Abstract
The serine/arginine-rich (SR) protein splicing factor 2/alternative splicing factor (SF2/ASF) has a role in splicing, stability, export and translation of messenger RNA. Here, we present the structure of the RNA recognition motif (RRM) 2 from SF2/ASF, which has an RRM fold with a considerably extended loop 5 region, containing a two-stranded β-sheet. The loop 5 extension places the previously identified SR protein kinase 1 docking sequence largely within the RRM fold. We show that RRM2 binds to RNA in a new way, by using a tryptophan within a conserved SWQLKD motif that resides on helix α1, together with amino acids from strand β2 and a histidine on loop 5. The linker connecting RRM1 and RRM2 contains arginine residues, which provide a binding site for the mRNA export factor TAP, and when TAP binds to this region it displaces RNA bound to RRM2.
Introduction
The characteristic of canonical serine/arginine-rich (SR) proteins is a carboxy-terminal domain rich in serines and arginines, preceded by either single or tandem RNA recognition motifs (RRMs). The SR proteins recognize and bind to sequences within messenger RNA, known as exonic splice enhancers (ESEs; Smith et al, 2006), together with certain intronic sequences (Kanopka et al, 1996). These interactions have wide ranging downstream effects on mRNA, including splice site choices, stability, export and translation (Sanford et al, 2005a). Recognition of ESEs is controlled by the RRMs, whereas the RS domain acts as a protein interaction module and makes direct contacts with the pre-mRNA during assembly of the spliceosome (Hertel & Graveley, 2005).
The SR protein splicing factor 2/alternative splicing factor (SF2/ASF) has two RRMs connected by a glycine- and arginine-rich linker. Although each RRM binds to RNA independently, they act synergistically for optimal interaction with RNA (Caceres & Krainer, 1993; Zuo & Manley, 1993), and both RRMs contribute to the RNA-binding specificity of the protein (Tacke & Manley, 1995). RRM2 is unusual as crucial residues within the ribonucleoprotein RNP-1 and RNP-2 motifs normally associated with RNA binding are missing; therefore, how it binds to RNA is unclear. SF2/ASF, together with the SR proteins 9G8 and SRP20, interact with the TIP associated protein (TAP) mRNA export factor (Huang et al, 2003; Hargous et al, 2006). This interaction is regulated by the phosphorylation status of the RS domain and only hypophosphorylated forms, generated during splicing, bind to TAP (Huang et al, 2004; Lai & Tarn, 2004). The cytoplasmic rephosphorylation of SR proteins is controlled by two protein kinases: SR protein kinase 1 (SRPK1), which phosphorylates serines adjacent to RRM2; and Clk/Sty (Clk for CDC-like kinase), which phosphorylates more distal serines (Ngo et al, 2005). Here, we present the structure of SF2/ASF RRM2 and identify amino acids involved in binding RNA and TAP.
Results And Discussion
Structural overview
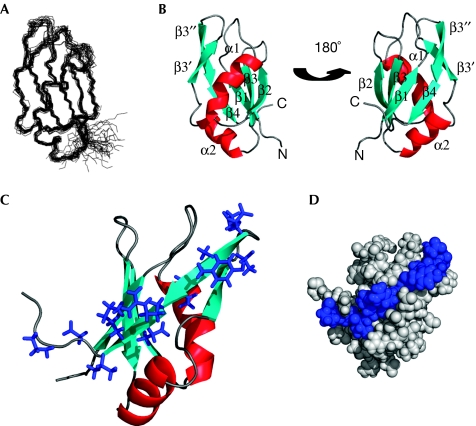
The solution structure of SF2/ASF amino acids (aa) 107–215, encompassing RRM2, has been determined by nuclear magnetic resonance (NMR) spectroscopy (Fig 1; Table 1). The fold is similar to typical RRMs (Maris et al, 2005) and contains and a four-stranded β-sheet packed against two helices. In common with certain RRMs, loop 5 is extended by a two-stranded β-sheet (β3′ and β3″), and this extension is longer than in most other RRMs with these elements. This is highlighted in supplementary Fig S1 online, in which the SF2/ASF fold is overlaid with structurally related proteins identified using the DALI server (Holm & Sander, 1993). In this overlay, only 2 of the 44 other folds contain a similarly large loop, 1UW4 and 2DNL. Residues from 107–120 and from 196–215 are disordered in the structure calculations, and increased mobility of this section is confirmed by 15N relaxation measurements (supplementary Fig S2 online).
