP1 fusion proteins demonstrated that subcellular localization of specific HP1 isoforms can be monitored by live-cell imaging (21).
The association of IE2 foci with HCMV DNA early after infection is strikingly reminiscent of its functional analogous protein, ICP4 of HSV-1, which has also been defined as a promiscuous DNA-binding protein (14). It was demonstrated by live-cell imaging that at the early stages of the replication cycle ICP4 forms dot-like accumulations which colocalize with viral DNA but are juxtaposed to ND10, with some of the ICP4 foci later developing into viral replication compartments (18). Furthermore, it was shown that ND10 components are able to actively relocate to sites of HSV-1 nucleoprotein complexes, which provides an explanation for the commonly observed association of viral DNA with ND10 (15, 17). Thus, our results obtained for the IE2 transactivator of HCMV further support the previously proposed concept that the formation of a viral nucleoprotein complex between the respective major transcriptional transactivator and viral DNA is likely to be a critical step common to the alpha- and betaherpesviruses (18).
The functional role of the intimate association between IE2 and viral DNA, however, remains to be determined. In addition to its role as a classical transcription factor binding directly or indirectly to several HCMV promoter sequences (6, 23, 27, 42, 43), there is increasing evidence for a chromatin-remodeling function of IE2. For instance, it was shown very recently that a repressive chromatin structure dependent on site-specific binding of IE2 to the cis repression sequence upstream of the IE1/2 transcriptional start site is established around the MIEP, and this was correlated with an interaction of IE2 with the histone deacetylase HDAC1 and the histone methyltransferases G9a and SUVAR(3-9)H1 (38). Paradoxically, however, IE2 not only binds to chromatin modifiers that induce repressive modifications but also to histone acetyltransferases known to open the chromatin structure, thus leading to transcriptional activation (10, 41). One plausible hypothesis on the role of the IE2 nucleoprotein complex may be that the immediate association of IE2 with viral DNA upon the start of the replication cycle may be necessary in order to establish a chromatin conformation that ensures efficient viral transcription. Experiments to address this hypothesis are in progress.
Acknowledgments
We thank B. Britt (Birmingham, AL), G. Hahn (Munich, Germany), U. Koszinowski (Munich, Germany), and B. Plachter (Mainz, Germany) for providing reagents.
This work was supported by SFB473, the IZKF Erlangen, BIGSS, and a Marie Curie Reintegration Grant of the European Commission Framework 6 Programme (contract no. MERG-CT-2004-513448).
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Adler, H., M. Messerle, and U. H. Koszinowski. 2003. Cloning of herpesviral genomes as bacterial artificial chromosomes. Rev. Med. Virol. 13:111-121. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J.-H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, J.-H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn, J.-H., W.-J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreoni, M., M. Faircloth, L. Vugler, and W. J. Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157-167. [DOI] [PubMed] [Google Scholar]
- 6.Arlt, H., D. Lang, S. Gebert, and T. Stamminger. 1994. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J. Virol. 68:4117-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bewley, C. A., A. M. Gronenborn, and G. M. Clore. 1998. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu. Rev. Biophys. Biomol. Struct. 27:105-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borst, E.-M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology. Lippincott-Raven Publishers, Philadelphia, PA.
