Abstract
Three forms of PPARs are expressed in the heart. In animal models, PPARγ agonist treatment improves lipotoxic cardiomyopathy; however, PPARγ agonist treatment of humans is associated with peripheral edema and increased heart failure. To directly assess effects of increased PPARγ on heart function, we created transgenic mice expressing PPARγ1 in the heart via the cardiac α–myosin heavy chain (α-MHC) promoter. PPARγ1-transgenic mice had increased cardiac expression of fatty acid oxidation genes and increased lipoprotein triglyceride (TG) uptake. Unlike in cardiac PPARα-transgenic mice, heart glucose transporter 4 (GLUT4) mRNA expression and glucose uptake were not decreased. PPARγ1-transgenic mice developed a dilated cardiomyopathy associated with increased lipid and glycogen stores, distorted architecture of the mitochondrial inner matrix, and disrupted cristae. Thus, while PPARγ agonists appear to have multiple beneficial effects, their direct actions on the myocardium have the potential to lead to deterioration in heart function.
Introduction
PPARs play a central role in cellular and tissue metabolism (1) and affect inflammatory processes (2). Several of these receptors have been studied in the heart, one of the most energy-demanding organs in the body. Heart energy is primarily derived from oxidation of fatty acids (FAs), and this correlates with the relatively high levels of PPARα expression in the heart (3). In contrast, PPARγ is expressed at relatively low levels in the heart. Nonetheless, recent data have shown that tissue-specific loss of PPARγ alters heart function (4). Moreover, rosiglitazone still caused cardiac hypertrophy in these mice, indicating non-PPARγ effects of the drug and highlighting the difficulty of relying on pharmacologic agents to understand the biologic actions of specific transcription factors.
In other tissues, PPARγ regulates tissue triglyceride (TG) accumulation. Hepatocyte overexpression of PPARγ leads to steatosis (5); loss of adipose tissue PPARγ leads to defective production of mature adipocytes (6). Whether PPARγ in the heart can function in a similar manner is not known.
Increased accumulation of lipids leads to organ dysfunction, a process termed lipotoxicity. In the heart, defects in lipid metabolism, diabetes, and severe obesity can cause dilated cardiomyopathy (7). This disorder has been modeled in animals. Cardiac overexpression of PPARα increases FA oxidation, but it also leads to heart lipid accumulation (8). Severe obesity associated with defective leptin actions (9) and genetic alterations that increase heart lipid due to greater uptake (10) or increased FA trapping (11) also cause lipotoxic cardiomyopathies.
In some rodent models of lipotoxic dilated cardiomyopathy, PPARγ agonist treatment improves heart function (12, 13). It has been postulated that PPARγ agonists have salutary effects due to direct actions on the heart (14–16); this is surprising, since PPARγ causes lipid accumulation in other tissues. PPARγ agonists have a variety of systemic effects, and, most remarkably, they channel a greater proportion of plasma TG and FA to adipose tissue (17). Such an action alone could reduce lipid uptake by the heart and improve lipotoxicity.
To directly assess effects of increased PPARγ on heart function, lipid metabolism, and gene expression, we created transgenic mice expressing PPARγ1 via the cardiac α–myosin heavy chain (α-MHC) promoter. These mice have increased cardiac uptake of FA and no reduction in glucose uptake and develop dilated cardiomyopathy. Although PPARγ agonists did not increase cardiac gene expression in wild-type mice, these drugs increased mRNA levels of FA oxidation and uptake genes and exacerbated heart dysfunction in PPARγ-transgenic animals.
Results
Creation of cardiac-specific PPARγ1–transgenic mice.