Figure 1.
Structure of SF2/ASF RNA-recognition motif 2. (A) Superposition of the 20 lowest energy structures of SF2/ASF RRM2 (amino acids 107–215). (B) Ribbon representation of SF2/ASF RRM2 shown in two orientations. (C) SRPK1 docking motif (blue side chains) highlighted on the RRM2 structure. (D) Space-filled model of RRM2 showing the SRPK1 docking motif in blue. RRM2, RNA-recognition motif 2; SF2/ASF, splicing factor 2/alternative splicing factor; SRPK1, serine/arginine-rich protein kinase 1.
Table 1.
Statistics for an ensemble of the 20 lowest energy structures of SF2/ASF amino acids 107–215
| NOE distances | |
|---|---|
| Intraresidue | 349 |
| Sequential | 248 |
| Medium range | 111 |
| Long range (Δ>4) | 254 |
| Dihedral restraints | 76 |
| Hydrogen bond restraints | 48 |
| NOE violations >0.2 Å | 3 |
| Dihedral restraint violation >2.5° | 0 |
| R.m.s. deviation from mean structure (Å)* | |
| Backbone heavy atoms | 0.6 |
| All heavy atoms | 1.3 |
| Ramachandran analysis* | |
| Most favoured regions (%) | 83.5 |
| Additionally allowed regions (%) | 16 |
| Generously allowed regions (%) | 0.6 |
| Disallowed regions (%) | 0.4 |
*Amino acids 121–195. SF2/ASF, splicing factor 2/alternative slicing factor.
SRPK1 docking sequence
Several database entries, for example, GenBank accession number AAH33785, incorrectly annotate the boundaries of SF2/ASF RRM2 as aa 124–185, which has significant consequences. In particular, a docking motif, which contributes to the interaction between SRPK1 and SF2/ASF was defined between aa 184 and 197, and was assumed to lie between RRM2 and the RS domain (Ngo et al, 2005). A C-terminal truncation lacking these residues in SF2/ASF (ΔCT; aa 1–179) or an internal deletion (ΔDOCK; deletion of aa 184–197) caused a marked reduction in the SRPK1 interaction (Ngo et al, 2005). The SF2/ASF structure shows that the SPRK1 docking motif (Fig 1C,D) extends significantly into the RRM fold, which finishes at D195. Therefore, in the ΔDOCK and ΔCT mutants, strands β3″ and β4 are deleted, and in the ΔCT mutant strands β3′ and loop 5 are also deleted. This removes a major section of the folded portion of RRM2, including four residues from the hydrophobic core (Fig 2B), which is likely to disrupt the fold. Therefore, the extent of contribution of the docking motif to the interaction with SRPK1 cannot be determined reliably on the basis of these truncations. Notably, R191A/K193A mutations within the docking sequence in SF2/ASF only led to a modest reduction in the SRPK1 co-precipitating. Furthermore, high concentrations (10 mM) of a peptide derived from the docking sequence were required to inhibit SF2/ASF phosphorylation by SRPK1 in vitro (Ngo et al, 2005). Together, this suggests that other regions of RRM2 contribute to the interaction with SRPK1. The β-sheet surface, which includes the docking motif, might provide further contacts with SRPK1, in a manner analogous to other RRM modules such as Y14, which mediates interactions with MAGOH through the β-sheet surface (Fribourg et al, 2003; Lau et al, 2003; Shi & Xu, 2003).
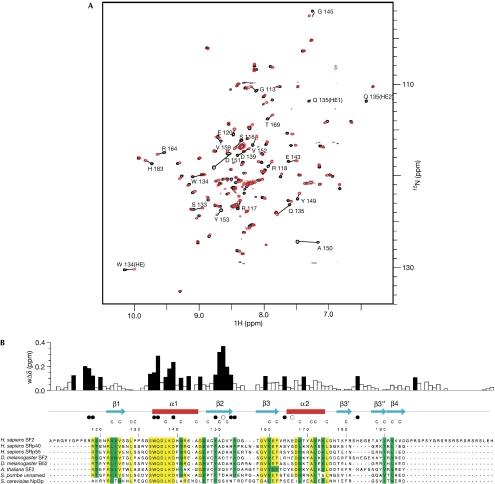
Figure 2.
