Abstract
Highly quantitative biomarkers of neurodegenerative disease remain an important need in the urgent quest for disease-modifying therapies. For Huntington's disease (HD), a genetic test is available (trait marker), but necessary state markers are still in development. In this report, we describe a large battery of transcriptomic tests explored as state biomarker candidates. In an attempt to exploit the known neuroinflammatory and transcriptional perturbations of disease, we measured relevant mRNAs in peripheral blood cells. The performance of these potential markers was weak overall, with only one mRNA, immediate early response 3 (IER3), showing a modest but significant increase of 32% in HD samples compared with controls. No statistically significant differences were found for any other mRNAs tested, including a panel of 12 RNA biomarkers identified in a previous report [Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, et al. (2005) Proc Natl Acad Sci USA 102:11023–11028]. The present results may nonetheless inform the future design and testing of HD biomarker strategies.
Keywords: state biomarker, RNA biomarker, gene expression profiling, polyglutamine disease, neurodegenerative disease
In 1993, the Huntington's Disease (HD) Collaborative Research Group identified a simple mutation, a CAG repeat expansion in exon 1 of the HD (or IT15) gene, to be the cause of HD. An intense ongoing effort has attempted to elucidate the cellular mechanisms through which this gene's protein product, huntingtin, evokes such a devastating neurodegenerative disorder. Despite its clear etiology at the genetic level, however, we are still far from understanding the multiple processes that determine the symptoms and fatal outcome of HD.
At the pathological level, postmortem studies of HD brain have revealed the pronounced death of medium spiny GABAergic projection neurons within the caudate nucleus and putamen. Although the most pronounced pathology is observed in the basal ganglia, cell death also occurs early in the cerebral cortex. In parallel with neuronal loss, there is a sustained activation of inflammatory processes, which manifest histologically as astrogliosis and microgliosis in the neostriatum, cortex, globus pallidus, and the adjoining white matter of HD brains (1). Microinflammation is a common feature of many neurodegenerative disorders including Alzheimer's disease, Parkinson's disease, dementia with Lewy bodies, amyotrophic lateral sclerosis, and prion diseases (reviewed in ref. 2). At the molecular level, both type I and type II immune pathway involvement has been observed (3), and the complement system has been implicated in the elimination of toxic proteinaceous aggregates (4). On the other hand, inflammatory responses mediated by glial cells have also been hypothesized to contribute to the dysfunction and death of neurons in neurodegenerative disorders, including HD (5, 6), and, as such, inflammation could be considered as an integral component of the pathogenic process.
The first symptoms of HD typically appear at an age between 30 and 40 years, after which patients die within ≈15–20 years. Early symptoms often comprise psychiatric abnormalities including depression, anxiety, sleep disorders, and irritability together with frontal and subcortical cognitive deficits. These disease manifestations are typically followed by motor symptoms including choreic movements, dystonia, and rigidity. Peripherally, weight loss is a common feature of HD, and there is increasing evidence of neuroendocrine dysfunction in HD (7, 8).
No therapy has been shown to delay disease onset or slow progression in humans. Thus, there is an urgent need for clinical trials to identify and validate such treatments. Studies in HD model systems suggest that identifying disease-modifying compounds is possible (9, 10), but screening these agents in human trials remains challenging. In large part, this difficulty is due to the slowness of disease progression relative to the time frame in which a trial is conducted. The current standard for the assessment of the clinical stage of HD patients is the United Huntington's Disease Rating Scale (UHDRS) (11, 12), a method limited in sensitivity and prone to subjectivity (13). The identification of robust HD state biomarkers would thus provide valuable assistance in objectively and sensitively monitoring disease onset and progression, which are the prerequisites for the validation of new therapeutic strategies. The search for biomarkers of HD and other neurodegenerative diseases encompasses many technologies, including neuroimaging, proteomic, metabonomic, and genomic approaches (reviewed in ref. 14).
The utility of HD state biomarkers for clinical trials depends not only on the sensitivity of the measure but also on the practicality of the sampling and testing, because repeated measures are required. Focusing on the analysis of samples that can be collected through rapid and minimally invasive procedures, such as the collection of peripheral blood, is thus a sound strategy from this perspective. The implementation of widely available and financially accessible detection technologies, such as assay by PCR, is also a desirable feature of a clinical biomarker test.
