Abstract
The features of modern dog breeds that increase the ease of mapping common diseases, such as reduced heterogeneity and extensive linkage disequilibrium, may also increase the difficulty associated with fine mapping and identifying causative mutations. One way to address this problem is by combining data from multiple breeds segregating the same trait after initial linkage has been determined. The multibreed approach increases the number of potentially informative recombination events and reduces the size of the critical haplotype by taking advantage of shortened linkage disequilibrium distances found across breeds. In order to identify breeds that likely share a trait inherited from the same ancestral source, we have used cluster analysis to divide 132 breeds of dog into five primary breed groups. We then use the multibreed approach to fine-map Collie eye anomaly (cea), a complex disorder of ocular development that was initially mapped to a 3.9-cM region on canine chromosome 37. Combined genotypes from affected individuals from four breeds of a single breed group significantly narrowed the candidate gene region to a 103-kb interval spanning only four genes. Sequence analysis revealed that all affected dogs share a homozygous deletion of 7.8 kb in the NHEJ1 gene. This intronic deletion spans a highly conserved binding domain to which several developmentally important proteins bind. This work both establishes that the primary cea mutation arose as a single disease allele in a common ancestor of herding breeds as well as highlights the value of comparative population analysis for refining regions of linkage.
Dog breeds are uniquely suited for genetic analysis of traits that have proven problematic for study in human families (Sutter and Ostrander 2004; Lindblad-Toh et al. 2005; Parker and Ostrander 2005). Each dog breed is a closed population, and breed membership requires that both parents be registered members of the same breed. Population bottlenecks resulting from small numbers of founders, over-representation of popular sires, and fluctuations in breed popularity contribute to reduced heterogeneity and increase the average length of linkage disequilibrium (LD) within dog breeds.
The same factors that make the dog system ideal for mapping complex traits also create a challenge for moving beyond locus identification to specifying the genetic sequence variant(s) responsible for phenotypes of interest. Much of the canine genome is contained in large continuous segments shared among all or most members a single breed (Sutter et al. 2004; Lindblad-Toh et al. 2005). As a result, many successful linkage studies have been followed by struggles to narrow the initial disease interval, which usually extends over several megabases and may contain >100 genes. However, if multiple breeds with allelic mutations can be identified, then a combination of data from these breeds could be used to narrow the interval of interest to a manageable size (Goldstein et al. 2006), as has been demonstrated by the successful identification of a gene for progressive rod–cone degeneration (prcd) (Zangerl et al. 2006) and the merle coat color mutation (Clark et al. 2006). However, it is rarely feasible to perform genetic crosses to determine allelism. Hence a genetic method for identifying breeds that likely share ancestral mutations is required.
We have used clustering analysis to group 132 domestic dog breeds into five groups. We have also identified subclusters within some of the larger groups showing additional levels of relatedness among some breeds. This classification scheme is based on genotypic data generated from 96 microsatellite markers that were initially used in a cluster analysis of 85 dog breeds (Parker et al. 2004). In this study, we demonstrate the utility of the breed classification system for multibreed analyses by applying it to the problem of fine-mapping the Collie eye anomaly (cea) locus.
Collie eye anomaly is a complex trait in which the pattern of chorioretinal and scleral development is variously disturbed. The primary aspect of the phenotype, termed choroidal hypoplasia, presents as a localized defect of choroidal development in the temporal quadrant of the ocular fundus. This lesion, similar to the human macular coloboma, segregates to a very close approximation as an autosomal recessive trait. The locus (cea) that is associated with this trait was previously mapped to a 3.9-cM region of canine chromosome 37 (CFA37) (Lowe et al. 2003).
In this study, we developed a dense set of markers for CFA37 that allowed us to assemble and compare haplotypes among affected dogs from multiple breeds hypothesized to share an ancestral haplotype based on the cluster analysis. This reduced the disease interval to 103 kb and only four genes. We show that a large deletion within an intron of one of these genes was present in all cea-affected dogs and absent in unaffected dogs of multiple breeds. Thus, the results presented here not only define the presumptive underlying mutation of the primary cea defect, but also demonstrate the utility and power of linkage disequilibrium mapping among breeds with a shared history.
Results
Population structure
To better understand the relationships among dog breeds, we have analyzed data from 638 dogs representing 132 distinct breeds or breed varieties using cluster analysis (Supplemental Table 1). The data set includes >79% of American Kennel Club (AKC) recognized breeds and represents 92% of all dogs registered by the AKC based on statistics from 2005 (http://www.akc.org/reg/dogreg_stats.cfm).
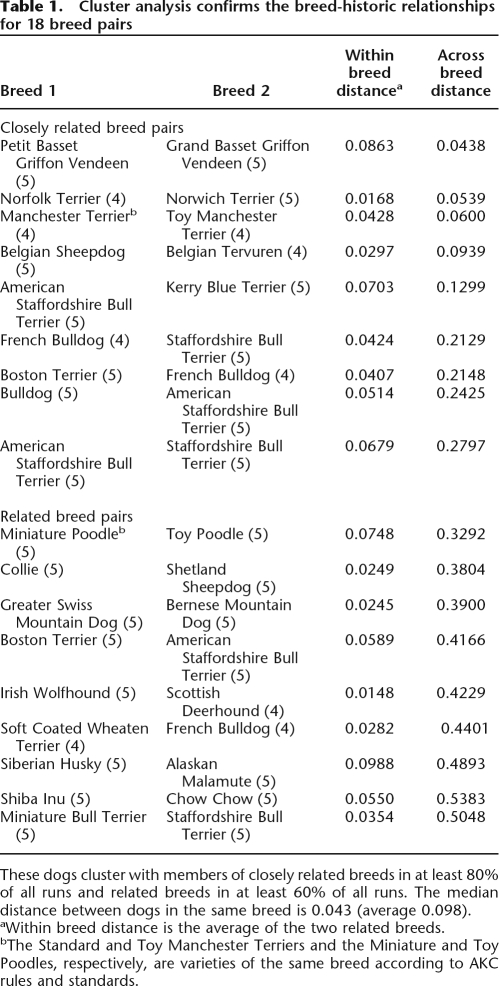
Using the computer program STRUCTURE (Pritchard et al. 2000; Falush et al. 2003), we first determined if each breed formed a unique, breed-specific cluster (Supplemental Fig. 1). At the maximum number of populations (K = the total number of breeds), STRUCTURE indicated that 114 of 132 breeds form distinct, single clusters composed in each case of dogs of only one breed (Supplemental Fig. 1). In addition, nine closely related pairs of breeds were observed that cluster in at least 80% of all runs (Table 1). These pairs can be identified in Supplemental Figure 1 as red squares off the diagonal. Nine additional pairs of breeds, three that had been previously noted (Parker et al. 2004) and six new pairs, were found to cluster in 60%–79% of all runs and are referred to as related breeds (Table 1). These pairs can be identified as orange squares off the diagonal in Supplemental Figure 1. When removed from the full data set and analyzed as a single pair at K = 2, four of the six closely related breed pairs and all of the related breed pairs each divide into two distinct breed groups in all runs (Supplemental Fig. 2). The only exceptions were the Petit and Grand Basset Griffon Vendeens (PBGV and GBGV, respectively), which were assigned to separate breed groups <20% of the time, and the Belgian Sheepdog and Tervuren, described previously (Parker et al. 2004). The Miniature and Toy Poodles, two size varieties of a single breed, separate into two populations although not strictly along variety lines. Fourteen breeds display within-breed clustering distances outside the 95% confidence interval, with the top six forming a single cluster <75% of the time (Supplemental Table 1).
Table 1.
Cluster analysis confirms the breed-historic relationships for 18 breed pairs
These dogs cluster with members of closely related breeds in at least 80% of all runs and related breeds in at least 60% of all runs. The median distance between dogs in the same breed is 0.043 (average 0.098).
aWithin breed distance is the average of the two related breeds.
bThe Standard and Toy Manchester Terriers and the Miniature and Toy Poodles, respectively, are varieties of the same breed according to AKC rules and standards.