Interaction of SF2/ASF RNA-recognition motif 2 with RNA. (A) Chemical shift perturbation following the addition of 5′-ACGA RNA to 15N-labelled RRM2. The heteronuclear single quantum correlation (HSQC) spectrum of free protein (black) and after the addition of 1.75 molar equivalents of RNA (red) are shown. Amino acids showing marked changes are indicated. (B) Magnitude of weighted amide chemical shift perturbations (wΔδ) following the addition of 5′-ACGA RNA to 15N-labelled RRM2. Mapping of these perturbations to secondary structural elements are shown above the SF2/ASF sequence alignment with related proteins. Residues with chemical shift changes of more than 0.1 p.p.m. are shown by black bars. The side chain Hɛ resonance of W134 also shows a sizeable shift of more than 0.1 p.p.m., and the side chain Hɛ of Q135 is unobservable, following the first addition of ligand, which indicates exchange broadening owing to a large chemical shift change. Filled circles below the secondary structural elements represent amino acids directly involved in RNA binding identified by mutagenesis, and open circles represent amino acids in which mutagenesis does not affect RNA binding. Residues that form the core of the RRM fold are denoted by the letter C above the sequences. Within the sequence alignments, residues highlighted in yellow are strongly conserved and residues in green have conservative substitutions. RRM, RNA recognition motif; SF2/ASF, splicing factor 2/alternative splicing factor.
RNA binding
To investigate how SF2/ASF RRM2 binds to RNA, we followed chemical shift perturbations with an SF2/ASF consensus binding motif 5′-CACACGA, defined as a high-scoring site using an ESE score matrix (Cartegni & Krainer, 2002; Smith et al, 2006). In titrations with this sequence, peaks in the heteronuclear single quantum correlation (HSQC) spectrum of RRM2 shifted gradually with increasing RNA concentration, indicating predominantly fast exchange. Although some peaks could be followed up to a stoichiometry approaching 1:1, other peaks became rapidly attenuated, with some crosspeaks attenuated below detection level (supplementary Fig S3A online). This suggests that binding is approaching intermediate exchange for shift changes of approximately 200 Hz, which, with the assumption of a diffusion limited on rate of 1 × 108 M−1 s−1, indicates a dissociation constant in the low micromolar range. In addition, a common reduction in the intensity of all peaks was observed so that the HSQC spectrum became weak as a 1:1 stoichiometry was approached, suggesting aggregation was occurring. This might have been triggered by the binding of two molecules of RRM2 with a single RNA molecule.
To avoid these effects, the titration was carried out with a sub-sequence of the SF2/ASF consensus, 5′-ACGA (Fig 2A). In this titration, virtually all peaks could be followed beyond a 1:1 stoichiometry, and general attenuation of peaks was reduced to a loss of approximately 20% at 1:1 stoichiometry. Notably, 5′-ACGA influenced chemical shifts for the same amino acids as those affected by 5′-CACACGA RNA (supplementary Fig S3A online). The magnitudes of chemical shift changes observed in the titration of RRM2 with 5′-ACGA are shown in Fig 2B, and a fit of data for chemical shift changes against ligand concentration gave an estimated binding constant of 60±30 μM for this interaction (supplementary Fig S3B online). The amino acids showing the largest chemical shifts include conserved residues S133, W134, Q135, K138 and D139 in helix α1, and Y149–Y153 from strand β2; these amino acids cluster on one surface of RRM2 (Fig 3B). Several other amino acids discussed below show chemical shifts of more than 0.1 p.p.m., suggesting an involvement in RNA binding.
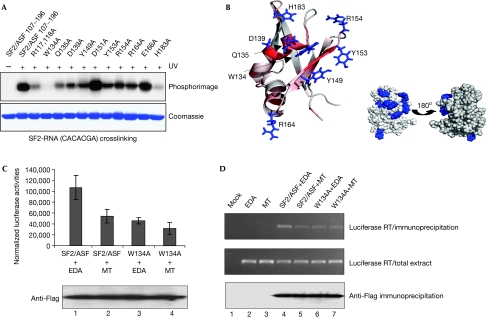
Figure 3.