Although the identification of a valid HD biomarker does not necessarily depend on its connection to a central disease mechanism, it is attractive to explore molecular and cellular pathways previously implicated in disease. Transcription has been identified as a potential target of mutant huntingtin's toxicity, and previous transcriptomic studies of tissues from HD patients and HD model systems have demonstrated significant and progressive disease-related effects on gene expression (15–18). Here, we report a multipartite strategy to identify an RNA biomarker for HD in peripheral blood motivated by previous evidence for transcriptional dysregulation and immune system involvement in the pathogenesis of HD.
Results
Given that neuroinflammation is an established and progressive facet of HD pathology, we hypothesized that inflammation-related transcriptional changes might also be measured in HD blood. To address this question, we first performed microarray gene expression profiling analyses of lymphocyte samples from 12 moderate-stage HD patients and 10 controls (Table 1) using U133 Human Genome 2.0 Plus arrays (Affymetrix, Santa Clara, CA). Although gender-related differences could be identified by using the standard criterion of RMA limma FDR P < 0.05 (Table 2), no HD-related changes met statistically robust detection on a single gene testing basis (same criterion, Table 3). Analysis using a different normalization method and cutoff criteria (as per ref. 17) gave qualitatively similar results [see Materials and Methods and supporting information (SI) Tables 4 and 5]. Because our previous experience indicated that real changes are sometimes missed by these criteria, we nonetheless chose selected mRNAs showing top-ranked differential expression measures to carry into further testing (Table 3). Represented among those chosen were genes involved in immune response, cell cycle, and cell death pathways.
Table 1.
Human samples for gene expression analyses
| HD states | Female | Age range (mean age) | Male | Age range (mean age) | Total |
|---|---|---|---|---|---|
| QPCR assays for candidate biomarkers (whole-blood RNA) | |||||
| Control | 17 | 25–65 (44.9) | 13 | 29–64 (48.6) | 30 |
| Presymptomatic* | 2 | 26–27 (26.5) | 3 | 31–35 (33.0) | 5 |
| Early | 2 | 30–31 (30.5) | 2 | 23–34 (30.0) | 4 |
| Moderate | 17 | 25–64 (47.5) | 9 | 38–74 (52.4) | 26 |
| Advanced | 0 | 5 | 45–64 (56.8) | 5 | |
| Lymphocyte RNA samples for high-density microarrays | |||||
| Control | 5 | 30–68 (49.6) | 5 | 38–60 (47.2) | 10 |
| HD moderate | 8 | 36–61 (46.8) | 4 | 51–62 (56.8) | 12 |
| Human immune profiling low-density arrays (whole-blood RNA) | |||||
| Control | 5 | 35–46 (42.0) | 4 | 45–59 (50.5) | 9 |
| HD moderate | 6 | 35–45 (41.5) | 5 | 43–59 (50.4) | 11 |
Complementary analyses including presymptomatic cases can be found in SI Figs. 4–7.
*Results presented in text for manifest HD (excluding HD presymptomatic).
Table 2.
Probe sets detecting gender effects in high-density microarray analysis
| Probe set ID | P (nominal) | FDR P | log2 FC | Gene symbol | Gene title |
|---|---|---|---|---|---|
| 201909_at | 4.38E-12 | 2.99E-08 | 5.35 | RPS4Y1 | Ribosomal protein S4, Y-linked 1 |
| 205000_at | 3.24E-14 | 2.96E-10 | 4.30 | DDX3Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked |
| 206700_s_at | 2.33E-11 | 1.38E-07 | 4.01 | SMCY | Smcy homolog, Y-linked (mouse) |
| 223646_s_at | 2.53E-11 | 1.38E-07 | 2.73 | CYorf15B | Chromosome Y open reading frame 15B |
| 223645_s_at | 2.51E-12 | 1.96E-08 | 2.39 | CYorf15B | Chromosome Y open reading frame 15B |
| 214131_at | 3.39E-09 | 1.43E-05 | 2.29 | CYorf15B | Chromosome Y open reading frame 15B |
| 228492_at | 6.01E-11 | 2.74E-07 | 2.13 | USP9Y | Ubiquitin specific peptidase 9, Y-linked (fat facets-like, Drosophila) |
| 204409_s_at | 3.44E-11 | 1.71E-07 | 2.04 | EIF1AY | Eukaryotic translation initiation factor 1A, Y-linked |
| 205001_s_at | 7.53E-09 | 2.94E-05 | 2.02 | DDX3Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked |
| 206624_at | 7.54E-07 | 1.96E-03 | 1.51 | USP9Y | Ubiquitin specific peptidase 9, Y-linked (fat facets-like, Drosophila) |
| 232618_at | 1.95E-07 | 5.93E-04 | 1.49 | CYorf15A | Chromosome Y open reading frame 15A |
| 236694_at | 2.18E-05 | 4.58E-02 | 0.97 | CYorf15A | Chromosome Y open reading frame 15A |
| 204410_at | 7.60E-06 | 1.73E-02 | 0.88 | EIF1AY | Eukaryotic translation initiation factor 1A, Y-linked |
| 208067_x_at | 3.19E-08 | 1.16E-04 | 0.61 | UTY | Ubiquitously transcribed tetratricopeptide repeat gene, Y-linked |
| 244482_at | 2.19E-06 | 5.44E-03 | 0.51 | EIF1AY | Eukaryotic translation initiation factor 1A, Y-linked |
| 230760_at | 2.26E-07 | 6.49E-04 | 0.45 | ZFY | Zinc finger protein, Y-linked |
| 1570360_s_at | 1.22E-07 | 3.92E-04 | 0.37 | DDX3Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked |
| 211149_at | 2.81E-06 | 6.67E-03 | 0.33 | UTY | Ubiquitously transcribed tetratricopeptide repeat gene, Y-linked |
| 216342_x_at | 2.17E-05 | 4.58E-02 | −0.32 | LOC390183//LOC442162 | Similar to 40S ribosomal protein S4, X isoform |
| 213876_x_at | 5.19E-07 | 1.42E-03 | −0.75 | U2AF1L2 | U2 small nuclear RNA auxiliary factor 1-like 2 |
| 224589_at | 3.45E-08 | 1.18E-04 | −3.01 | XIST | X (inactive)-specific transcript |
| 224590_at | 1.98E-17 | 5.42E-13 | −5.20 | XIST | X (inactive)-specific transcript |
| 214218_s_at | 4.46E-17 | 8.12E-13 | −5.46 | XIST | X (inactive)-specific transcript |
| 221728_x_at | 2.18E-18 | 1.19E-13 | −5.72 | XIST | X (inactive)-specific transcript |
| 227671_at | 4.74E-15 | 5.19E-11 | −6.23 | XIST | X (inactive)-specific transcript |
| 224588_at | 4.01E-15 | 5.19E-11 | −7.15 | XIST | X (inactive)-specific transcript |
Table 3.
Candidate RNA biomarkers identified from human lymphocyte microarray data
| Pathway | Gene title | Gene symbol | Probe set | log2 FC | P (nominal)* |
|---|---|---|---|---|---|
| Immune response | Major histocompatibility complex, class II, DQ α 1 | HLA-DQA1 | 212671_s_at | −1.114 | 0.0091 |
| Immune response | Tumor necrosis factor (TNF superfamily, member 2) | TNF-a | 207113_s_at | 1.229 | 0.0437 |
| Immune response | Interleukin 8 | IL8 | 211506_s_at | 1.038 | 0.0636 |
| 202859_x_at | 1.018 | 0.2921 | |||
| Immune response | Aquaporin 9 | AQP9 | 205568_at | 0.562 | 0.1662 |
| Immune response | Interleukin 1, β | IL1B | 205067_at | 1.954 | 0.0370 |
| 39402_at | 1.747 | 0.0491 | |||
| Immune response | Tumor necrosis factor receptor superfamily, member 17 | TNFRSF17 | 206641_at | −0.411 | 0.0690 |
| Cell cycle/cell death | BCL2-like 1 | BCL2L1 | 1569067_at | 0.258 | 0.0002 |