We next examined clustering across breeds to determine if hierarchical relationships could be discerned. Again we utilized the program STRUCTURE, this time allowing the breeds to assort into two to 50 clusters by incrementally increasing the number of populations (K) in each subsequent run. To determine the best value of K that describes the breed structure, we examined both likelihood measures taken from the program STRUCTURE and measures of consistency across multiple runs. The likelihood of each run increased with successively higher values of K. This is expected based on the analysis of individual breed clusters. Calculations of ΔK based on variation in the likelihood across multiple runs does not indicate a single best K value (Evanno et al. 2005; Supplemental Fig. 3). Therefore, the consistency of the individual runs was deemed more informative for assessing population structure across the breeds. A distribution of standard deviations across multiple runs of structure at each value of K from 2 to 20 was used to identify the most informative value of K (Supplemental Fig. 4). This distribution is lowest at K = 2, where a separation of the Eastern or Asian breeds from the general population of modern European breeds is observed. Variation between runs increases at K = 3 and peaks at K = 4 and K = 6, with a significant decrease at K = 5 (Supplemental Fig. 4). The variation between runs at K = 5 is significantly lower than that found at K = 4, K = 6, K = 7, and K = 8 (p = 2.14 × 10−10, 2.2 × 10−16, 2.6 × 10−4, and 2.6 × 10−3, respectively). As K increases, the mean and median standard deviation (SD) decreases gradually toward an ultimate breed-specific clustering solution.
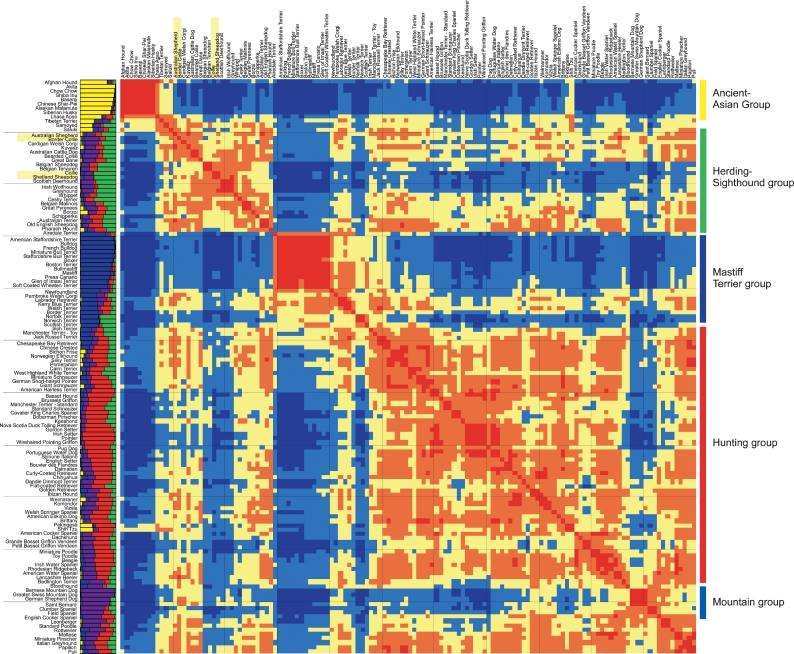
Based on these findings, there are five probable clusters of breeds present in the current data set (Fig. 1). Individually, the STRUCTURE results produced two different patterns of clusters at K = 5, with neither appearing more frequently than the other (Supplemental Fig. 5). To determine the single best clustering solution, all 20 runs were converted to distance matrices and averaged into one single matrix represented by the color map in Figure 1. By comparing all of the runs simultaneously, we observe a fifth, previously unrecognized cluster comprised of large mountain dogs and a subset of spaniels that are clearly more related to each other than to the majority of dogs from the other four clusters. The new “mountain” cluster, shown in purple in the structure graph on the left side of Figure 1, is anchored by the Bernese Mountain Dog and Greater Swiss Mountain Dog and includes other large dogs such as the German Shepherd and Saint Bernard (Fig. 1). The spaniels are divided between the mountain cluster and the hunting cluster, shown in red, which is the largest cluster. The hunting cluster is comprised of modern gun dogs such as the pointers, setters, and retrievers, as well as an assortment of hounds and companion dogs.
Figure 1.
Population structure of 132 domestic dog breeds. On the left of the heatmap graph is a structure graph with each bar representing a breed comprised of four to five individuals. The bars are divided into K colors, where K is the number of populations assumed. The length of the colored segment shows the breed’s estimated proportion of membership in that cluster averaged over 20 runs with K = 5. The heatmap graph shows consistency of clustering between breeds across the same 20 runs of structure. (Red blocks) Breeds that cluster together in >80% of runs; (orange) breeds that cluster together in 60%–80% of all runs; (yellow) breeds that cluster in 40%–60% of runs; (light blue) breeds that cluster in 20%–40% of runs; and (dark blue) breeds that cluster in <20% of all runs. Breeds are ordered identically along both the X- and Y-axes of the color map. Order was determined by a dendrogram based on the distances displayed (dendrogram not shown). The names of the breeds are shown to the left of the structure graph and above the heatmap. The four breeds used to map cea are highlighted in yellow. Group names are listed to the right with colored lines indicating the location of the group on the heatmap.
The other two clusters are the mastiff/terrier cluster, which first becomes apparent at K = 3, and the herding/sighthound cluster (Supplemental Fig. 5). Detailed examination of the color-map graph of breed relationships reveals not only the five primary clusters but smaller closely related groups within the larger clusters (Fig. 1). Many of the breeds within these subclusters appear mixed on the averaged structure graph, although they clearly group with one another based on comparisons across runs regardless of the actual cluster to which they are assigned in each.
At values of K > 5, there is clearly additional structure depicted in each run, as evidenced by increasing likelihood and the steady decrease in SD between runs (Supplemental Figs. 2, 3). Individuals tend to be clustered in one group rather then split between multiple groups, but the majority of these assignments are not consistent across multiple runs indicative of the admixed history of most breeds. As K approaches the total number of breeds, the dogs are divided into breed-specific groups giving no additional information about breed relationships. Therefore, further analyses at greater values of K were not considered.
Fine-mapping and mutation analysis
We applied the above findings to the problem of cea, a developmental defect found in several herding breeds. We previously localized the primary cea defect to the region between markers FH4306 and AHTh174 on canine chromosome 37 (CFA37; LOD = 22.17 at θ = 0.076) (Lowe et al. 2003). This interval is now known to correspond to the 1.7-Mb CanFam2 region on canine chromosome 37, between bases 27,726,776 and 29,432,968. This region corresponds to an ∼2-Mb region on human chromosome 2q35 (Chr2:218,528,849–220,575,553; Human, March 2006 assembly) that contains >40 known human genes. Likely candidate genes for cea would be those that affect development or induction of the ocular mesenchymal tissue (Latshaw et al. 1969), such as genes involved in cellular differentiation or migration. Due to both the size of the region and the number of candidate genes, we sought to reduce the interval by developing haplotypes composed of SNP and microsatellite markers in affected dogs from several breeds.
The four breeds in which the disease is most prevalent are Collie-like herding breeds that belong to the herding/sighthound cluster (Fig. 1). The Collie and Shetland Sheepdog form a related pair at the center of the cluster, while the Australian Shepherd and the Border Collie cluster together. Directed matings had already shown the disease to be allelic in the Collie, Border Collie, and Australian Shepherd (Lowe et al. 2003). Clustering results suggest that these three breeds, as well as the Shetland Sheepdog, are likely to share a recent common ancestor, and the causative mutation may thus be identical by descent (IBD) as well.
The genes AAMP and EPHA4 were identified as flanking the previously reported cea interval on the integrated map of CFA37 by Lowe et al. (2003). To design new markers for the fine-mapping of the cea interval, prior to the availability of the 7.5× whole genome sequence assembly (Lindblad-Toh et al. 2005), we assumed a similar gene order and content for this region of CFA37 and the homologous region of human chromosome 2 (HSA2). Interrogation of the TIGR 1.5× canine sequence database (http://www.ncbi.nlm.nih.gov/projects/genome/seq/BlastGen/BlastGen.cgi?pid=10726) using human sequence for genes located between AAMP and EPHA4 on HSA2 yielded homologous canine sequences and identified eight new microsatellite markers (Supplemental Table 2). Evaluation of these microsatellites suggested several variant haplotypes among cea-affected Collies, Shetland Sheepdogs, Border Collies, and Australian Shepherds. Most affected individuals shared common alleles at markers FH4619 and FH4620 (Supplemental Table 2). However, at least one affected dog was heterozygous at each of these markers, suggesting that higher-resolution genotyping of the interval with more stable polymorphisms would better define the ancestral recombination events.
