Abstract
We describe the temporal order of recruitment of transcription factors, cofactors and basal transcriptional components and the consequent biochemical events that lead to activation of the major histocompatibility class II (MHCII) DRA gene transcription by IFN-γ. We found that the gene is ‘poised’ for activation since both the activators and a fraction of the basal transcriptional machinery are pre-assembled at the enhancer and promoter prior to IFN-γ treatment. The class II transactivator is synthesized following IFN-γ treatment and it is recruited to the enhanceosome leading to the subsequent recruitment of the CBP and GCN5 coactivators. This is followed by histone acetylation and recruitment of the SWI/SNF chromatin remodeling complex. CIITA also recruits the CDK7 and CDK9 kinases and enhances the ability of CDK7 to phosphorylate Pol II at Ser5 leading to initiation of mRNA synthesis. Thus, the gene-specific class II transactivator selects the target genes for expression by coordinating a multiple set of biochemical activities ranging from chromatin alterations and pre-initiation complex assembly to promoter clearance.
Keywords: chromatin immunoprecipitation/CIITA/enhanceosome/RNA Pol II phosphorylation/transcriptional regulation
Introduction
Gene transcription is regulated at multiple steps including binding of transcription factors to the gene’s regulatory elements, recruitment and assembly of the basal transcriptional apparatus at the core promoter, and elongation of mRNA synthesis (Lee and Young, 2000; Lemon and Tjan, 2000). In some genes, localized chromatin alterations such as histone modifications and nucleosome remodeling (Narlikar et al., 2002) are required in order to allow these processes to take place in the context of an otherwise non-permissive chromatin environment. In signal-dependent transcription, the cascade of events that follow receptor triggering results in the formation of activatory circuits in the nucleus, which ensure a highly cell-specific transcriptional response. A classical example of signal-dependent transcriptional mechanisms is the IFN-γ-induced activation of the major histocompatibility complex class II (MHCII) genes.
The MHCII gene products are surface glycoproteins that determine antigen presentation to the T-cell receptor (TCR) of CD4+ T lymphocytes and play a critical role in the development of the thymus TCR repertoire and the peripheral immune response (Cresswell, 1994; Viret and Janeway, 1999). MHCII gene expression is regulated by developmental cues during differentiation of B-lymphocytes and by cytokines, mainly by IFN-γ, in several cell types (Reith and Mach, 2001; Ting and Trowsdale, 2002). Regulated MHCII gene transcription depends on the combined presence of promoter proximal conserved DNA regulatory elements known as S (also known as W, Z or H in the mouse), X, X2 and Y boxes that combinatorially form the class II enhancer (Glimcher and Kara, 1992). Studies on the bare lymphocyte syndrome (BLS), a hereditary immunodeficiency disease due to deficient MHCII gene expression, have led to the identification of the regulatory factors that are essential for MHCII expression. These genes encode the X box binding RFX complex and the MHCII transactivator CIITA (Waldburger et al., 2000). Other ubiquitously expressed proteins such as CREB and the NFY complex also bind to the well-conserved motifs S, X, Y of the MHCII enhancer (Reith and Mach, 2001). RFX, CREB and NFY interact and bind cooperatively to the enhancer DNA to assemble the MHCII enhanceosome (Masternak et al., 2000; Reith and Mach, 2001). Although essential, these transcription factors cannot support class II transcription in the absence of an additional factor, the MHCII-specific transcriptional co-activator CIITA. CIITA is a non-DNA binding factor, which is expressed under the same conditions as MHCII genes and which is constitutively present in professional antigen presenting cells (B cells, dendritic cells) or it is cytokine-inducible in most other cells (Harton and Ting, 2000; Waldburger et al., 2000; Reith and Mach, 2001). CIITA is recruited to its target genes (Masternak et al., 2000) via multiple protein–protein interactions with several components of the enhanceosome (DeSandro et al., 2000; Hake et al., 2000;Zhu et al., 2000). In addition, CIITA can interact with several components of the basal transcription machinery such as TBP, TAFII32, TAFII70, TFIIH and P-TEFb (Fontes et al., 1997b; Mahanta et al., 1997; Kanazawa et al., 2000) and thus it could affect pre-initiation complex assembly and/or function. Furthermore CIITA also binds to members of the histone acetyltransferase-type (HAT) general coactivators CBP/p300 (Kretsovali et al., 1998; Fontes et al., 1999) and pCAF (Spilianakis et al., 2000).
The in vivo relevance of the above protein–protein interactions and the way their functions are integrated and coordinated during induction by IFN-γ is not known. To this end, we have examined here the functional interplay between the enhanceosome and CIITA that leads to the assembly of cofactors required for IFN-γ-induced transcription of the human MHCII gene DRA. We show that seven proteins that constitute a minimal class II enhanceosome are necessary and sufficient for recruiting to the promoter not only CIITA, but also a subset of the general transcription factors and RNA Pol II. CIITA in turn functions by stabilizing the enhanceosome–cofactor assembly, inducing promoter chromatin acetylation and finally by facilitating Ser5 phosphorylation and promoter clearance of RNA Pol II, which determines the exact timing of IFN-γ-induced transcription.
Results
In vitro assembly and characterization of the MHC class II enhanceosome
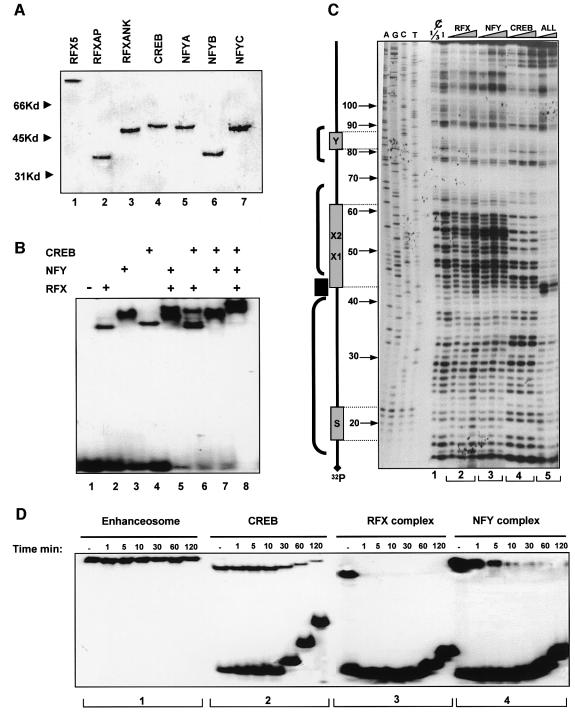
Previous studies performed using crude nuclear extracts as a source of transcription factors have suggested that CIITA is recruited to the class II enhanceosome by interacting with multiple enhanceosome components such as RFX, NFY and CREB (Masternak et al., 2000). To investigate whether the class II enhanceosome can recruit directly CIITA to the promoter in the absence of any other factor, we reconstituted the enhanceosome in vitro using recombinant factors and tested its ability to recruit CIITA. The three RFX subunits (RFX5, RFXAP and RFXANK), together with the three NFY subunits (NFYA, NFYB and NFYC) and CREB were expressed and purified to near homogeneity from Escherichia coli (Figure 1A). Equimolar amounts of each subunit of the RFX and NFY complex were co-renatured and the assembly and identity of the trimeric complexes was monitored by electrophoretic mobility shift assays (EMSAs) coupled with antibody supershift experiments (data not shown). Class II enhanceosome assembly was monitored by EMSAs and by DNase I footprinting experiments. Figure 1B shows that the MHCII DRA promoter (run alone in lane 1) forms distinct complexes with RFX, NFY and CREB (lanes 2–4). Pairwise combination of the factors revealed the assembly of class II promoter complexes containing both of the factors as well as individual DNA-bound proteins (lanes 5–7). Remarkably, when all factors were allowed to interact with the promoter a new complex was formed containing all three proteins (Figure 1B, lane 8). The cooperative nature of the MHCII enhanceosome assembly was monitored by quantitative DNase I footprinting experiments. Figure 1C shows that increasing amounts of recombinant RFX or NFY only slightly protected the promoter from DNase I cleavage at the X and Y boxes respectively (Figure 1C, lane sections 2 and 3, numbered at the base of the gel). In contrast, CREB alone bound stably to the X2 subregion (Figure 1C, section 4). Remarkably, when the two lower concentrations of RFX, NFY and CREB were allowed to interact with the promoter simultaneously, a strong footprint that extended over the X-Y-S region was observed, indicative of enhanceosome assembly (Figure 1C, section 5). Importantly, enhanceosome assembly is highly cooperative since these activators are incapable of strong protection of DNA when tested alone (compare sections 2, 3, 4 with 5). Figure 1D compares the DNA binding stability of the enhanceosome factors by off-rate competition experiments. In agreement with earlier observations using nuclear extracts (Reith et al., 1994a,b) or recombinant RFX and NFY (Caretti et al., 2000) individual enhanceosome subunits bind very unstably to DNA (Figure 1D, sections 3 and 4 with apparent half life <1 min for RFX and 1–5 min for NFY). CREB showed higher stability (Figure 1D, section 2 with half life ∼40 min). Strikingly, the enhanceosome showed very high stability (Figure 1D, section 1, half life >2 h) as compared with individual CREB, RFX or NFY subcomplexes. Since CREB binds relatively stably on its own, we assume that it contributes significantly to the overall stability of the enhanceosome. A similar effect was reported for the X2BP–RFX subcomplex using nuclear extracts (Reith et al., 1994a).
