Abstract
The patterning and development of multicellular organisms requires a precisely controlled balance between cell proliferation, differentiation and death. The regulation of apoptosis is an important aspect to achieve this balance, by eliminating unnecessary or mis-specified cells which, otherwise, may have harmful effects on the whole organism. Apoptosis is also important for the morphogenetic processes that occur during development and that lead to the sculpting of organs and other body structures. Here, we review recent progress in understanding how apoptosis is regulated during development, focusing on studies using Drosophila or C. elegans as model organisms.
Introduction
Apoptosis is the most widely studied form of programmed cell death and is characterized by a variety of morphological and biochemical aspects, such as the condensation of the nucleus and cytoplasm, the activation of proteases (caspases) and nucleases, that respectively degrade cellular proteins and DNA, and the fragmentation of apoptotic cells into membrane-bound bodies that are rapidly phagocytosed by neighboring cells. These aspects of apoptotic cell death make it distinct from necrosis, a form of cell death resulting from overwhelming cellular injury, in which cells swell and lyse, releasing their cytoplasmic contents into the extracellular space [1,2,3].
The nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster have been instrumental in defining the genetic and molecular pathways that regulate apoptosis during development. In brief, execution of apoptosis in these organisms requires the activation of caspases, a class of cysteine proteases that are constitutively expressed in virtually all cells as inactive zymogens. Upon death inducing signals, the inactive caspases are cleaved at specific aspartic acid residues, resulting in the removal of an inhibitory N-terminal domain and production of a large and a small subunit. These subunits then associate as a hetero-tetramer (2 large/2 small) to form the active protease that cleaves many cellular targets, leading to the apoptotic death of the cell.
The regulation of caspases occurs mainly by two distinct general mechanisms, employing regulatory cascades designed to either activate or inhibit caspases (Figure 1). In the “classic pathway”, derived from studies in C. elegans and mammalian systems, activation of the initiator caspase (caspase-9) results from the formation the apoptosome, a multi-protein complex containing CED-4/Apaf-1 and Cytochrome C (in mammals). This pathway is also regulated by the Bcl-2 family of proteins, which can facilitate or prevent the release of Cytochrome C from mitochondria to the cytoplasm. A second pathway, largely derived from work in insects, is an inhibitory one, where caspase activation is blocked by Inhibitor of Apoptosis Proteins (IAPs). In this case, the activation of apoptosis requires the action of IAP antagonists, such as reaper, hid and grim in Drosophila [4–6]. It appears that both pathways are used in coordination to control the activation of caspases, but the relevant contribution of either branch varies according to the particular cell type and signaling paradigm [3,7–10].
Figure 1.
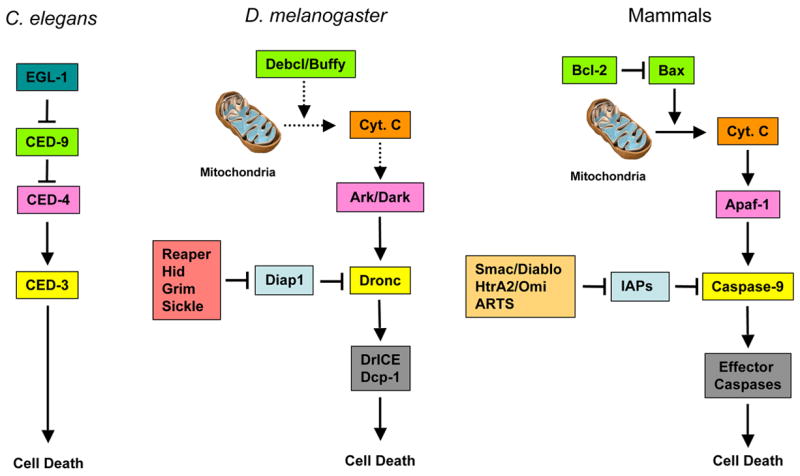
The core apoptotic pathways in C. elegans, Drosophila and mammals. The functional homologues between species are represented with boxes with the same background color. For the sake of simplicity, we did not include the pathways leading to the activation of caspase-2 and caspase-8 (extrinsic pathway).
