Abstract
Ubiquitin (Ub) regulates diverse functions in eukaryotes through its attachment to other proteins. The defining step in this protein modification pathway is the attack of a substrate lysine residue on Ub bound through its C-terminus to the active site cysteine residue of a Ub-conjugating enzyme (E2) or certain Ub ligases (E3s). So far, these E2 and E3 cysteine residues are the only enzyme groups known to participate in the catalysis of conjugation. Here we show that a strictly conserved E2 asparagine residue is critical for catalysis of E2- and E2/RING E3-dependent isopeptide bond formation, but dispensable for upstream and downstream reactions of Ub thiol ester formation. In constrast, the strictly conserved histidine and proline residues immediately upstream of the asparagine are dispensable for catalysis of isopeptide bond formation. We propose that the conserved asparagine side chain stabilizes the oxyanion intermediate formed during lysine attack. The E2 asparagine is the first non-covalent catalytic group to be proposed in any Ub conjugation factor.
Keywords: catalytic mechanism/E2/E3/isopeptide/ubiquitin
Introduction
The covalent conjugation of ubiquitin (Ub) to proteins regulates a host of eukaryotic cell functions, including the progression of the cell cycle, antigen presentation, the induction of the inflammatory response and DNA damage tolerance (Hershko and Ciechanover, 1998; Ulrich, 2002). The role of Ub in these processes is that of a signal, which frequently, but not always, targets the substrate protein for degradation by 26S proteasomes. Ub–substrate ligation is accomplished through the actions of a cascade of conjugating factors (Hershko and Ciechanover, 1998; Pickart, 2001). Ub-activating enzyme (E1) first uses ATP to drive the formation of a thiol ester between an E1 cysteine residue and the C-terminus of Ub (G76). Ub is then transferred to the active site cysteine of a Ub-conjugating enzyme (E2). The Ub in this thiol ester (or a subsequent one, see below) is then attacked by the ε-amino group of a substrate lysine residue to form an isopeptide bond. This final step is usually facilitated by a Ub–protein ligase (E3) that binds the E2–Ub thiol ester and the substrate at distinct sites (Huang et al., 1999; Zheng et al., 2000, 2002; Pickart, 2001; Brzovic et al., 2003; Verdecia et al., 2003). The existence of multiple E2 and E3 enzymes facilitates the selective targeting of individual substrates for ubiquitylation (Hershko and Ciechanover, 1998; Pickart, 2001).
Most known E3 enzymes belong to two protein families. RING E3s utilize a globular zinc-binding domain to recruit the E2–Ub intermediate (Deshaies, 1999; Joazeiro and Weissman, 2000), whereas HECT (homologous to the E6AP C-terminus) domain E3s accomplish this using a different globular domain (Huibregtse et al., 1995; Huang et al., 1999). The two families are also mechanistically distinct, in that each uses a different source of activated Ub in the isopeptide bond formation step. In RING E3-dependent reactions, the substrate lysine directly attacks the RING-bound E2–Ub intermediate (Deshaies, 1999), while in HECT E3-dependent reactions, Ub must be transferred to a conserved cysteine of the E3 before it can be attacked by the substrate lysine (Huibregtse et al., 1995; Scheffner et al., 1995).
E3 enzymes play a well-established role in substrate selection and are generally necessary for substrate ubiquitylation in vivo (Hershko and Ciechanover, 1998; Laney and Hochstrasser, 1999). However, the extent to which E2s or E3s accomplish chemical catalysis remains uncertain. Efficient isopeptide bond formation is expected to depend on a general base to deprotonate the attacking lysine, along with a group(s) to stabilize the developing negative charge on the (former) carbonyl oxygen of Ub-G76 (Rawlings and Barrett, 1994; Pickart, 2001). However, crystal and solution structures of E2s and E2/E3 complexes have failed to reveal credible candidates for these groups (for example see Tong et al., 1997; Huang et al., 1999; Zheng et al., 2000, 2002; Van Demark et al., 2001), leading to the view that E3-induced proximity of the substrate and the E2–Ub intermediate is the principal mode of catalysis (reviewed in Pickart, 2001; Van Demark and Hill, 2002). Here we report that a strictly conserved E2 asparagine residue is necessary for efficient catalysis of isopeptide bond formation. We propose that this asparagine side chain stabilizes the oxyanion intermediate that is formed when lysine attacks the E2–Ub thiol ester.
Results
Conserved E2 asparagine residue is dispensable for E2 folding and E2–Ub thiol ester formation
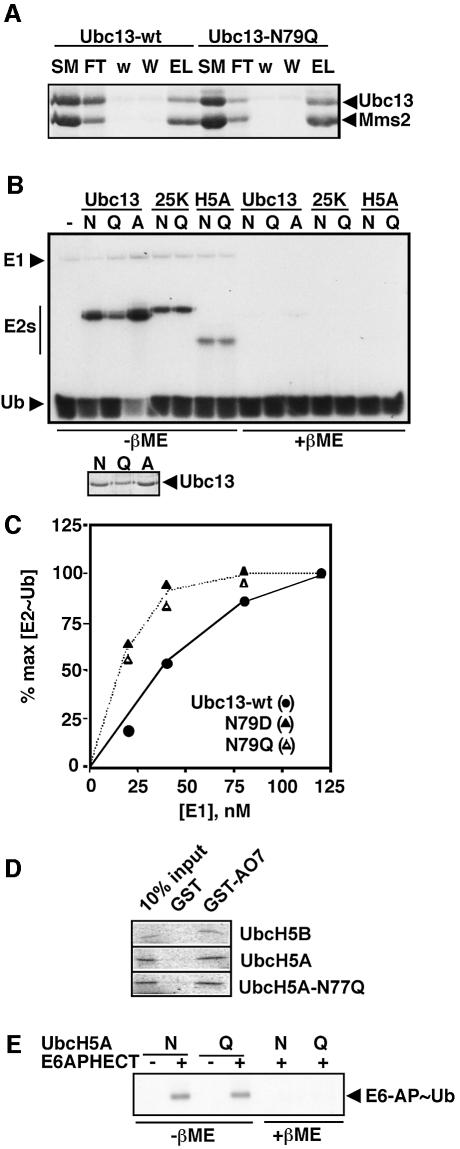
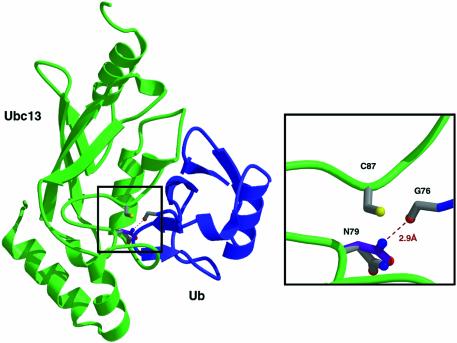
Structures are now available for many E2s, several E3 RING domains and two E3 HECT domains (reviewed in Van Demark and Hill, 2002; Verdecia et al., 2003). Each of these structures reveals a remarkably sterile catalytic landscape (Pickart, 2001; Van Demark and Hill, 2002). Conspicuously absent are groups that could deprotonate an attacking lysine (or cysteine) residue, or hydrogen-bond to the developing negative charge on the Ub carbonyl oxygen (Figure 1A). Nonetheless, we were intrigued by the absolute conservation of an asparagine residue positioned just upstream of the E2 active site cysteine (Figure 1B). Existing structures show that this side chain is fully hydrogen-bonded and pointed away from the E2 cysteine, as shown for Ubc13 in Figure 1C. This asparagine has been suggested to play a role in stabilizing E2 structure (Tong et al., 1997; Pickart, 2001; Bernier-Villamor et al., 2002), but its atomic contacts are not fully conserved. In many E2s, including Ubc13, the asparagine side chain hydrogen-bonds to the peptide backbone (Figure 1C). In other cases, the asparagine hydrogen-bonds to the side chain of a strictly conserved, buried histidine positioned two residues upstream (Worthylake et al., 1998; Hamilton et al., 2001). In addition, none of the residues in the HPN motif (Figure 1B) is conserved in ubiquitin E2 variant (UEV) proteins, which display an E2-like fold (Moraes et al., 2001; Van Demark et al., 2001). We decided to test whether the conserved asparagine residue plays a role in catalysis.