| 206665_s_at | 0.357 | 0.1303 | |||
| 212312_at | 0.338 | 0.1563 | |||
| 215037_s_at | 0.375 | 0.1595 | |||
| Cell cycle/cell death | MAX dimerization protein 1 | MDX1 | 226275_at | 0.801 | 0.0544 |
| 228846_at | 0.469 | 0.1181 | |||
| Cell cycle/cell death | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, α | NFKBIA | 201502_s_at | 0.923 | 0.0690 |
| 231699_at | 0.132 | 0.0966 | |||
| Cell cycle/cell death | Disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | DAB2 | 201279_s_at | 0.596 | 0.0748 |
| 201278_at | 0.460 | 0.0821 | |||
| 210757_x_at | 0.478 | 0.1286 | |||
| 201280_s_at | 0.508 | 0.1766 | |||
| Cell cycle/cell death | Absent in melanoma 2 | AIM2 | 206513_at | −0.541 | 0.0534 |
| Cell cycle/cell death | Immediate early response 3 | IER3 | 201631_s_at | 1.428 | 0.0197 |
| Cell cycle/cell death | Purinergic receptor P2X, ligand-gated ion channel, 1 | P2RX1 | 210401_at | 0.590 | 0.0059 |
| 1569346_a_at | 0.193 | 0.1413 | |||
| Cell cycle/cell death | Peptidylprolyl isomerase F (cyclophilin F) | PPIF | 201490_s_at | 0.514 | 0.0436 |
| 201489_at | 0.616 | 0.0589 | |||
| Cell cycle/cell death | Dual-specificity phosphatase 1 | DUSP1 | 201044_x_at | 0.560 | 0.1112 |
| 226578_s_at | 0.268 | 0.1120 | |||
| Cell cycle/cell death | Protein phosphatase 1, regulatory (inhibitor) subunit 15A | PPP1R15A | 37028_at | 0.982 | 0.0325 |
| 202014_at | 0.904 | 0.0362 | |||
| Transcription | Early growth response 1 | EGR1 | 201694_s_at | 1.259 | 0.1181 |
| 227404_s_at | 1.119 | 0.1690 | |||
| 201693_s_at | 0.587 | 0.2592 | |||
| Miscellaneous | MutL homolog 3 (Escherichia coli) | MLH3 | 214525_x_at | 0.657 | 0.0488 |
| 217216_x_at | 0.484 | 0.1026 | |||
| 204838_s_at | 0.706 | 0.1199 | |||
| Miscellaneous | Trophoblast-derived noncoding RNA | TncRNA | 224566_at | 0.841 | 0.0044 |
| 224565_at | 0.429 | 0.0080 | |||
| 214657_s_at | 1.011 | 0.0101 | |||
| 238320_at | 1.010 | 0.0151 | |||
| 234989_at | 0.487 | 0.2052 |
*FDR P > 0.05 for all.
To gain another view into possible immune-related changes, we subjected 11 mid-stage HD and 9 control whole-blood RNA samples (Table 1) to a panel of 90 PCR-based immune marker assays (see Materials and Methods and SI Table 6). In this analysis, three assays (for IL-1b, COX-2, and TGF-b mRNAs) showed differences between HD and control samples and were carried into a second round of testing.
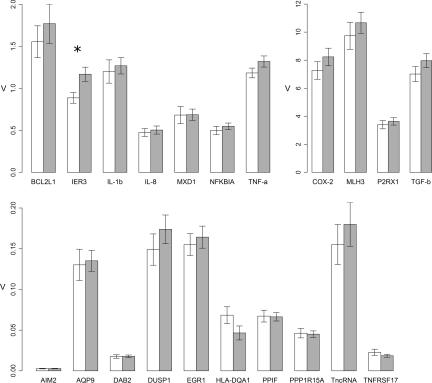
Quantitative PCR (QPCR) assays to measure RNA targets identified by both lymphocyte microarray and immune marker panel screens were then applied to a larger cohort of whole-blood samples from individuals representing presymptomatic, early, moderate, and advanced HD (see Table 1 and Materials and Methods). Fig. 1 shows the relative expression values for manifest HD versus controls from this enlarged sample set (for additional analyses, see SI Fig. 4). Only one RNA showed a mean expression difference; the expression of IER3 showed a 32% increase in HD versus controls.
Fig. 1.
Relative expression values of candidate biomarker RNAs identified by lymphocyte microarray or whole-blood immune panel gene expression screens. Error bars represent SEM. white, control samples; gray, manifest HD samples (classified as early, moderate, and advanced HD in Table 1). *, nominal P value <0.05.
