Abstract
Epstein-Barr virus (EBV) can infect various cell types but limits its classical growth-transforming function to B lymphocytes, the cells in which it persists in vivo. Transformation initiates with the activation of Wp, a promoter present as tandemly repeated copies in the viral genome. Assays with short Wp reporter constructs have identified two promoter-activating regions, one of which (UAS2) appears to be lineage independent, while the other (UAS1) was B-cell specific and contained two putative binding sites for the B-cell-specific activator protein BSAP/Pax5. To address the physiologic relevance of these findings, we first used chromosome immunoprecipitation assays and found that BSAP is indeed bound to Wp sequences on the EBV genome in transformed cells. Thereafter, we constructed recombinant EBVs carrying two Wp copies, both wild type, with UAS1 or UAS2 deleted, or mutated in the BSAP binding sites. All the viruses delivered their genomes to the B-cell nucleus equally well. However, the BSAP binding mutant (and the virus with UAS1 deleted) showed no detectable activity in B cells, whether measured by early Wp transcription, expression of EBV latent proteins, or outgrowth of transformed cells. This was a B-cell-specific defect since, on entry into epithelial cells, an environment where Wp is not the latent promoter of choice, all the Wp mutant viruses initiated infection as efficiently as wild-type virus. We infer that EBV ensures the B-cell specificity of its growth-transforming function by exploiting BSAP/Pax5 as a lineage-specific activator of the transforming program.
Epstein-Barr virus (EBV), an orally transmitted gamma-1 herpesvirus widespread in human populations, replicates in a permissive, probably epithelial, cell type in the oropharynx and then establishes latency in B lymphocytes. To assist the establishment of latency, EBV has acquired a unique set of latent-cycle genes whose expression drives B-cell growth, thereby allowing the transient expansion of infected cells in the naive host. Such expansions are usually contained by the emerging host T-cell response, but in T-cell-compromised patients, uncontrolled expansion can lead to fatal lymphoproliferative disease. The virus's growth-transforming function can be studied in vitro, where the infection of resting B cells leads to permanent B lymphoblastoid cell lines (LCLs) expressing the full set of latent-cycle proteins, the nuclear antigens EBNA1, -2, -3A, -3B, -3C and -LP, and the latent membrane proteins LMP1 and -2 (reviewed in reference 31).
Though EBV is markedly B lymphotropic in vitro, experimental infections of certain non-B cell types, in particular epithelial cells and some T- and NK cell lines, can be obtained. These infections are often abortive or, in already established lines, lead to persistent infections with restricted patterns of latent gene expression (14, 17, 26, 44, 45) that mirror those seen in EBV-associated tumors of epithelial, T-, or NK cell origin (31). Thus, EBV limits the use of the full growth-transforming program to B lymphocytes, the lineage in which it has evolved to disseminate and persist. The present work set out to determine how this lineage specificity is achieved.
B-cell transformation is initiated through the activation of a promoter, Wp, present in tandem repeats of the BamHI W fragment of the viral genome. This leads to the expression initially of EBNA2 and EBNA-LP and subsequently of all six EBNAs as an alternative pan-EBNA promoter, Cp (situated upstream in the adjacent BamHI C fragment), gradually becomes dominant and Wp activity declines (2, 5, 42). Infections of non-B cells select for a third promoter, Qp (in the downstream BamHI Q fragment), leading to the expression of the virus genome maintenance protein EBNA1 in the absence of the other EBNAs (16, 23, 45). The factors that determine Wp activation in B cells are poorly understood. Transient transfection of reporter constructs into established cell lines showed that Wp is more active in B cells than non-B cells and contains lineage-independent and B-cell-specific regulatory regions with various binding sites for transcription factors (6, 22, 41), only one of which, the B-cell-specific activator protein BSAP/Pax5, is restricted to the B-cell lineage (1). However, the relevance of these findings to the physiologic controls governing Wp usage when an entire EBV genome enters a resting B cell remains to be determined.
MATERIALS AND METHODS
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were carried out for BSAP binding as previously described (41).
Luciferase reporter assays.
All luciferase reporter constructs used in this work were generated from Wp440/GL2 (6), which contains Wp sequences from −440 to +173 (relative to the RNA start site) cloned between the BglII and HindIII sites of pGL2 Basic (Promega). WpΔ1 was created by deleting the sequences between −264 and −87 by digestion with AvrII and SnaBI, blunt-ending with Klenow DNA polymerase, and recircularizing the vector fragment. WpΔ2 was created by deleting the sequences between −440 and −264 by digestion with NheI (present in the multiple cloning site of pGL2 Basic upstream of the BglII site) and AvrII and recircularizing the vector in the presence of an adaptor oligonucleotide which restored both the original BglII and AvrII sites. WpΔ1+Δ2 was created by deleting the UAS1 sequence from WpΔ2 as described above. A further derivative, WpBSAPm, in which both the BSAP B and D binding sites are inactivated in the context of an otherwise wild-type Wp440 sequence, was generated by using a Quickchange site-directed mutagenesis kit (Stratagene). Briefly, this was done by replacing the sequence 5′ ATGGTT 3′ (−234 to −229) with 5′ AGTACT 3′ within the BSAP site D sequence (simultaneously introducing a ScaI restriction site) of Wp440Bm, a Wp derivative in which BSAP site B has already been mutated (41). Luciferase reporter assays were performed as described previously (6).
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed essentially as described previously (8). Briefly, cells were treated with formaldehyde to cross-link protein-DNA interactions and total chromatin was isolated. Immunoprecipitation reactions were then performed with anti-BSAP antibody (C-20X polyclonal; Santa Cruz Biotechnology), anti-EBNA1 antibody (Ch.EBNA1) (8), or an anti-mouse immunoglobulin gamma (IgG) control antibody (Jackson Immunoresearch). Immunocomplexes were collected with protein A-Sepharose beads, the protein removed, and the DNA eluted and purified. The amounts of cellular (CD19) and viral (Wp, oriP, and Qp) DNA sequences in each immunoprecipitate were then quantified by PCR. Quantitative PCR (Q-PCR) assays were performed in 25-μl volumes containing 5 μl of input DNA, 1× universal Taqman PCR mastermix (Applied Biosystems), and the primer/probe combinations and concentrations shown in Table 1. Samples were cycled for 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s.