Transcription is important
Many regulatory steps of the apoptotic process occur at the post-translational levels, either by protein-protein interactions and protein cleavage or by ubiquitin-dependent protein degradation [11]. Although all proteins required for the execution of apoptosis are constitutively expressed in virtually all cells, it has also been known for a long time that the induction of apoptosis often requires de novo transcription and protein synthesis [2,12,13].
Work in C. elegans has highlighted the importance of transcriptional regulation of the core apoptotic genes. The transcriptional upregulation of egl-1 promotes cell death by antagonizing the interaction between CED-9 (Bcl-2 like) and CED-4 (Apaf-1 like) and allowing the release of CED-4 from mitochondria [14,15]. In the hermaphrodite specific neurons (HSN), the transcription factor TRA-1 directly binds the egl-1 promoter to repress egl-1 transcription and induce apoptosis of these cells in the males [16]. Recent work showed that the Hox gene mab-5 is also required for the regulation of egl-1, and induction of apoptosis in the P11 and P12 cell lineages that generate neurons in the posterior ventral nerve cord. In the P11 lineage, MAB-5 forms a complex with the Pbx homologue CEH-20 to directly regulate egl-1 transcription [17].
But egl-1 is not the only cell death gene for which regulation at the transcriptional level is important. The correct timing of onset of death of the tail-spike cell requires transcriptional up-regulation of the ced-3 caspase [18]. In this case, EGL-1 and CED-9 play only a minor role, and cell death occurs shortly after the onset of ced-3 expression. The transcription factor PAL-1 (homologue of the mammalian tumor suppressor Cdx2) plays an important role by binding to the ced-3 promoter to direct ced-3 expression [18]. Also, egl-38 and pax-2, members of the Pax2/5/8 class of transcription factors, act as modulators of ced-9 transcription both in somatic and germline cell death [19].
One of the strongest evidence that new gene transcription is required for activation of apoptosis during normal development has come from Drosophila, where Reaper-family proteins are transcriptionally activated in doomed cells prior to their death [4]. More recently, it was also shown that expression of the apical caspase Dronc and the effector caspase Drice are upregulated in response to ecdysone [20,21], a hormone which induces apoptosis to eliminate the larval tissues during the process of metamorphosis. The transcriptional levels of thread (which encodes Drosophila Inhibitor of Apoptosis Protein 1 – Diap1) are also regulated by the Hippo signaling pathway, through the phosphorylation and inactivation of the transcriptional activator Yorkie [22] (see below). It appears that these recently discovered phenomena contribute to determine the general susceptibility of a cell towards apoptosis, but that additional regulation is needed for the acute initiation of cell death.
Fat signals to the Hippo pathway
Genetic screens in Drosophila, designed to identify mutants with excessive cell proliferation and hyperplastic growth, led to the identification of several genes acting in the Hippo signaling pathway [23,24]. These include two serine/threonine kinases, Hippo (Hpo) and Warts (Wts)/Lats, two adaptor proteins, Salvador (Sav) and “Mob as tumor suppressor” (Mats), and the transcriptional activator Yorkie (Yki). Wts phosphorylation and activation by Hpo is facilitated by Sav and Mats. Activated Wts phosphorylates and inactivates Yorkie (Yki). Loss of Hpo, Wts, or Sav function or overexpression of Yki lead to up-regulation of cyclin E and diap-1 and an increase in cell proliferation and inhibition of apoptosis.
Recent studies link the membrane protein Fat with the Hpo pathway [25,26,27, 28]. Fat is a large protocadherin previously shown to function in the establishment of planar cell polarity, proximal-distal patterning of appendages, and to act as a tumour suppressor, restricting the growth of imaginal discs during larval development. Clones of fat mutant cells, as in clones of mutants of the Hpo pathway, overgrow due to increased proliferation and suppression of apoptosis and cyclin E and diap1, which are upregulated in Hpo pathway mutants, are also upregulated in fat mutants.