Fig. 1. In vitro assembly of the class II enhanceosome. (A) SDS–PAGE analysis of bacterially synthesized Ni–NTA purified, subunits of FRX, CREB and NFY. His-tagged proteins were run on a 10% acrylamide gel and were stained by Coomassie Blue. (B) EMSA was performed using the indicated recombinant factors and a 32P-labeled XY oligonucleotide. Reactions were run on a 5% acrylamide gel. (C) DNase I footprinting without (¢) and following binding of increasing amounts (see Materials and methods) of the indicated factors, assembled as above, to the DRA promoter fragment extending from –120 to +13 bp. Reactions were run on a 6% sequencing gel. Protected regions (brackets) and a hypersensitive site (HS) are indicated on the left based on the coordinations derived by the sequencing lanes (AGCT) run on the left. ALL indicates the use of RFX, NFY complexes and CREB together. (D) Off-rate experiments using complexes of the radioactive DRA-XY oligonucleotide and similar amounts of the indicated factors used for EMSA. Reactions shown in sections 1–4 were allowed to proceed for 20 min were chased by a 100-fold excess of cold DNA competitor and loaded serially after the indicated time on a 5% acrylamide gel.
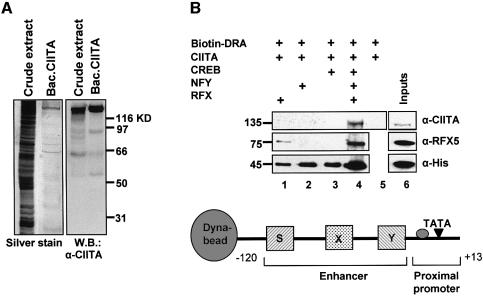
To investigate the ability of the enhanceosome to recruit directly CIITA we carried out in vitro recruitment experiments using immobilized promoter templates and recombinant factors. A biotinylated fragment extending from –120 to +13 bp of the DRA promoter (Figure 2B, bottom) was attached to paramagnetic streptavidin beads (Dynal) followed by enhanceosome assembly. Next, the templates were incubated with recombinant and purified CIITA protein (Figure 2A) and after washing the bound proteins were detected by western blotting. Figure 2B depicts the immunodetection of CIITA and RFX5 using specific antibodies as well as detection of the comigrating RFX/Ank, NFY/A, NFY/C and CREB at 45 kDa using anti-His antisera. Figure 2B shows that CIITA alone (lane 5 top) cannot bind to naked DNA. Similarly RFX, NFY or CREB cannot recruit CIITA when bound individually to the DRA promoter (lanes 1–3). However, the class II enhanceosome assembled from all three factors (RFX, NFY and CREB) recruited CIITA efficiently (lane 4). Thus seven proteins (three subunits of RFX, three of NFY and CREB) constitute the minimal enhanceosome that is necessary and sufficient for CIITA recruitment to DRA promoter.
Fig. 2. CIITA is recruited in an enhanceosome-dependent manner. (A) CIITA protein was produced by a baculovirus vector in Sf9 cells, further purified with an Ni–NTA column and run on an acrylamide gel. Purity was checked by silver stain (left panel) and western blotting to an anti-CIITA antibody (right panel). (B) The –120 to +13 DRA fragment was biotinylated and bound to Dyna-beads. The indicated His-tagged enhanceosome factors were added either individually or in combinations along with CIITA. The presence of the retained proteins relative to their inputs (lane 6) was detected by immunoblotting with the antibodies indicated on the right: anti-CIITA and anti-RFX5 detect specifically CIITA (140 kDa) and RFX5 (75 kDa) whereas anti-histidine (α-His) antibody detects RFXANK, CREB, NFYA and NFYC proteins that comigrate at 45 kDa on an 8% polyacrylamide gel. Lane 6 shows 20% of the input proteins.
In vivo recruitment of coactivators and chromatin modifiers to the promoter
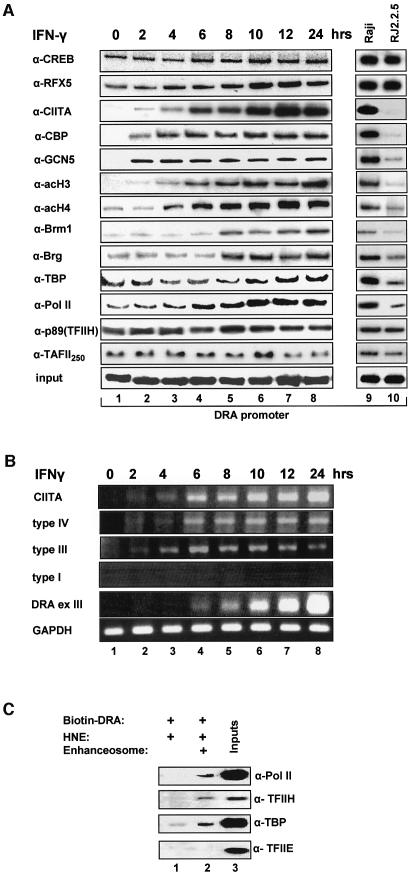
To investigate recruitment of CIITA to the endogenous class II promoters we carried out chromatin immunoprecipitation experiments using chromatin prepared from HeLa cells induced with IFN-γ for different amounts of time. Figure 3A shows that in uninduced cells the DRA promoter is occupied by various enhanceosome components such as CREB and RFX5 (top two rows), although the DRA gene is not expressed (Figure 3B, lane 1). This observation is in agreement with previous in vivo footprinting data indicating partial protection of class II promoters in uninduced HeLa cells (Kara and Glimcher, 1993). Surprisingly, we found that a subset of the basal transcriptional machinery including TBP, TAFII 250, RNA Pol II and the p89 subunit of TFIIH were also bound to the DRA promoter in the uninduced and non-expressing cells (Figure 3A). These results suggested that the class II enhanceosome could recruit these basal factors even in the absence of CIITA. To investigate this possibility, we carried out in vitro recruitment experiments using the immobilized DRA promoter with or without the enhanceosome and HeLa nuclear extracts (HNEs). Figure 3C shows that when HNEs that contain low levels of endogenous enhanceosome factors were allowed to react with the DNA very low levels of these basal factors were recruited to the promoter (lane 1). However, templates bearing the recombinant enhanceosome efficiently recruited TBP, Pol II and TFIIH (p89) but not TFIIE (lane 2). Thus, the class II enhanceosome can direct recruitment of key components of the basal machinery to the promoter in the absence of CIITA, a result consistent with the chromatin immunoprecipitation experiments described above.