TABLE 1.
Real-time PCR primer/probe combinations used to amplify immunoprecipitates from ChIP assays
| Target | Primer or probe and sequencea | Concn | EBV sequence coordinates |
|---|---|---|---|
| CD19 | 5′ primer, 5′-TGGAGAATGGGGCCTGAG | 300 nM | |
| 3′ primer, 5′-AGGTGGCATGGTGGTCAGAC | 300 nM | ||
| Probe, 5′-CAGCATCCCCTGCGCGAAGCT | 200 nM | ||
| Wp | 5′ primer, 5′-GAATGGGCGCCATTTTGTC | 300 nM | 14079-14098 |
| 3′ primer, 5′-GGCTTATTCCTCTTTTCCCCTCTA | 300 nM | 14200-14177 | |
| Probe, 5′-AGATAGCAGCAGCGCAGCCAACCATA | 200 nM | 14175-14150 | |
| oriP | 5′ primer, 5′-ATGTAAATAAAACCGTGACAGCTCAT | 1 μM | 8957-8982 |
| 3′ primer, 5′-TTACCCAACGGGAAGCATATG | 1 μM | 9038-9058 | |
| Probe, 5′-TGGGAGATATCGCTGTTCCTTAGGACCC | 200 nM | 8987-9014 | |
| Qp | 5′ primer, 5′-TTGAAAAGGCGCGGGATA | 1 μM | 62420-62437 |
| 3′ primer, 5′-TCCCAGCTGCCCAAAATG | 1 μM | 62503-62486 | |
| Probe, 5′-TAAGGATAGCATGTATTACCCGCCATCCG | 200 nM | 62477-62449 |
The TaqMan probes were labeled with a 6-carboxyfluorescein phosphoramidite reporter dye at the 5′ end and 6-carboxytetramethyl-rhodamine at the 3′ end.
Recombinant viruses.
Recombinant EBV genomes were created by using the EBV bacterial artificial chromosome (BAC) cloning system (11; data not shown).
B-cell infection.
B cells were positively selected from buffy coats by using CD19 Dynabeads (Invitrogen) followed by detachment with CD19 Detachabead (Invitrogen) according to the manufacturer's protocol. The purified B cells were incubated overnight at 37°C with a recombinant virus preparation, with or without EBNA2KO virus, at known multiplicities of infection (MOI). The delivery of recombinant EBV genomes was assessed by fluorescence in situ hybridization (FISH) using a fluorescein-labeled EBV DNA cosmid probe (36). The level of EBV antigen expression was determined by immunofluorescence staining using JF186 (EBNA-LP), PE2 (EBNA2), and CS1-4 (LMP1) antibodies followed by a Cy3-conjugated anti-mouse IgG secondary antibody (Stratech). EBV transcripts were quantified by quantitative reverse transcriptase PCR (Q-RT-PCR) using previously described primer/probe combinations (7) on RNA isolated from 1 × 106 infected cells per time point. Cell proliferation was examined by FACS analysis of 5- (and 6-) carboxyfluorescein diacetate (CFSE)-labeled B cells infected with each recombinant virus, with or without EBNA2KO virus coinfection, and 8-week transformation assays as described previously (36).
Epithelial cell infection.
AdAH cells were infected by using the transfer infection method (37) and examined by immunofluorescence cell staining with R4 (EBNA1), CS1-4 (LMP1) and BZ1 (BZLF1) antibodies followed by a suitable Cy3-conjugated secondary IgG antibody.
RESULTS
BSAP binding sites within Wp.
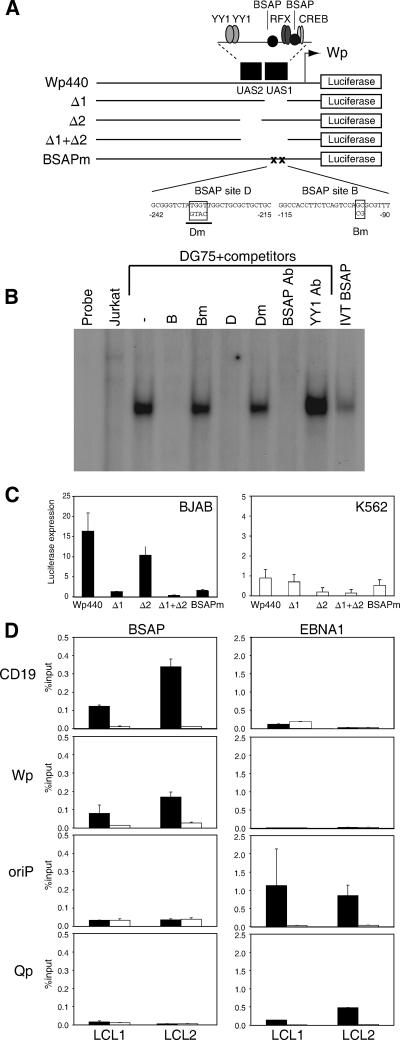
Based on reporter gene assays using progressive 5′ deletions of the 440-bp sequence upstream of the transcription start site, Wp contains a lineage-independent activating region, UAS2, with two YY1 binding sites, and a B-cell-specific region, UAS1, with RFX-, CREB-, and two BSAP-binding sites (Fig. 1A). We created a new set of reporter constructs in which these two regulatory regions had been deleted, either individually (Δ1 and Δ2) or together (Δ1+Δ2), or in which both BSAP sites had been mutated (BSAPm) using nucleotide substitutions previously shown to abrogate BSAP binding to site B (41) and shown here to abrogate BSAP binding to site D (Fig. 1B); note that the BSAP site D mutation introduced a ScaI restriction site (underlined in Fig. 1A) that could serve as a marker during recombinant virus construction. Transient transfection assays confirmed that the wild-type Wp440 construct was more active in B-cell lines (BJAB and DG75) than in non-B-cell lines (K562 and Jurkat) and that the Δ1 deletion markedly reduced reporter activity in B cells, while the Δ2 deletion had the greater effect in a non-B-cell environment (Fig. 1C and data not shown). Importantly, in all cases the effects of the Δ1 deletion could be reproduced by mutation of the BSAP sites within an otherwise wild-type construct.