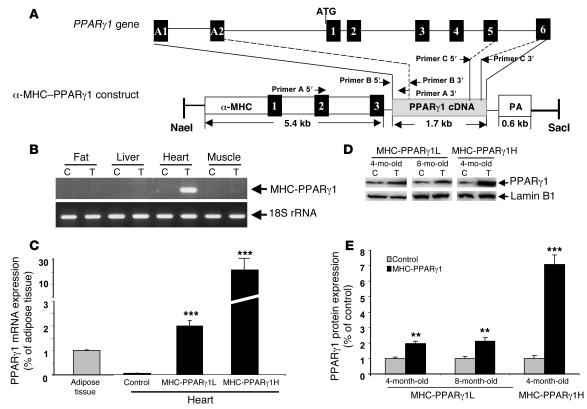
Mice overexpressing PPARγ in cardiomyocytes via the α-MHC promoter were designated MHC-PPARγ1 mice (Figure 1A). PCR screening of the offspring identified 3 founders harboring the MHC-PPARγ1 transgene. Offspring from 2 of these founders, designated MHC-PPARγ1L (low-expressing) and MHC-PPARγ1H (high-expressing), were used in this report. Male mice were used in the experiments unless indicated otherwise. The PPARγ1 transgene was expressed specifically in the heart; no expression was detected in the liver, skeletal muscle, or fat (Figure 1B). In control mouse hearts, PPARγ1 mRNA levels were low; the ratio of PPARγ to 18S rRNA was much lower in control heart than adipose tissue. In contrast, MHC-PPARγ1L mice had PPARγ to 18S rRNA levels approximately twice those found in adipose tissue (Figure 1C). MHC-PPARγ1H hearts had PPARγ mRNA levels approximately 10-fold higher than those in MHC-PPARγ1L hearts. PPARγ protein levels were 1.9-fold (MHC-PPARγ1L) and 7.2-fold (MHC-PPARγ1H) higher in the cardiac ventricles of transgenic mice than littermate controls (Figure 1, D and E).
Figure 1. Construct and gene expression in PPARγ1-transgenic mice.
(A) Diagram of PPARγ1 genomic locus and PPARγ1 construct design. The α-MHC promoter was used to drive PPARγ1 cDNA expression. Black boxes indicate exons that are numbered. PCR primers are indicated. Boxes A1 and A2 indicate PPARγ1-specific exons. (B) Cardiac-specific MHC-PPARγ1 expression. RT-PCR analysis of RNA was performed in 3-month-old MHC-PPARγ1L male mice using primer A: 5ι end specific to α-MHC promoter exon 2 and 3′ end specific to PPARγ1 cDNA nucleotides 173–192. (C) Heart expression of total PPARγ mRNA (both endogenous gene and transgene) in MHC-PPARγ1L and MHC-PPARγ1H male mice was quantified by qRT-PCR. Primer C was used for PCR amplification. Data are shown as mean ratio (±SD; n = 5 in each group) corrected for 18S rRNA. ***P < 0.001 for MHC-PPARγ1 mice versus control mice. (D) Nuclear protein (30 μg) from heart tissues of 4- and 8-month-old MHC-PPARγ1L and 4-month-old MHC-PPARγ1H mice and their littermate controls was analyzed by Western blot using polyclonal PPARγ antibody. Western blot for lamin B1 is shown as a control. (E) Bands were quantified using Molecular Analysis Software. Data were normalized to values for littermate controls (set at 1.0), and normalized units of expression are shown (±SD; n = 3 in each group). **P < 0.01; ***P < 0.001 for MHC-PPARγ1 versus control mice. C, littermate control; Tg, transgenic MHC-PPARγ1 mouse; PA, poly(A) site.
Transgenic mice from each of the lines bred normally, and glucose, body weight, and plasma lipid levels were similar to those of nontransgenic mice (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI30335DS1).
PPARγ activity in hearts of MHC-PPARγ1 mice.
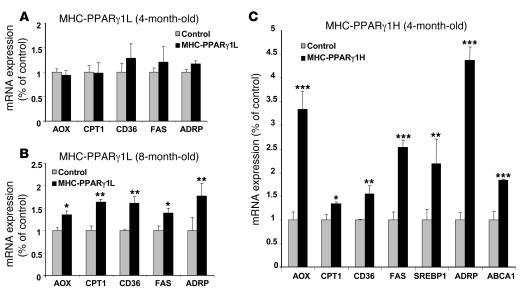
We estimated the degree of PPARγ activity in the MHC-PPARγ1 mice by assessing expression of downstream genes. Surprisingly, despite the adipose-like PPARγ1 expression level, hearts from young MHC-PPARγ1L mice (4 months old) had an expression of downstream genes similar to that of controls (Figure 2A). Eight-month-old MHC-PPARγ1L mice, however, had greater expression of acyl-CoA oxidase (AOX) (1.35-fold), carnitine palmitoyl transferase–1 (CPT1) (1.63-fold), FA translocase (CD36) (1.61-fold), FA synthase (FAS) (1.44-fold), and adipose differentiation–related protein (ADRP) (1.81-fold) (Figure 2B).