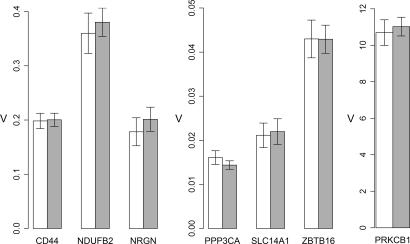
Exploiting our previous data on gene expression in the HD brain (18), we next investigated whether genes differentially expressed in brain tissues would also show HD-related changes in peripheral blood. Among mRNAs with statistical cutoff criterion of P < 0.001 for differential expression in HD caudate (32 cases, pathologic grade 0–2) versus control (32 samples, age- and gender-matched), we selected seven RNAs that could be reliably detected in 1- to 5-ng whole-blood RNA samples. Despite their dysregulation in HD brain, none of the seven interrogated candidates: CD44 antigen (CD44), NADH dehydrogenase (ubiquinone) 1 β-subcomplex, 2 (NDUFB2), neurogranin (NRGN), protein phosphatase 3, catalytic subunit, α-isoform (PPP3CA), protein kinase C β1 (PRKCB1), solute carrier family 14 (urea transporter), member 1 (SLC14A1), and zinc finger and BTB domain containing 16 (ZBTB16), showed differential expression in peripheral blood samples from individuals with HD (Fig. 2 and SI Fig. 5).
Fig. 2.
Relative expression values of candidate biomarker RNAs identified by HD brain gene expression data. Error bars represent SEM. white, control samples; gray, manifest HD samples (classified as early, moderate, and advanced HD in Table 1).
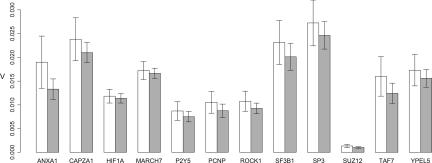
We also examined the ability of a previously identified set of RNA biomarkers (17) to discriminate between HD and control states in our sample cohort. PCR amplification efficacy of the previously published assays was tested, showing excellent results (efficiencies between 90% and 100%, data not shown). From this standard curve, input amounts of total blood RNA were chosen based on the linear range of each assay. As depicted in Fig. 3, however, these marker RNAs did not show differential expression across HD and control samples. To exclude the possibility that the nondetection of expression differences was due to a difference in the RNA preparation procedure, all assays were repeated with a new set of blood samples subjected to two different incubation periods in the RNA-stabilizing reagent, and these RNA extractions were carried out by another subset of investigators in a different laboratory. Despite these modifications, no differences were observed between samples from HD and control individuals (SI Table 7 and SI Fig. 6). Moreover, biological replicate samples subjected to independent processing showed minimal differences in expression measures (<10% for all assays; SI Fig. 7). Thus, we conclude that biological, and not technical, factors underlie the differences between our results and those previously reported.
Fig. 3.
Relative expression values of genes identified by ref. 17. Error bars represent SEM. white, control samples; gray, manifest HD samples (classified as early, moderate, and advanced HD in Table 1).
Discussion
New disease-modifying therapeutic agents are desperately needed for Huntington's and other neurodegenerative diseases. To optimize the process for testing new candidate therapies, however, clinical trials will need to be able to implement sensitive and reliable disease biomarker measures. Blood is an obvious tissue of choice for molecular biomarker development because of its easy access and rapid renewability. In the present study, we explored the possibility that mRNA readouts from human blood could serve as reliable indicators of HD clinical status.
Although a previous study showed that transcriptomic blood biomarkers are robust in tracking HD progression (17), our findings do not support this conclusion. Using a similar sample size and the same QPCR primer pairs, we saw no differential expression between manifest HD and control samples in any of 12 previously reported marker RNAs. Among the many additional RNA assays tested, only one detected a difference between manifest HD and control samples, and this difference was relatively small (32% increase in IER3). Although IER3 could be considered an interesting candidate marker for a variety of reasons (including a trend to track with HD status, as shown in SI Fig. 4), our expectation is that the small fold change observed in its expression offers little potential to provide sensitive disease monitoring. Nevertheless, IER3 is the top candidate RNA marker in the present study, and it would be reasonable to consider additional analyses of its expression, particularly in longitudinal HD patient samples.
We were surprised that the examination of immune cell and inflammatory pathway RNAs in peripheral blood cells appeared to be of such little use in detecting or tracking HD. Neuroinflammation is a seemingly uniform and progressive feature of HD brain pathology (5, 6), and one might have expected the corresponding compartment of immune cells in the periphery to indicate this effect. On the other hand, high interindividual variabilities in blood RNA levels have been observed (19). Specific non-HD conditions known to influence blood gene expression, such as anemia, infectious disease, and psychoactive drugs, were ruled out as causes for high interindividual variability in our study, however. Moreover, variability imparted by extreme values in a minority of cases is unlikely to account for the finding that the difference between mean values of the HD and control groups is close to zero (i.e., there is not even a clear trend to be distinguished between HD and control values).