FIG. 1.
BSAP binding sites in Wp. (A) Map of the Wp440 reporter construct showing the UAS1 and UAS2 regulatory sequences (black boxes) containing binding sites for known transcription factors, and the Wp mutants Δ1 (with UAS1 deleted), Δ2 (with UAS2 deleted), Δ1+Δ2 (double deletion), and BSAPm (mutations introduced into BSAP sites B and D as shown). (B) Electrophoretic mobility shift assay with a BSAP binding H2B2.1 probe. Radiolabeled probe was incubated alone (probe), with T cell (Jurkat) or B cell (DG75) nuclear extract, or with in vitro-translated BSAP (IVT BSAP) protein. Competition assays were carried out with Wp BSAP site binding sequences (B and D) and with their mutated derivatives (Bm and Dm). Supershift assays were performed with a BSAP-specific antibody (BSAP Ab) and an irrelevant YY1 antibody (YY1 Ab). (C) Wp reporter activity assayed by quantifying luciferase expression (arbitrary units) in BJAB and K562 cell lines transiently transfected with the constructs shown in A. (D) ChIP assays to examine BSAP and EBNA1 binding activities in two different LCLs. Immunoprecipitates were analyzed by Q-PCR for cellular (CD19) and EBV (Wp UAS1, oriP, and Qp) DNA sequences, and in each case, the results obtained (black bars) are shown relative to those obtained with a control antibody (open bars). Assays were performed in duplicate, and the amount of DNA present in immunoprecipitates is expressed as a percentage of input DNA values.
We then used ChIP assays to ask whether an interaction could be detected between BSAP and Wp sequences in the viral genome of EBV-infected cells. To this end, a BSAP-specific antibody was used to precipitate protein-DNA complexes from chromatin preparations of two different LCLs; in parallel, we used an EBNA1-specific antibody, previously shown to detect EBNA1 bound to its two sites on the EBV genome, oriP and Qp (8), and an irrelevant control antibody. In each case, the immunoprecipitated DNA was amplified alongside the original input DNA in a series of Q-PCR assays using one of four primer pairs specific for Wp UAS1, for the BSAP-regulated CD19 gene promoter as a positive control for BSAP binding (24), and for oriP and Qp as positive controls for EBNA1 binding (8). The anti-BSAP immunoprecipitates showed a clear enrichment of both CD19 DNA and Wp UAS1 sequences compared to the levels in the control antibody and no enrichment of oriP and Qp sequences. Conversely, anti-EBNA1 immunoprecipitates were preferentially enriched in oriP and Qp sequences but not in Wp or CD19 DNA (Fig. 1D). These results were reproducible and provide the first evidence that BSAP does indeed bind to Wp sequences on the resident EBV genome in a latently infected B cell.
Recombinant viruses carrying Wp mutations.
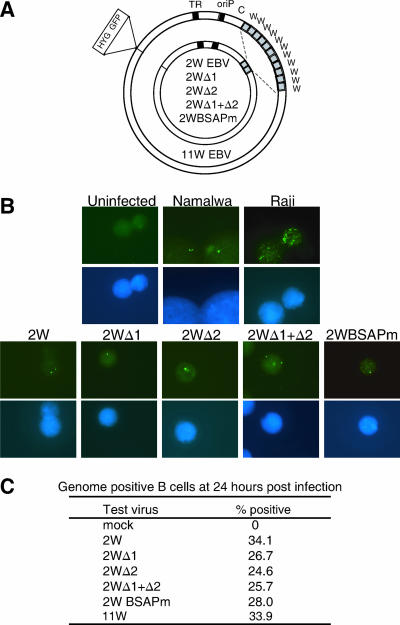
We then engineered a series of recombinant EBVs based on BAC clone 2089, containing the B95.8 EBV genome (11) and carrying the different Wp sequences shown in Fig. 1A. Naturally occurring EBV isolates contain different numbers of copies of the BamHI W fragment (3), up to the 11 copies present in BAC clone 2089 (here called 11W). The manipulation of Wp sequences in the context of the viral genome required us to reduce the number of BamHI W copies to a minimum; note that this inevitably reduced the achievable size of one of the transforming proteins, EBNA-LP, since the W1 and W2 exons within BamHI W encode the N-terminal repeat domain of the EBNA-LP protein (35, 38). However, earlier work had shown that a virus with only two copies retains the transforming function (21) and so, using the 11W virus as a template, we generated a set of recombinants containing just two BamHI W repeats (Fig. 2A), in which both Wp sequences were either wild type (2W), had UAS1 (Δ1), UAS2 (Δ2), or UAS1 and UAS2 (Δ1+Δ2) deleted, or were mutated in both their BSAP sites (BSAPm) (data not shown).
FIG. 2.
Delivery of recombinant viral genomes to the nucleus. (A) Schematic representation of the 11W recombinant 2089 and the panel of 2W wild-type and 2W mutant recombinant genomes, all of which carry the genes for hygromycin (HYG) resistance and green fluorescent protein (GFP); also shown are the BamHI W repeats (gray boxes), terminal repeats (TR), and origin of latent replication (oriP). (B) Detection of viral genomes (green spots) within B-cell nuclei (blue DAPI stains) by FISH using a fluorescein-labeled EBV DNA cosmid probe. Upper panels show hybridization to uninfected B cells and to the EBV-positive Namalwa and Raji cell lines. Lower panels show EBV genomes detected in primary B cells 24 h p.i. with the indicated 2W recombinant viruses (50 MOI). (C) Data showing the percentages of B cells acquiring detectable EBV genomes within 24 h of exposure to the indicated 2W recombinant viruses and to the 11W virus (all at 50 MOI).