Mutagenesis defines SF2/ASF residues required for RNA binding. (A) UV crosslinking with RNA 5′-CACACGA and SF2/ASF RRM2 (aa 107–215) and point mutants. The top panel shows a phosphorimage of the Coomassie blue-stained gel shown in the bottom panel. (B) Side chains of amino acids required for RNA binding are shown in blue (left). The ribbon representation of the structure is also coloured according to the magnitude of chemical shifts seen on the addition of RNA 5′-ACGA (red=large). The amino acids required for RNA binding (blue) map to a localized patch on the space-filled model of the structure (right). (C) W134A mutation in RRM2 reduces translational activation in vivo. The pLCS-EDA reporter carries an SF2-binding site sequence and is translationally activated by overexpression of SF2/ASF. The pLCS-EDAmt carries a mutated SF2-binding site, which shows poor activation by SF2/ASF (Sanford et al, 2004, 2005b). pLCS-EDA (EDA) or pLCS-EDAmt (MT) plasmids were co-transfected with Flag-SF2/ASF (aa 11–196) or the W134A mutant into 293T cells and luciferase activity was measured after 24 h. Transfection efficiencies were normalized by co-transfection of a cytomegalovirus (CMV)-driven β-galactosidase expression vector; results represent averages from transfections carried out in triplicate. Flag-tagged SF2/ASF proteins were detected by western blotting (lower panel). (D) W134A mutation in RRM2 reduces the interaction with pLCS-EDA messenger RNA in vivo. 293T cells were transfected with the indicated plasmids, immunoprecipitated with anti-Flag–agarose, RNA extracted and assayed by RT–PCR. The top panel shows the results of RT–PCR analysis from immunoprecipitates, the middle panel results from total extracts before immunoprecipitation and the bottom panel a western blot of immunoprecipitates using Flag antibodies. RRM, RNA recognition motif; RT–PCR, reverse transcriptase–PCR; SF2/ASF, splicing factor 2/alternative splicing factor.
As chemical shifts can arise from indirect effects, we investigated the contribution of selected amino acids to RNA binding by using UV crosslinking (Fig 3A). We were unable to detect a UV crosslink between 5′-ACGA and RRM2, perhaps because RNA titration data indicate that it binds less tightly than 5′-CACACGA; therefore we used 5′-CACACGA. Mutation of surface-exposed residues in helix α1 reduced RNA binding for Q135A and D139A, and led to a complete loss of binding for W134A, indicating that W134A is essential for interaction with RNA. The HSQC spectrum for W134A showed no significant perturbations, indicating that this mutation had not disrupted the protein fold (supplementary Fig S4 online). Interestingly, the amino acids SWQLKD from helix α1 were originally identified as part of an invariant signature motif for SR proteins containing a second RRM (Birney et al, 1993), their strong conservation no doubt being influenced by their importance for RNA binding.
Some residues in the β2 strand showed large chemical shifts, including Y149, A150 and D151. Mutagenesis of Y149 showed a significant reduction in RNA binding, indicating its involvement, whereas mutagenesis of D151, which shows a large chemical shift, had no effect on the RNA crosslink (Fig 3A). This suggests that the chemical shift on D151 is an indirect effect caused by the surrounding residues binding to RNA. Notably, Y149, which is surface exposed, is conserved in SF2/ASF-related proteins as either tyrosine or phenylalanine, whereas many other strongly conserved hydrophobic residues—with the exception of W134—contribute to the hydrophobic core of SF2/ASF (Fig 2B). This level of conservation implies that Y/F149 is used by SF2/ASF-related proteins for RNA binding and might be involved in stacking interactions with RNA. The amino acids Y153 and R154 form part of loop 3, and mutation of either of these residues reduces the UV crosslink with RNA. R164 is also required for optimal RNA binding, whereas the nearby residue E166 is not required. Finally, H183, which forms part of loop 5, has a crucial role in RNA binding, as its mutation markedly reduces the ability of RRM2 to crosslink with RNA. Although most of the residues required for efficient interaction with RNA in human SF2/ASF are conserved in human SRP40 and SRP55 proteins, these latter proteins have no equivalent of H183; furthermore, loop 5 is truncated in these larger SR proteins. Therefore, the loop 5 extension in SF2/ASF probably acts to correctly position H183 for RNA binding.