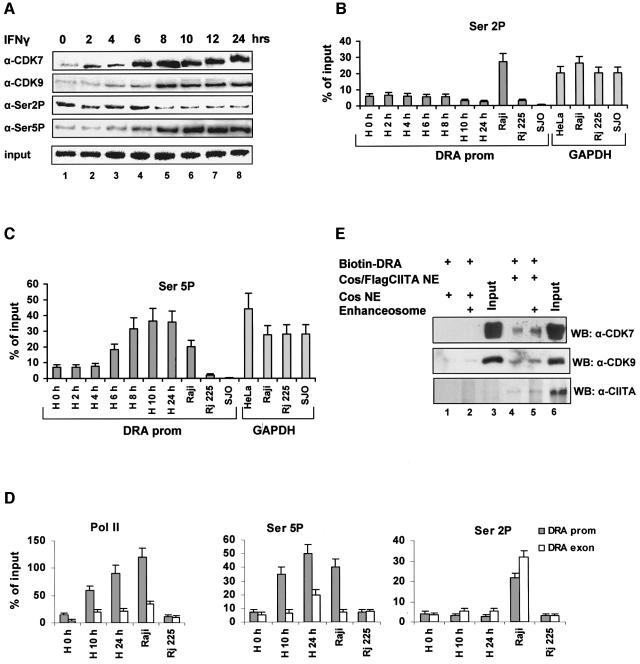
Fig. 3. Dynamic cofactor recruitment on the DRA promoter in vivo during IFN-γ induction. (A) Temporal order of factor recruitment on the promoter upon IFN-γ addition to HeLa cells (lanes 1–8) run in parallel with constitutively expressing Raji cells (lane 9) and their isogenic CIITA defective RJ225 cells (lane 10). Chromatin immunoprecipitation experiments were performed with antibodies against the indicated proteins or against acetylated histone 3 (α-H3) and histone 4 (α-H4) and semi-quantitative radioactive PCR reactions were carried out on the immunoprecipitated or 1% of initial material (input) using primers specific for the proximal promoter of the MHC class II gene. Results are representative of 2–5 experiments. (B) Activation of the class II gene during IFN-γ induction. HeLa cells were treated with IFN-γ for the indicated time points. Levels of mRNA were detected following semi-quantitative RT-PCR using primers spanning the CIITA coding region between amino acids 960–1110 (CIITA), CIITA promoter types IV, III or I, and CIITA common exon 2 and GAPDH coding region as indicated on the left. (C) In vitro recruitment of general transcription factors by the assembled enhanceosome. HNE (50 µg) were added to the biotinylated DRA promoter in the presence (+) or the absence of the enhanceosome (RFX, CREB and NFY) that was formed as in Figure 1. Detection of the recruited cofactors relative to the input (that is equal to 30% of the amount used for the recruitment) contained in the native HNEs were detected by immunoblotting with the antibodies shown on the right.
Two hours following IFN-γ treatment the enhanceosome recruited newly synthesized CIITA, whose IFN-γ-induced expression is driven by the type III and IV promoters but not by the type I promoter (Figure 3B, lane 2). The amount of CIITA recruited to the promoter is proportional to the amount of CIITA synthesized upon IFN-γ treatment (Figure 3A and B). At the same time the enhanceosome also recruits CBP and GCN5 to the promoter although their expression is not regulated by IFN-γ (data not shown). Presumably, the recruitment of these HAT proteins to the class II promoter is mediated via their interactions with CIITA (Kretsovali et al., 1998; Fontes et al., 1999). The recruitment of CBP, GCN5 and CIITA to the promoter is followed by increasing histones H3 and H4 acetylation (Figure 3A, lanes 3–8) in agreement with previous observations (Beresford and Boss, 2001). Importantly, histone acetylation precedes the recruitment of the SWI/SNF chromatin remodeling complexes bearing Brg-1or Brm-1. Figure 3B shows that the first DRA transcripts appear at 6 h following induction when the recruitment of CIITA, GCN5, CBP, Pol II and histone acetylation have reached their nearly peak activities (lane 4). The transcripts continue to accumulate throughout the entire time course (Figure 3B) and this increase correlates with increased enhanceosome stability, CIITA and SWI/SNF recruitment (Figure 2A). To compare the IFN-γ inducible with the constitutive class II expression state, we performed side by side chromatin immunoprecipitation experiments using chromatin prepared from MHCII expressing Raji B lymphoid cells and from the isogenic non-expressing and CIITA negative RJ225 cells. Figure 3A shows (compare lane 1 with 10 and lane 8 with 9), that for all the factors tested here, the IFN-γ-induced HeLa cell is fully comparable to the class II-expressing Raji cell whereas the uniduced HeLa cells resemble the CIITA-deficient RJ225 cells.
As a specificity control for all chromatin immunoprecipitation experiments we performed PCR reactions using the same immunoprecipitated DNA templates and primers amplifying the human IFN-β promoter, which is not expressed under these conditions. In all cases examined, the IFN-β signals were invariably close to background (see Supplementary figure 1 available at The EMBO Journal Online).