The precise molecular mechanisms linking Fat and the Hpo pathway are not known yet, but it seems Fat can regulate the Hpo pathway in more than one way. First, Fat can regulate Wts protein stability via the unconventional myosin Dachs, which directly binds Wts [25]. Second, Fat can interact with the Hpo pathway through the regulation of Expanded (Ex) [26–28]. Ex and the related protein Merlin (Mer) are members of the FERM (4.1, Ezrin, Radixin, Moesin) domain superfamily of proteins and act as upstream regulators of the Hpo pathway [29]. Fat co-localizes with Ex at the apical adherens junctions and is required for Ex, but not Mer or Moesin, apical localization [26–28]. It is not know how Fat regulates Ex apical localization or how Ex and Mer influence Hippo signaling, but it has been suggested that Fat might bring Ex together with Hpo and thereby activate this pathway [27]. Attempts to coimmunoprecipitate Fat and Exp failed, but Fat is a large (560 kDa) protein, which may be difficult to detect by coimmunoprecipitation assay [27]. On the other hand, Ex and Mer regulate endocytosis, and the steady state levels of several membrane receptors, such as Fat, Notch, EGFR, Patched and Smoothened are altered in Ex/Mer double mutants [30]. Hence, it is also possible that regulation of Hpo signaling by Ex/Mer occurs in an indirect manner, via the regulation of endocytosis of Fat. Additional experiments are required to address this possibility and the exact biochemical interactions between Fat, Ex and the Hpo pathway. Interestingly, cells mutant for vps25, a component of the ESCRT (Endosomal Sorting Complex Required for Transport), have higher levels of Hpo signaling [31], which points to the importance of endocytosis for the regulation of the Hpo pathway.
Another relevant finding was the identification of the microRNA bantam (ban) as a target of Yki and the Hpo pathway [32,33]. ban was previously identified as a developmentally regulated microRNA that induces cell proliferation and prevents apoptosis through the downregulation of the apoptosis-inducing gene hid [34]. In addition to ban other microRNAs, such as miR-14, miR-278 and the miR-2 family, play important roles as regulators of cell death genes [35,36].
Compensatory proliferation and cell competition
In developing tissues, cells that undergo apoptosis in response to stress or injury can induce proliferation of neighboring progenitor cells [37,38,39]. This regenerative phenomenon, termed “compensatory proliferation”, permits the organism to compensate for the loss of large numbers of cells and to develop normally patterned and sized organs in these situations. Significantly, doomed Drosophila imaginal disc cells produce proteins with known mitogenic and morphogenetic activities, such as Wnts and BMPs [38,39]. Furthermore, Wnt signaling is required for cell proliferation in this system [39]. Given the role of Wnt signaling for self-renewal of stem cells and in tumorigenesis in mammals, a similar mechanism may operate in vertebrates and there is considerable interest in elucidating this process in more detail.
New findings clearly establish the central role of the apical caspase Dronc in the induction of ‘compensatory proliferation” [40]. Dronc mutants, but not Drice mutants, suppress compensatory proliferation induced by γ-irradiation or by expression of apoptotic proteins. These findings establish a bifurcation downstream of Dronc in the regulation of apoptosis and “compensatory proliferation”. One branch leading to Drice cleavage and execution of apoptosis, and a second branch, independent of Drice, leading to the induction of “compensatory proliferation” [40]. Interestingly, p53 appears to be an important component of this second branch downstream of Dronc [41]. p53 is required for the induction of compensatory proliferation and is transcriptionally activated in “undead cells” by a mechanism that requires Dronc function [41]. Further investigations will certainly focus on the mechanism by which Dronc activates p53 transcription.
Cells can also undergo “compensatory apoptosis”, when rapidly proliferating cells induce apoptosis of their neighbors. For example, Drosophila cells expressing high levels of the proto-oncogene Myc proliferate faster and induce apoptosis in neighboring cells with lower levels of Myc [42,43]. This property is known as “cell competition” and is thought to occur due to the competition between fast-proliferating (Myc positive) and neighboring cells for growth and survival signals. Decapentaplegic (Dpp) seems to be an important survival signal in this context, since experimental activation of Dpp signaling in the out-competed cells (low Myc) can rescue these cells from apoptosis [43]. In addition, Dpp as also been associated with “morphogenetic apoptosis” [44], which occurs when there is an experimentally induced disruption of a Dpp signaling gradient between juxtaposed cells. Sharp boundaries of Dpp signaling during normal development can induce reaper-dependent apoptosis to sculpt the joint in the Drosophila leg [45] (see also “Patterning signals and proliferation in Drosophila imaginal discs” by Nick Baker in this issue).