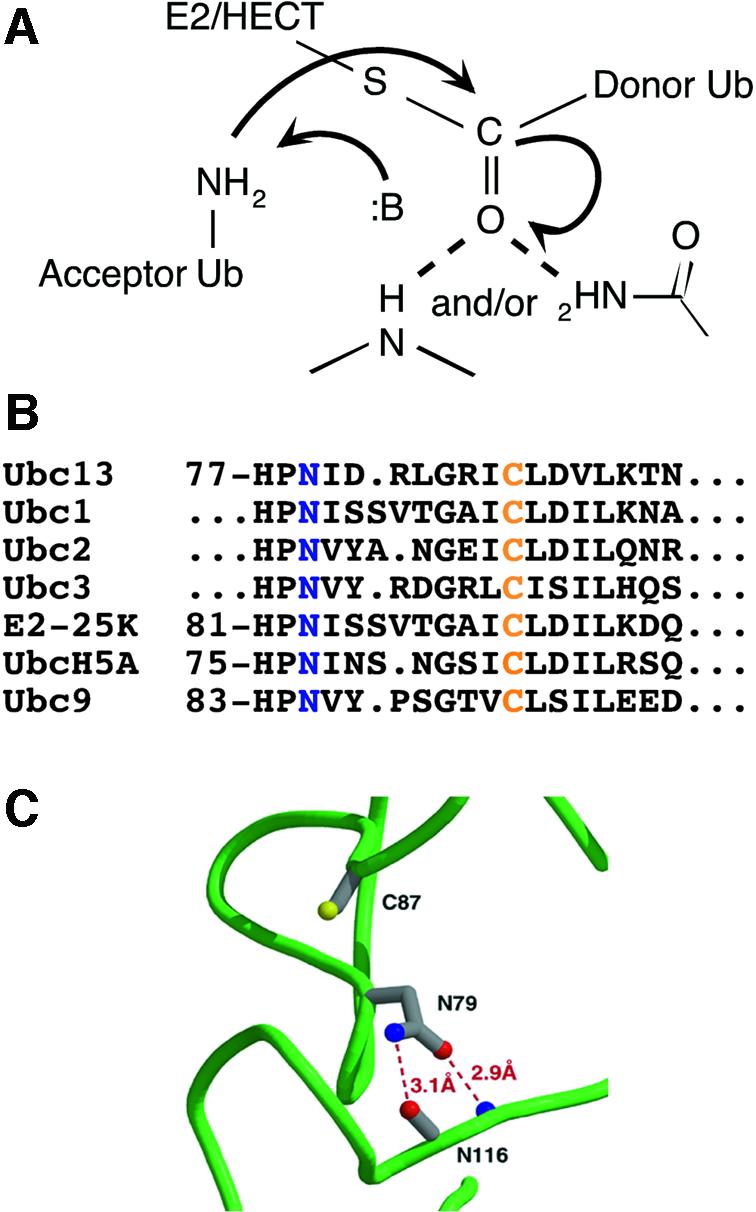
Fig. 1. Chemical and structural issues in the catalysis of Ub conjugation. (A) Model for catalysis of isopeptide bond formation in Ub chain synthesis (:B denotes a general base). (B) E2 active site sequences (blue, conserved asparagine; orange, active site cysteine). (C) Representative interactions of conserved asparagine in unliganded E2s, taken from the crystal structure of Ubc13 (Van Demark et al., 2001).
The E2 enzyme known as Ubc13 partners with a UEV protein (Mms2) and a RING E3 (Rad5) to modify the DNA polymerase cofactor PCNA with a K63-linked polyubiquitin (polyUb) chain, thus promoting DNA damage tolerance (Hofmann and Pickart, 1999; Ulrich and Jentsch, 2000; Hoege et al., 2002; Ulrich, 2002). The Mms2/Ubc13 heterodimer also catalyzes the synthesis of free K63-linked polyUb chains in a Rad5-independent manner in vitro (Hofmann and Pickart, 1999). In this reaction, Mms2 plays an E3-like role by binding a free Ub molecule (the acceptor) in a manner that positions K63, but not other lysines of Ub, to attack the E2–Ub thiol ester bound at a nearby site (Van Demark et al., 2001; McKenna et al., 2003). We used this well-studied reaction for our initial functional characterization of the conserved E2 asparagine residue.
Yeast Ubc13-N79Q, -N79D and -N79A were all highly expressed and soluble in Escherichia coli (data not shown). Several physical and functional criteria suggest that the mutant Ubc13s were properly folded. The circular dichroism (CD) spectrum of Ubc13-N79Q was virtually identical to that of the wild-type enzyme (Supplementary figure 1, available at The EMBO Journal Online). Preliminary 15N-HSQC NMR studies of Ubc13-N79Q also suggest that its fold is very similar to that of wild-type Ubc13 (D.Fushman, personal communication), and NMR studies by Klevit and colleagues suggest that an orthologous mutation in UbcH5C (N77S) causes little perturbation of folding (R.Klevit, personal communication). Each mutant E2 interacted with Mms2 in a manner that was indistinguishable from wild-type Ubc13, as shown for Ubc13-N79Q in Figure 2A. Moreover, none of the mutations affected the yield of Ubc13–Ub thiol ester produced in the presence of E1 and MgATP (Figure 2B). Quantitative kinetic studies of Ub transfer from E1 to Ubc13 showed that the transfer rate was unimpaired by the N79D and N79Q mutations and that the mutant proteins bound slightly better than wild-type Ubc13 to the E1–Ub thiol ester (Figure 2C). (It will be shown below that mutations at the orthologous position do not impair the interaction of a different E2 with two of its cognate E3s.) Thus, despite its absolute conservation in the E2 enzyme family, this asparagine appears to be dispensable for Ubc13 folding and for the catalytic events that produce the Ubc13–Ub thiol ester. The dispensability of the conserved asparagine for Ub transfer from E1 to E2 may reflect the presence of groups in the E1 active site that stabilize the oxyanion which forms as the E2 cysteine attacks the E1–Ub thiol ester (Pickart, 2001).