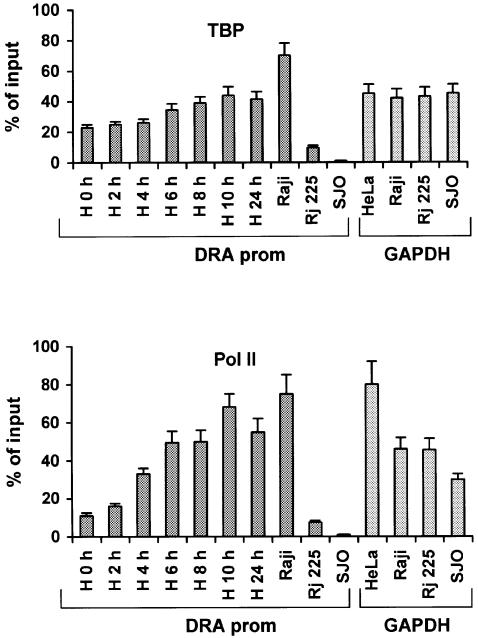
Our observation that a significant amount of TBP and Pol II is bound to the promoter before IFN-γ induction was surprising and therefore we measured these amounts by chromatin immunoprecipitation experiments followed by quantitative real-time PCR analysis. The amounts of TBP and Pol II bound to the DRA promoter in uninduced and IFN-γ-induced HeLa cells were calculated in side-by-side PCR experiments using primers amplifying the promoter of the constitutively expressed GAPDH gene (Figure 4). Furthermore, these amounts were also compared with the amounts of TBP- and Pol II-immunoprecipitated DRA promoter from Raji B lymphoid cells that constitutively express the class II genes, the isogenic RJ225 CIITA negative mutant cells or from SJO cells that lack one of the essential enhanceosome components (RFX5-deficient cells; Waldburger et al., 2000). The upper panel of Figure 4 shows that the amount of TBP bound to the silent IFN-γ-uninduced promoter was 51% of that bound to the highly expressed GAPDH gene promoter and these amounts rose to 99% following IFN-γ induction. The amount of TBP bound to the GAPDH gene promoter displayed small and random variation during the time course of IFN-γ induction (not shown). As expected, the constitutively expressed promoter in Raji B cells showed high levels of TBP occupancy (Figure 4). However, the amount of TBP bound to cells deficient for CIITA or RFX5 was 17% and <1% of the GAPDH levels, respectively. Thus, TBP cannot be recruited to the DRA promoter in cells lacking the RFX5 enhanceosome component. This result is in agreement with our observation that the preformed enhanceosome recruits TBP in the absence of CIITA. A similar result was obtained when we calculated the relative amounts of Pol II recruited to the DRA or GAPDH promoters. As seen in the lower part of Figure 4, a significant amount of Pol II occupies the promoter even before IFN-γ induction and this amount increases during the time course of induction in HeLa cells. We calculated that Pol II recruitment is increased ∼6-fold upon IFN-γ induction to levels similar to those recruited in Raji cells or to those recruited at the GAPDH promoter in all cell types tested. Importantly, the amount of Pol II recruited to the uninduced HeLa cells is comparable with that of CIITA-deficient cells but still they are ∼10-fold higher than those found in RFX5-deficient cells. Thus, TBP and Pol II are both recruited in an enhanceosome-dependent manner in the transcriptionally silent DRA gene in HeLa cells.
Fig. 4. Quantitative analysis of TBP (top) and RNA–Pol II (bottom) promoter recruitment. Immunoprecipitated chromatin from: uninduced (H0 h) or induced HeLa for the indicated time periods, class II expressing B lymphoid cells (Raji), CIITA deficient (RJ225) or RFX5 deficient (SJO) was amplified with DRA or GAPDH promoter specific primers and analyzed quantitatively. GAPDH values during IFN-γ induction of HeLa cells showed small and random temporal variation and were averaged. Results are percentages of the input chromatin and represent averages from at least three experiments from two independently prepared chromatin samples with shown standard error.
CIITA enhances Pol II C-terminal domain Ser5 phosphorylation
The presence of Pol II in the unstimulated promoter and its increasing recruitment upon activation prompted us to examine the phosphorylation status of the C-terminal domain (CTD) which determines the transition from transcriptional initiation to elongation (Dahmus, 1996). Figure 5A (top row, lanes 1–8) shows that CDK7, a component of the holo-TFIIH complex that phosphorylates RNA Pol II CTD, is increasingly recruited to the promoter and this recruitment perfectly correlates with that of CIITA (Figure 3A, third row, lanes 1–8). Furthermore, CDK9, a component of the PTEF-b complex (Price, 2000) that is also a kinase of RNA Pol II CTD, is also recruited to the class II promoter with a slightly delayed kinetics relative to CDK7. Using antibodies that specifically recognize phosphorylated RNA Pol II CTD (Figure 5A), we showed that transcription of the DRA gene, which appears at 6 h post-induction (Figure 3B), perfectly correlates with an increase in Ser5 but not Ser2 phosphorylation at the promoter (Figure 5A). These observations were further supported by quantitative real-time PCR analysis (Figure 5B and C). As seen in Figure 5C, an increase of phosphorylated Ser5 appears first at 6 h and reaches a maximum between 10 and 24 h of induction, thus correlating with DRA transcription. The amounts of Ser5 phosphorylated Pol II at the induced DRA promoter are similar to those detected at the GAPDH gene promoter (Figure 5C). Interestingly, we found that there is a perfect correlation of Ser5 phosphorylation at the DRA promoter between IFN-γ-induced HeLa cells and constitutively active Raji cells. However, the levels of Ser2 phosphorylation in Raji cells were higher relative to either induced or uniduced HeLa cells. As a control we showed that the levels of phosphorylated Ser2 or Ser5 in the RFX5-deficient cells (SJO) in which there is no Pol II bound to the promoter were undetectable (Figure 5B and C). Finally, as a quality control we showed that the amount of CREB bound to the DRA promoter in all cell types was similar (Supplementary figure 2). Of note, the amounts of CREB protein bound to the promoter in SJO cells (lacking RFX5) was reduced, a result consistent with the cooperative nature of enhanceosome assembly (Supplementary figure 2).
Fig. 5. Dynamic recruitment of RNA Pol II kinases and CTD serine phosphorylation upon IFN-γ-induced transcription. (A) Chromatin immunoprecipitation analysis of CDK7, CDK9 and RNA Pol II-serine2 (Ser2P) or -serine5 (Ser5P) phosphorylation during IFN-γ induction of HeLa cells. (B and C) Real-time PCR analysis of the Ser2 (Ser2P) or Ser5 (Ser5P) phosphorylated Pol II on the class II promoter in IFN-γ-induced HeLa and B lymphoid (Raji, RJ225 and SJO) cells. Control experiments were performed on the GAPDH promoter (right panels). (D) Real-time PCR analysis of the DRA promoter (gray bars) or exon 5 (open bars) associated Pol II, Ser2P or Ser5P as labeled. HeLa were uninduced (H0h) or IFN-γ-induced for 10 or 24 h and compared with Raji and RJ225 B lymphoid cell types. (E) Nuclear extracts from control (lanes 1 and 2) or CIITA expressing cells (lanes 4 and 5) were incubated with the DRA promoter without (lanes 1 and 4) or with (lanes 2 and 5) the addition of the enhanceosome factors used in Figure 2. The recruitment of CDK7 and CDK9 and CIITA relative to their input (20%) was monitored by specific antibodies.
To further understand the role of differential CTD serine phosphorylation in the transcription of the DRA gene, we compared the promoter and exon 5 levels of phospho-serine 2 and 5 recruited Pol II under various conditions. Both promoter and exon associated Pol II increased proportionally upon induction of HeLa cells and resemble the pattern of phosphoserine 5 CTD (Figure 5D, compare left and middle graphs). Ser5 phosphorylation is mainly promoter linked whereas serine 2 phosphorylation showed to be equally distributed between promoter and exons (Figure 5D, right graph) or even higher at the exons at a late time point after IFN induction. However, the levels of Ser2P Pol II found associated with either the promoter or the exon remained lower in induced HeLa relative to Raji cells.
Since there is a perfect correlation between recruitment of CDKs and CIITA to the DRA promoter, we investigated the role of the latter in CDK recruitment: the enhanceosome was assembled in vitro and the amount of kinases recruited in the presence of control or CIITA containing nuclear extracts was examined. As shown in Figure 5E, both the endogenous and the recombinant enhanceosome did not recruit either CDK7 or CDK9 (lanes 1 and 2) in the absence of CIITA. However, in the presence of CIITA the enhanceosome recruited CDK7 and CDK9 efficiently (lanes 4 and 5).