As UV crosslinking is sensitive to the availability and accessibility of suitable functional groups, we investigated the effects of W134A and H183A mutations on RNA binding by using NMR titrations (supplementary Fig S4 online). These studies showed that both these mutations impair the ability of RRM2 to bind to RNA, and W134A showed the most profound reduction, which is consistent with the UV-crosslinking results. The mutation R117,118A, which shows significant chemical shifts with RNA (Fig 2A), reduces the ability of SF2/ASF to crosslink with RNA and bind to RNA in NMR titrations (Fig 3A; supplementary Fig S4 online). However, the HSQC spectrum for R117/118A showed several chemical shift perturbations compared with the wild-type protein, which suggests that the otherwise unstructured region around R117,118 makes transient interactions with the folded RRM2 domain. Loss of these transient interactions could account for weakened RNA binding rather than the loss of direct interactions between R117,118 and RNA. However, the involvement of a linker connecting RRM motifs in RNA binding is not unprecedented; for example, the linker between the RRMs of nucleolin interacts with five nucleotides (Allain et al, 2000). Together, the amino acids of RRM2 showing large chemical shifts on RNA binding and reduced RNA crosslinking activity when mutagenized cluster on the same face of the protein (Fig 3B) and define a new mode for RNA interaction with an RRM.
To confirm the importance of W134 for SF2/ASF function in vivo, we examined whether this mutation affected the ability of SF2/ASF to bind to and activate expression of a reporter mRNA (rmRNA) containing an SF2/ASF-binding site (Sanford et al, 2005b; Fig 3C). SF2/ASF (aa 11–196) was able to activate expression of this rmRNA in vivo, and activated an rmRNA with a mutated SF2/ASF-binding site poorly. By contrast, the W134A mutant was unable to activate expression of wild-type or mutant rmRNAs, which is consistent with the observation that a WQD-AAA mutant was not functional in this assay (Sanford et al, 2005b). To investigate whether this was due to a reduced interaction with rmRNA, we immunoprecipitated SF2/ASF and assessed the amount of bound rmRNA (Fig 3D). Wild-type SF2/ASF bound rmRNA efficiently but interacted weakly with mutated rmRNA. By contrast, W134A showed a weaker interaction with both wild-type and mutant rmRNAs despite the presence of RRM1. This is consistent with studies showing that RRM1 and RRM2 are required for optimal interaction with SF2/ASF-binding sites on mRNA (Caceres & Krainer, 1993; Zuo & Manley, 1993) and that the substrate specificity of SF2/ASF results from the combined action of both RRMs (Tacke & Manley, 1995). However, SF2/ASF carrying a WQD-AAA RRM2 mutation still retains some RNA-binding activity in a messenger ribonucleoprotein capture assay presumably through RRM1 and the linker (Sanford et al, 2005b).
SF2/ASF interaction with TAP
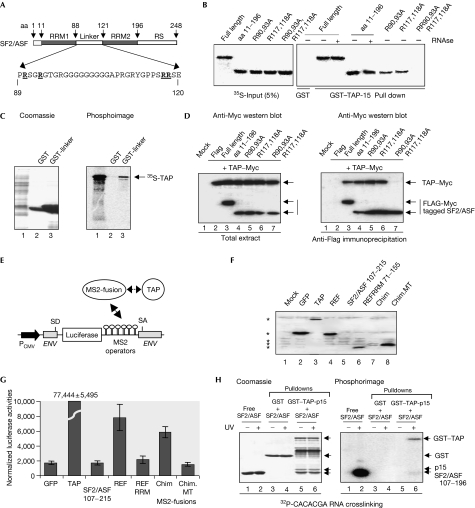
Both 9G8 and SRp20 use arginines within peptides adjacent to RRMs to bind to TAP (Hargous et al, 2006). Furthermore, the SF2/ASF RS domain is not required for the interaction with TAP (Huang et al, 2003); therefore candidate peptides for binding TAP in SF2/ASF are aa 1–11 preceding RRM1, containing a single arginine, and the linker connecting RRM1 and RRM2, containing multiple arginines (Fig 4A). SF2/ASF (aa 11–196) bound to TAP and also to full-length SF2/ASF, indicating that aa 1–11 were not involved (Fig 4B). Therefore, we mutated arginines within the linker region connecting RRM1 and RRM2, and assayed their ability to bind to glutathione-S-transferase (GST)–TAP-p15 (Fig 4B). The mutation R90,93A reduced the amount of SF2/ASF bound to GST–TAP-p15, as did mutation of R117/118A, and the combination of these mutations completely blocked TAP binding. Given that mutation of R117,118A influenced the NMR spectrum for RRM2 (supplementary Fig S4 online), we wanted to exclude the possibility that indirect alterations to the RRM2 structure might influence the interaction with TAP. Therefore, we confirmed that the isolated linker (aa 89–120) was sufficient for the interaction with TAP (Fig 4C). Furthermore, we showed that R90,93,117,118 were required for the interaction between SF2/ASF (aa 11–196) and TAP in vivo by using a co-immunoprecipitation assay (Fig 4D). We conclude that TAP binding to SF2/ASF involves R90,93,117,118 within the linker region connecting RRM1 and RRM2.