Fig. 2. E2 asparagine mutations do not impair E2 folding, E2–Ub formation or HECT E3–Ub formation. (A) Binding of wild-type Ubc13 (Ubc13-wt) and Ubc13-N79Q proteins to H10-Mms2 immobilized on Ni beads (Coomassie-stained gel). Successive lanes show proportional loading of the starting material (SM), unbound fraction (FT), washes with 0.1 M (w) and 0.4 M NaCl (W), and EDTA strip (EL). There is no binding of Ubc13 to the beads in the absence of Mms2 (data not shown). (B) Steady-state level of E2–[125I]Ub (autoradiograph). Assays were quenched as indicated (E2–Ub is labile to β-mercaptoethanol, βME). Small panel, Coomassie staining of Ubc13, showing that differences in E2–Ub levels reflect variations in [Ubc13]. (C) The level of Ubc13–Ub formed after 2 min of reaction (representing the initial rate) was determined at varying [E1]. (D) Binding of in vitro-translated E2s to immobilized GST–AO7 RING domain (autoradiograph). UbcH5B and GST are positive and negative controls, respectively. (E) E6-AP-HECT–Ub thiol ester formation (autoradioagraph). Assays with the indicated version of UbcH5A and [125I]Ub were quenched with or without β-mercaptoethanol as indicated.
Conserved asparagine is vital for E2-catalyzed Ub conjugation
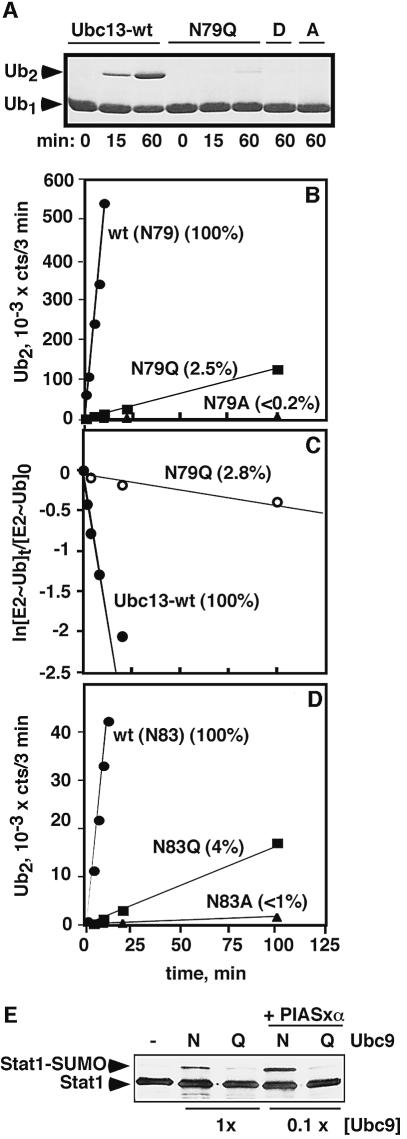
Mutation of N79 had a strikingly different effect in assays of isopeptide bond formation. The Mms2–Ubc13-N79Q complex was severely deficient in catalyzing the synthesis of K63-linked diubiquitin (Ub2), while the Ubc13-N79D and Ubc13-N79A complexes lacked detectable activity (Figure 3A). Quantitative assays revealed rate reductions of 40- and >500-fold, respectively, for the conservative (N79Q) and non-conservative (N79A) mutations (Figure 3B). The acceptor Ub apparently interacts with surfaces of the Mms2/Ubc13 complex that are distant from the N79 side chain (Van Demark et al., 2001; McKenna et al., 2003), so the mutations are unlikely to inhibit acceptor Ub binding. However, the low affinity of Ub for the wild-type Mms2/Ubc13 complex (Hofmann and Pickart, 2001), combined with the low activity of the mutant complex (Figure 3A and B), make it difficult to measure Ub binding directly. To confirm that the asparagine mutations affect the chemical step we replaced the acceptor Ub with free lysine, which is an inefficient but detectable E2 substrate (Pickart and Rose, 1985). Unlike polyUb chain synthesis, which requires the acceptor Ub binding site contributed by Mms2 (Hofmann and Pickart, 1999; Van Demark et al., 2001), Ubc13-catalyzed lysine ubiquitylation is Mms2-independent (data not shown), indicating that there are few binding interactions with this minimal substrate. The N79Q mutation in Ubc13 nevertheless reduced the rate of Ub transfer to free lysine by 36-fold (Figure 3C), essentially identical to the 40-fold inhibition of transfer to Ub-K63 catalyzed by Mms2/Ubc13 (Figure 3B). Thus, N79 plays a prominent role in the catalysis of isopeptide bond formation catalyzed by Ubc13 (also see Figure 4B).
Fig. 3. E2 asparagine mutations impair E2/UEV- and E2-catalyzed isopeptide bond synthesis. (A) Qualitative assay of K63-Ub2 synthesis catalyzed by Mms2/Ubc13 (Coomassie-stained gel). (B) Quantitative assay of K63-Ub2 synthesis by Mms2/Ubc13. [125I]Ub was employed; the Ub2 product was quantitated by band excision and gamma counting. Percent values refer to relative slopes of the lines. (C) The Ubc13–Ub thiol ester was pre-formed in a short pulse incubation with E1 and [125I]Ub. Lysine was then added to initiate the chase (see Materials and methods). The plot shows the decay of the respective E2–Ub thiol esters. (D) E2-25K-catalyzed synthesis of K48-Ub2 [assayed as in (B)]. (E) Stat1 sumoylation catalyzed by Ubc9 (–, no E1/E2; N, Ubc9-wt; Q, Ubc9-N85Q). A western blot was developed with Stat1 antibodies. In the lanes labeled 1×, the [Ubc9] was 0.2 µM. In the lanes labeled 0.1×, [Ubc9] was 20 nM. PIASxα (0.3 µM) was included in the 0.1× assays. Similar results were obtained in an assay with in vitro-translated Stat1 (data not shown).
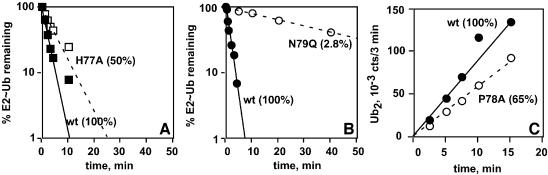
Fig. 4. Conserved E2 histidine and proline are dispensable for catalysis of isopeptide bond formation. (A and B) Pulse–chase assays of Ub2 synthesis were conducted with the indicated Ubc13 proteins. The assays were carried out as in Figure 3C, except that wild-type Ub (117 µM) served as the acceptor during the chase, producing K63-Ub2 as the product. (A) Comparison of Ubc13-wt and Ubc13-H77A; (B) (a separate experiment) compares Ubc13-wt and Ubc13-N79Q. (C) Steady-state K63-Ub2 synthesis. Assays comparing Ubc13-wt and Ubc13-P78A were carried out as in Figure 3B.