To compare the efficiencies with which these different viruses could enter B cells, we used standard methods of FISH with an EBV cosmid probe capable of detecting individual EBV genomes in cell nuclei (10). As illustrated by the control results shown in Fig. 2B, uninfected B cells gave no detectable signals, whereas the Namalwa-BL reference line had the expected two genome copies per cell, seen as fluorescent green spots by microscopy, and the Raji-BL line had around 50 copies per cell (Fig. 2B). In a typical experiment, when resting B cells were exposed to a standard dose (50 MOI) of each of the recombinant viruses and analyzed by FISH 24 h later, nuclear viral genomes (typically 1 to 2 per infected cell) were detectable in each case (Fig. 2B) and roughly equal percentages of cells (25 to 35%) had become genome positive (Fig. 2C).
Effects of Wp mutations on B-cell-transforming infection.
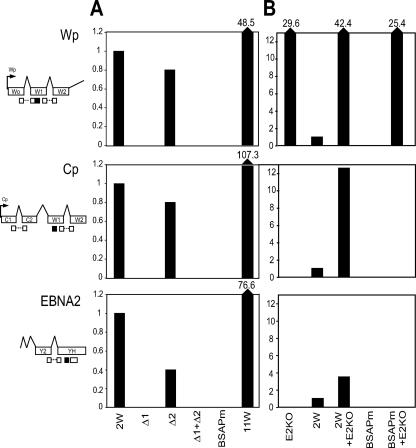
We then used a series of Q-RT-PCR assays with transcript-specific EBV primer-probe combinations (7) to track the initial burst of Wp transcription that occurs within the first 1 to 2 days postinfection (p.i.) and drives the expression of EBNA2 and EBNA-LP mRNAs and is then followed by the activation of transcription from Cp. Cells exposed to the 2W wild-type virus showed easily detectable Wp-initiated transcripts and appropriately spliced EBNA2 mRNAs at day 1 p.i. and Cp-initiated transcripts by day 2 (Fig. 3A). Cells exposed to the Δ2 virus gave similar values, albeit with somewhat slower activation of EBNA2 transcription in this particular experiment. However, we never observed detectable Wp or Cp activity or EBNA2 transcription at any time up to day 10 p.i. with either the Δ1 deletion, the Δ1+Δ2 double deletion or, most significantly, the BSAPm virus (Fig. 3A and data not shown).
FIG. 3.
EBV latent gene transcription in primary B cells infected with recombinant viruses. Wp and EBNA2 transcripts (assayed at 24 h p.i.) and Cp transcripts (at 48 h p.i.) were quantified by Q-RT-PCR in B cells exposed to the indicated viruses (50 MOI) alone (A) or in combination with the EBNA2KO virus (E2KO) (B). The exon structures of the transcripts are shown on the left together with the relative positions of the primers (open boxes) and probes (black boxes) used in the Q-RT-PCR assays. Values are expressed relative to those seen in cells infected with the 2W wild-type virus alone.
The Q-RT-PCR results indicated that the overall levels of viral transcripts seen from the 2W wild-type virus within the first 2 days p.i. were substantially lower than those seen with an 11W virus (Fig. 3A). While this might partly reflect an influence of Wp copy number per se on transcript levels immediately p.i., another possible factor was the inability of the 2W virus to make a sufficiently large EBNA-LP. To control for this, we carried out coinfections with a recombinant EBNA2 gene knockout (EBNA2KO) helper virus (4) which, though nontransforming, retains 11 wild-type BamHI W copies and can provide a full-length EBNA-LP in trans. Fig. 3B shows the transcript levels observed in cells infected with the 2W wild-type or BSAPm viruses used at an MOI of 50, either alone as above or with an equivalent MOI of the helper virus. Note that the EBNA2KO virus alone produces high levels of Wp transcription (expressed relative to that seen with 2W wild-type virus alone) but does not progress to Cp activation. Coinfection of the 2W wild-type virus with the EBNA2KO virus leads to a further increase in total Wp transcription and to levels of Cp and EBNA2 transcripts that are markedly higher than those seen with 2W virus alone; we infer that these Cp transcripts, like those of EBNA2, are expressed from the 2W virus. By contrast, coinfection with the EBNA2KO virus did not rescue any detectable expression from the BSAPm virus genome, whether monitored by an enhancement of overall Wp activity or by the appearance of either Cp-initiated or EBNA2 transcripts (Fig. 3B). When tested in parallel, the Δ2 virus behaved like the 2W wild type, whereas the Δ1 and Δ1+Δ2 viruses were as inactive as the BSAPm virus (data not shown).
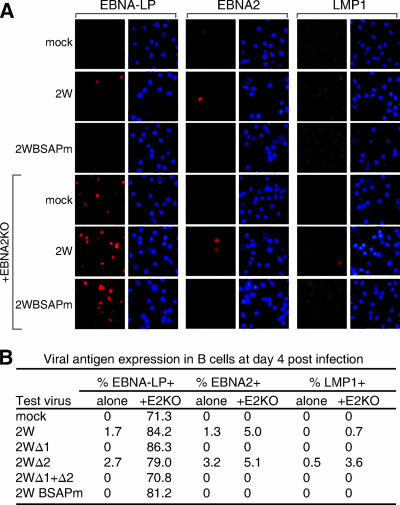
In further experiments, cells exposed to 2W wild-type or mutant viruses (with or without the EBNA2KO helper) were harvested at day 4 p.i. and the progress of infection was monitored by immunofluorescence staining for EBNA-LP, EBNA2, and LMP1. Representative EBV antigen stains (red) and 4′,6′-diamidino-2-phenylindole (DAPI) counterstains (blue) of the same fields are shown in Fig. 4A, and the percentages of antigen-positive cells seen in one such experiment are given in Fig. 4B. Infection with 2W wild-type or Δ2 virus alone produced small numbers of EBNA-LP- and EBNA2-positive cells on day 4; LMP1 staining was first detected either on day 4, as seen here in the 2W Δ2 virus infection, or slightly later (data not shown). However, no EBNA-LP, EBNA2, or LMP1 staining was observed in cultures infected with Δ1, Δ1+Δ2, and BSAPm virus, even with the analysis extended up to 10 days p.i. In line with the Q-RT-PCR data, the EBNA2KO helper virus alone induced detectable EBNA-LP expression in the majority of cells, but never EBNA2 or LMP1. This helper virus caused a small but significant increase in the number of cells expressing EBNA2 and LMP1 in response to 2W and Δ2 virus infection, seen consistently in several experiments. However, even in the presence of the helper virus, we never observed EBNA2- or LMP1-positive cells following Δ1, Δ1+Δ2, or BSAPm virus infection.