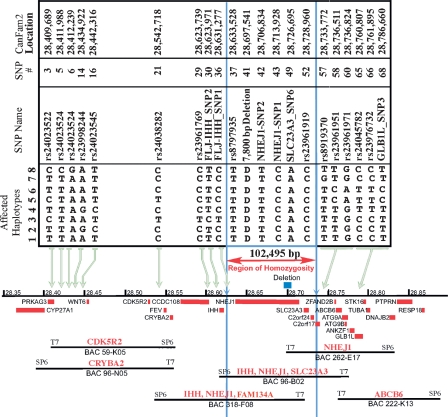
Additional markers were discovered from sequence reads based on eight genes (ABCB6, FAM134A, GLB1L, IHH, NHEJ1, PRKAG3, SLC23A3, WNT6) predicted to be present in the region between AAMP and EPHA4. Canine sequence for several other genes predicted to be in the candidate region was also examined but yielded no polymorphisms among the panel of dogs tested (the full list of primers is available at http://research.nhgri.nih.gov/dog_genome/). Twenty-nine SNPs, four insertion/deletion polymorphisms, and one simple sequence repeat were identified and genotyped in 29 affected, unaffected, and carrier dogs. Initially, the genotypes of 14 affected dogs representing four related breeds revealed a common haplotype bounded by the closest flanking markers, IHH-Indel1 and rs23961951 (SNPs 28 and 58) (Supplemental Table 3), for which heterozygosity was detected among affected chromosomes. Release of the 7.5× Boxer sequence assembly (CanFam1) and a SNP database (Lindblad-Toh et al. 2005) allowed additional SNPs to be identified and tested. Heterozygosity among cea-affected chromosomes in SNPs rs23961769 and FLJ-IHH_SNP1 proximally, and rs8919370 distally, reduced the shared haplotype to 102,495 bp (corresponding to chr37:28,631,277–28,733,772 on CanFam2) and eliminated all exons of IHH from the conserved interval (Fig. 2). A subset of the haplotypes with critical recombinants identified is shown in Figure 2. A full list of the markers, primer sequences, genotypes, and haplotypes in the region can be found in Supplemental Tables 2 and 3.
Figure 2.
Alignment of SNP haplotypes and a canine BAC contig to the CanFam2 genomic sequence for the cea interval on canine chromosome 37. Eight haplotypes, spanning >376 kb on CFA37, represent cea-transmitting chromosomes segregating in four different breeds and define a linkage disequilibrium interval common to all known cea-affected breeds. Haplotype 1 was observed in all cea-affected Rough and Smooth Collies and Australian Shepherds tested, and in some but not all Border Collies. Haplotypes 2 through 7 were identified in specific Border Collies, and haplotype 8 was only seen in European Shetland Sheepdogs. All haplotypes are identical, and all affected dogs are homozygous for SNPs rs8797935 through rs23961919 (the region bounded by vertical blue lines). (Blue box above the chromosome, the black line in the center of the figure) The position of the 7799-bp deletion in intron 4 of NHEJ1. (Red boxes below the chromosome line) Genes in the region. BACs are tiled below the genes and are identified by their number. The gene-specific probes used to pick the BACs and align them to the chromosome are listed in red above the BAC.
Interrogation of the RPC181 BAC library yielded several clones positive for genes anticipated to be within the cea candidate region. Location of the end sequences of these BACs allowed alignment against CanFam1 and then CanFam2 when these assemblies became available, and construction of a canine BAC contig across the cea interval. PCR analysis of this BAC contig with SNPs and other markers confirmed that marker and gene order in this BAC contig was in agreement with that reported in the 7.5× sequence assembly (Fig. 2). This analysis further confirmed that IHH exon 1 was present in the BAC contig, although represented by a gap in CanFam1. Eventually, the entire cea LD interval, defined by the SNP haplotype shared by all tested cea-affected chromosomes, was shown to be included in a single BAC clone, 96B02 (Fig. 2).
The shared haplotype encompasses the coding region of four genes: nonhomologous end joining factor 1 (NHEJ1), solute carrier family 23 member 3 (SLC23A3), and two hypothetical proteins, FAM134A and C2orf24; plus the non-coding region 5′ to Indian Hedgehog (IHH) exon 1. Sequence analysis of all exons and flanking regions within the shared haplotype revealed no variants within a coding region that would provide a likely candidate for the causative mutation. One potentially significant intronic variant was detected, however, in all affected chromosomes tested: a 7799-bp deletion within intron 4 of the gene NHEJ1, corresponding to nucleotides 28,697,542–28,705,340 on canine chromosome 37 (CFA37) in CanFam2. Two additional noncoding SNPs (NHEJ1_1, NHEJ1_2) in the same gene were also detected (see Supplemental Table 3).
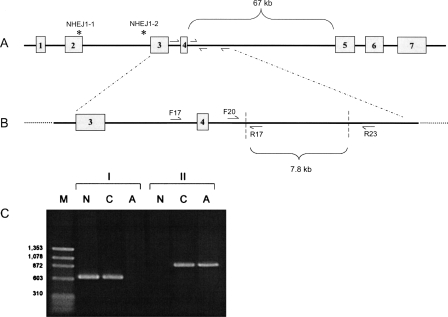
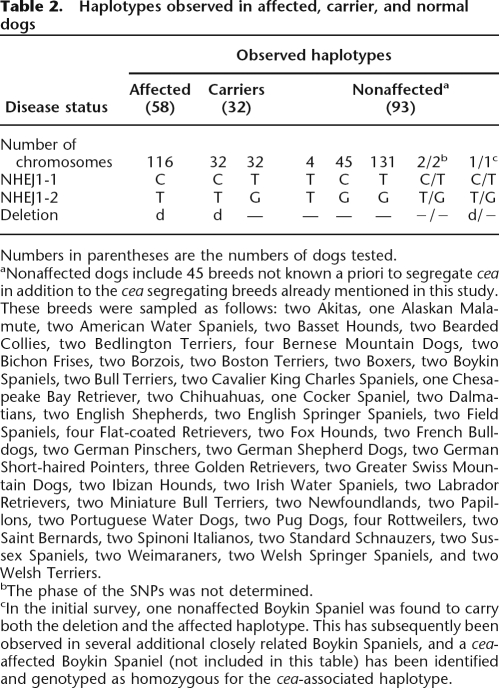
To estimate the occurrence of this deletion in the greater dog population, a two-step PCR test was used to evaluate a set of samples representing multiple breeds. The test utilizes primers placed inside and outside the deletion to quickly identify chromosomes with and without the mutation (Fig. 3). Ninety dogs (58 cea-affected, 32 obligate heterozygotes) representing four breeds segregating cea were genotyped for the 7799-bp deletion, as well as for the closely linked NHEJ1_1 and NHEJ1_2 SNPs. These 90 dogs represented several extended cea-informative pedigrees, both purebred and experimental mixed-breed. An additional 93 dogs from 45 breeds not known to segregate cea were similarly scanned. The deletion was present in both chromosomes of all 58 affected dogs and in one and only one chromosome of each of the 32 obligate carriers from the extended pedigrees (Table 2). All dogs carrying the 7799-bp deletion also carried the affected haplotype at SNPs NHEJ1_1 and NHEJ1_2.
Figure 3.
A two-step PCR protocol for genotyping the cea-associated deletion. (A) The NHEJ1 gene with the positions of the two linked SNPs designated by * and primer locations within intron four. (B) Expanded representation of the cea-associated deletion region (outlined by diagonal dashed lines), with flanking and internal primers. (C) Electrophoretogram demonstrating PCR results using primers shown in B on DNA from Normal (N), Carrier (C), and Affected (A) dogs. (Lane 1, M) A marker lane with sizes as indicated. Set I of PCR products (lanes 2–4) presents amplification results using primers NHEJ1-F17 and NHEJ1-R17. Set II (lanes 5–7) presents results using primers NHEJ1-F20 and NHEJ1-R23.
Table 2.