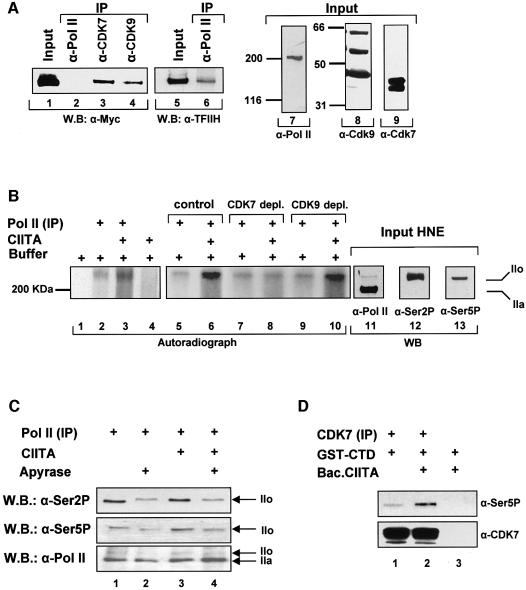
The CIITA-dependent recruitment of kinases led us to investigate whether CIITA interacts with CDKs. Figure 6A shows that antibodies against the endogenous CDK7 or CDK9, can precipitate myc-tagged CIITA (lanes 3 and 4) that was expressed into COS cells following transfection. In contrast, an antibody against Pol II could not precipitate CIITA (lane 2), although it did precipitate the p89 subunit of TFIIH (lanes 5 and 6). The western blots of Figure 6A (lanes 7, 8 and 9) depict the inputs and specificity of the antibodies used for immunoprecipitation.
Fig. 6. CIITA promotes serine 5 phosphorylation of Pol II. (A) CIITA interacts in vivo with CDK7 and CDK9. Whole cell extracts containing Myc-tagged CIITA were separated on a 8% SDS–PAGE and immunoblotted with an antibody against the myc epitope (α-Myc) or the p89 subunit of TFIIH (α-TFIIH) either before (Input) or following immunoprecipitation (IP) with antibodies against RNA Pol II, CDK7 and CDK9 as indicated. Input of the extracts (10%) were separated on 6 and 10% gels and immunoblotted with anti-Pol II and anti-CDK7, -CDK9 antibodies respectively. (B) RNA Pol II was immunoprecipitated from HNEs (lanes 2, 3 and 5–10) and equal amounts were phosphorylated in vitro with [γ-32P] ATP in the presence (lanes 3, 6, 8 and 10) or the absence (lanes 2, 5, 7 and 9) of CIITA. Reactions were separated on 6% SDS–PAGE together with input HNEs (lanes 11–13). The radioactive part of the gel was subject to fixation drying and autoradiography. The non-radioactive part was transferred onto nitrocellulose for subsequent immunoblotting with antibodies against Pol II and the Ser2 (Ser2P) and the Ser5 (Ser5P) phosphorylated forms of the CTD of Pol II. IIo and IIa correspond to the phosphorylated and non-phosphorylated forms of Pol II. (C) In vitro phosphorylation reactions of Pol II were performed in the presence of CIITA and/or apyrase as indicated. Phosphorylated forms of RNA Pol II were detected by immunoblotting using antibodies H5 and H14 specific for the phosphorylated Ser2P and Ser5P of Pol II. (D) A GST fusion of the CTD of Pol II (GST–CTD) was incubated with immunoprecipitated CDK7 (lanes 1 and 2) in the presence (lane 2) or the absence (lane 1) of 100 ng of baculovirus CIITA. Ser 5 phosphorylated RNA Pol II CTD was detected by H14 antibody and CDK7 was detected by specific antibody.
The experiments described above suggested that CIITA could affect the phosphorylation status of Pol II by recruiting the CDK7 and CDK9 kinases. To investigate this possibility directly we examined the effect of CIITA on RNA Pol II phosphorylation in vitro. The Pol II holoenzyme complex was immunoprecipitated using a specific Pol II antibody and Pol II phosphorylation was determined by adding [γ-32P]ATP and recombinant CIITA protein in the reaction. Figure 6B shows that the addition of baculovirus-expressed CIITA increased the in vitro phosphorylation of Pol II (compare lanes 2 and 3) by the associated kinases. To examine the relative contribution of CDK7 and CDK9 to this effect, we used extracts immunodepleted for these kinases (Supplementary figure 3). Depletion of CDK7 rendered CIITA unable to enhance Pol II phosphorylation (compare lanes 7 and 8 with 5 and 6) whereas CDK9 depletion had no effect (lanes 9 and 10). These results strongly suggest that CDK7 or an associated unknown kinase is the main Pol II kinase potentiated by CIITA. The western blots of Figure 6B (lanes 11–13) depict the inputs and identify the phosphorylated Pol II species as RNA Pol II form IIo.
To determine the Ser residue of the RNA Pol II CTD that was preferentially phosphorylated we performed cold in vitro phosphorylation assays followed by western blot analysis using antibodies against phosphorylated Ser2 or Ser5 CTD. Figure 6C shows that CIITA specifically increased phosphorylation of Ser5 but not Ser2 in the absence (lane 3) but not in the presence of apyrase (lane 4). Importantly, bacterially expressed CTD was used as a phosphorylation substrate for CDK7 since previous depletion experiments indicated that it is the kinase that is affected by CIITA. Immunoprecipitated CDK7 phosphorylated Ser5 of CTD (Figure 6D, lane 1). In the presence of CIITA Ser5 phosphorylation by CDK7 was strongly increased (Figure 6D, compare lanes 1 and 2). Thus, one of the actions of CIITA is to recruit both Pol II kinases and increase the Ser5 phosphorylation by CDK7.
Discussion
Previous studies have shown that assembly of the transcriptional apparatus in response to environmental cues is governed by distinct mechanisms in different genes. In general, the transcriptional activators bind sequentially to their regulatory elements and this leads to a defined order of recruitment of both chromatin-modifying activities and general transcription factors (Cosma, 2002). These observations led to the hypothesis that these differences are associated with different biological needs for each gene product. In this manuscript we describe the order of events taking place during activation of an MHCII gene in response to IFN-γ. We found that in contrast to the previously described cases of signal-regulated gene transcription by virus (Agalioti et al., 2000), estrogens (Shang, et al., 2000), phosphate deprivation (Gregory et al., 1999) or by differentiating signals (Soutoglou and Talianidis, 2002) the activators responsible for the expression of the DRA gene are prebound to the enhancer forming an enhanceosome. The enhanceosome then serves as a docking surface for the subsequent recruitment of a subset of the basal transcription machinery that remains associated to the core promoter even under conditions where the gene is transcriptionally silent. In other words the class II genes are ‘poised’ for activation. Transcriptional activation in response to IFN-γ requires the synthesis of CIITA, which is rapidly recruited to class II promoters by the enhanceosome. We showed that this recruitment correlates with the subsequent recruitment of chromatin-modifying activities such as HATs and ATP-dependent remodeling machines. Surprisingly, CIITA also recruits CDK7 and CDK9 thus causing RNA Pol II CTD phosphorylation and transcriptional elongation. Collectively, our data describe a novel strategy by which a gene-specific transcriptional coactivator orchestrates the MHCII gene expression program.