FIG. 4.
EBV latent antigen expression in B cells infected with recombinant viruses. (A) Immunofluorescence staining for EBNA-LP, EBNA2, and LMP1 in primary B cells either unexposed (mock) or exposed to 2W wild-type or BSAPm virus (50 MOI), in each case with or without the EBNA2KO virus (50 MOI), and stained 4 days p.i.; blue DAPI staining shows all nuclei in the field. (B) Summary of results showing percentages of cells positive for EBNA-LP, EBNA2, and LMP1 at day 4 p.i.
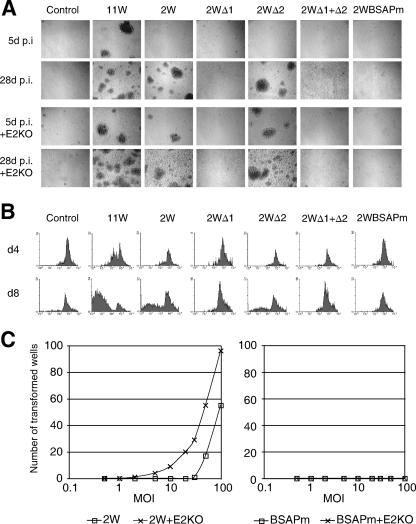
The ultimate test of a virus's B-cell-transforming activity requires the analysis of cultures for morphological change and for cell proliferation leading to successful LCL outgrowth. Cultures infected with the 2W wild-type and Δ2 recombinants did not contain visible foci at day 5 p.i., but foci clearly did develop later and were easily detectable by day 28 (Fig. 5A); these same viruses did induce foci by day 5, however, in coinfections with the EBNA2KO helper. Note that the original 11W wild-type virus, included here for comparison, formed foci by day 5 with and without the helper. By contrast, no foci developed in cultures infected with Δ1, Δ1+Δ2, or BSAPm viruses, whether alone or with the helper. To quantitate the early proliferative events, B cells were stained with CFSE prior to infection with each of the above viruses plus the EBNA2KO helper. FACS profiles of CFSE levels on days 4 and 8 p.i. (Fig. 5B) confirmed proliferation induced by the 2W wild-type and Δ2 viruses, albeit less extensive than in 11W-infected cultures, whereas no proliferation was detectable using the other mutants; note that the helper virus (control, Fig. 5B) was also incapable of driving cells into cycle. Finally, we titrated the 2W wild-type and BSAPm viruses in long-term transformation assays, infecting B cells with these agents at doses of 0.5 to 100 MOI (with and without 50 MOI of helper virus), plated the cells at 104 cells/well in 96-well plates, and after 8 weeks, scored all wells with active growth leading, where subcultured, to LCL establishment. The highest dose of 2W wild-type virus induced transformation in every well if given with the helper virus and in >50% of wells if given alone (Fig. 5C); LCLs were obtained down to 2W virus doses of 2 and 30 MOI in the presence and absence of the helper, respectively. The Δ2 virus (data not shown) gave equivalent yields. However, the BSAPm virus (and the Δ1 and Δ1+Δ2 viruses, data not shown) never yielded LCLs either in this type of assay or in other high-MOI infections seeding cells at higher densities to optimize the chances of outgrowth.
FIG. 5.
Transformation efficiencies of recombinant viruses. (A) Bright-field images of B-cell cultures 5 and 28 days p.i. with the indicated recombinant viruses (50 MOI), with or without coinfection with the EBNA2KO virus (50 MOI). (B) FACS analysis of cell proliferation using CFSE-labeled B cells infected as described above in the presence of the EBNA2KO virus and cultured for 4 and 8 days p.i.; cell proliferation is seen as a decrease in CFSE staining. d4, day 4; d8, day 8. (C) Number of wells from a 96-well plate showing transformation within 8 weeks following exposure to the 2W wild-type and BSAPm viruses at a range of MOIs, with or without EBNA2KO virus (E2KO; 50 MOI).
Effects of Wp mutations on nontransforming infection of epithelial cells.
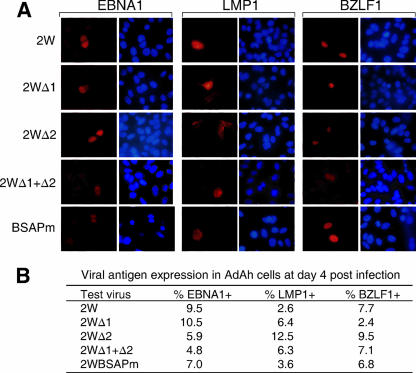
Finally, we compared the same viruses for their ability to initiate epithelial cell infection by using the recently developed protocol of transfer from the surface of virus-loaded resting B cells (37). Note that epithelial cell infections lead to the activation of Qp rather than Wp/Cp and to the expression of EBNA1 in the absence of the other EBNAs; in addition, some infected cells express LMP1 and/or the immediate-early lytic cycle antigen BZLF1 (23, 37). Cells of the AdAh epithelial cell line were cocultivated for 10 min with B cells that had been recently exposed to 25 MOI of 2W wild-type virus or the 2W mutants and then cultured for 4 days before being stained for EBNA1, LMP1, and BZLF1. Representative examples of staining are shown in Fig. 6A, and the percentages of antigen-positive cells seen in one such experiment are shown in Fig. 6B. All viruses showed active epithelial cell infection in this and other experiments, with no consistent differences between the recombinants in the levels of EBNA1, LMP1, or BZLF1 staining observed. Significantly, there was no detectable Wp or Cp activity in these cells and no detectable staining for EBNA2 (data not shown). This strongly suggests that the Wp mutant viruses that fail to transform B cells are perfectly competent to initiate infections in a cell environment that does not naturally involve Wp activation.