The assays of Ubc13 conjugation activity (Figure 3A–C) were carried out at 37°C, whereas CD spectra (Supplementary figure 1) were collected at 25°C. To exclude the possibility that the folding of the mutant Ubc13 proteins was selectively altered at the higher temperature, we assayed Ub2 synthesis at a reduced temperature (21°C). The mutant enzymes remained inactive at the lower temperature (Supplementary figure 2), arguing against a temperature-dependent conformational perturbation. Moreover, interactions with E1 (Figure 2) and HECT E3s (see below) were assayed at the higher temperature. Our finding that these interactions were unimpaired by the asparagine mutations (see below) provides further evidence against non-specific conformational perturbations.
To establish conservation of the asparagine residue’s function, we mutated the orthologous residue (N83) in E2-25K, an enzyme that synthesizes free polyUb chains linked through Ub-K48 (Chen et al., 1991). As with Ubc13, mutation to glutamine did not alter the rate or extent of Ub transfer from E1 to E2-25K (Figure 2B; data not shown). In contrast, the rate of synthesis of K48-linked Ub2 was reduced 25- and >100-fold, respectively, by the N83Q and N83A mutations (Figure 3D). These effects are quantitatively similar to the effects of analogous mutations in Ubc13 (Figure 3B), confirming that this residue plays a conserved and important role in E2-catalyzed isopeptide bond formation.
Conserved histidine and proline side chains are dispensable for E2-catalyzed Ub conjugation
Upstream histidine and proline residues, as well as the asparagine residue corresponding to N79 of Ubc13, are completely conserved in known E2s (Figure 1B). Although histidine is frequently employed as a general base in enzyme-catalyzed reactions, the conserved E2 histidine is largely buried, arguing against such a catalytic role (Pickart, 2001). The proline residue is surface exposed. To determine whether these strictly conserved residues could be functionally distinguished from N79, we individually mutated H77 and P78 to alanine in Ubc13. Both mutant enzymes were highly expressed, but largely insoluble (>90%; data not shown). However, we were able to purify small amounts of the soluble mutant proteins.
Because Ubc13-H77A displayed a modest defect in E2–Ub formation (∼5-fold rate reduction at subsaturating [E1]; data not shown), we probed the catalytic competence of the pre-formed mutant thiol ester in a pulse–chase assay. As shown in Figure 4A, the rate of Ub2 synthesis observed in a single turnover of the mutant thiol ester was only 2-fold slower than the rate seen for the wild-type intermediate, indicating that the reactive center for isopeptide bond formation is essentially unaffected by the H77A mutation. In contrast, the rate of Ub2 synthesis was below the limit of detectability when Ubc13-N79A was assayed in this manner (data not shown). The conservative N79Q mutation caused 36-fold inhibition (Figure 4B), in excellent agreement with the quantitative effect seen in two other assays (Figure 3B and C). Ubc13-P78A was assayed in steady-state Ub2 synthesis; here the reduction in rate was even less than 2-fold (Figure 4C). Thus, the effects of mutating N79 are not mimicked by altering nearby conserved residues. The marked and specific effects of the N79 mutations rather suggest that this side chain plays a role in catalysis.
Conserved asparagine is important for RING E3/E2-dependent Ub conjugation
Substrate ubiquitylation usually requires an E3 that brings its cognate E2 and substrate into close proximity, but it remains uncertain whether E3s play a role in chemical catalysis. The uncertainty is greatest for E3s of the RING family. These E3s use a globular zinc-binding domain to recruit the E2–Ub intermediate (Deshaies, 1999; Joazeiro and Weissman, 2000), but no E3 side chain comes closer than ∼15 Å to the E2 active site (Zheng et al., 2000, 2002; Brzovic et al., 2003). We therefore suspected that the E2 asparagine might be important for RING E3-dependent ubiquitylation.
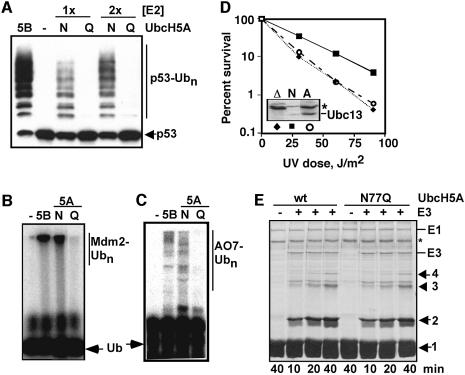
We tested this hypothesis using Mdm2, a RING E3 that partners with E2s of the UbcH5 subfamily to regulate the stability of the p53 tumor suppressor protein (Fang et al., 2000). As expected, UbcH5A-N77Q behaved identically to wild-type UbcH5A in assays of E2–Ub formation (Figure 2B). The mutant E2 displayed a severe defect, however, in assays of the Mdm2-dependent conjugation of Ub to p53 (Figure 5A). A similar effect was seen in assays of Mdm2 autoubiquitylation (Figure 5B), which regulates the stability of Mdm2 in vivo (Fang et al., 2000). Note that it was previously shown that >90% of the Ub conjugated in the autoubiquitylation assay (Figure 5B) is linked to Mdm2 itself (Fang et al., 2000); we confirmed this expectation by stripping the blot in Figure 5A and re-probing it with an Mdm2 antibody (data not shown). With Mdm2, as with many RING E3s, stable E2 binding is not detectable using standard approaches (Lorick et al., 1999). The RING domain protein AO7 is an exception; fragments of AO7 that include the RING and surrounding regions display easily observable binding to members of the UbcH5 subfamily of E2s (Lorick et al., 1999). We found that wild-type and mutant UbcH5A bound to a glutathione S-transferase (GST)-fused AO7 RING construct with indistinguishable efficiencies (Figure 2D). However, the mutant UbcH5A displayed a severe defect in AO7-catalyzed ubiquitylation (Figure 5C). The results obtained with AO7 indicate that the E2 mutation affects the functionality of the UbcH5A/AO7 complex, rather than its concentration. A similar situation is likely to apply with Mdm2 (Figure 5A and B). These results are consistent with the proposed catalytic role of the asparagine. The Mms2/Ubc13-N79A complex is likely to be similarly defective in polyubiquitylation dependent on the Rad5 RING E3, because expression of Ubc13-N79A failed to restore DNA damage tolerance to a ubc13Δ yeast strain, despite robust expression (Figure 5D). These results show that the conserved asparagine of Ubc13 is important for this E2’s in vivo function.