We do not have specific data to explain the inability of a previously identified blood RNA biomarker set to discriminate between HD and control samples in our cohort. We have, however, ruled out known biological causes, specific technical issues with RNA preparation or QPCR, and insufficient sample size. The present findings suggest that some patient populations may not be amenable to monitoring by using these indicators and thus emphasize a need for further validation of these biomarkers before their widespread use in clinical trials.
Biomarker identification strategies beyond those based on blood RNA also provide hope that universally applicable tests can be developed. These range from structural imaging, to novel clinical and cognitive testing batteries, to proteomic and metabonomic assays. A combinatorial use of these indicators may in fact be the most rational strategy for sensitive and universal disease assessment (20). Thus, there is still room for optimism that these emerging new approaches will be able to improve on standard clinical readouts for the monitoring of future therapeutic trials.
Materials and Methods
Human Subjects.
Subjects were recruited through the HD Multidisciplinary Clinic of the National Hospital for Neurology and Neurosurgery, London, U.K. Blood samples were collected from patients who had tested positive for the HD genetic mutation. Control subjects were partners and spouses of HD patients, at-risk individuals who tested negative for the HD mutation, and healthy individuals from the general population. Premanifest HD mutation carriers were identified according to the absence of diagnostic motor abnormalities on the Unified Huntington's Disease Rating Scale (12). Patients with motor abnormalities were defined as having early, moderate, or advanced disease by using the total functional capacity (TFC) scale (13–7, early; 6–3, moderate) (21) assessed by experienced clinical raters.
All participants gave informed written consent, and the study protocol received the approval of local and national ethics review boards. Subjects with concomitant CNS disorders, significant medical comorbidity, known liver dysfunction, recent alcohol or substance abuse, and those taking medications or supplements suspected or known to interfere with the experimental methods used were excluded.
Sample Collection.
All samples were collected between 2 and 4 pm. For microarray expression profiling, blood samples were collected in EDTA Vacutainer tubes (BD, Oxford, U.K.) and fractionated within 2 h by density gradient centrifugation using a standard technique to obtain mononuclear cells, predominantly lymphocytes (>90%) (22).
Blood samples for PCR analyses of whole-blood gene expression were collected directly into PAXgene RNA tubes (Qiagen, Valencia, CA), incubated at room temperature for 2 h, and then frozen to −80°C before extraction. To investigate possible effects of room-temperature incubation time and freezing on RNA quality and gene expression profiles, we obtained samples from an additional set of patients and controls; for these samples, 2 PAXgene RNA tubes of blood were collected from each individual, after which one was incubated at room temperature for 2 h and to −80°C and the other was extracted after 24 h without freezing (see SI Table 7).
Samples included in the reported analyses are summarized in Table 1.
Microarray Gene Expression Profiling.
Lymphocyte samples from 12 moderate stage HD patients and 10 controls were processed for gene expression analysis by using Affymetrix U133 Human Genome 2.0 Plus arrays. Four micrograms of total RNA were converted into biotinylated cRNA, and hybridization, washing, and staining were conducted according to the GeneChip Expression Analysis protocol (Affymetrix). Data were analyzed by using the Bioconductor software package (www.bioconductor.org). Quality control for microarrays used the method of ref. 23, with a median NUSE cutoff of 1.05. Gene expression was quantified by robust multiarray analysis (24, 25) using the affy package (26), and differential gene expression was determined from empirical Bayes moderated t-statistics calculated with the limma package (27). An alternate analysis with normalization performed with the MAS5 algorithm for normalization and a classic two-sided t test for statistical testing is presented in SI Tables 4 and 5.
RNA Isolation.
For microarrays, RNA was isolated by using TRIzol (Invitrogen, Carlsbad, CA) following the manufacturer's protocol, followed by RNeasy column cleanup and ethanol precipitation.
For PCR analyses, RNA was isolated by using the PAXgene Blood RNA kit (PreAnalytiX, Hombrechtikon, Switzerland) according to the manufacturer's protocol (omitting the optional final incubation at 65°C) and stored at −80°C. To remove residual salt contamination, RNAs were ethanol-precipitated in the presence of glycogen (3.5 μg per sample). RNA quality was assessed on a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). Only highly intact RNAs with a RNA integrity number of ≥7 were used for reverse transcription.
Reverse Transcription and Real-Time PCR Analyses.