Figure 4.
TAP binds to the SF2/ASF linker between RRM1 and RRM2. (A) Schematic of SF2/ASF and linker region sequence. Mutated arginine residues are underlined. (B) Pull-down assays with GST–TAP-p15, and 35S-SF2/ASF truncations and point mutants. (C) Pull-down assays with GST–SF2/ASF linker and 35S-TAP. (D) Co-immunoprecipitation of SF2/ASF mutants with TAP. SF2/ASF has Flag and Myc tags; TAP is Myc tagged. Total extracts (left) and Flag immunoprecipitates (right) were probed with Myc antibody. (E) Schematic of tethered messenger RNA export assay. (F) Western blot detection of Myc-tagged MS2 fusions as indicated by using Myc antibodies. Specific bands are highlighted with asterisks. Chim pLCS-EDAmt (MT) is the same construct carrying R90,93,117,118A mutations. (G) Normalized luciferase activities for MS2 fusions indicated in the tethered export assay. (H) UV crosslinking assay with RNA, GST–TAP-p15 and SF2/ASF (107–196). The right panel shows a phosphorimage of the Coomassie-stained gel (left panel). For all lanes, 32P-labelled RNA was added to the samples at the start together with SF2/ASF (107–196). Lanes 1+2 contain free SF2/ASF; lanes 3–6 show GST pull-downs subsequently eluted from beads and then UV crosslinked as indicated. Chim, REF-RRM-SF2/ASF linker peptide chimaera; ENV, HIV-1 envelope gene; GFP, green fluorescent protein; GST, glutathione-S-transferase; MS2, bacteriophage MS2 coat protein; PCMV, cytomegalovirus promoter; RRM, RNA recognition motif; SA, splice acceptor; SD, splice donor; SF2/ASF, splicing factor 2/alternative splicing factor; TAP, TIP associated protein.
To examine whether the TAP-binding peptide from SF2/ASF was a transferable signal, we assayed its ability to restore activity to a mutated form of the mRNA export adaptor REF2-I, which lacks the main TAP-binding site, in a tethered export assay. SR proteins do not function in this assay, presumably because direct tethering to the unspliced mRNA means that they evade splicing-dependent dephosphorylation, which is required for the interaction with TAP. However, the TAP-binding domains of 9G8 and SRp20 can restore the activity of an MS2–REF2-I fusion, which lacks its principal TAP-binding domain (Hargous et al, 2006). Similarly, the chimaera of REF2-I RRM and SF2/ASF linker peptide was active in this assay, despite poor expression (Fig 4F,G). By contrast, a chimaera with mutated arginine residues was not active, indicating that arginine residues involved in binding to TAP are required for the mRNA export activity of the REF2-I–SF2/ASF chimaera.
That R117/118 are necessary for efficient interaction with RNA and TAP (Fig 3A) raises the possibility that the TAP interaction with SF2/ASF might be mutually exclusive with RNA binding. Therefore, we analysed the ability of SF2/ASF RRM2 (107–196) to bind to RNA alone and in complex with GST–TAP-p15. In isolation, SF2/ASF (107–196) showed a strong interaction with RNA (Fig 4H). However, when a complex was formed between GST–TAP-p15 and SF2/ASF (107–196), the amount of RNA crosslinked to SF2/ASF was markedly reduced, whereas TAP retained the ability to bind to RNA. Thus, TAP binding displaces RNA bound to SF2/ASF (107–196). Given the synergy between RRM1 and RRM2 for binding to RNA (Caceres & Krainer, 1993), we suggest that TAP binding to the linker might markedly alter the ability of SF2 RRM1 and RRM2 to interact with RNA, and might even destabilize binding completely. However, we were unable to test this directly in this assay because of the poor solubility of SF2/ASF (aa 11–196).