Fig. 5. Effects of E2 asparagine mutations on E3-dependent isopeptide bond synthesis. (A) Mdm2-dependent ubiquitylation of p53 was assayed with UbcH5B (5B, positive control), no E2 (–, negative control), or two different concentrations (1×, 2×) of wild-type UbcH5A (N) or UbcH5A-N77Q (Q). A western blot was developed with p53 antibodies. (B) Mdm2 autoubiquitylation was monitored with radiolabeled Ub [autoradiograph; annotation is the same as in (A)]. The blot shown in (A) was also stripped and re-probed with an Mdm2 antibody to confirm that autoubiquitylation was impaired by the N77Q mutation (data not shown). (C) AO7-catalyzed autoubiquitylation was assayed [autoradiograph, annotation is the same as in (A)]. (D) Ubc13-N79A cannot support DNA damage tolerance in vivo. A yeast mms2Δubc13Δ strain (Hofmann and Pickart, 1999) was transformed with empty vectors (diamonds), plasmids expressing Mms2 and HA-tagged wild-type Ubc13 (squares) or plasmids expressing Mms2 and HA-tagged Ubc13-N79A (circles). Inset, Ubc13-N79A is highly expressed (western blot developed with HA antibodies). Asterisk, cross-reacting band (loading control; the wild-type lane is underloaded). (E) KIAA10 (HECT) E3-dependent synthesis of K48-linked polyUb chains (Coomassie-stained gel). E3 was omitted from lanes 1 and 5 (negative controls). Numbers denote chains consisting of the indicated number of Ub units. Asterisk denotes bovine serum albumin (carrier protein).
Conserved asparagine is dispensable for E2–E3 Ub transfer
In HECT E3-dependent reactions, recruitment of the E2–Ub thiol ester is followed by Ub transfer to a conserved E3 cysteine residue (Huibregtse et al., 1995; Scheffner et al., 1995; Huang et al., 1999). Here the HECT E3–Ub intermediate is the proximal Ub donor, so groups needed to catalyze isopeptide bond formation should reside in the E3 (versus E2) active site. However, the E2 asparagine still might facilitate attack of the HECT cysteine on the E2–Ub intermediate (Kursula et al., 2002). In contrast to prediction from this model, UbcH5A-N77Q behaved identically to wild-type UbcH5A in assays of thiol ester formation with a representative family member, E6-AP (Figure 2E). As expected from this result, polyUb chain synthesis catalyzed by the HECT-containing C-domain of KIAA10 (You and Pickart, 2001) was unaffected by the N77Q mutation in UbcH5A (Figure 5E). [In this assay E3–Ub thiol ester formation is the rate-limiting step in chain synthesis (data not shown.)] Similar results were obtained in assays of chain assembly catalyzed by the E6-AP HECT domain and in studies of Nedd4-dependent autoubiquitylation (data not shown). These results show that the conserved E2 asparagine is dispensable for Ub transfer from UbcH5A to HECT E3s. They also provide further confirmation that the mutant UbcH5A is correctly folded.
Conserved asparagine is important for conjugation of a Ub-like protein
Ub defines a family of proteins, known as Ub-like proteins (UbLs), that share a similar fold and a similar mechanism of signaling through covalent conjugation (Hochstrasser, 2000; Jentsch and Pyrowolakis, 2000). The ligation of each UbL to its specific substrates relies on a dedicated E2 enzyme with an asparagine residue positioned identically to the conserved asparagine of the Ub E2s (Figure 1B). Ubc9, the E2 enzyme for SUMO, functions in cell cycle progression and nucleocytoplasmic transport (Hochstrasser, 2000; Jentsch and Pyrowolakis, 2000; Melchior, 2000). Consistent with the properties of Ub E2s reported above, human Ubc9-N85Q catalyzes the sumoylation of Stat1, a mammalian SUMO substrate (Rogers et al., 2003), at a markedly reduced rate (Figure 3E). The Ubc9-dependent sumoylation of Stat1 is stimulated by PIASxα, one of a family of RING-like SUMO E3s (Johnson and Gupta, 2001; Kahyo et al., 2001; Rogers et al., 2003). Inclusion of PIASxα in Stat1 sumoylation assays did not overcome the defect conferred by the N85Q mutation in Ubc9 (Figure 3E, last two lanes), similar to findings with Ubc13 (Figure 5D). As expected, Ubc9-N85Q was fully competent in SUMO thiol ester formation (data not shown). These findings suggest that the conserved asparagine plays an important role in Ubc9 catalysis. Consistent with this model, human Ubc9-N85D cannot fulfill the essential function of Ubc9 in Saccharomyces cerevisiae and the N85A mutation causes a severe growth defect (C.Lima, personal communication).
Proposed role of conserved asparagine residue: oxyanion stabilization
Our findings (Figures 2 and 5E) argue strongly against a generalized structural role for the conserved E2 asparagine. The selective requirement for this side chain in isopeptide bond formation (Figures 3 and 5A–D) instead suggests a role in the catalysis of this reaction. Many cysteine proteases, including those that remove Ub/UbLs from substrates, use a glutamine or asparagine side chain to stabilize the oxyanion intermediate formed during the attack of cysteine or water on an amide or thiol ester bond (Figure 1A) (Rawlings and Barrett, 1994; Johnston et al., 1997, 1999; Mossessova and Lima, 2000; Hu et al., 2002). This is an attractive role for the conserved E2 asparagine, but it would necessitate a change in side chain orientation relative to the conformation seen in existing E2 structures.
Substrate-induced changes in the orientations of catalytic groups are well documented. Specific precedent, including an altered conformation of an oxyanion-stabilizing asparagine, is provided by the Ub deconjugating enzyme known as Hausp (Hu et al., 2002). In the case of the Ubc1 E2, NMR studies have shown that the conserved asparagine experiences a marked change in environment during E2–Ub formation (Hamilton et al., 2001), which could be explained by a change in side chain orientation. We used existing models of the Ubc1–Ub thiol ester (Hamilton et al., 2001) and the Mms2/Ubc13 complex (Van Demark et al., 2001) to model N79 of Ubc13 in a conformation appropriate for oxyanion stabilization. We were able to model the asparagine side chain so that its nitrogen was positioned within hydrogen-bonding distance (2.9 Å) of the Ub-G76 carbonyl (Figure 6). We propose that the conserved asparagine residue directly stabilizes the oxyanion intermediate formed during lysine attack.
Fig. 6. Model for oxyanion stabilization by conserved E2 asparagine residue. Ubc1 in the model of the Ubc1–Ub thiol ester (Hamilton et al., 2001) was replaced by Ubc13 and the N79 side chain was modeled in a conformation suitable for oxyanion stabilization.
Discussion
A strictly conserved E2 asparagine residue (Figure 1B and C) was previously proposed to stabilize E2 folding (reviewed in Tong et al., 1997; Pickart, 2001; Bernier-Villamor et al., 2002; Van Demark and Hill, 2002). However, we found that even non-conservative changes at this site did not reduce the steady-state level of E2–Ub thiol ester (Figure 2B), impair the kinetics of E2–Ub formation (Figure 2C), reduce binding to a RING domain (Figure 2D) or alter the efficiency of Ub transfer to the active site cysteine of a HECT E3 (Figures 2E and 5E). Competence of the mutant E2s in upstream and downstream (E1 and E3) interactions argues against an important role for the asparagine in E2 folding.