Reverse transcription was performed by using the High Capacity cDNA Archive kit (Applied Biosystems), following the manufacturer's protocol and using 2 μg of each RNA sample in a reaction volume of 100 μl. After an initial 10 min at 25°C, reverse transcription was conducted at 37°C for 120 min. The resulting cDNA was stored at −20°C. Quantitative real-time PCR experiments were performed with a 7900HT Fast Real-Time PCR System from Applied Biosystems. Each assay sample was run in triplicates alongside its reference assay (on the same plate). Samples comprised those listed in Table 1.
TaqMan Gene Expression Assays (Applied Biosystems) were used according to the manufacturer's recommendations. Cycling parameters comprised an initial polymerase activation step (10 min at 95°C), followed by 2-step cycling for 40 cycles (15 sec at 95°C and 60 sec at 60°C). Ten-microliter reactions were composed of TaqMan Fast Universal Master Mix without AmpErase UNG and cDNA inputs equivalent to the following amounts of total RNA: 1 ng (AIM2, ACTB, AQP9, CD44, DAB2, DUSP1, HLA-DQA1, MXD1, NFKBIA, PPIF, PPP1R15A, PPP3CA, NRGN, RPLP, SLC14A1, TncRNA, ZBTB16; 2 ng (COX-2, IL-1b, IL-8, TNF-a, BCL-XL, TGF-b, and HPRT); 5 ng (IER3, P2RX1, EGR1, MLH3, TNFRSF17, ACTB, PRKCB1, NDUFB2, and HPRT). Ct values were normalized to RPLP (1 or 2 ng) or HPRT (5 ng), respectively.
A subset of samples (Table 1) was run on TaqMan Low-Density Array Human Immune Profiling Arrays. cDNA samples equivalent to 100 ng of total RNA were loaded in TaqMan Universal Master Mix without AmpErase UNG in each fill port. For Immune Profiling Array assays, 28S ribosomal RNA served as the reference.
SYBR green assays (for ANXA1, MARCH7, CAPZA1, HIF1A, SUZ12, P2Y5, PCNP, ROCK1, SF3B1, SP3, TAF7, and YPEL5) were performed by using oligonucleotide primer pairs described in ref. 17. Ten-microliter reactions were composed of 300 nM primer, Power SYBR green Master Mix (Applied Biosystems), and cDNA inputs equivalent to 1 ng of total RNA. Cycling parameter comprised an initial polymerase activation step (10 min at 95°C), followed by a 2-step cycling for 40 cycles (15 sec at 95°C and 1 min at 60°C). Relative expression values were normalized to β-actin or 28S ribosomal RNAs; use of either reference yielded comparable results.
Statistical Analyses.
Primer efficiencies were evaluated by running standard curves with input amounts ranging between 10 pg and 10 ng by using the following equation: Ex=10(−1/slope) −1. The relative expression values (V) of target genes were calculated by the ΔCt method corrected for amplification efficiencies as described (28).
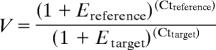 |
Using ANOVA, we tested for differences in mean relative expression values between HD manifest and control groups (or, alternatively, among three groups: controls, presymptomatic/early, and moderate/advanced, or four groups: controls, pre symptomatic, early, moderate/advanced; see SI Figs. 4–6). A preliminary analysis explored gender and age as additional factors, but no significant age effect was detected for any of the genes analyzed by QPCR (significance level 0.05), and only one gene (MLH3) showed a significant difference between women and men that was independent of HD status.
Supplementary Material
Acknowledgments
We are grateful to the HD patients and their families who contributed samples for this study. We also thank Maria Rey and the staff of the Lausanne DNA Array Facility for technical assistance. This work was supported by the HighQ Foundation, the Ecole Polytechnique Fédérale de Lausanne, and the Swiss National Science Foundation. S.J.T. is a U.K. Department of Health National Clinician Scientist.
Abbreviation
- HD
Huntington's disease.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.R. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE8762).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703652104/DC1.