Haplotypes observed in affected, carrier, and normal dogs
Numbers in parentheses are the numbers of dogs tested.
aNonaffected dogs include 45 breeds not known a priori to segregate cea in addition to the cea segregating breeds already mentioned in this study. These breeds were sampled as follows: two Akitas, one Alaskan Malamute, two American Water Spaniels, two Basset Hounds, two Bearded Collies, two Bedlington Terriers, four Bernese Mountain Dogs, two Bichon Frises, two Borzois, two Boston Terriers, two Boxers, two Boykin Spaniels, two Bull Terriers, two Cavalier King Charles Spaniels, one Chesapeake Bay Retriever, two Chihuahuas, one Cocker Spaniel, two Dalmatians, two English Shepherds, two English Springer Spaniels, two Field Spaniels, four Flat-coated Retrievers, two Fox Hounds, two French Bulldogs, two German Pinschers, two German Shepherd Dogs, two German Short-haired Pointers, three Golden Retrievers, two Greater Swiss Mountain Dogs, two Ibizan Hounds, two Irish Water Spaniels, two Labrador Retrievers, two Miniature Bull Terriers, two Newfoundlands, two Papillons, two Portuguese Water Dogs, two Pug Dogs, four Rottweilers, two Saint Bernards, two Spinoni Italianos, two Standard Schnauzers, two Sussex Spaniels, two Weimaraners, two Welsh Springer Spaniels, and two Welsh Terriers.
bThe phase of the SNPs was not determined.
cIn the initial survey, one nonaffected Boykin Spaniel was found to carry both the deletion and the affected haplotype. This has subsequently been observed in several additional closely related Boykin Spaniels, and a cea-affected Boykin Spaniel (not included in this table) has been identified and genotyped as homozygous for the cea-associated haplotype.
Among the 93 dogs from breeds that did not segregate cea, two dogs (an Alaskan Malamute and a Dalmatian) carried the alleles associated with cea at SNPs NHEJ1_1 and NHEJ1_2 but not the 7799-bp deletion. One dog, a Boykin Spaniel, was heterozygous for both the deletion and the NHEJ1_1 / NHEJ1_2 haplotype and was presumed a carrier of the disorder. Sixteen additional Boykin Spaniels closely related to this individual were subsequently tested, and five were found to be heterozygous for the mutation and haplotype, demonstrating that the recognized cea-affected haplotype was segregating in this breed. Most recently, a small family of Boykin Spaniels segregating the cea-affected phenotype has been identified and genotyped to confirm that the CFA37 haplotype cosegregates with the disorder in the Boykin Spaniel (see Supplemental Table 3).
Dogs from several additional breeds in which a cea or cea-like phenotype was reported to segregate, but were not included in the initial mapping analysis, were subsequently tested for the presence of the 7799-bp deletion. In the Lancashire Heeler, Nova Scotia Duck Tolling Retriever, and Longhaired Whippet breeds, all dogs diagnosed as cea-affected were homozygous for the 7799-bp deletion; cea-nonaffected obligate carriers were heterozygous for the deletion; and it cosegregated with cea in informative pedigrees (see Supplemental Table 3). Two dogs from the Berger des Pyrenees breed that were clinically diagnosed as affected with colobomas were tested but did not have the 7799-bp deletion. A small pedigree of Soft Coated Wheaten Terriers that segregates a phenotype including lesions resembling cea (Van der Woerdt et al. 1995) was also tested, but none of the dogs carried the deletion.
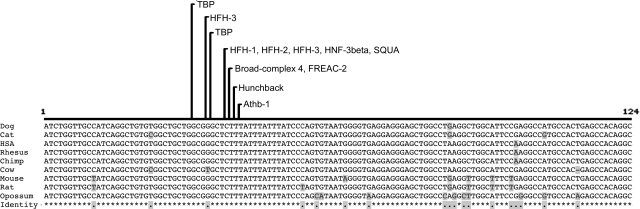
The entire 7799-bp deletion comprises nucleotides 28,697,542–28,705,340 on chromosome 37 based on the CanFam2 assembly (http://genome.ucsc.edu/). It is located within the 67-kb intron 4 of the gene NHEJ1, ∼460 bp from exon 5. Comparative genome analysis of this non-coding region among dog, human, mouse, and rat genomes revealed islands of high sequence conservation (PhastCons Conserved Elements) (Siepel et al. 2005; see http://genome.ucsc.edu/cgi-bin/hgTables). The largest such conserved element (Score: 692, LOD = 210) comprises a 323-bp interval corresponding to chr37:28,702,317–28,702,639. The homologous region was readily identified in the genomes of all nine mammals currently available, including that of the opossum (Monodelphis domestica, a marsupial). Multiple sequence alignment of these nine mammalian homologs identified a core 124-bp element (chr37:28,702,470–28,702,599) that was the most highly conserved sequence. Within this element, there are conserved binding sites for several DNA-binding proteins (Fig. 4).
Figure 4.
Comparative alignment of the 124-bp highly conserved region within the canine cea-associated deletion for nine diverse mammalian sequences. The most strongly conserved region includes a cluster of recognition domains for several DNA-binding proteins, conserved in all nine species. These sites are listed above the alignment with lines pointing to the starting position. Locations and sequence information for these conserved binding domains are listed in Supplemental Table 6. The sequence locations for alignment are dog, chr37:28,702,470–28,702,593 rc; human, chr2:219,715,427–219,715,550 rc; chimp, chr2b:225,076,398–225,076,521 rc; rhesus, chr12:82,998,197–82,998,320 rc; mouse, chr1:74,973,801–74,973,924 rc; rat, chr9:74,386,991–74,387,114 rc; cat, scaffold_100612:247,076–247,199; cow, chr2:65,088,541–65,088,663 rc; opossum, chr7:175,293,269–175,293,392.
Discussion
Collie eye anomaly is a hereditary canine ocular disorder characterized by regional hypoplasia of the choroid, the highly vascularized layer of the eye underlying the retina. The characteristic ophthalmoscopically detectable defect in the ocular fundus is located temporal to the optic nerve (Roberts 1960; Roberts and Dellaporta 1965; Roberts et al. 1966) and corresponds to lesions termed “macular colobomas” when recognized in humans (Alur and Brooks 2004; Gregory-Evans et al. 2004; Chang et al. 2006). Colobomas and staphylomas of the optic nerve head and/or adjacent tissues may also occur as part of the cea extended phenotype. Also, although less frequently, tortuous retinal vessels, multiple retinal folds, retinal detachment, and/or retinal neovascularization may be observed. Most commonly, dogs affected with cea belong to one of the herding breeds with Collie ancestry. We previously mapped the primary cea locus to a 3.9-cM interval on CFA37 flanked by markers FH4306 and AHTh174, a region projected to encompass >40 genes (Lowe et al. 2003).
The addition of several microsatellite markers and comparison of assembled haplotypes between affected and unaffected dogs initially strongly suggested that the cea locus was restricted to the 691-kb region between FH4617 and FH4622 (Supplemental Table 2). However, no additional recombination events were found within the initial mapping families to confirm this reduced interval, and no disease-associated mutations were observed to segregate within a selected set of putative candidate genes tested.
The founding dogs of the mapping families came from three different affected breeds—the Collie, Border Collie, and Australian Shepherd. All of these breeds fall into one breed cluster and were considered likely to have acquired their mutation from the same ancestral source. A fourth affected breed, the Shetland Sheepdog, was predicted to possess the same mutation based solely on its close genetic relationship to the Collie. With the family information exhausted, we decided to use a comparative mapping strategy, including multiple breeds with shared ancestry, to reduce the region of interest to the coding regions of four candidate genes. This reduction in haplotype allowed for the identification of a disease-associated deletion within the gene NHEJ1. The deletion, though intronic, includes several conserved elements, most notably a 124-bp segment that is highly conserved among all available mammalian genomes, including that of the opossum, and contains binding sites for multiple regulatory proteins. It is the interaction of just such a protein with the conserved region that we postulate is responsible for the cea defect. Either NHEJ1 or IHH could be the target gene regulated by such an interaction. IHH lies just 1250 bp outside the minimum shared interval and is a member of a family of morphogens that regulate cell proliferation, differentiation, and cell–cell communication in developing embryos, which makes it a particularly attractive candidate gene for such a scenario.