We propose that the side chain of the conserved E2 asparagine stabilizes the oxyanion formed during the attack of lysine on the E2–Ub intermediate. Multiple features of our data support this model. First, E2-catalyzed Ub conjugation reactions are impaired moderately (25- to 40-fold inhibition) or severely (100- to 500-fold inhibition) by conservative or non-conservative mutations, respectively, of the asparagine (Figure 3A–D). In addition, mutation of the orthologous asparagine in Ubc9 inhibits the conjugation of SUMO to Stat1 (Figure 3E). E2- and RING E3-dependent ubiquitylation reactions share a reaction center in which the substrate lysine directly attacks the E2–Ub thiol ester. Thus, our finding that mutating the conserved asparagine in UbcH5A severely inhibits ubiquitylation catalyzed by two RING E3s (Figure 5A–C), without impeding E2/RING interactions (Figure 2D), is also in agreement with prediction from our model. Conjugation catalyzed by another RING E3, the BRCA1/BARD1 heterodimer (Brzovic et al., 2001), is severely inhibited by the N77S mutation in UbcH5C (R.Klevit, personal communication). The finding that non-conservative mutations of this residue in Ubc13 and Ubc9 confer a loss-of-function phenotype in yeast is further consistent with the proposal of an important catalytic role (Figure 5D; C.Lima, personal communication). The rate reduction we observe for non-conservative mutations (Figure 3) is of the magnitude expected for loss of oxyanion stabilization, which is thought to accelerate serine/thiol protease-catalyzed reactions by factors of 100- to 300-fold (Bryan et al., 1986; Carter and Wells, 1988). Finally, molecular modeling presented here suggests that the E2 asparagine side chain can adopt a conformation that is suitable for oxyanion stabilization (Figure 6).
Although our data suggest that the asparagine mutant proteins are properly folded, we cannot definitively exclude the possibility that the mutations lead to a subtle structural perturbation. In this case, the high specificity of the observed effects could be explained if the structural alteration repositions an unidentified catalytic element, for example a main chain nitrogen that hydrogen-bonds to the oxyanion. Alternatively, there could be changes in the orientation of the covalently bound Ub or in the positioning of the acceptor lysine residue of the substrate. However, the competence of the asparagine mutants in HECT E3 reactions argues against the first possibility, while the similar inhibition seen with a variety of substrates is difficult to reconcile with the second one. Although the proposed model (Figure 6) is the simplest one that can accommodate all of the data, a definitive resolution of the role of the conserved asparagine awaits atomic-resolution structural data for specific reaction intermediates.
The proposed repositioning of the asparagine (Figure 6) may be coupled to active site changes that occur upon Ub thiol ester formation, and could be further modulated by substrate or E3 binding. Recently, Brzovic et al. (2003) reported that the specific association of an E2 and E3 can occur in the absence of catalytic competence. Thus, the conformations of key residues not directly associated with binding, such as the E2 asparagine discovered in the present work, may be important for the formation of catalytically active complexes. The crystal structure of a Hausp–Ub thiohemiacetal adduct (a mimic of the Hausp–Ub thiol ester) shows how the covalent binding of Ub can drive the formation of a functional oxyanion hole in an enzyme that catalyzes the reverse reaction to that studied here (Hu et al., 2002).
The E3–Ub thiol ester that is strictly required in certain E3-dependent reactions is formed by attack of the HECT cysteine on the E2–Ub thiol ester. In contrast to the strong inhibition seen when isopeptide bond formation involves direct attack of lysine on the E2–Ub intermediate (Figures 3 and 5A–D), there was no inhibition when the same E2 (UbcH5A) participates in HECT E3-dependent conjugation (Figure 5E). This result dramatically demonstrates the absence of the E2–Ub intermediate in the reaction center for isopeptide bond formation by HECT E3s, but it also rules out a role for the E2 asparagine in catalyzing E3–Ub formation from E2–Ub. Nonetheless, efficient attack of the E3 cysteine is expected to depend on oxyanion stabilization (Kursula et al., 2002).
Our results can be reconciled with this requirement if a different oxyanion site facilitates attack by the E3 thiol on the E2–Ub intermediate. Although serine/thiol proteases employ a single site to catalyze the attack of distinct nucleophiles, one of the attacking groups is water, which has minimal interactions with the enzyme. The situation is very different in the transfer of Ub from E2–Ub to a substrate lysine versus a HECT cysteine. Here each acceptor is part of a protein that probably contacts the E2 and/or E3 at distinct sites (Huang et al., 1999; Van Demark et al., 2001; Bernier-Villamor et al., 2002; Verdecia et al., 2003). However, if the HECT cysteine and the substrate lysine approach Ub-G76 by different routes, then groups stabilizing the oxyanion formed during one reaction may not perform this task efficiently in the other reaction. Thiolase, which catalyzes sequential acyl transfers to two different thiol acceptors, utilizes a distinct oxyanion site for each reaction (Kursula et al., 2002). Since the E2 asparagine seems to stabilize the oxyanion formed during lysine attack, we suggest that an oxyanion hole for thiol (E3) attack could reside either in the HECT active site or elsewhere in the E2 active site. We note that a main chain-based oxyanion site in the E2 would have escaped detection in studies to date. The proposed asparagine-based oxyanion hole for lysine attack could also include a second hydrogen bond from an unidentified main chain nitrogen (Rawlings and Barrett, 1994; Hu et al., 2002). It remains unclear whether E2s and/or E3s use a general base to deprotonate the attacking lysine.
The E2 asparagine is the first candidate for a non-nucleophilic catalytic element in any Ub conjugation factor. The location of this element in the E2 explains how RING E3s are able to facilitate specific substrate ubiquitylation even though they appear to lack a bona fide active site: according to our model, the E2 enzyme contributes not only thermodynamically activated Ub, but also a catalytic group to facilitate its appropriate discharge. The existence of such a mechanism may facilitate the design of inhibitors that target the chemistry of the conjugation reaction.
Materials and methods
Plasmids and proteins
Existing expression plasmids were mutated by standard PCR methods and verified by sequencing. Yeast H10-Mms2 was purified on Ni beads (Hofmann and Pickart, 1999); yeast Ubc13 (Raasi and Pickart, 2003), human UbcH5A (You and Pickart, 2001), the E6-AP HECT domain (Lorick et al., 1999) and the C-domain of the human KIAA10 HECT E3 (You and Pickart, 2001) were all produced as GST fusion proteins, adsorbed onto GSH beads, and released by thrombin cleavage. Thrombin was removed by absorbing the protein of interest onto anion exchange resin (Ubc13, KIAA10-CD, E6-AP) or by absorbing thrombin onto cation exchange resin (UbcH5A). Human E2-25K was purified in an untagged form (Haldeman et al., 1997). All E2-25K proteins carried the benign C170S mutation (Haldeman et al., 1997). Mdm2, Nedd4 and the AO7-RING construct were expressed in a GST-fused form, purified on GSH beads, and assayed as intact fusion proteins (Lorick et al., 1999; Fang et al., 2000). H6-E1 was expressed in insect cells and purified on Ni beads (Raasi and Pickart, 2003). Sumo-1, its activating enzyme (Aos1/Uba2) and Ubc9 were expressed and purified by established procedures (Zhang et al., 2002). Wild-type and mutant Ubs were expressed in E.coli and purified as described previously (Haldeman et al., 1997). For some experiments, wild-type Ub was labeled with Na125I to a specific radioactivity of ∼8000 c.p.m./pmol (Haldeman et al., 1997).