Caspases without death
While it has long been know that certain caspases have non-apoptotic functions, a number of recent studies have illustrated the role of “apoptotic” caspases in cell differentiation or cellular morphogenesis [46,47]. For example, caspase activity is required for the removal of the bulk cytoplasm from developing spermatids in Drosophila [48]. Although caspase activation in this system does not lead to the death of the entire cell, sperm individualization resembles apoptosis in the sense that many cellular structures are degraded. Notably, the activation of apoptotic effector caspases in this system strictly requires a testis-specific form of Cytochrome C [48, 9]. This contrasts with the finding that Cytochrome C appears to only accelerate developmental apoptosis in the Drosophila retina [10]
Apoptotic proteins can also help alter the shape of neurons by dendrite pruning, the local degeneration of dendrites that occurs during Drosophila metamorphosis. Dendrite pruning requires the degradation of Diap1 mediated by Ubcd1 (an E2 ubiquitin-conjugating enzyme) and activation of Dronc [49,50]. Like in spermatid individualization, the apoptotic machinery is also used in a spatially restricted way to destroy only parts of a cell. The precise regulatory mechanisms to achieve this local activation of caspases are still poorly understood. Diap1, Dronc and Dark (Apaf1 homologue) have also a non-apoptotic role during border cell migration in the Drosophila ovary [51].
Another example of the non-apoptotic use of caspases is the specification of neural precursor cells in Drosophila [Kuranaga and Miura, 2007]. The Drosophila IKK-related kinase (DmIKKε) was identified as a new regulator of Diap1 protein degradation [52]. DmIKKe directly binds and phosphorylates Diap1, promoting Diap1 degradation. Loss of function of DmIKKε by RNAi led to the upregulation of Diap1 protein levels, and resulted in the formation of extra macrochaetae due to reduced caspase activity. However, cell death during development was not affected by the loss of DmIKKε function [52]. Regulation of Diap1 by DmIKKε is also important for organization of the actin cytoskeleton and changes in cell morphology [53]. Collectively, these studies raise the interesting question of how the potentially lethal activity of apoptotic proteins is restricted to specific compartments within a cell.
Conclusions
Although in a few select cases the signaling pathways regulating cell death and their intersection with the core apoptotic program are known in considerable detail, in general we still do not know much about how a particular cell chooses between life and death during normal development. Given the terminal nature of cell death and the potentially catastrophic consequences of mistakes, it should not come as a surprise that many distinct layers of control are used to tightly regulate the activity of caspases. This includes transcriptional regulation of caspases, activation of caspases by apoptosome formation, degradation of pro-caspases (such as Dronc) by IAPs and the ubiquitin-proteasome system, activation of caspases through inhibition of IAPs by Reaper-family proteins, regulation of core cell death proteins by phosphorylation, micro-RNAs, and by changing their subcellular localization. Furthermore, it has become increasingly clear that apoptotic cells can actively communicate with their cellular environment, and not just to attract phagocytes, but also to stimulate cell proliferation and tissue regeneration. Finally, core components of the cell death program, including apoptotic effector caspases, were found to have non-apoptotic roles in cellular remodeling, differentiation and cell specification. With each advance many more new questions have emerged that will undoubtedly keep the field busy for years to come.