Figure 2. Expression of PPARγ target genes in the hearts of MHC-PPARγ1 mice.
(A and B) Four- and 8-month-old MHC-PPARγ1L male mice; (C) 4-month-old PPARγ1H male mice. Data are shown as mean ratio (±SD; n = 5–9 in each group) corrected for 18S rRNA and normalized to those for littermate controls (set as 1.0). *P < 0.05, **P < 0.01, ***P < 0.001 for MHC-PPARγ1 mice versus littermate controls.
In contrast, multiple genes were markedly upregulated in the hearts of 4-month-old MHC-PPARγ1H mice (Figure 2C), including genes involved in lipid oxidation, uptake, synthesis, and storage, such as AOX, CPT1, CD36, FAS, ADRP, and SREBP1. Expression of ATP-binding cassette transport A1 (ABCA1), which mediates reverse cholesterol transport to apoA-I and is induced by PPARγ (18), was 1.73-fold higher in 4-month-old MHC-PPARγ1 H mice (Figure 2C).
PPAR gene expression in control and MHC-PPARγ mice with aging.
To determine why the MHC-PPARγL transgene did not alter downstream gene expression in the young mice, we assessed expression of PPAR family members in control and transgenic mice with aging. As noted by others (19), PPARα expression decreased as the mice aged (Supplemental Figure 1A). Moreover, expression of PPARα was reduced by 29% and 60% in the 2- and 9-month-old MHC-PPARγL transgenic mice, respectively, compared with their littermate controls. Neither endogenous and transgenic PPARγ1 nor PPARδ expression differed between 2- and 9-month-old mouse hearts (Supplemental Figure 1, B and C). Thus, changes in PPARα expression might modify the downstream effects of the MHC-PPARγL transgene.
Cardiac lipid content and uptake in MHC-PPARγ1 mice.
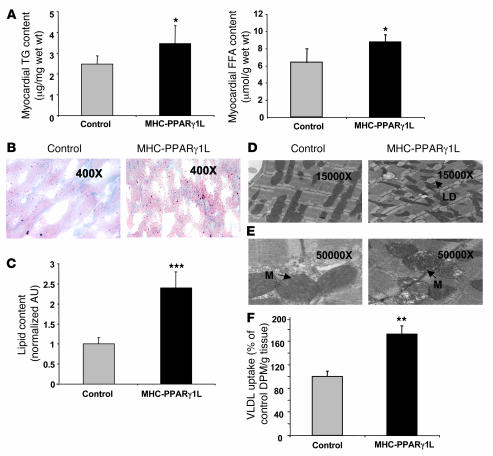
Greater CD36 expression in the MHC-PPARγ1 mice could increase uptake of plasma free and lipoprotein-derived FA. Heart TG and FA content were increased in 8-month-old MHC-PPARγ1L mice (Figure 3A). Oil red O staining revealed greater accumulation of intracellular neutral lipids in hearts from MHC-PPARγ1L mice compared with littermate controls (Figure 3, B and C). Electron microscopy showed more lipid droplets within the sarcoplasm of cardiomyocytes, with distortion of the mitochondrial contours in MHC-PPARγ1L mice (Figure 3D). In some areas the cristae were disrupted (Figure 3E).
Figure 3. Increased heart lipid accumulation in MHC-PPARγ1 mice.