In conclusion, SF2/ASF RRM2 has a distinctive fold, involving an extended loop 5, placing most of the SRPK1 kinase-docking sequence within the β-sheet surface. The RRM2 fold uses new elements to interact with RNA together with amino acids from the linker, and arginine residues within the linker connecting RRM1 and RRM2 provide a docking site for TAP. TAP binding triggers displacement of RNA from RRM2, which might destabilize the interaction between full-length SF2/ASF and RNA, given the dependency of SF2/ASF on both RRMs for stable specific RNA–protein interactions.
Methods
SF2/ASF complementary DNA truncations were generated by PCR and subcloned into the vector pET24b. Details of oligonucleotides are available on request. Quikchange mutagenesis and GST pull-down assays were carried out as described previously (Hargous et al, 2006). RNA UV crosslinking was carried out with 32P-end-labelled RNA oligonucleotides (Dharmacon, Lafayette, CO, USA). The tethered mRNA export assay was carried out as described previously (Hargous et al, 2006). Immunoprecipitations used Flag–agarose (Sigma) and reverse transcriptase–PCR (RT–PCR) analysis on pLCS-EDA, and pLCS-EDAMT (Sanford et al, 2005a, 2005b) used PCR primers recognizing luciferase. The number of PCR cycles was kept within the linear range (25 cycles) by assessing a control RT–PCR reaction at various points up to 40 cycles. Samples of SF2/ASF RRM2 for NMR analysis were prepared as described previously (Tintaru et al, 2007). Details on NMR spectroscopy and structure calculations can be found in the supplementary information online.
Data deposition. The coordinates for SF2/ASF RRM2 have been deposited in Protein Data Bank, accession number 2O3D.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v8/n8/extref/7401031-s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank J. Caceres for providing plasmids. This work was supported by the Biotechnology and Biological Sciences Research Council.
References
- Allain FH, Gilbert DE, Bouvet P, Feigon J (2000) Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J Mol Biol 303: 227–241 [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 21: 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Krainer AR (1993) Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J 12: 4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30: 377–384 [DOI] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E (2003) A novel mode of RBD-protein recognition in the Y14–Mago complex. Nat Struct Biol 10: 433–439 [DOI] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH (2006) Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J 25: 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ, Graveley BR (2005) RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem Sci 30: 115–118 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stevenin J, Steitz JA (2003) SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Molecular Cell 11: 837–843 [DOI] [PubMed] [Google Scholar]
- Huang Y, Yario TA, Steitz JA (2004) A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci USA 101: 9666–9670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka A, Muhlemann O, Akusjarvi G (1996) Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381: 535–538 [DOI] [PubMed] [Google Scholar]
- Lai MC, Tarn WY (2004) Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J Biol Chem 279: 31745–31749 [DOI] [PubMed] [Google Scholar]
- Lau CK, Diem MD, Dreyfuss G, Van Duyne GD (2003) Structure of the Y14-Magoh core of the exon junction complex. Curr Biol 13: 933–941 [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH (2005) The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131 [DOI] [PubMed] [Google Scholar]
- Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G (2005) Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell 20: 77–89 [DOI] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Caceres JF (2004) A novel role for shuttling SR proteins in mRNA translation. Genes Dev 18: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Ellis J, Caceres JF (2005a) Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem Soc Trans 33: 443–446 [DOI] [PubMed] [Google Scholar]
- Sanford JR, Ellis JD, Cazalla D, Caceres JF (2005b) Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci USA 102: 15042–15047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xu RM (2003) Crystal structure of the Drosophila Mago nashi–Y14 complex. Genes Dev 17: 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR (2006) An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 15: 2490–2508 [DOI] [PubMed] [Google Scholar]
- Tacke R, Manley JL (1995) The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J 14: 3540–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintaru AM, Hautbergue GM, Hounslow AM, Lian LY, Wilson SA, Craven CJ (2007) Assignment of 1H, 13C and 15N resonances for ASF/SF2 RNA recognition motif 2. J Biomol NMR 38: 193. [DOI] [PubMed] [Google Scholar]
- Zuo P, Manley JL (1993) Functional domains of the human splicing factor ASF/SF2. EMBO J 12: 4727–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information