The class II enhanceosome is assembled via multiple cooperative interactions between its protein constituents and the DNA (Masternak et al., 2000). Previous reports employing nuclear extracts (Reith et al., 1994a,b; Moreno et al., 1995; Fontes et al., 1997a; Louis-Plence et al., 1997) or a subset of recombinant enhanceosome components (RFX and NFY) (Caretti et al., 2000) have shown cooperative DNA binding between RFX, NFY and X2BP. We have reconstituted the enhanceosome in vitro using all seven subunits of the class II binding factors (RFX, CREB and NFY) and found that it can recruit CIITA. This is the first report of CIITA recruitment by a purely recombinant class II gene enhanceosome.
Our chromatin immunoprecipitation experiments revealed that the enhanceosome and a subset of the basal transcriptional machinery that is already bound to the promoter before induction is ‘energized’ by the presence of newly synthesized CIITA recruited upon IFN-γ treatment. CIITA recruitment leads to a further stabilization of the transcription complex by integrating and coordinating a new set of protein–protein interactions. Thus, CIITA coordinates several alterations of cofactor recruitment to the promoter that precede or are concomitant with the onset of transcription.
An extensive cofactor recruitment analysis by chromatin immunoprecipitations indicated that the molecular architecture of the transcription complexes of IFN-γ-induced HeLa cells is qualitatively similar to the constitutively expressing Raji cells. On the other hand, uninduced HeLa cells resemble the CIITA negative B lymphoblastoid cells RJ225 with the exception of TBP that shows high levels in the former cell type.
CIITA orchestrates recruitment of the HATs CBP and GCN5 correlating with chromatin acetylation that precedes initiation of transcription. Therefore, the synergy between CIITA and CBP and/or PCAF/GCN5 that was shown in transfection systems (Kretsovali et al., 1998; Fontes et al., 1999; Spilianakis et al., 2000) reflects a physiological role of HAT-type coactivators in class II gene transcription. Although the HAT activity of various coactivators—as assayed by transient experiments—seems to be partly dispensable for MHCII gene expression, the present data in combination with results that correlate transcriptional inactivation after IFN-γ removal (Beresford and Boss, 2001) or in regulatory mutant cell lines with reduced acetylation (Masternak and Reith, 2002), support a causal relationship between promoter localized histone acetylation and transcription. Our experiments also showed that both Brg-1 and Brm-1 containing chromatin remodeling machines are recruited to the promoter, subsequently to the wave of acetylation and their presence temporally correlates with a role in transcriptional elongation. Thus, the need for SWI–SNF-mediated remodeling is not only restricted to the expression of CIITA itself (Mudhasani and Fontes, 2002) but it is also directly required for MHCII gene transcription.
Although promoter occupancy by factors of the basal machinery did not change significantly over time after IFN-γ induction, occupancy by Pol II showed a strong increase that temporally parallels both CIITA recruitment and gene transcription. Phosphorylation of the CTD of Pol II is a critical process that controls promoter clearance and elongation (Dahmus, 1996). The cyclin H–CDK7 complex (which belongs to TFIIH) is responsible for Ser5 phosphorylation whereas cyclin T–CDK9 (P-TEF) is responsible for Ser2 phosphorylation. CIITA interacts with and recruits both RNA Pol II kinases CDK7 and CDK9. Interestingly, CDK7 recruitment is directly proportional to the recruitment of CIITA. As a consequence, CTD phosphorylation at Ser5 is the event that closely correlates with DRA gene transcription onset that occurs 6 h post induction. In another system of signal-induced transcription, the IL-8 gene, induction by TNF leads to both Ser5 and Ser2 phosphorylation of promoter-bound Pol II (Nissen and Yamamoto, 2000; Barboric et al., 2001). Apparently the kinase specificity for distinct CTD serines is subject to gene-, cell- and regulatory-type specific rules. Previous work with Saccharomyces cerevisiae has demonstrated that RNA Pol II is Ser5 phosphorylated at the promoters and Ser2 phosphorylated within transcribed regions (Komarnitsky et al., 2000). On the DRA gene, RNA Pol II and its Ser5-phosphorylated form are more concentrated at the promoter as compared with the coding regions. In contrast, Ser2-phosphorylated RNA Pol II is evenly distributed between promoter and coding regions or even more concentrated at the coding regions. These data are in agreement with reports from studies on the DHFR and γ-actin genes in mammalian cells (Cheng and Sharp, 2003). Ser2 phosphorylation in IFN-γ-induced epithelial-like cells is low relative to Raji B lymphoid cells that constitutively transcribe the DRA gene. Induced HeLa express ∼5× less DRA mRNA than Raji cells (not shown) yet their Pol II levels do not differ significantly. Therefore, we conclude that Raji cells have a higher rate or density of elongating polymerase relative to HeLa. Alternatively, paused Pol II at the DRA promoter, similarly to the heat shock (Lis, 1998), c-myc (Krumm et al., 1992) and c-fos (Pinaud and Mirkovitch, 1998) genes, might occur in higher proportions in HeLa cells relative to the Raji.
Strikingly, CIITA augments serine 5 phosphorylation of Pol II in vitro. Such an effect may be due to its ability to increase enzyme activity or stabilize enzyme–substrate interaction. We provide evidence that CIITA potentiates the CDK7-driven phosphorylation of RNA Pol II CTD employing either depleted extracts or immunoprecipitated CDK7 and recombinant CTD which was phosphorylated at Ser5.
In summary, our data describe a novel mechanism of controlling gene transcription in which a mammalian gene is poised for activation and awaits to be ‘energized’ by the incoming gene-specific class II transactivator. The latter is able to promote Ser5 phosphorylation of RNA Pol II which is the critical event that determines the timing of transcription onset through promoter clearance and transcription elongation.
Materials and methods
Cell culture and reagents
HeLa and COS-1 cells were grown in DMEM supplemented with 10% bovine fetal serum. B lymphoid cell lines were maintained in RPMI. IFN-γ (R&D) was used at 100–200 U/ml. Antibodies against acetylated H3 and H4 were from Upstate Biotechnology. Monoclonal antibodies H5 and H14 were from Covance. All other antibodies were from Santa Cruz Biotechnology.
Recombinant proteins
His-tagged recombinant proteins were expressed in E.coli HB101 cells from the pRSET vector and were purified on an Ni–NTA column. A detailed protocol is included in Supplementary data section. Baculo-CIITA was produced using the Bac-to-Bac Baculovirus expression system (Gibco-BRL) according to manufacturers instructions. The protein was then purified on Ni–NTA agarose beads under non-denaturing conditions.
Electrophoretic mobility shift assays
Binding reactions contained radiolabeled (0.3 × 106 c.p.m./reaction) of the XY fragment XY sense 5′-GATCCCCTAGCAACAGATGCGT CATCTCAAAATATTTTT.
For the off-rate EMSA experiments 3 × 106 c.p.m. radiolabeled DRA enhancer DNA fragment and the appropriate amounts of each protein complex were used: RFX5, 2 ng; RFXAP, 1 ng; RFXANK, 1 ng; NFYA, 1.25 ng; NFYB, 1.25 ng; NFYC, 1.25 ng; CREB, 1.25 ng. Binding reactions were left to set on ice for 20 min and then 100× excess cold competitor of the same DRA DNA fragment was added to the reaction for the indicated time periods and run in 6% polyacrylamide gel. The first lane of each off-rate EMSA experiment for the enhanceosome, CREB, RFX complex and NFY complex are binding reactions using no cold competitor. The abnormal kinetics in some lanes is due to loading after increasing time periods.