References
- 1.Sapp E, Kegel KB, Aronin N, Hashikawa T, Uchiyama Y, Tohyama K, Bhide PG, Vonsattel JP, DiFiglia M. J Neuropathol Exp Neurol. 2001;60:161–172. doi: 10.1093/jnen/60.2.161. [DOI] [PubMed] [Google Scholar]
- 2.Hunot S, Hirsch EC. Ann Neurol. 2003;53(Suppl 3):49–58. doi: 10.1002/ana.10481. [DOI] [PubMed] [Google Scholar]
- 3.Singhrao SK, Neal JW, Morgan BP, Gasque P. Exp Neurol. 1999;159:362–376. doi: 10.1006/exnr.1999.7170. [DOI] [PubMed] [Google Scholar]
- 4.Bonifati DM, Kishore U. Mol Immunol. 2007;44:999–1010. doi: 10.1016/j.molimm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Leblhuber F, Walli J, Jellinger K, Tilz GP, Widner B, Laccone F, Fuchs D. Clin Chem Lab Med. 1998;36:747–750. doi: 10.1515/CCLM.1998.132. [DOI] [PubMed] [Google Scholar]
- 6.Pavese N, Gerhard A, Tai YF, Ho AK, Turkheimer F, Barker RA, Brooks DJ, Piccini P. Neurology. 2006;66:1638–1643. doi: 10.1212/01.wnl.0000222734.56412.17. [DOI] [PubMed] [Google Scholar]
- 7.Heuser IJ, Chase TN, Mouradian MM. Biol Psychiatry. 1991;30:943–952. doi: 10.1016/0006-3223(91)90007-9. [DOI] [PubMed] [Google Scholar]
- 8.Petersen A, Bjorkqvist M. Eur J Neurosci. 2006;24:961–967. doi: 10.1111/j.1460-9568.2006.04985.x. [DOI] [PubMed] [Google Scholar]
- 9.Beal MF, Ferrante RJ. Nat Rev Neurosci. 2004;5:373–384. doi: 10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- 10.Butler R, Bates GP. Nat Rev Neurosci. 2006;7:784–796. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- 11.Shoulson I, Fahn S. Neurology. 1979;29:1–3. doi: 10.1212/wnl.29.1.1. [DOI] [PubMed] [Google Scholar]
- 12.The Huntington Study Group. Mov Disord. 1996;11:136–142. doi: 10.1002/mds.870110204. [DOI] [PubMed] [Google Scholar]
- 13.Paulsen JS, Hayden M, Stout JC, Langbehn DR, Aylward E, Ross CA, Guttman M, Nance M, Kieburtz K, Oakes D, et al. Arch Neurol. 2006;63:883–890. doi: 10.1001/archneur.63.6.883. [DOI] [PubMed] [Google Scholar]
- 14.Henley SMD, Bates GP, Tabrizi SJ. Curr Opin Neurol. 2005;18:698–705. doi: 10.1097/01.wco.0000186842.51129.cb. [DOI] [PubMed] [Google Scholar]
- 15.Luthi-Carter R, Hanson SA, Strand AD, Bergstrom DA, Chun W, Peters NL, Woods AM, Chan EY, Kooperberg C, Krainc D, et al. Hum Mol Genet. 2002;11:1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 16.Strand AD, Aragaki AK, Shaw D, Bird T, Holton J, Turner C, Tapscott SJ, Tabrizi SJ, Schapira AH, Kooperberg C, et al. Hum Mol Genet. 2005;14:1863–1876. doi: 10.1093/hmg/ddi192. [DOI] [PubMed] [Google Scholar]
- 17.Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, et al. Proc Natl Acad Sci USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, et al. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 19.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Proc Natl Acad Sci USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wild EJ, Tabrizi SJ. Lancet Neurol. 2006;5:724–725. doi: 10.1016/S1474-4422(06)70531-2. [DOI] [PubMed] [Google Scholar]
- 21.Shoulson I. Neurology. 1981;31:1333–1335. doi: 10.1212/wnl.31.10.1333. [DOI] [PubMed] [Google Scholar]
- 22.Boyum A. Scand J Clin Lab Invest Suppl. 1968;97:9–29. [PubMed] [Google Scholar]
- 23.Brettschneider J, Collin F, Bolstad BM, Speed TP. Technometrics. 2007 in press. [Google Scholar]
- 24.Bolstad BM, Irizarry RA, Astrand M, Speed TP. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 25.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Gautier L, Cope L, Bolstad BM, Irizarry RA. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 27.Smyth GK. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. article 3. [DOI] [PubMed] [Google Scholar]
- 28.Zucker B, Luthi-Carter R, Kama JA, Dunah AW, Stern EA, Fox JH, Standaert DG, Young AB, Augood SJ. Hum Mol Genet. 2005;14:179–189. doi: 10.1093/hmg/ddi014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.