The apparent absence of any sequence homologous to the cea-associated deletion in all the currently available genomes of nonmammalian species suggests strongly that the mechanism destroyed by the cea-associated deletion has evolutionary significance for differences in the patterning of ocular development unique to the mammalian eye.
This work establishes that the primary cea mutation arose as a single disease allele and was transmitted to multiple herding breeds through outcrosses in early dog breed development. The breeds used in this comparative study are closely related both by genetic analysis, as shown in the cluster analysis, and historically, as herding dogs. Although cea-affected dogs are most consistently seen in the herding breeds with Collie ancestry, lesions resembling Collie eye anomaly are infrequently seen in a much wider range of breeds. In the most recent compilation of such data (American College of Veterinary Ophthalmologists 2007; see Supplemental Table 4), nine breeds and varieties are reported as known to cosegregate the cea phenotype and the cea-associated haplotype reported herein. Dogs from a further 18 breeds are infrequently reported to present with choroidal hypoplasia (macular coloboma) and optic nerve head coloboma/staphyloma. Sporadic isolated case reports of colobomas are further tabulated for an additional 77 breeds.
The deletion described here is found only in dogs affected with or carrying cea and can be identified using a simple PCR-based test. Aside from the standard use of a genetic test to control breeding and reduce the prevalence of the disease, the presence of this mutation can be used to assist with diagnoses as well. Dogs presenting with unidentified choroidal hypoplasias or colobomas can be tested for the deletion to determine if the condition is indicative of cea. This may prove particularly useful in breeds that are not typically associated with cea. The test may also be used to identify cea in merle dogs where pale eye coloring can lead to false diagnosis.
Because carriers of the cea-associated haplotype were found by chance in dogs from a hunting breed, the Boykin Spaniel, not previously known to be affected with cea, efforts were made to identify and genotype a cea-affected Boykin Spaniel. Finding that the affected Boykin Spaniels were homozygous for the CFA37 haplotype described herein confirmed that this breed segregates cea as well as the mutation and haplotype. We also tested three other breeds (Lancashire Heeler, Longhaired Whippet, and Nova Scotia Duck Tolling Retriever) in which a cea-like phenotype was reported to segregate and demonstrated concordance between their genotypes for the 7799-bp deletion in intron 4 of NHEJ1 and their cea phenotypic status. The Lancashire Heeler was developed by crossing a herding breed and a terrier and shows a close relationship to the Border Collie according to our analysis, although it clusters with the hunting dogs. The Longhaired Whippet is a recently developed breed that may have had long-coated herding dogs, such as the Shetland Sheepdog, among its founders. The Nova Scotia Duck Tolling Retriever (NSDTR) is a gun dog breed that reportedly includes “farm collies” among its ancestors, although it does not cluster with the herding breeds. Both the Boykin Spaniel and the NSDTR are hunting breeds that were developed in North America from a mixture of European breeds and mongrels, which may have included local farm Collies. Without deliberate selection for any of the traits associated with herding dogs, the only signature of herding ancestry in these two sporting breeds is the cea defect commonly found in Collie-like dogs. This finding highlights the inter-relatedness of all dog breeds and suggests that although breeds are strictly segregated in modern times, all dogs stem from one or more common founding populations, and the relationships among established breeds are not always evident.
In addition to the cea-affected breeds, we tested a small pedigree of Soft Coated Wheaten Terriers (SCWT), which exhibited a phenotype broadly similar to cea. In the SCWT, the eye disorder presents with the choroidal hypoplasia and colobomas that are reminiscent of cea, but mild anterior segment dysgenesis, which is not typical of cea, is also observed in the SCWT syndrome. Interestingly, neither the affected nor unaffected dogs had the cea deletion, and markers in the region did not segregate with the disease (genotypes available at http://research.nhgri.nih.gov/dog_genome/). The SCWT is an old terrier breed from Ireland and has more in common genetically with guarding breeds, such as the bulldog, than with herding or sporting breeds, as evidenced by the cluster analysis presented here (Fig. 1). We hypothesize that the disorder found in the SCWT, although similar in appearance to cea, is caused by a mutation in another gene in the pathway leading to development of the choroid.
Applied broadly, these results suggest that the search for causative mutations associated with canine diseases will be most successful when (1) the disease has been identified in multiple breeds of common ancestry; and (2) the disease alleles are IBD in affected individuals. Multiple instances of mutations shared across breeds have been observed to date. For instance, the same mutations in the MC1R, TYRP1, and AGRP genes have been found to segregate with coat color in multiple dog breeds (Newton et al. 2000; Schmutz et al. 2002; Berryere et al. 2005). In addition, recent identification of SILV, the gene responsible for merle or mottled coloring in dogs, revealed the identical causative mutation, a SINE insertion, in multiple breeds of dog with a variety of genetic and historical backgrounds (Clark et al. 2006). These studies suggest that the mutations associated with each phenotype arose a limited number of times, in some cases only once, were introduced into new breeds through out-crossing, and were maintained either through direct selection for a desired trait or through close proximity to a region under selection. Similarly, many disease alleles, although not specifically selected for, appear to have developed relatively few times and can be found in related breeds via a common ancestor. For example, a haplotype of microsatellites associated with the multi-drug-resistance mutation in ABCB1 was found to be IBD in nine distinct dog breeds, four of which cluster together in our analysis while the other five share historical or anecdotal relationships to these breeds (Neff et al. 2004).
In dogs, as in humans, there are multiple loci for most common diseases including deafness, progressive retinal disease, heart disease, and epilepsy (for review, see Petersen-Jones 2005; Rak and Distl 2005; Chandler 2006; Parker et al. 2006). Examining identical diseases in unrelated breeds will likely reveal independent genetic origins for diseases with similar phenotypes. By sampling from breeds with predicted common ancestry, researchers can enrich their studies for affected individuals that share a common disease-causing mutation.
In summary, a classification system based on genetic markers, such as the breed clusters described here, allows for quick identification of related breeds. Canine breed history is complex and we cannot know what selective forces were strongest during the development of a particular breed or how those have changed over time. Clustering analysis at the current level does not identify each historical introgression; however, it does provide a starting point for selecting breeds with common ancestry. The breeds contained within a single cluster share not only a large portion of their genome, but as the cea analysis demonstrates, may be more likely than randomly selected breeds to share deleterious mutations inherited from a common ancestor. As geneticists working in the human system struggle to understand the genetic basis of complex traits, it is clear the dog has much to offer. Subsets of dog breeds have been identified with an increased risk for nearly every disease that plagues humans. Using comparative genomics, we can link not only the genomes but the phenotypes and geographic boundaries that define populations to categorize the breeds, trace their history, and identify traits shared between them. This offers a unique opportunity for those interested in studying the genetics of isolated populations, regardless of species.
Methods
Canine pedigrees and diagnostic methods
Naturally occurring and experimental pedigrees derived from affected purebred Collies, Border Collies, and Australian Shepherds in which cea segregates were sampled as described previously (Lowe et al. 2003). Other affected purebred dogs were collected from private owners. All diagnoses were made by clinical ophthalmoscopic examination (G.M. Acland), and in selected cases confirmed by gross examination of the fixed posterior segment and/or ocular histopathology. For the purposes of this study, any dog with ophthalmoscopic evidence of choroidal hypoplasia (one or both eyes) was classified as affected.
DNA isolation
For cluster analyses, samples from 224 purebred dogs representing 132 breeds were obtained by buccal (cheek) swabs (76%) and/or blood samples (24%) from American Kennel Club (AKC) sanctioned dog shows, specialty events, and mail-in donations. Particular effort was made to sample the many breed groups that were under-represented in previous studies of breed relatedness (Zajc and Sampson 1999; Koskinen and Bredbacka 2000; Irion et al. 2003; Parker et al. 2004). AKC registration number and detailed pedigree information were requested for all dogs, as participation was limited to dogs that were unrelated through the grandparent generation. In the case of nine dogs for which pedigrees were not submitted, verification of pedigree was provided by breed club representatives or collection managers.
Buccal swab samples were collected according to AKC guidelines (http://www.akc.org/) using cytology brushes (Medical Packaging Corp.). DNA was extracted from buccal swabs using QiaAmp DNA extraction kits following the manufacturer’s protocol (QIAGEN). DNA was extracted from blood samples using a phenol/chloroform protocol as described previously (Comstock et al. 2002).