FIG. 6.
EBV latent antigen expression in AdAH epithelial cells infected with recombinant viruses by transfer infection. (A) Immunofluorescence staining for EBNA1, LMP1, and BZLF1 in AdAH cells cultured for 4 days p.i. with the indicated viruses (50 MOI). (B) Summary of results showing percentages of cells positive for EBNA1, LMP1, and BZLF1 at day 4 p.i.
DISCUSSION
This work was prompted by the marked contrast in EBV promoter usage, and hence in gene expression, observed after the entry of the virus into B cells versus non-B cells. De novo B-cell infection naturally leads to Wp-mediated activation and growth transformation both in vitro and, from the evidence of newly infected cells in tonsillar B-cell populations (19, 25), also in vivo. By contrast, experimental infections of epithelial, T-, or NK cells (14, 17, 26, 44, 45) and EBV-positive tumors of epithelial, T-, or NK cell origin (31) are all characterized by restricted forms of latency with selective expression of EBNA1 from Qp and suppression of the classical growth-transforming program. We therefore hypothesized that the lineage specificity of EBV's transforming function was determined by factors within the B-cell environment that act on the incoming viral genome to drive Wp activation.
Previous studies of Wp regulation have relied almost entirely on transient transfection assays with short reporter constructs. While an initial report of activation through a putative NF-κB site (39) has not been confirmed, other positive regulatory regions were detected upstream of the Wp start site (18, 30, 32). These were later defined more clearly as a lineage-independent region, UAS2, containing two YY1 sites and responsible for most of the low baseline activity shown by Wp reporters in non-B cells, and a B-cell-specific region, UAS1, containing CREB, RFX, and two BSAP binding sites and responsible for the much-higher reporter activity in B cells (6, 22, 41). The physiologic relevance of these findings was always in doubt, however, since data from EBV reporter constructs often do not accurately reflect promoter regulation in the more complex setting of the natural virus genome (12). As a first step to address the question of physiologic relevance, we looked for evidence that the BSAP protein, which reporter assays had identified as a potential regulator of Wp activity, was in fact bound to Wp UAS1 sequences on the viral episome in EBV-transformed LCL cells. ChIP assays, carried out using the known BSAP binding sites in the cellular CD19 promoter as a positive control (24), showed that this was indeed the case, with significant enrichment of Wp UAS1 sequences in the anti-BSAP immunoprecipitates (Fig. 1).
We then set out to construct EBV recombinants in which either the UAS1, UAS2, or UAS1 and UAS2 regions of Wp had been deleted or the BSAP-binding sites in UAS1 had been mutated. The difficulties inherent in using virus recombinants to analyze a multimerized promoter such as Wp had been illustrated by the earlier work of Yoo et al. (43). They attempted to complement the nontransforming EBV strain P3HR1 that has EBNA2 deleted by using a combination of two constructs, one straddling the EBNA2 deletion, and the other containing a BamHI W fragment with a 200-bp deletion within Wp that removes both YY1 sites in UAS2 and BSAP binding site D in UAS1. While the resident EBV genome in all 253 LCLs established by complementation had acquired the EBNA2 gene, in only one case was the mutant Wp sequence present as the most 5′ copy and this still had wild-type Wp copies downstream (43). Although such poor representation in the resultant LCLs suggested that the mutant Wp had been selected against in the transformation assay, the complexity of the system precluded any firm conclusions being made.
With this as background, we turned to the BAC system to generate homogeneous recombinant virus preparations of known genomic structure (11). To facilitate mutant virus construction, we opted for a virus backbone with just two BamHI W copies since it was already known that such a virus was transforming (21). We in fact found that the 2W wild-type virus, though capable of transformation, is less active in this respect than the prototype EBV recombinant 2089, which has 11 wild-type W copies. This appears to reflect, at least in part, the fact that the 2W virus encodes an EBNA-LP with a suboptimal number of W1W2 repeat domains, since transformation could be improved by coinfection with a derivative of 2089 that has EBNA2 deleted, a helper virus that lacks all transforming function but provides a full-length EBNA-LP in trans. We infer that, while an EBNA-LP with just two W1W2 repeat domains is clearly able to enhance EBNA2's transactivating function in in vitro reporter assays (28, 29), there may be other activities of EBNA-LP which are important to the early events of transformation and which require more W1W2 copies for optimal function. Though this helper virus with EBNA2 deleted was useful in the present context, it has to be stressed that its use did not affect the essential pattern of results obtained from Wp mutant viruses but merely served to magnify the contrast between transforming and nontransforming mutants.
Our results show that the deletion of UAS1, which in transient reporter assays in B-cell lines typically reduces Wp activity by around 10-fold (Fig. 1), completely inactivates the transforming function when analyzed in the context of the viral genome (Fig. 3 to 5). By contrast, the deletion of UAS2, which typically causes a slight reduction of Wp reporter activity in B-cell lines (but a marked reduction in non-B cells), has no detectable effect on the virus's transforming function. Most importantly, mutation of the two BSAP binding sites in UAS1 was functionally equivalent to a full UAS1 deletion; the BSAPm virus had likewise lost all detectable transforming activity (Fig. 3 to 5). From the formal titration assays (Fig. 5), we can say that the transforming function of the BSAPm virus is at least 100-fold lower than that of the 2W wild type. However, it is likely that the difference is much greater than this, since BSAP site mutation completely abrogated (i) any expression of Wp-initiated (as well as Cp-initiated and EBNA2) transcripts in Q-RT-PCR assays that are capable of detecting a single Wp-expressing cell in a background of 105 negative cells (7); (ii) any expression of EBNA-LP, EBNA2, and LMP1 in infected cells detectable by immunofluorescence staining; (iii) any virus-induced B-cell proliferation in the early culture period detectable by CFSE staining; and (iv) any visible signs of transformation and of LCL outgrowth both in formal titration assays and also in bulk cultures set up after the exposure of B cells to the highest deliverable virus dose.