Enzyme assays
Assays employed 0.1 µM E1 and were conducted at 37°C unless otherwise stated. Assays of E2–Ub formation used 4 µM [125I]Ub and 1–2 µM E2 (Haldeman et al., 1997). The assay of HECT–Ub formation (Figure 2E) employed 0.2 µM E2 and 0.4 µM E6-AP-HECT. Assays of Ub2 synthesis catalyzed by the Mms2/Ubc13 complex (Hofmann and Pickart, 2001) or E2-25K (Haldeman et al., 1997) employed Ub-D77 (∼500 µM) and [125I]Ub (3–10 µM) as substrates with 0.3–3 µM E2 (except in Figure 3A and Supplementary figure 2, where Ub-D77 and unlabeled Ub-K63R were ∼200 µM each). Where added, Mms2 was stoichiometric to Ubc13. Higher concentrations of mutant E2s were used to accurately measure slow rates; normalized data are presented in Figure 3B and D. Single-turnover assays of [125I]Ub transfer to lysine by Ubc13 used an established pulse–chase method (Pickart and Rose, 1985) with N-acetyl lysine methyl ester (125 mM) added to initiate the chase. This reaction was abolished by adding 8 M urea together with lysine, indicating that it is catalyzed (data not shown). Ubiquitylation of 125 mM lysine by wild-type Ubc13-Ub was ∼30-fold faster than thiol ester hydrolysis (native conditions, data not shown). Pulse–chase assays with Ub (117 µM) as acceptor were carried out as described previously (Hofmann and Pickart, 2001). The synthesis of K48-linked polyUb chains catalyzed by KIAA10 C-domain/UbcH5A was assayed using Ub-K29R (Raasi and Pickart, 2003). All assays of chain assembly were performed at subsaturating acceptor Ub concentration.
For assays of Ub conjugation to p53, extract from doxycline-treated p53-inducible Saos-2 cells (Ryan et al., 2000) was incubated with 2 pmol GST–Mdm2 that had been pre-coupled to GSH beads, followed by washing to remove lysate proteins and unbound p53. The bead-bound Mdm2/p53 complex was incubated for 30 min (30°C, 30 µl, pH 8) with ∼0.1 µM E2 and 40 µM Ub. Samples were analyzed by western blotting (8% gels), using DO-1 antibody to detect p53. An independent assay of Mdm2 autoubiquitylation (without added p53; Fang et al., 2000) employed [32P]Ub (Scheffner et al., 1994). Stat1 sumoylation was assayed under established conditions and detected by western blotting of Stat1 (Rogers et al., 2003). The assay of AO7-dependent ubiquitylation, which has been described previously (Lorick et al., 1999), employed 0.4 µM E2.
E2 binding assays
The binding of UbcH5 E2s to the GST–AO7 RING construct was assayed as described previously (Lorick et al., 1999). pGEM-UbcH5B, pET3a-UbcH5A (Scheffner et al., 1994) and pET3a-UbcH5A-N77Q were transcribed and translated using T7 polymerase in wheat germ lysate (Promega). Active GST–AO7 RING (or GST as a negative control), 20 pmol in crude bacterial lysate, was bound to 50 µl glutathione beads, washed and incubated with in vitro-translated E2 (105 c.p.m.) in 200 µl binding buffer (Lorick et al., 1999). After washing, bound E2 was eluted with SDS sample buffer, resolved by SDS–PAGE, and quantified using a PhosphorImager. The binding of Ubc13 to H10-Mms2 immobilized on nickel beads was monitored as described previously (Van Demark et al., 2001).
UV sensitivity
The N79A mutation was introduced into a centromeric plasmid carrying the S.cerevisiae UBC13 promoter region and gene (including the intron) fused to a C-terminal HA tag (Hofmann and Pickart, 1999). Plasmid specifying Ubc13-N79A or the wild-type protein was co-transformed with a plasmid specifying H10-Mms2 into a ubc13Δmms2Δ strain of S.cerevisiae and UV survival was measured (Hofmann and Pickart, 1999).
Molecular modeling
A model of the Ubc13-Ub thiol ester was produced based on the model of the Ubc1-Ub thiol ester (1FXT) (Hamilton et al., 2001) and the Mms2–Ubc13 structure (1JAT) (Van Demark et al., 2001). The N79 side chain in Ubc13 was rotated toward C87. The C-terminus of Ub was manually docked into the Ubc13 active site, putting the carbonyl oxygen of Ub-G76 within hydrogen bonding distance of Nδ of Asn79. Energy minimization was then done using GROMOS from the Swiss-PDB viewer (Guex and Peitsch, 1997) and PROCHECK (Laskowski et al., 1993) was used to verify the geometry of the final model.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to C.Lima, R.Klevit and D.Fushman for permission to cite unpublished data. We thank A.Davis for performing CD spectroscopy, A.Hu for assistance in studies of Ubc13-P78A, M.Wang and S.Zhu for assistance in preliminary studies, M.Ajua-Alemanji for technical assistance, R.Cohen for a critical reading of the manuscript, and H.Gilbert and M.Wogulis for helpful discussions. This study was funded by grants to C.M.P. from the NIH (GM60372) and to M.J.M. from the NIH (GM60980) and the American Cancer Society. C.P.W. is an Investigator of the Howard Hughes Medical Institute.