- 10.Bryant, L. A., P. Mixon, M. Davidson, A. J. Bannister, T. Kouzarides, and J. H. Sinclair. 2000. The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF. J. Virol. 74:7230-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantrell, S. R., and W. A. Bresnahan. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80:6188-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiou, C.-J., J. Zong, I. Waheed, and G. S. Hayward. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J. Virol. 67:6201-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demarchi, J. M. 1981. Human cytomegalovirus DNA: restriction enzyme cleavage maps and map locations for immediate-early, early, and late RNAs. Virology 114:23-38. [DOI] [PubMed] [Google Scholar]
- 14.DiDonato, J. A., J. R. Spitzner, and M. T. Muller. 1991. A predictive model for DNA recognition by the herpes simplex virus protein ICP4. J. Mol. Biol. 219:451-470. [DOI] [PubMed] [Google Scholar]
- 15.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., G. Sourvinos, C. Leiper, J. B. Clements, and A. Orr. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 78:1903-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn, G., M. Jarosch, J. B. Wang, C. Berbes, and M. A. McVoy. 2003. Tn7-mediated introduction of DNA sequences into bacmid-cloned cytomegalovirus genomes for rapid recombinant virus construction. J. Virol. Methods 107:185-194. [DOI] [PubMed] [Google Scholar]
- 21.Hayakawa, T., T. Haraguchi, H. Masumoto, and Y. Hiraoka. 2003. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 116:3327-3338. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann, H., S. Flöss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, L., and M. F. Stinski. 1995. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J. Virol. 69:7612-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 27.Lang, D., and T. Stamminger. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J. Virol. 67:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang, D., and T. Stamminger. 1994. Minor groove contacts are essential for an interaction of the human cytomegalovirus IE2 protein with its DNA target. Nucleic Acids Res. 22:3331-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lischka, P., Z. Toth, M. Thomas, R. Mueller, and T. Stamminger. 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 26:1631-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorz, K., H. Hofmann, A. Berndt, N. Tavalai, R. Mueller, U. Schlotzer-Schrehardt, and T. Stamminger. 2006. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 80:5423-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macias, M. P., and M. F. Stinski. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc. Natl. Acad. Sci. USA 90:707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonough, S. H., and D. H. Spector. 1983. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology 125:31-46. [DOI] [PubMed] [Google Scholar]
- 36.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 37.Preston, C. M., and M. J. Nicholl. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J Gen. Virol. 87:1113-1121. [DOI] [PubMed] [Google Scholar]
- 38.Reeves, M., J. Murphy, R. Greaves, J. Fairley, A. Brehm, and J. Sinclair. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J. Virol. 80:9998-10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz, R., B. Helmich, and D. H. Spector. 1996. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J. Virol. 70:6955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz, R., M. H. Sommer, A. Scully, and D. H. Spector. 1994. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J. Virol. 68:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 69:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 45.Stenberg, R. M., A. S. Depto, J. Fortney, and J. A. Nelson. 1989. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J. Virol. 63:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenberg, R. M., D. R. Thomsen, and M. F. Stinski. 1984. Structural analysis of the major immediate early gene of human cytomegalovirus. J. Virol. 49:190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stenberg, R. M., P. R. Witte, and M. F. Stinski. 1985. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J. Virol. 56:665-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sternsdorf, T., K. Jensen, and H. Will. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139:1621-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waheed, I., C.-J. Chiou, J.-H. Ahn, and G. S. Hayward. 1998. Binding of the human cytomegalovirus 80-kDa immediate-early protein (IE2) to minor groove A/T-rich sequences bounded by CG dinucleotides is regulated by protein oligomerization and phosphorylation. Virology 252:235-257. [DOI] [PubMed] [Google Scholar]
- 52.Wathen, M. W., D. R. Thomsen, and M. F. Stinski. 1981. Temporal regulation of human cytomegalovirus transcription at immediate early and early times after infection. J. Virol. 38:446-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkinson, G. W. G., C. Kelly, J. H. Sinclair, and C. Rickards. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79:1233-1245. [DOI] [PubMed] [Google Scholar]
- 55.Winkler, M., T. aus dem Siepen, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodhall, D. L., I. J. Groves, M. B. Reeves, G. Wilkinson, and J. H. Sinclair. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652-37660. [DOI] [PubMed] [Google Scholar]
- 57.Xu, Y., S. A. Cei, H. A. Rodriguez, K. S. Colletti, and G. S. Pari. 2004. Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within oriLyt. J. Virol. 78:11664-11677. [DOI] [PMC free article] [PubMed] [Google Scholar]