(A) Heart TG (left panel) and FFA content (right panel) was significantly increased in MHC-PPARγ1L transgenic mice (n = 7 for each group). (B) Oil red O staining showed an abundance of neutral lipid droplets randomly scattered throughout the cytoplasm of cardiomyocytes in MHC-PPARγ1L female mice (right panel) after 24-hour fasting (original magnification, ×400). (C) Oil red O staining was quantified using Molecular Analysis Software (n = 3 in each group). Data were normalized to values for littermate controls (set as 1.0). (D). Electron micrographs (original magnification, ×15,000) of myocardial tissue showed a large increase in lipid droplets within the sarcoplasm of cardiomyocytes in MHC-PPARγ1L male mice (right panel) compared with littermates (left panel). All of these lipid droplets were located adjacent to mitochondria, with distortion of the mitochondrial contours. (E) Electron micrographs (original magnification, ×50,000) detailed distorted architecture of the mitochondrial inner matrix with electron lucent foci (arrows) in MHC-PPARγ1L mice, and in some areas the cristae were disrupted. (F) [14C-TG]VLDL uptake into heart of MHC-PPARγ1L male mice and littermate control mice at 8 months of age (n = 6 per group). LD, lipid droplet; M, mitochondria. Data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 compared with littermate controls. DPM, decays per minute.
Cardiac uptake of VLDL-TG in 8-month-old MHC-PPARγ1L hearts was 74% greater than that in littermate control hearts (P < 0.01; Figure 3F), which corresponds with increased expression of lipid uptake genes and cardiac lipid storage. Liver and skeletal muscle lipid uptake were not altered by the PPARγ1 transgene (data not shown).
Glucose transporter 4 expression and glucose uptake in MHC-PPARγ1 mice.
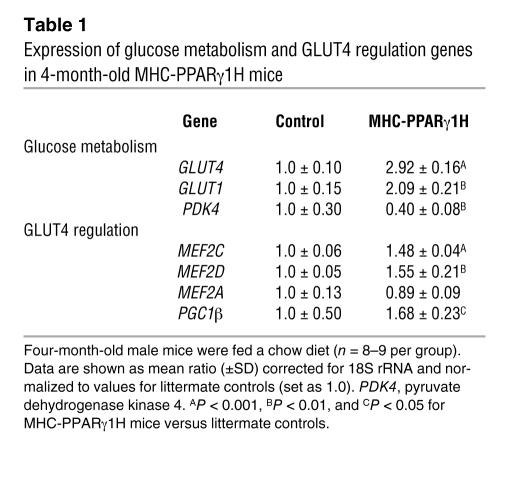
Unlike in MHC-PPARα transgenic mice (8), glucose transporter 4 (GLUT4) and GLUT1 mRNA levels were unchanged in the MHC-PPARγ1L mice (data not shown) and were upregulated in MHC-PPARγ1H mice (Table 1). Pyruvate dehydrogenase kinase 4, the gene regulating glucose oxidation, was also downregulated in the MHC-PPARγ1H mice (Table 1).
Table 1 .
Expression of glucose metabolism and GLUT4 regulation genes in 4-month-old MHC-PPARγ1H mice
The GLUT4 gene undergoes a complex program of gene regulation. The GLUT4–myocyte enhancer factor 2 (GLUT4-MEF2) site is required for metabolic regulation of GLUT4 transcription (20–22). MEF2A, MEF2C, and MEF2D isoforms, but not MEF2B isoforms, are expressed in the heart. PPARγ coactivator 1 (PGC1) also interacts with MEF2C to upregulate endogenous GLUT4 expression and glucose uptake in cultured muscle cells (23). GLUT4 expression in MHC-PPARγ1H mice was associated with increased expression of MEF2C and MEF2D (1.48-fold, P < 0.001, and 1.55-fold, P = 0.009, respectively), but with no change in MEF2A. PGC1β gene expression was also upregulated (1.68-fold; P = 0.01) in the hearts of the MHC-PPARγ1H mice (Table 1).