Acknowledgments
We thank Hyung Don Ryoo and César S. Mendes for critically reading this manuscript. Hermann Steller is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Tittel JN, Steller H. A comparison of programmed cell death between species. Genome Biol. 2000;1:REVIEWS0003. doi: 10.1186/gb-2000-1-3-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 3.Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 4.Song Z, Steller H. Death by design: mechanism and control of apoptosis. Trends Cell Biol. 1999;9:M49–52. [PubMed] [Google Scholar]
- 5.Kaufmann SH, Hengartner MO. Programmed cell death: alive and well in the new millennium. Trends Cell Biol. 2001;11:526–534. doi: 10.1016/s0962-8924(01)02173-0. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y. Mechanical aspects of apoptosome assembly. Curr Opin Cell Biol. 2006;18:677–684. doi: 10.1016/j.ceb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhou L, Steller H. Distinct pathways mediate UV-induced apoptosis in Drosophila embryos. Dev Cell. 2003;4:599–605. doi: 10.1016/s1534-5807(03)00085-6. [DOI] [PubMed] [Google Scholar]
- 9.Arama E, Bader M, Srivastava M, Bergmann A, Steller H. The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. Embo J. 2006;25:232–243. doi: 10.1038/sj.emboj.7600920. • A testis specific form of Cytochrome C is strictly required for activation of caspases during terminal differentiation of spermatids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendes CS, Arama E, Brown S, Scherr H, Srivastava M, Bergmann A, Steller H, Mollereau B. Cytochrome c-d regulates developmental apoptosis in the Drosophila retina. EMBO Rep. 2006;7:933–939. doi: 10.1038/sj.embor.7400773. • Cytochrome C is required for "on time" death of the extra lattice cells in the Drosophila eye. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ditzel M, Meier P. Ubiquitylation in apoptosis: DIAP1's (N-)en(d)igma. Cell Death Differ. 2005;12:1208–1212. doi: 10.1038/sj.cdd.4401711. [DOI] [PubMed] [Google Scholar]
- 12.Lockshin RA. Programmed cell death. Activation of lysis by a mechanism involving the synthesis of protein. J Insect Physiol. 1969;15:1505–1516. doi: 10.1016/0022-1910(69)90172-3. [DOI] [PubMed] [Google Scholar]
- 13.Oppenheim RW, Prevette D, Tytell M, Homma S. Naturally occurring and induced neuronal death in the chick embryo in vivo requires protein and RNA synthesis: evidence for the role of cell death genes. Dev Biol. 1990;138:104–113. doi: 10.1016/0012-1606(90)90180-q. [DOI] [PubMed] [Google Scholar]
- 14.Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 15.Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, Horvitz HR. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- 16.Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Strauss TJ, Potts MB, Cameron S. Direct regulation of egl-1 and of programmed cell death by the Hox protein MAB-5 and by CEH-20, a C. elegans homolog of Pbx1. Development. 2006;133:641–650. doi: 10.1242/dev.02234. [DOI] [PubMed] [Google Scholar]
- 18.Maurer CW, Chiorazzi M, Shaham S. Timing of developmental cell death onset controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development. 2007;134:1357–1368. doi: 10.1242/dev.02818. •• The transcriptional activation of ced-3 is under PAL-1 control and is required for the timely death of the tail-spike cell. [DOI] [PubMed] [Google Scholar]
- 19.Park D, Jia H, Rajakumar V, Chamberlin HM. Pax2/5/8 proteins promote cell survival in C. elegans. Development. 2006;133:4193–4202. doi: 10.1242/dev.02614. [DOI] [PubMed] [Google Scholar]
- 20.Cakouros D, Daish TJ, Kumar S. Ecdysone receptor directly binds the promoter of the Drosophila caspase dronc, regulating its expression in specific tissues. J Cell Biol. 2004;165:631–640. doi: 10.1083/jcb.200311057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilpatrick ZE, Cakouros D, Kumar S. Ecdysone-mediated up-regulation of the effector caspase DRICE is required for hormone-dependent apoptosis in Drosophila cells. J Biol Chem. 2005;280:11981–11986. doi: 10.1074/jbc.M413971200. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. •• The transcription factor Yorkie is regulated by the Hippo signaling pathway. Yorkie is phosphorylated and inactivated by Warts. Yorkie is required for Diap 1 transcription. [DOI] [PubMed] [Google Scholar]
- 23.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Hariharan IK, Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu Rev Genet. 2006;40:335–361. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- 25.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. •• Together with 26, 27 and 28. The authors identified the protochaderin Fat as an upstream regulator of the Hippo signaling pathway. Discs overgrown and Dachs are part of the Fat pathway that regulates Warts protein levels. [DOI] [PubMed] [Google Scholar]
- 26.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. •• see anotation 25. [DOI] [PubMed] [Google Scholar]
- 27.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. •• see anotation 25. [DOI] [PubMed] [Google Scholar]
- 28.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. •• see anotation 25. [DOI] [PubMed] [Google Scholar]
- 29.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. • The tumour supressors Merlin and Expanded are upstream regulators of the Hippo signaling pathway. [DOI] [PubMed] [Google Scholar]