References
- Bernier-Villamor V., Sampson,D.A., Matunis,M.J. and Lima,C.D. (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell, 108, 345–356. [DOI] [PubMed] [Google Scholar]
- Bryan P., Pantolizno,M.W., Quill,S.G., Hsiao,H.-Y. and Poulos,T. (1986) Site-directed mutagenesis and the role of the oxyanion hole in subtilisin. Proc. Natl Acad. Sci. USA, 83, 3743–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic P.S., Rajagopal,P., Hoyt,D.W., King,M.-C. and Klevit,R.E. (2001) Structure of a BRCA1–BARD1 heterodimeric RING–RING complex. Nat. Struct. Biol., 8, 833–837. [DOI] [PubMed] [Google Scholar]
- Brzovic P.S., Keeffe,J.R., Nishikawa,H., Mayamoto,K., Fox,D., Fukuda,M., Ohta,T. and Klevit,R. (2003) Binding and recognition in the assembly of an active BRCA1–BARD1 ubiquitin ligase complex. Proc. Natl Acad. Sci. USA, 100, 5646–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P. and Wells,J.A. (1988) Dissecting the catalytic triad of a serine protease. Nature, 332, 564–568. [DOI] [PubMed] [Google Scholar]
- Chen Z., Niles,E.G. and Pickart,C.M. (1991) Isolation of a cDNA encoding a mammalian multi-ubiquitinating enzyme (E2-25K) and overexpression of the functional enzyme in E. coli. J. Biol. Chem., 266, 15698–15704. [PubMed] [Google Scholar]
- Deshaies R.J. (1999) SCF and cullin/RING H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol., 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Fang S., Jensen,J.P., Ludwig,R.L., Vousden,K.H. and Weissman,A.M. (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem., 275, 8945–8951. [DOI] [PubMed] [Google Scholar]
- Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Haldeman M.T., Xia,G., Kasperek,E.M. and Pickart,C.M. (1997) Structure and function of ubiquitin conjugating enzyme E2-25K: the tail is a core-dependent activity element. Biochemistry, 36, 10526–10537. [DOI] [PubMed] [Google Scholar]
- Hamilton K.S., Ellison,M.J., Barber,K.R., Williams,R.S., Huzil,J.T., Mckenna,S., Ptak,C., Glover,M. and Shaw,G.S. (2001) Structure of a conjugating enzyme–ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure, 9, 897–904. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (2000) Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol., 2, E153–E157. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander,B., Moldovan,G.-L., Pyrowolakis,G. and Jentsch,S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature, 419, 135–141. [DOI] [PubMed] [Google Scholar]
- Hofmann R.M. and Pickart,C.M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell, 96, 645–653. [DOI] [PubMed] [Google Scholar]
- Hofmann R.M. and Pickart,C.M. (2001) In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem., 276, 27936–27943. [DOI] [PubMed] [Google Scholar]
- Hu M., Li,P., Li,M., Li,W., Yao,T., Wu,J.-W., Gu,W., Cohen,R.E. and Shi,Y. (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell, 111, 1041–1054. [DOI] [PubMed] [Google Scholar]
- Huang L., Kinnucan,E., Wang,G., Beaudenon,S., Howley,P.M., Huibregtse,J.M. and Pavletich,N.P. (1999) Structure of an E6AP–UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science, 286, 1321–1326. [DOI] [PubMed] [Google Scholar]
- Huibregtse J.M., Scheffner,M., Beaudenon,S. and Howley,P.M. (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin–protein ligase. Proc. Natl Acad. Sci. USA, 92, 2563–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S. and Pyrowolakis,G. (2000) Ubiquitin and its kin: how close are the family ties? Trends Cell Biol., 10, 335–342. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A.P. and Weissman,A.M. (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell, 102, 549–552. [DOI] [PubMed] [Google Scholar]
- Johnson E.S. and Gupta,A.A. (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell, 106, 735–744. [DOI] [PubMed] [Google Scholar]
- Johnston S.C., Larsen,C.N., Cook,W.J., Wilkinson,K.D. and Hill,C.P. (1997) Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 Å resolution. EMBO J., 16, 3787–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.C., Riddle,S.M., Cohen,R.E. and Hill,C.P. (1999) Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J., 18, 3877–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahyo T., Nishida,T. and Yasuda,H. (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell, 8, 713–718. [DOI] [PubMed] [Google Scholar]
- Kursula P., Ojala,J., Lambeir,A.-M. and Wierenga,R.K. (2002) The catalytic cycle of biosynthetic thiolase: a conformational journey of an acetyl group through four binding modes and two oxyanion holes. Biochemistry, 41, 15543–15556. [DOI] [PubMed] [Google Scholar]
- Laney J.D. and Hochstrasser,M. (1999) Substrate targeting in the ubiquitin system. Cell, 97, 427–430. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the sterochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lorick K.L., Jensen,J.P., Fang,S., Ong,A.M., Hatakeyama,S. and Weissman,A.M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA, 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna S., Moraes,T., Pastushok,L., Ptak,C., Xiao,W., Spyracopoulos,L. and Ellison,M.J. (2003) An NMR-based model of the ubiquitin-bound human ubiquitin conjugation complex Mms2–Ubc13. The structural basis for lysine 63 chain synthesis. J. Biol. Chem., 278, 13151–13158. [DOI] [PubMed] [Google Scholar]
- Melchior F. (2000) SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol., 16, 591–626. [DOI] [PubMed] [Google Scholar]
- Moraes T.F., Edwards,R.A., McKenna,S., Pastushok,L., Xiao,W., Glover,J.N.M. and Ellison,M.J. (2001) Crystal structure of the human ubiquitin conjugating complex, hMms2–hUbc13. Nat. Struct. Biol., 8, 669–673. [DOI] [PubMed] [Google Scholar]
- Mossessova E. and Lima,C.D. (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell, 5, 865–876. [DOI] [PubMed] [Google Scholar]
- Pickart C. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem., 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. and Rose,I.A. (1985) Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem., 260, 1573–1581. [PubMed] [Google Scholar]
- Raasi S. and Pickart,C.M. (2003) Rad23 ubiquitin-associated domains (UBA) inhibit 26S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem., 278, 8951–8959. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D. and Barrett,A.J. (1994) Families of cysteine peptidases. Methods Enzymol., 244, 461–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers R.S., Horvath,C.M. and Matunis,M.J. (2003) SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J. Biol. Chem., 278, 30091–30097. [DOI] [PubMed] [Google Scholar]
- Ryan K.M., Ernst,J.K., Rice,N.R. and Vousden,K.H. (2000) Role of NF-κB in p53-mediated programmed cell death. Nature, 404, 892–897. [DOI] [PubMed] [Google Scholar]
- Scheffner M., Huibregtse,J.M. and Howley,P.M. (1994) Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc. Natl Acad. Sci. USA, 91, 8797–8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M., Nuber,U. and Huibregtse,J.M. (1995) Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature, 373, 81–83. [DOI] [PubMed] [Google Scholar]
- Tong H., Hateboer,G., Perrakis,A., Bernards,R. and Sixma,T.K. (1997) Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system. J. Biol. Chem., 272, 21381–21387. [DOI] [PubMed] [Google Scholar]
- Ulrich H.D. (2002) Degradation or maintenance: actions of the ubiquitin system on eukaryotic chromatin. Eukaryot. Cell, 1, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H.D. and Jentsch,S. (2000) Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J., 19, 3388–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Demark A.P. and Hill,C.P. (2002) Structural basis of ubiquitylation. Curr. Opin. Struct. Biol., 12, 822–830. [DOI] [PubMed] [Google Scholar]
- Van Demark A.P., Hofmann,R.M., Tsui,C., Pickart,C.M. and Wolberger,C. (2001) Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell, 105, 711–720. [DOI] [PubMed] [Google Scholar]
- Verdecia M.A., Joazeiro,C.A.P., Wells,N.J., Ferrer,J.-L., Bowman,M.E., Hunter,T. and Noel,J.P. (2003) Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell, 11, 249–259. [DOI] [PubMed] [Google Scholar]
- Worthylake D.K., Prakash,S., Prakash,L. and Hill,C.P. (1998) Crystal structure of the Saccharomyces cerevisiae ubiquitin-conjugating enzyme Rad6 at 2.6 Å resolution. J. Biol. Chem., 273, 6271–6276. [DOI] [PubMed] [Google Scholar]
- You J. and Pickart,C.M. (2001) A hect domain E3 enzyme assembles novel polyubiquitin chains. J. Biol. Chem., 276, 19871–19878. [DOI] [PubMed] [Google Scholar]
- Zhang H., Saitoh,H. and Matunis,M.J. (2002) Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol., 22, 6498–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Wang,P., Jeffrey,P.D. and Pavletich,N.P. (2000) Structure of a c-Cbl–UbcH7 complex: RING domain function in ubiquitin–protein ligases. Cell, 102, 533–539. [DOI] [PubMed] [Google Scholar]
- Zheng N. et al. (2002) Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature, 416, 703–709. [DOI] [PubMed] [Google Scholar]