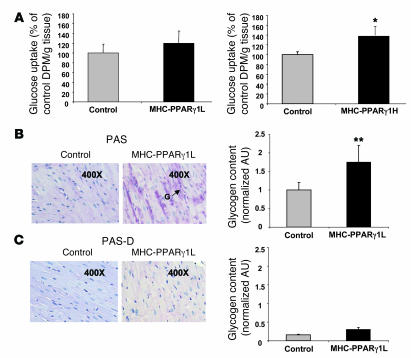
Myocardial glucose import in vivo was assessed using 2-deoxy-d-[3H]glucose. Heart uptake of glucose was unchanged in the MHC-PPARγ1L hearts (Figure 4A, left panel) and was increased by 37% in MHC-PPARγ1H mice compared with littermate controls (P < 0.05; Figure 4A, right panel). The PAS technique revealed greater cardiac glycogen storage in MHC-PPARγ1L mice compared with littermate controls (Figure 4B). PAS diastase (PAS-D) completely removed the glycogen in cardiac sections (Figure 4C). These data indicate that in the MHC-PPARγ mice, although cardiac lipid uptake was elevated, glucose uptake was not reduced.
Figure 4. Increased cardiac glucose uptake and glycogen storage in MHC-PPARγ1 male mice.
(A) 2-Deoxy-d-[3H]glucose uptake into heart of MHC-PPARγ1 and wild-type control mice. MHC-PPARγ1L mice were 8 months old (n = 7 in each group); MHC-PPARγ1H mice were 4 months old (n = 6 in each group). (B) PAS staining of heart tissue from 8-month-old MHC-PPARγ1L female mice (middle panel) and littermate controls (left panel) after 6-hour fasting (original magnification, ×400). The arrow shows focal increased staining. Glycogen content was quantified using Molecular Analysis software (n = 3–4 per group) and normalized to values for PAS-staining controls (set as 1.0) (right panel). G, glycogen. Data are shown as mean ± SD. *P < 0.05 compared with littermate controls; **P < 0.01. (C) The glycogen staining was removed by PAS diastase (PAS-D) digestion (left and middle panels), and the data were quantified using Molecular Analysis software (right panel).
Cardiac function in MHC-PPARγ1 mice.
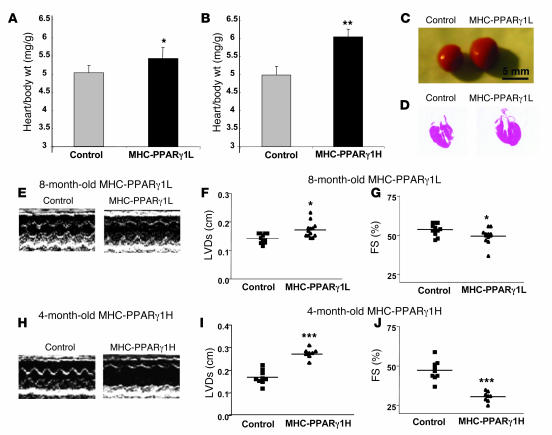
Both MHC-PPARγ1L and MHC-PPARγ1H hearts had dilated left ventricles and impaired systolic function. Heart to body weight ratios were increased in 8-month-old MHC-PPARγ1L and 4-month-old MHC-PPARγ1H mice (Figure 5, A and B). The dilated cardiomyopathy phenotype was grossly visible (Figure 5, C and D) and also seen by echocardiography (Figure 5, E–J). Left ventricular systolic dimension was increased (0.17 ± 0.03 versus 0.14 ± 0.01 cm; P = 0.021), and fractional shortening was reduced from 53% ± 3.6% to 48% ± 5.2% (P = 0.036) by age 8 months in PPARγ1L mice (Figure 5, F and G). The MHC-PPARγ1H mice had more severe cardiac dysfunction. Four-month-old PPARγ1H hearts had greater left ventricular systolic dimension (0.28 ± 0.02 versus 0.17 ± 0.03; P < 0.001) and a greater reduction in fractional shortening (from 47.3% ± 5.3% to 30.2% ± 3.2%; P < 0.001) (Figure 5, I and J).
Figure 5. Dilated cardiomyopathy in MHC-PPARγ1 mice.