- 30.Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. • The tumour supressors Merlin and Expanded regulate the endocytosis of several signaling receptors, including Fat, Notch, EGFR, Patched and Smoothened. [DOI] [PubMed] [Google Scholar]
- 31.Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. • Vps25 is a component of the ESCRT complex and is required for protein sorting at the early endossome. vps25 mutant cells dye by apoptosis and induce non autonomous cell proliferation and survival. Hippo signaling is upregulated in vps25 mutant cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. •• Together with 33, the microRNA bantam is a target of Yorkie and the Hippo pathway. bantam overexpression in sufficient to rescue survival of yorkie mutant cells. [DOI] [PubMed] [Google Scholar]
- 33.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. •• see anotation 32. [DOI] [PubMed] [Google Scholar]
- 34.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 35.Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:097–1108. doi: 10.1016/j.cell.2005.04.016. • Oligoribonucleotide-mediated depletion studies for several microRNA expressed during Drosophila development. The miR-2 family of microRNAs regulates the expression of the pro-apoptotic genes hid, grim and reaper. [DOI] [PubMed] [Google Scholar]
- 36.Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–6187. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 37.Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Garijo A, Martin FA, Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131:5591–5598. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- 39.Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 40.Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M. DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol. 2006;26:7258–7268. doi: 10.1128/MCB.00183-06. • The initiator caspase Dronc is required for apoptosis and compensatory proliferation. The effector caspase Drice is required for apoptosis but not for compensatory proliferation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells BS, Yoshida E, Johnston LA. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol. 2006;16:1606–1615. doi: 10.1016/j.cub2006.07.046. • Activation of p53 expression is downstream of Dronc and is required for compensatory proliferation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 43.Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 44.Adachi-Yamada T, O'Connor MB. Mechanisms for removal of developmentally abnormal cells: cell competition and morphogenetic apoptosis. J Biochem (Tokyo) 2004;136:13–17. doi: 10.1093/jb/mvh099. [DOI] [PubMed] [Google Scholar]
- 45.Manjon C, Sanchez-Herrero E, Suzanne M. Sharp boundaries of Dpp signalling trigger local cell death required for Drosophila leg morphogenesis. Nat Cell Biol. 2007;9:57–63. doi: 10.1038/ncb1518. •• Discontinuities of Dpp signaling activate JNK/reaper-dependent apoptosis, generating the joints of the Drosophila leg. First evidence that apoptosis induced by sharp boundaries of Dpp signaling is used during normal development in a morphogenetic process. [DOI] [PubMed] [Google Scholar]
- 46.Abraham MC, Shaham S. Death without caspases, caspases without death. Trends Cell Biol. 2004;14:184–193. doi: 10.1016/j.tcb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Kuranaga E, Miura M. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 2007 doi: 10.1016/j.tcb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 48.Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 49.Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. •• Together with 50. Regulation of Diap1 protein levels by the E2 ubiquitin-conjugating enzime ubcD1 is important for the process of dendrite pruning during Drosophila metamorphosis. The caspase Dronc is also involved in this process and caspase activity is exclusively observed in the dendrites to be pruned. Another example of a non-apoptotic function of caspases. [DOI] [PubMed] [Google Scholar]
- 50.Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci. 2006;9:1234–1236. doi: 10.1038/nn1774. •• see anotation 49. [DOI] [PubMed] [Google Scholar]
- 51.Geisbrecht ER, Montell DJ. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell. 2004;118:111–125. doi: 10.1016/j.cell.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 52.Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, Kobayashi M, Hayashi S, Miura M. Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell. 2006;126:583–596. doi: 10.1016/j.cell.2006.05.048. • Together with 53. Mutations in Drosophila IKK-related kinase were identified as suppressors of reaper killing activity in the eye. DmIKK-ε phophorylates and promotes Diap1 degradation. Knowdown of DmIKK-ε resulted in actin assembly and cell shape defects. DmIKK-ε regulates non-apoptotic functions of Diap1/Dronc. [DOI] [PubMed] [Google Scholar]
- 53.Oshima K, Takeda M, Kuranaga E, Ueda R, Aigaki T, Miura M, Hayashi S. IKK epsilon regulates F actin assembly and interacts with Drosophila IAP1 in cellular morphogenesis. Curr Biol. 2006;16:1531–1537. doi: 10.1016/j.cub.2006.06.032. • see anotation 52. [DOI] [PubMed] [Google Scholar]