In vitro DNase I footprinting
In vitro DNase I footprinting used C-terminally radiolabeled DNA fragments of the human DRA enhancer. A detailed protocol is included in the Supplementary data section.
Recruitment on immobilized template
The biotinylated DRA promoter was synthesized by PCR using specific DRA primers. Blocking was performed in BC-100 buffer (100 mM KCl, 20 mM HEPES pH 7.9, 20% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF) supplemented with 10% BSA for 1 h at 4°C in the presence of 2 µg of salmon sperm DNA. Then recombinant RFX, NFY and CREB were added at concentrations established on DNase footprinting (RFX5, 80 ng; RFXAP, 40 ng; RFXANK, 40 ng; NFYA, 50 ng; NFYB, 50 ng; NFYC, 50 ng; CREB, 100 ng) and incubated for an additional hour. Finally baculoviral CIITA (∼100 ng as determined by silver staining) was added for 1 h. Washing was performed three times in BC-100 supplemented with 0.01% Triton.
RT–PCR
RNA was prepared with the Trizol reagent (Gibco-BRL). Reverse transcription–PCR reactions were made using MMLV Promega enzyme. PCR conditions for all the reactions included an initial 5 min step at 94°C, followed by 24 cycles of 30 s at 94°C, 30 s at 64°C, 30 s at 72°C, and a final step of 5 min at 72°C. PCR products were analyzed in a 2% agarose gel.
The sequences of RT–PCR primers for CIITA are the following: CIITA type III.sense, 5′-ATGCGTTGCCTGGCTCCACGCC-3′; CIITA type IV.sense, 5′-CCAGAGCTGGCGGGAGGGAG-3′; CIITA type I.sense, 5′-GCCATCAGCCCAGCCTGGTGC-3′.
The above primers were used along with the following common reverse primer: CIITA.antisense, 5′-CAGGCAGCTCAACGAGGAAC TGGAG-3′.
Sequences of RT–PCR primers for the DRA gene are: DRA.exIIIsense, 5′-GAGTTTGATGCTCCAAGCCCTCTCCCA-3′; DRA.exIIIantisense, 5′-CAGAGGCCCCCTGCGTTCTGCTGCATT-3′.
Sequences for RT–PCR primers for GAPDH gene are: GAPDH.sense, 5′-CCCCCTCTGCTGATGCCC-3′; GAPDH.antisense, 5′-CCCCGCG GCCATCACG-3′.
Chromatin immunoprecipitations
A detailed protocol for chromatin immunoprecipitation assays is provided in the Supplementary data section. For radioactive PCR analysis, DNA was resuspended in 100 µl TE buffer pH 7.5 and 10 µl were used in a radioactive PCR reaction using [32P]dATP and [32P]dCTP supplemented with 0.1 mM cold dNTPs, 3 units of Taq polymerase, 0.25 mM MgCl2, 1× PCR reaction buffer and 400 ng of each primer. PCR conditions included an initial step of 5 min at 94°C, 24 cycles of 30 s at 94°C, 30 s at 64°C, 30 s at 72°C and a final step of 2 min at 72°C.
Real-time PCR analysis was performed using the Opticon monitor (MJ Research) and the intercalating fluorescent dye SYBR Green I. The fraction of immunoprecipitated promoter DNA was calculated from a standard curve/relative to standard input chromatin. The immunoprecipitated values obtained with different antibodies using the class II promoter were compared with the values of the reference gene GAPDH which displays high levels of expression and is not regulated by IFN-γ. The IFN-β promoter was used as a negative control. Chromatin immunoprecipitations were repeated at least three times from different chromatin material and radioactive or real-time PCR analysis was repeated three to five times.
The primers used were the following:
DRA promoter: DRAsense, 5′-GTTGTCCTGTTTGTTTAAGAAC-3′; DRAantisense, 5′-GCTCTTTTGGGAGTCAG-3′.
DRAexon 5: DRAsense, 5′-GAAAGCAGTCATCTTCAGCGTT-3′; DRAantisense 5′-ATTATCACCATGCAATGCCTCT-3′.
IFN-β promoter: IFN-βsense, 5′-GCTTTCCTTTGCTTTCTCCCAA GTC-3′; IFN-βantisense, 5′-CCTTTCTCCATGGGTATGGCC-3′.
GAPDH-promoter: GAPDHsense, 5′-TGAGCAGACCGGTGTCAC TA-3′; GAPDHantisense, 5′-AGGACTTTGGGAACGACTGA-3′.
In vitro phosphorylation assays
HNEs (∼600 µg) were immunoprecipitated with 30 µl (200 ng/µl, Santa Cruz) of anti-Pol II antibody overnight at 4°C, supplemented with 20 µl of Protein A slurry preblocked with 500 µl of BC100, 5% BSA, 0.01% Triton X-100, for 1 h at 4°C. The immune complexes were pelleted, resuspended in 10 µl of BC-100 and used for phosphorylation experiments. Phosphorylation reaction was set up in a buffer containing 5 mM HEPES pH 7.8, 20 mM Tris–HCl pH 7.9, 7 mM MgCl2, 60 mM KCl, 12% (v/v) glycerol, 2% (w/v) polyethylene glycol 8000, 2 mM 2-mercaptoethanol, 0.1 mM EDTA, 6 µg of BSA, 3 µCi [γ-32P]ATP for radioactive reactions and 15 µM cold ATP. Reactions were performed for 1 h at 25°C, in a total volume of 25 µl. For detection of phosphorylated forms of Pol II by western blotting we used 0.2 mM ATP and the monoclonal antibodies H14 and H5 that recognize the phosphoserine 5 and phosphoserine 2 version of RNA Pol II–CTD respectively (Covance-Babco). For the experiments employing CDK7 and CDK9-depleted extracts, two rounds of treatment with 30 µl of anti-CDK7 or 30 µl of anti-CDK9 antibodies preceded the immunoprecipitation of RNA Pol II. For the experiments using RNA Pol II CTD as a template, 500 ng of a GST–CTD fusion protein (kindly provided by Dr D.Reinberg) was phosphorylated by CDK7 immunoprecipitated from 100 µg of HNEs in a buffer containing 50 mM Tris pH 7.5, 4 mM MgCl2, 5 mM DTT, 5 mM MnCl2, 0.1 mM ZnSO4 and 0.2 mM ATP.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank G.Vretzos for expert assistance with cell culture and E.Tzortzakaki and S.Lomvardas for help with some experiments. We also thank D.Reinberg for GST–CTD and I.Talianidis for antibodies. This work was supported by grants from the Greek General Secretariat for Science and Technology (PENED 2284).