DNA from cea-affected dogs for fine-mapping studies was isolated from whole blood or splenic tissue, using standard protocols (Maniatis et al. 1982). All procedures were performed in adherence to the ARVO Resolution for the Use of Animals in Ophthalmic and Vision Research. All DNA was handled in accordance with the Fred Hutchinson Cancer Research Center and NIH Animal Care and Use Committee approved protocols.
Microsatellite genotyping for cluster analysis
Data from 414 dogs representing 85 distinct breeds that had been genotyped by us previously were included in this study (Parker et al. 2004). Five unrelated dogs from each of 35 breeds and four dogs from a further 12 breeds, for a total of 47 new breeds, were additionally genotyped using 96 microsatellite markers as described (Parker et al. 2004). DNA samples were arrayed in 96-well plates. A positive control was included on each plate to ensure consistent allele binning. PCR was carried out using 2.5–5 ng of genomic DNA as template and a standard protocol (http://research.nhgri.nih.gov/dog_genome/). PCR amplicons were labeled by the addition of 0.25 pmol of an M13 primer covalently linked with either 6FAM, VIC, NED, or PET fluorescent dyes (ABI) to each reaction. For each DNA sample, two to four amplicons spanning separate microsatellites and labeled with different dyes were multiplexed following completion of PCR. Samples were denatured in Hi-Di formamide with 15 pmol of GeneScan-500LIZ size standard (ABI) according to the manufacturer’s protocols. All samples were loaded on an ABI 3730xl capillary electrophoresis instrument for allele separation. Genotypes were called using GeneMapper 4.0 (ABI). All calls were checked manually, and each plate was scanned for the appearance of new alleles outside existing bins.
Statistical analysis of population clusters
Clustering of 132 breeds was performed using the program STRUCTURE (Pritchard et al. 2000; Falush et al. 2003) at 100,000 iterations of the Gibbs sampler after a burn-in of 20,000 iterations. The correlated allele frequency model was used with asymmetric admixture allowed. Each run was repeated 20 times at values of K from 2 to 5, 10 times at values of K from 6 to 20, and five times at values of K from 21 to 50. Runs were completed both with and without population information to assess consistency of clustering.
Two methods were used to compare independent runs of STRUCTURE. At values of K ≤ 5, populations were manually matched by breed membership and then averaged over all runs. Consistency was determined by the presence of multiple breeds with at least 80% of their genome assigned to only one of the K clusters. Manually matching breeds introduces a bias toward the breed chosen as anchor for each cluster and thus implies greater purity within these breeds that may or may not be realistic. To reduce this bias and to assess the stability of runs at higher values of K, similarity was determined by calculating an average standard deviation across multiple runs. In order to bypass the need to manually order populations, Euclidean distance matrices between individuals and populations were calculated from the cluster results for each run. The Euclidean distance gives a model-free linear distance between individuals based strictly on changes in the cluster assignments of each. The standard deviation of these distances based on multiple runs at the same value of K was calculated for each population pair, and the distribution of standard deviations was assessed as a measure of overall consistency (Supplemental Fig. 4). A one-tailed t-test was used to assess the significance in the distribution of standard deviations at each value of K. Calculations and significance tests were carried out using the statistical package R (R Development Core Team 2006). Relationships between the breeds are displayed as a heatmap based on average distances between the breeds as calculated above. Heatmaps were created using the gplots graphing package in R (http://cran.r-project.org/src/contrib/Descriptions/gplots.html).
To determine the clustering integrity of individual breeds, the entire data set was divided into subsets of 10–11 breeds each, and all possible pairs of subsets were run five times with K equal to n, n + 1, and n + 2, where n equals the number of breeds in the combined subsets (20, 21, or 22). All runs were compared by calculating the Euclidean distance between individual dogs based on their clustering assignment at each run and then averaging that distance over all runs in which both individuals of the pair were included. The average distance between a pair of dogs within a single breed is 0.09 (median 0.04), while the average distance between any two dogs of two different breeds is 1.3 (median 1.3; Wilcoxon test for significant difference in the distributions p < 2 × 10−16) (Supplemental Fig. 1B). The outer limit of the 95% confidence interval describing within-breed clustering is equivalent to individuals clustering together in ∼85% of runs. Breeds in which all individuals cluster with all members of another breed in at least 80% of runs are therefore considered to be a closely related pair. Breeds that cluster together in >60% but <80% of runs are considered a related pair (Table 1).
Canine BAC library screening for fine-mapping of cea
The RPC181 canine 8.1-fold BAC library (BACPAC Resource Center, Children’s Hospital Oakland Research Institute, Oakland, CA; http://bacpac.chori.org/mcanine81.htm) was screened to identify BAC clones containing the genes CRYBA2, CDK5R2, NHEJ1, SLC23A3, and ABCB6. Canine partial sequences for these genes were acquired from the Institute for Genomic Research (TIGR) database and used to design primers for canine gene-specific probes (Supplemental Table 5). Amplification of the correct gene product was confirmed by direct sequencing. Amplified products were labeled with [α-32P]dCTP and hybridized to high-density gridded BAC clone filters according to a standard protocol (Li et al. 1999). DNA from identified BAC clones was extracted using the alkaline lysis method (Bimboim and Doly 1979). PCR reactions using probe-specific primers and sequencing of resulting amplicons was used to confirm that the identified BAC clones contained the expected genes.
End sequence was obtained for eight positive BAC clones by the sequencing center at TIGR. SNP data and end sequences were used to align the BACs with the canine 7.5× sequence (Lindblad-Toh et al. 2005) and create a contig covering the region of linkage.
Sequence alignment and analysis
Sequence alignment of multiple genomes was performed using BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) and BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Conserved domains were analyzed using the Web program Consite (http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite/) to look for regulatory sites.
SNP genotyping for fine-mapping
Polymorphisms for fine mapping the cea interval were identified using two approaches. Initially, a set of informative polymorphisms was developed de novo by resequencing selected dogs. This was done prior to the availability of the 7.5× canine genome sequence assembly (Lindblad-Toh et al. 2005) and subsequently in order to identify additional gene-specific markers in the region. PCR primers were designed from gene sequences in the cea disease interval in a region that corresponds to human chromosome 2q35 (Supplemental Table 3). Prior to the availability of the whole genome sequence assembly, amplicons were selected based on alignment of the canine 1.5× sequence with the human genome (Kirkness et al. 2003; Hitte et al. 2005). To identify informative polymorphisms and build haplotypes, each gene region was amplified initially in eight dogs; four affected dogs, three carriers, and one unaffected dog. Regions that contained SNPs were then sequenced using DNA from 46 additional dogs including 32 affected dogs, 13 carriers, and two additional unaffected animals. All sequence reads were aligned using Phred, Phrap, and Consed packages (Ewing et al. 1998; Gordon et al. 1998). SNPs were identified from aligned sequences using PolyPhred (Nickerson et al. 1997).
Once the canine genome sequence alignments (CanFam1 and CanFam2) became publicly available (http://genome. ucsc.edu/ and http://www.ncbi.nlm.nih.gov/Genomes/), an additional set of SNPs from the cea interval was selected and tested for informativeness in several cea-affected and heterozygous dogs (Supplemental Table 3). Canine SNPs can be found at http://www.broad.mit.edu/mammals/dog/snp/ and http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=Display&DB=snp.
Haplotypes were assigned manually by ordering SNPs according to the genomic position. The haplotypes of homozygous dogs were determined first. Heterozygous dogs were assigned phase based on the most likely haplotypes previously identified. Where available, phase was verified with family data.
Population screening for a cea-associated deletion
Once a deletion and two tightly linked SNPs were identified in the initial study group of cea-affected dogs, a set of DNA samples from an additional 34 cea-affected dogs, 33 obligate cea-heterozygotes, and 94 phenotypically unaffected individuals representing 45 breeds was tested for the presence of these alleles. PCR amplification and sequencing were performed using standard protocols as described previously (Lowe et al. 2003; Zangerl et al. 2006). Conditions are available at http://research.nhgri.nih.gov/dog_genome/. The deletion was tracked by genotyping using two sets of primers: NHEJ1-F17, 5′-TCTCACAGGCAGAAAGCTCA-3′, with NHEJ1-R17, 5′-CCATTCATTCCTTTGCCAGT-3′, to amplify within the deletion; and NHEJ1-F20, 5′-TGGGCTGGTGAACATTTGTA-3′, with NHEJ1-R23, 5′-CCTTTTTGTTTGCCCTCAGA-3′, to amplify across the deletion.