We consider it very likely that these results genuinely reflect the consequences of Wp mutation and are not caused by other changes accidentally introduced into the recombinant EBV genomes during their construction or transfer into 293 cells. We saw no evidence of any unscheduled changes by restriction enzyme and sequence analysis and, for each recombinant described, we have tested virus preparations from two independently established 293 producer cell clones; in each case, the two isolates gave identical results (data not shown). Most importantly, the recombinants that were so dramatically different in their B-cell-transforming activities were all equally competent at initiating infection in epithelial cells (Fig. 6), i.e., in an environment where the natural course of infection involves the activation of the Qp promoter and not of Wp/Cp.
This work throws light on a poorly understood aspect of EBV biology, the relationship between growth-transforming infection and the B-cell environment. The acquisition of the growth-transforming function has been a defining step in the evolution of EBV and the other lymphocryptoviruses. It appears to provide these viruses with an efficient means of colonizing their cellular reservoir of persistence, the B-cell system, both through driving the expansion of latently infected cells and perhaps also through enhancing the cells' survival potential postexpansion (40). However, because these viruses can infect other cell types in vivo, it is clearly important to prevent the activation of the transforming program in an inappropriate lineage. We suggest that EBV, and by inference its lymphocryptovirus relatives in which Wp sequences are highly conserved (33; A. Bell, unpublished observations), has achieved this by exploiting the B-cell-specific protein BSAP as a key regulator of the promoter that initiates transformation. During lymphocyte ontogeny, BSAP is first expressed at the multipotent pro-B-cell stage and actually drives the final commitment to the B-cell lineage (27); thereafter, it remains expressed in mature B cells and is only extinguished with the final differentiation to a plasma cell (1). It is therefore interesting that both pro- and pre-B cells have been found to be susceptible to EBV transformation in vitro (13, 15, 20). By contrast, plasma cells are not transformable, and indeed, the plasmacytic differentiation of LCL cells has been linked to lytic cycle induction and the suppression of Wp/Cp activity (9, 34). The identification of the Wp/BSAP connection is therefore helpful in explaining a number of aspects of EBV's interaction with the B-cell lineage. Several other aspects of EBV latency in vivo remain poorly understood, however, but may likewise involve the influence of particular cellular factors on the virus transcription program.
Acknowledgments
We thank Charles Chau and Paul Lieberman (Wistar Institute, Philadelphia, PA) for generously providing the Ch.EBNA1 antibody and Lori Frappier (University of Toronto, Canada) for kindly providing the R4 antibody.
This work was funded by Cancer Research UK (CR-UK) grant number C910/A3891 and Association for International Cancer Research (AICR) grant number 04-397.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Adams, B., P. Dorfler, A. Aguzzi, Z. Kozmik, P. Urbanek, I. Maurer-Fogy, and M. Busslinger. 1992. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 6:1589-1607. [DOI] [PubMed] [Google Scholar]
- 2.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. [DOI] [PubMed] [Google Scholar]
- 3.Allan, G. J., and D. T. Rowe. 1989. Size and stability of the Epstein-Barr virus major internal repeat (IR-1) in Burkitt's lymphoma and lymphoblastoid cell lines. Virology 173:489-498. [DOI] [PubMed] [Google Scholar]
- 4.Altmann, M., and W. Hammerschmidt. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 3:e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altmann, M., D. Pich, R. Ruiss, J. Wang, B. Sugden, and W. Hammerschmidt. 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc. Natl. Acad. Sci. USA 103:14188-14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, A., J. Skinner, H. Kirby, and A. Rickinson. 1998. Characterisation of regulatory sequences at the Epstein-Barr virus BamHI W promoter. Virology 252:149-161. [DOI] [PubMed] [Google Scholar]
- 7.Bell, A. I., K. Groves, G. L. Kelly, D. Croom-Carter, E. Hui, A. T. Chan, and A. B. Rickinson. 2006. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J. Gen. Virol. 87:2885-2890. [DOI] [PubMed] [Google Scholar]
- 8.Chau, C. M., and P. M. Lieberman. 2004. Dynamic chromatin boundaries delineate a latency control region of Epstein-Barr virus. J. Virol. 78:12308-12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford, D. H., and I. Ando. 1986. EB virus induction is associated with B-cell maturation. Immunology 59:405-409. [PMC free article] [PubMed] [Google Scholar]
- 10.Delecluse, H. J., S. Bartnizke, W. Hammerschmidt, J. Bullerdiek, and G. W. Bornkamm. 1993. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J. Virol. 67:1292-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, T. J., P. J. Farrell, and S. Swaminathan. 1996. Molecular genetic analysis of Epstein-Barr virus Cp promoter function. J. Virol. 70:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, S. M., J. N. Hurley, J. M. McCune, H. G. Kunkel, and R. A. Good. 1980. Pre-B cells and other possible precursor lymphoid cell lines derived from patients with X-linked agammaglobulinemia. J. Exp. Med. 152:1519-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara, S., and Y. Ono. 1995. Isolation of Epstein-Barr virus-infected clones of the human T-cell line MT-2: use of recombinant viruses with a positive selection marker. J. Virol. 69:3900-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansson, M., K. Falk, and I. Ernberg. 1983. Epstein-Barr virus transformation of human pre-B cells. J. Exp. Med. 158:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai, S., J. Nishikawa, and K. Takada. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 72:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isobe, Y., K. Sugimoto, L. Yang, K. Tamayose, M. Egashira, T. Kaneko, K. Takada, and K. Oshimi. 2004. Epstein-Barr virus infection of human natural killer cell lines and peripheral blood natural killer cells. Cancer Res. 64:2167-2174. [DOI] [PubMed] [Google Scholar]