References
- Agalioti T., Lomvardas,S., Parekh,B., Yie,J., Maniatis,T. and Thanos,D. (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-α promoter. Cell, 103, 667–678. [DOI] [PubMed] [Google Scholar]
- Barboric M., Nissen,R.M., Kanazawa,S., Jabrane-Ferrat,N. and Peterlin,B.M. (2001) NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell, 8, 327–337. [DOI] [PubMed] [Google Scholar]
- Beresford G.W. and Boss,J.M. (2001) CIITA coordinates multiple histone acetylation modifications at the HLA–DRA promoter. Nat. Immunol., 2, 652–657. [DOI] [PubMed] [Google Scholar]
- Caretti G., Cocchiarella,F., Sidoli,C., Villard,J., Peretti,M., Reith,W. and Mantovani,R. (2000) Dissection of functional NF-Y-RFX cooperative interactions on the MHC class II Ea promoter. J. Mol. Biol., 302, 539–552. [DOI] [PubMed] [Google Scholar]
- Cheng C. and Sharp,P. (2003) RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol., 23, 1961–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma M.P. (2002) Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell, 10, 227–236. [DOI] [PubMed] [Google Scholar]
- Cresswell P. (1994) Antigen presentation. Getting peptides into MHC class II molecules. Curr. Biol., 4, 541–543. [DOI] [PubMed] [Google Scholar]
- Dahmus M.E. (1996) Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem., 271, 19009–19012. [DOI] [PubMed] [Google Scholar]
- DeSandro A.M., Nagarajan,U.M. and Boss,J.M. (2000) Associations and interactions between bare lymphocyte syndrome factors. Mol. Cell. Biol., 20, 6587–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes J., Jabrane-Ferrat,N. and Peterlin,B. (1997a) Assembly of functional regulatory complexes on MHC class II promoters in vivo. J. Mol. Biol., 270, 336–345. [DOI] [PubMed] [Google Scholar]
- Fontes J.D., Jiang,B. and Peterlin,B.M. (1997b) The class II trans-activator CIITA interacts with the TBP-associated factor TAFII32. Nucleic Acids Res., 25, 2522–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes J.D., Kanazawa,S., Jean,D. and Peterlin,B.M. (1999) Interactions between the Class II transactivator and CREB binding protein increase transcription of major histocompatibility complex Class II genes. Mol. Cell. Biol., 19, 941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimcher L.H. and Kara,C.J. (1992) Sequences and factors: a guide to MHC class-II transcription. Annu. Rev. Immunol., 10, 13–49. [DOI] [PubMed] [Google Scholar]
- Gregory P.D., Schmid,A., Zavari,M., Munsterkotter,M. and Horz,W. (1999) Chromatin remodelling at the PHO8 promoter requires SWI–SNF and SAGA at a step subsequent to activator binding. EMBO J., 18, 6407–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S., Masternak,K., Kammerbauer,C., Jansen,C., Reith,W. and Steimle,V. (2000) CIITA leucine-rich repeats control nuclear localization in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome and MHC class II gene transcription. Mol. Cell. Biol., 20, 7716–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harton J. and Ting,J.P. (2000) Class II transactivator: mastering the art of major histocompatibility complex expression. Mol. Cell. Biol., 20, 6185–6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa S., Okamoto,T. and Peterlin,B.M. (2000) Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity, 12, 61–70. [DOI] [PubMed] [Google Scholar]
- Kara C.J. and Glimcher,L.H. (1993) Developmental and cytokine-mediated regulation of MHC class II gene promoter occupancy in vivo. J. Immunol., 150, 4934–4942. [PubMed] [Google Scholar]
- Komarnitsky P., Cho,E. and Buratowski,S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev., 14, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsovali A., Agalioti,T., Spilianakis,C., Tzortzakaki,E., Merika,M. and Papamatheakis,J. (1998) Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol. Cell. Biol., 18, 6777–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A., Meulia,T., Brunvand,M. and Groudine,M. (1992) The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev., 6, 2201–2213. [DOI] [PubMed] [Google Scholar]
- Lee T.I. and Young,R.A. (2000) Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet., 34, 77–137. [DOI] [PubMed] [Google Scholar]
- Lemon B. and Tjan,R. (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev., 14, 2551–2569. [DOI] [PubMed] [Google Scholar]
- Lis J. (1998) Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol., 63, 347–356. [DOI] [PubMed] [Google Scholar]
- Louis-Plence P., Moreno,C.S. and Boss,J.M. (1997) Formation of a regulatory factor X/X2 box-binding protein/nuclear factor-Y multiprotein complex on the conserved regulatory regions of HLA class II genes. J. Immunol., 159, 3899–3909. [PubMed] [Google Scholar]
- Mahanta S.K., Scholl,T., Yang,F.C. and Strominger,J.L. (1997) Transactivation by CIITA, the type II bare lymphocyte syndrome-associated factor, requires participation of multiple regions of the TATA box binding protein. Proc. Natl Acad. Sci. USA, 94, 6324–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masternak K. and Reith,W. (2002) Promoter-specific functions of CIITA and the MHC class II enhanceosome in transcriptional activation. EMBO J., 21, 1379–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masternak K., Muhlethaler-Mottet,A., Villard,J., Zufferey,M., Steimle,V. and Reith,W. (2000) CIITA is a transcriptional coactivator that is recruited to the MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev., 14, 1156–1166. [PMC free article] [PubMed] [Google Scholar]
- Moreno C.S., Emery,P., West,J.E., Durand,B., Reith,W., Mach,B. and Boss,J.M. (1995) Purified X2 binding protein (X2BP) cooperatively binds the class II MHC X box region in the presence of purified RFX, the X box factor deficient in the bare lymphocyte syndrome. J. Immunol., 155, 4313–4321. [PubMed] [Google Scholar]
- Mudhasani R. and Fontes,J.D. (2002) The class II transactivator requires brahma-related gene 1 to activate transcription of major histocompatibility complex class II genes. Mol. Cell. Biol., 22, 5019–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar G.J., Fan,H.-Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Nissen R. and Yamamoto,K. (2000) The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev., 14, 2314–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud S. and Mirkovitch,J. (1998) Regulation of c-fos expression by RNA polymerase elongation competence. J. Mol. Biol., 280, 785–798. [DOI] [PubMed] [Google Scholar]
- Price D. (2000) P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol., 20, 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith W. and Mach,B. (2001) The bare lymphocyte syndrome and the regulation of MHC expresion. Annu. Rev. Immunol., 19, 331–373. [DOI] [PubMed] [Google Scholar]
- Reith W., Kobr,M., Emery,P., Durand,B., Siegrist,C.A. and Mach,B. (1994a) Cooperative binding between factors RFX and X2bp to the X and X2 boxes of MHC class II promoters. J. Biol. Chem., 269, 20020–20025. [PubMed] [Google Scholar]
- Reith W., Siegrist,C., Durand,B., Barras,E. and Mach,B. (1994b) Function of major histocompatibility complex class II promoters requires cooperative binding between factors RFX and NF-Y. Proc. Natl Acad. Sci. USA, 91, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Co-factor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Soutoglou E. and Talianidis,I. (2002) Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science, 295, 1901–1904. [DOI] [PubMed] [Google Scholar]
- Spilianakis C., Papamatheakis,J. and Kretsovali,A. (2000) Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol. Cell. Biol., 20, 8489–8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting J.P. and Trowsdale,J. (2002) Genetic control of MHC class II expression. Cell, 109, S21–S33. [DOI] [PubMed] [Google Scholar]
- Viret C. and Janeway,C.J. (1999) MHC and T cell development. Rev. Immunogenet., 1, 91–104. [PubMed] [Google Scholar]
- Waldburger J.M., Masternak,K., Muhlethaler-Mottet,A., Villard,J., Perreti,M., Landman,S. and Reith,W. (2000) Lessons from the bare lymphocyte syndrome: molecular mechanisms regulating MHC class II expression. Immunol. Rev., 178, 148–165. [DOI] [PubMed] [Google Scholar]
- Zhu X.-S., Linhof,M.W., Li,G., Chin,K.-C., Maity,S.N. and Ting,J.P. (2000) Transcriptional scaffold: CIITA interacts with NF-Y, RFX and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol. Cell. Biol., 20, 6051–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]