Acknowledgments
We gratefully acknowledge Sue Pearce Kelling and Jennifer Johnson for technical assistance, Aaron Sethman and Gabriel Renaud from the NHGRI Bioinformatics Core for programming assistance, Nathan B. Sutter and Pascale Quignon for helpful discussions about analysis, Keith Murphy for helpful discussions regarding Boykin Spaniels, and many dog owners and breeders who provided samples for this study. This work was funded by NIH grant EY06855, The Foundation Fighting Blindness, the American Border Collie Association, and by the Intramural Program of the National Human Genome Research Institute.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6772807
References
- Alur R., Brooks B., Brooks B. Clinical and genetic analysis of coloboma: A review. Asian J. Exp. Sci. 2004;20:1–15. [Google Scholar]
- American College of Veterinary Ophthalmologists . Ocular disorders proven or suspected to be inherited in purebred dogs. Genetics Committee Report; Meridian, ID: 2007. [Google Scholar]
- Berryere T.G., Kerns J.A., Barsh G.S., Schmutz S.M., Kerns J.A., Barsh G.S., Schmutz S.M., Barsh G.S., Schmutz S.M., Schmutz S.M. Association of an Agouti allele with fawn or sable coat color in domestic dogs. Mamm. Genome. 2005;16:262–272. doi: 10.1007/s00335-004-2445-6. [DOI] [PubMed] [Google Scholar]
- Bimboim H.C., Doly J., Doly J. A rapid alkaline procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler K. Canine epilepsy: What can we learn from human seizure disorders? Vet. J. 2006;172:207–217. doi: 10.1016/j.tvjl.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Chang L., Blain D., Bertuzzi S., Brooks B.P., Blain D., Bertuzzi S., Brooks B.P., Bertuzzi S., Brooks B.P., Brooks B.P. Uveal coloboma: Clinical and basic science update. Curr. Opin. Ophthalmol. 2006;17:447–470. doi: 10.1097/01.icu.0000243020.82380.f6. [DOI] [PubMed] [Google Scholar]
- Clark L.A., Wahl J.M., Rees C.A., Murphy K.E., Wahl J.M., Rees C.A., Murphy K.E., Rees C.A., Murphy K.E., Murphy K.E. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc. Natl. Acad. Sci. 2006;103:1376–1381. doi: 10.1073/pnas.0506940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock K.E., Georgiadis N., Pecon-Slattery J., Roca A.L., Ostrander E.A., O’Brien S.J., Wasser S.K., Georgiadis N., Pecon-Slattery J., Roca A.L., Ostrander E.A., O’Brien S.J., Wasser S.K., Pecon-Slattery J., Roca A.L., Ostrander E.A., O’Brien S.J., Wasser S.K., Roca A.L., Ostrander E.A., O’Brien S.J., Wasser S.K., Ostrander E.A., O’Brien S.J., Wasser S.K., O’Brien S.J., Wasser S.K., Wasser S.K. Patterns of molecular genetic variation among African elephant populations. Mol. Ecol. 2002;11:2489–2498. doi: 10.1046/j.1365-294x.2002.01615.x. [DOI] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J., Regnaut S., Goudet J., Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P., Hillier L., Wendl M.C., Green P., Wendl M.C., Green P., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J.K., Stephens M., Pritchard J.K., Pritchard J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein O., Zangerl B., Pearce-Kelling S., Sidjanin D.J., Kijas J.W., Felix J., Acland G.M., Aguirre G.D., Zangerl B., Pearce-Kelling S., Sidjanin D.J., Kijas J.W., Felix J., Acland G.M., Aguirre G.D., Pearce-Kelling S., Sidjanin D.J., Kijas J.W., Felix J., Acland G.M., Aguirre G.D., Sidjanin D.J., Kijas J.W., Felix J., Acland G.M., Aguirre G.D., Kijas J.W., Felix J., Acland G.M., Aguirre G.D., Felix J., Acland G.M., Aguirre G.D., Acland G.M., Aguirre G.D., Aguirre G.D. Linkage disequilibrium mapping in domestic dog breeds narrows the progressive rod–cone degeneration interval and identifies ancestral disease-transmitting chromosome. Genomics. 2006;88:541–550. doi: 10.1016/j.ygeno.2006.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P., Abajian C., Green P., Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Gregory-Evans C.Y., Williams M.J., Halford S., Gregory-Evans K., Williams M.J., Halford S., Gregory-Evans K., Halford S., Gregory-Evans K., Gregory-Evans K. Ocular coloboma: A reassessment in the age of molecular neuroscience. J. Med. Genet. 2004;41:881–891. doi: 10.1136/jmg.2004.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitte C., Madeoy J., Kirkness E.F., Priat C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Madeoy J., Kirkness E.F., Priat C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Kirkness E.F., Priat C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Priat C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Lorentzen T.D., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Senger F., Thomas D., Derrien T., Ramirez C., Scott C., Thomas D., Derrien T., Ramirez C., Scott C., Derrien T., Ramirez C., Scott C., Ramirez C., Scott C., Scott C., et al. Facilitating genome navigation: Survey sequencing and dense radiation-hybrid gene mapping. Nat. Rev. Genet. 2005;6:643–648. doi: 10.1038/nrg1658. [DOI] [PubMed] [Google Scholar]
- Irion D.N., Schaffer A.L., Famula T.R., Eggleston M.L., Hughes S.S., Pedersen N.C., Schaffer A.L., Famula T.R., Eggleston M.L., Hughes S.S., Pedersen N.C., Famula T.R., Eggleston M.L., Hughes S.S., Pedersen N.C., Eggleston M.L., Hughes S.S., Pedersen N.C., Hughes S.S., Pedersen N.C., Pedersen N.C. Analysis of genetic variation in 28 dog breed populations with 100 microsatellite markers. J. Hered. 2003;94:81–87. doi: 10.1093/jhered/esg004. [DOI] [PubMed] [Google Scholar]
- Kirkness E.F., Bafna V., Halpern A.L., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Bafna V., Halpern A.L., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Halpern A.L., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Levy S., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Remington K., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Rusch D.B., Delcher A.L., Pop M., Wang W., Fraser C.M., Delcher A.L., Pop M., Wang W., Fraser C.M., Pop M., Wang W., Fraser C.M., Wang W., Fraser C.M., Fraser C.M., et al. The dog genome: Survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Koskinen M.T., Bredbacka P., Bredbacka P. Assessment of the population structure of five Finnish dog breeds with microsatellites. Anim. Genet. 2000;31:310–317. doi: 10.1046/j.1365-2052.2000.00669.x. [DOI] [PubMed] [Google Scholar]
- Latshaw W.K., Wyman M., Venzke W.G., Wyman M., Venzke W.G., Venzke W.G. Embryologic development of an anomaly of ocular fundus in the Collie dog. Am. J. Vet. Res. 1969;30:211–217. [PubMed] [Google Scholar]