- 18.Jansson, A., M. Masucci, and L. Rymo. 1992. Methylation of discrete sites within the enhancer region regulates the activity of the Epstein-Barr virus BamHI W promoter in Burkitt lymphoma lines. J. Virol. 66:62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph, A. M., G. J. Babcock, and D. A. Thorley-Lawson. 2000. Cells expressing the Epstein-Barr virus growth program are present in and restricted to the naive B-cell subset of healthy tonsils. J. Virol. 74:9964-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katamine, S., M. Otsu, K. Tada, S. Tsuchiya, T. Sato, N. Ishida, T. Honjo, and Y. Ono. 1984. Epstein-Barr virus transforms precursor B cells even before immunoglobulin gene rearrangements. Nature 309:369-372. [DOI] [PubMed] [Google Scholar]
- 21.Kempkes, B., D. Pich, R. Zeidler, and W. Hammerschmidt. 1995. Immortalization of human primary B lymphocytes in vitro with DNA. Proc. Natl. Acad. Sci. USA 92:5875-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirby, H., A. Rickinson, and A. Bell. 2000. The activity of the Epstein-Barr virus BamHI W promoter in B cells is dependent on the binding of CREB/ATF factors. J. Gen. Virol. 81:1057-1066. [DOI] [PubMed] [Google Scholar]
- 23.Knox, P. G., Q. X. Li, A. B. Rickinson, and L. S. Young. 1996. In vitro production of stable Epstein-Barr virus-positive epithelial cell clones which resemble the virus: cell interaction observed in nasopharyngeal carcinoma. Virology 215:40-50. [DOI] [PubMed] [Google Scholar]
- 24.Kozmik, Z., S. Wang, P. Dorfler, B. Adams, and M. Busslinger. 1992. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol. Cell. Biol. 12:2662-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurth, J., T. Spieker, J. Wustrow, G. J. Strickler, L. M. Hansmann, K. Rajewsky, and R. Kuppers. 2000. EBV-infected B cells in infectious mononucleosis: viral strategies for spreading in the B cell compartment and establishing latency. Immunity 13:485-495. [DOI] [PubMed] [Google Scholar]
- 26.Li, Q. X., L. S. Young, G. Niedobitek, C. W. Dawson, M. Birkenbach, F. Wang, and A. B. Rickinson. 1992. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature 356:347-350. [DOI] [PubMed] [Google Scholar]
- 27.Matthias, P., and A. G. Rolink. 2005. Transcriptional networks in developing and mature B cells. Nat. Rev. Immunol. 5:497-508. [DOI] [PubMed] [Google Scholar]
- 28.McCann, E. M., G. L. Kelly, A. B. Rickinson, and A. I. Bell. 2001. Genetic analysis of the Epstein-Barr virus-coded leader protein EBNA-LP as a co-activator of EBNA2 function. J. Gen. Virol. 82:3067-3079. [DOI] [PubMed] [Google Scholar]
- 29.Peng, R., J. Tan, and P. D. Ling. 2000. Conserved regions in the Epstein-Barr virus leader protein define distinct domains required for nuclear localization and transcriptional cooperation with EBNA2. J. Virol. 74:9953-9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puglielli, M. T., N. Desai, and S. H. Speck. 1997. Regulation of EBNA gene transcription in lymphoblastoid cell lines: characterization of sequences downstream of BCR2 (Cp). J. Virol. 71:120-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 32.Ricksten, A., A. Olsson, T. Andersson, and L. Rymo. 1988. The 5′ flanking region of the gene for the Epstein-Barr virus-encoded nuclear antigen 2 contains a cell type specific cis-acting regulatory element that activates transcription in transfected B cells. Nucleic Acids Res. 16:8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivailler, P., H. Jiang, Y. G. Cho, C. Quink, and F. Wang. 2002. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J. Virol. 76:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rochford, R., M. V. Hobbs, J. L. Garnier, N. R. Cooper, and M. J. Cannon. 1993. Plasmacytoid differentiation of Epstein-Barr virus-transformed B cells in vivo is associated with reduced expression of viral latent genes. Proc. Natl. Acad. Sci. USA 90:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sample, J., M. Hummel, D. Braun, M. Birkenbach, and E. Kieff. 1986. Nucleotide sequences of mRNAs encoding Epstein-Barr virus nuclear proteins: a probable transcriptional initiation site. Proc. Natl. Acad. Sci. USA 83:5096-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon-Lowe, C., G. Baldwin, R. Feederle, A. Bell, A. Rickinson, and H. J. Delecluse. 2005. Epstein-Barr virus-induced B-cell transformation: quantitating events from virus binding to cell outgrowth. J. Gen. Virol. 86:3009-3019. [DOI] [PubMed] [Google Scholar]
- 37.Shannon-Lowe, C. D., B. Neuhierl, G. Baldwin, A. B. Rickinson, and H. J. Delecluse. 2006. Resting B cells as a transfer vehicle for Epstein-Barr virus infection of epithelial cells. Proc. Natl. Acad. Sci. USA 103:7065-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speck, S. H., A. Pfitzner, and J. L. Strominger. 1986. An Epstein-Barr virus transcript from a latently infected, growth-transformed B-cell line encodes a highly repetitive polypeptide. Proc. Natl. Acad. Sci. USA 83:9298-9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugano, N., W. P. Chen, M. L. Roberts, and N. R. Cooper. 1997. Epstein-Barr virus binding to CD21 activates the initial viral promoter via NF-kappa B induction. J. Exp. Med. 186:731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 1:75-82. [DOI] [PubMed] [Google Scholar]
- 41.Tierney, R., H. Kirby, J. Nagra, A. Rickinson, and A. Bell. 2000. The Epstein-Barr virus promoter initiating B-cell transformation is activated by RFX proteins and the B-cell-specific activator protein BSAP/Pax5. J. Virol. 74:10458-10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woisetschlaeger, M., C. N. Yandava, L. A. Furmanski, J. L. Strominger, and S. H. Speck. 1990. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. USA 87:1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo, L. I., J. Woloszynek, S. Templeton, and S. H. Speck. 2002. Deletion of Epstein-Barr virus regulatory sequences upstream of the EBNA gene promoter Wp1 is unfavorable for B-cell immortalization. J. Virol. 76:11763-11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshiyama, H., S. Imai, N. Shimizu, and K. Takada. 1997. Epstein-Barr virus infection of human gastric carcinoma cells: implication of the existence of a new virus receptor different from CD21. J. Virol. 71:5688-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshiyama, H., N. Shimizu, and K. Takada. 1995. Persistent Epstein-Barr virus infection in a human T-cell line: unique program of latent virus expression. EMBO J. 14:3706-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]