- Li R., Mignot E., Faraco J., Kadotani H., Cantanese J., Zhao B., Lin X., Hinton L., Ostrander E.A., Patterson D.F., Mignot E., Faraco J., Kadotani H., Cantanese J., Zhao B., Lin X., Hinton L., Ostrander E.A., Patterson D.F., Faraco J., Kadotani H., Cantanese J., Zhao B., Lin X., Hinton L., Ostrander E.A., Patterson D.F., Kadotani H., Cantanese J., Zhao B., Lin X., Hinton L., Ostrander E.A., Patterson D.F., Cantanese J., Zhao B., Lin X., Hinton L., Ostrander E.A., Patterson D.F., Zhao B., Lin X., Hinton L., Ostrander E.A., Patterson D.F., Lin X., Hinton L., Ostrander E.A., Patterson D.F., Hinton L., Ostrander E.A., Patterson D.F., Ostrander E.A., Patterson D.F., Patterson D.F., et al. Construction and characterization of an eightfold redundant dog genomic bacterial artificial chromosome library. Genomics. 1999;58:9–17. doi: 10.1006/geno.1999.5772. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Chang J.L., Kulbokas E.J., Zody M.C., Kulbokas E.J., Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lowe J.K., Kukekova A.V., Kirkness E.F., Langlois M.C., Aguirre G.D., Acland G.M., Ostrander E.A., Kukekova A.V., Kirkness E.F., Langlois M.C., Aguirre G.D., Acland G.M., Ostrander E.A., Kirkness E.F., Langlois M.C., Aguirre G.D., Acland G.M., Ostrander E.A., Langlois M.C., Aguirre G.D., Acland G.M., Ostrander E.A., Aguirre G.D., Acland G.M., Ostrander E.A., Acland G.M., Ostrander E.A., Ostrander E.A. Linkage mapping of the primary disease locus for Collie eye anomaly. Genomics. 2003;82:86–95. doi: 10.1016/s0888-7543(03)00078-8. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E.F., Sambrook J., Fritsch E.F., Sambrook J., Sambrook J. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1982. [Google Scholar]
- Neff M.W., Robertson K.R., Wong A.K., Safra N., Broman K.W., Slatkin M., Mealey K.L., Pedersen N.C., Robertson K.R., Wong A.K., Safra N., Broman K.W., Slatkin M., Mealey K.L., Pedersen N.C., Wong A.K., Safra N., Broman K.W., Slatkin M., Mealey K.L., Pedersen N.C., Safra N., Broman K.W., Slatkin M., Mealey K.L., Pedersen N.C., Broman K.W., Slatkin M., Mealey K.L., Pedersen N.C., Slatkin M., Mealey K.L., Pedersen N.C., Mealey K.L., Pedersen N.C., Pedersen N.C. Breed distribution and history of canine mdr1-1Δ, a pharmacogenetic mutation that marks the emergence of breeds from the Collie lineage. Proc. Natl. Acad. Sci. 2004;101:11725–11730. doi: 10.1073/pnas.0402374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton J.M., Wilkie A.L., He L., Jordan S.A., Metallinos D.L., Holmes N.G., Jackson I.J., Barsh G.S., Wilkie A.L., He L., Jordan S.A., Metallinos D.L., Holmes N.G., Jackson I.J., Barsh G.S., He L., Jordan S.A., Metallinos D.L., Holmes N.G., Jackson I.J., Barsh G.S., Jordan S.A., Metallinos D.L., Holmes N.G., Jackson I.J., Barsh G.S., Metallinos D.L., Holmes N.G., Jackson I.J., Barsh G.S., Holmes N.G., Jackson I.J., Barsh G.S., Jackson I.J., Barsh G.S., Barsh G.S. Melanocortin 1 receptor variation in the domestic dog. Mamm. Genome. 2000;11:24–30. doi: 10.1007/s003350010005. [DOI] [PubMed] [Google Scholar]
- Nickerson D.A., Tobe V.O., Taylor S.L., Tobe V.O., Taylor S.L., Taylor S.L. PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker H.G., Ostrander E.A., Ostrander E.A. Canine genomics and genetics: Running with the pack. PLoS Genet. 2005;1:e58. doi: 10.1371/journal.pgen.0010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker H.G., Kim L.V., Sutter N.B., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Kim L.V., Sutter N.B., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Sutter N.B., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L., DeFrance H.B., Ostrander E.A., Kruglyak L., Ostrander E.A., Kruglyak L., Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Parker H.G., Meurs K.M., Ostrander E.A., Meurs K.M., Ostrander E.A., Ostrander E.A. Finding cardiovascular disease genes in the dog. J. Vet. Cardiol. 2006;8:115–127. doi: 10.1016/j.jvc.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Jones S. Advances in the molecular understanding of canine retinal diseases. J. Small Anim. Pract. 2005;46:371–380. doi: 10.1111/j.1748-5827.2005.tb00333.x. [DOI] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P., Stephens M., Donnelly P., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. Foundation for Statistical Computing; Vienna, Austria: 2006. [Google Scholar]
- Rak S.G., Distl O., Distl O. Congenital sensorineural deafness in dogs: A molecular genetic approach toward unravelling the responsible genes. Vet. J. 2005;169:188–196. doi: 10.1016/j.tvjl.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Roberts S.R. Congenital posterior ectasia of the sclera in Collie dogs. Am. J. Ophthalmol. 1960;50:451–465. doi: 10.1016/0002-9394(60)90684-x. [DOI] [PubMed] [Google Scholar]
- Roberts S.R., Dellaporta A., Dellaporta A. Congenital posterior ectasia of the sclera in Collie dogs: Part I. Clinical features. Am. J. Ophthalmol. 1965;59:180–186. [PubMed] [Google Scholar]
- Roberts S.R., Dellaporta A., Winter F.C., Dellaporta A., Winter F.C., Winter F.C. The Collie ectasia syndrome. Pathology of eyes of young and adult dogs. Am. J. Ophthalmol. 1966;62:728–752. [PubMed] [Google Scholar]
- Schmutz S.M., Berryere T.G., Goldfinch A.D., Berryere T.G., Goldfinch A.D., Goldfinch A.D. TYRP1 and MC1R genotypes and their effects on coat color in dogs. Mamm. Genome. 2002;13:380–387. doi: 10.1007/s00335-001-2147-2. [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter N.B., Ostrander E.A., Ostrander E.A. Dog star rising: The canine genetic system. Nat. Rev. Genet. 2004;5:900–910. doi: 10.1038/nrg1492. [DOI] [PubMed] [Google Scholar]
- Sutter N.B., Eberle M.A., Parker H.G., Pullar B.J., Kirkness E.F., Kruglyak L., Ostrander E.A., Eberle M.A., Parker H.G., Pullar B.J., Kirkness E.F., Kruglyak L., Ostrander E.A., Parker H.G., Pullar B.J., Kirkness E.F., Kruglyak L., Ostrander E.A., Pullar B.J., Kirkness E.F., Kruglyak L., Ostrander E.A., Kirkness E.F., Kruglyak L., Ostrander E.A., Kruglyak L., Ostrander E.A., Ostrander E.A. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Woerdt A., Stades F.C., Van der Linde-Sipman J.S., Boeve M.H., Stades F.C., Van der Linde-Sipman J.S., Boeve M.H., Van der Linde-Sipman J.S., Boeve M.H., Boeve M.H. Multiple ocular anomalies in two related litters of soft-coated Wheaten Terriers. Vet. Comp. Ophthalmol. 1995;5:78–82. [Google Scholar]
- Zajc I., Sampson J., Sampson J. Utility of canine microsatellites in revealing the relationships of pure bred dogs. J. Hered. 1999;90:104–107. doi: 10.1093/jhered/90.1.104. [DOI] [PubMed] [Google Scholar]
- Zangerl B., Goldstein O., Philp A.R., Lindauer S.J., Pearce-Kelling S.E., Mullins R.F., Graphodatsky A.S., Ripoll D., Felix J.S., Stone E.M., Goldstein O., Philp A.R., Lindauer S.J., Pearce-Kelling S.E., Mullins R.F., Graphodatsky A.S., Ripoll D., Felix J.S., Stone E.M., Philp A.R., Lindauer S.J., Pearce-Kelling S.E., Mullins R.F., Graphodatsky A.S., Ripoll D., Felix J.S., Stone E.M., Lindauer S.J., Pearce-Kelling S.E., Mullins R.F., Graphodatsky A.S., Ripoll D., Felix J.S., Stone E.M., Pearce-Kelling S.E., Mullins R.F., Graphodatsky A.S., Ripoll D., Felix J.S., Stone E.M., Mullins R.F., Graphodatsky A.S., Ripoll D., Felix J.S., Stone E.M., Graphodatsky A.S., Ripoll D., Felix J.S., Stone E.M., Ripoll D., Felix J.S., Stone E.M., Felix J.S., Stone E.M., Stone E.M., et al. Identical mutation in a novel retinal gene causes progressive rod-cone degeneration in dogs and retinitis pigmentosa in humans. Genomics. 2006;88:551–563. doi: 10.1016/j.ygeno.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]