(A and B) The heart to body weight ratio was increased in both MHC-PPARγ1L (n = 11–13) and MHC-PPARγ1H male mice (n = 8–9). (C) Representative photographs of hearts of control and MHC-PPARγ1L male mice. (D) Histological section of H&E-stained cardiac tissue in control and MHC-PPARγ1L male mice. (E and H) Representative echocardiographic images of left ventricle motion in MHC-PPARγ1L and MHC-PPARγ1H mice. Echocardiography showed increased left ventricular systolic dimension (F and I) and reduced fractional shortening (G and J) in both MHC-PPARγ1L and MHC-PPARγ1H mice. FS, fractional shortening; LVDs, left ventricular end-systolic dimension. Data are shown as mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 versus littermate controls.
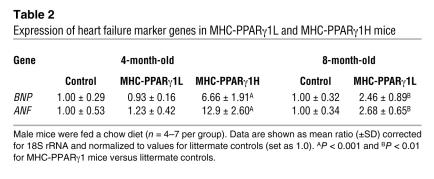
Expression of heart failure marker genes brain-type natriuretic peptide (BNP) and atrial natriuretic factor (ANF), was increased in MHC-PPARγ1 mice (Table 2). These increases were seen by 4 months in the PPARγ1H line but not until 8 months in the PPARγ1L line.
Table 2 .
Expression of heart failure marker genes in MHC-PPARγ1L and MHC-PPARγ1H mice
Effects of rosiglitazone in wild-type and MHC-PPARγ1 mice.
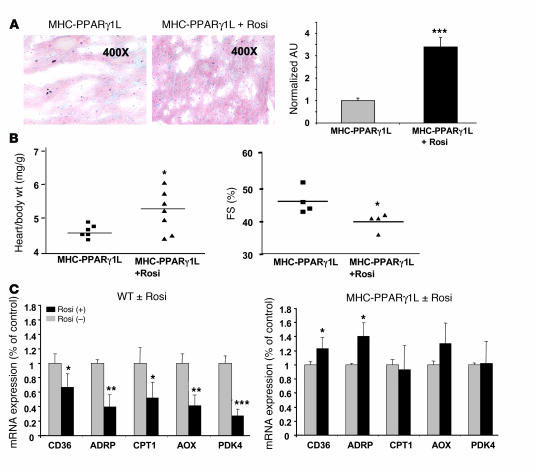
In contrast to other lipotoxic heart models (13, 16, 24), rosiglitazone treatment of 8-month-old MHC-PPARγ1L mice led to further deterioration of cardiac function: increased lipid accumulation, larger hearts, and decreased fractional shortening (Figure 6, A and B). This was associated with increased cardiac gene expression of CD36 and ADRP in rosiglitazone-treated MHC-PPARγ1L mice (Figure 6C). When wild-type mice were treated with rosiglitazone, cardiac expression of usual PPARγ downstream genes was paradoxically reduced in the 8-month-old mice (Figure 6C). Adipose tissue and muscle had increased CD36 expression after rosiglitazone treatment (Supplemental Figure 2).
Figure 6. MHC-PPARγ1L and C57BL/6 wild-type mice with or without rosiglitazone treatment.
(A) Oil red O heart histology of 8-month-old MHC-PPARγ1L female mice with (middle panel) or without (left panel) rosiglitazone treatment. Staining was quantified and normalized to values for littermate controls (set as 1.0) (right panel). Data are shown as mean (± SD; n = 3–4 in each group). (B) Heart to body weight ratio (left panel) and echocardiographic measurements of fractional shortening (right panel) in 8-month-old MHC-PPARγ1L male mice with or without rosiglitazone treatment. (C) qRT-PCR analysis of cardiac gene expression in 8-month-old male C57BL/6 wild-type (left panel) and MHC-PPARγ1L mice (right panel) with or without rosiglitazone treatment (n = 6–7 per group). *P < 0.05, **P < 0.01, and ***P < 0.001 versus the group without rosiglitazone treatment group. Rosi, rosiglitazone, WT, wild-type C57BL/6 mice.
Pathways leading to cardiac dysfunction in MHC-PPARγ1 mice.
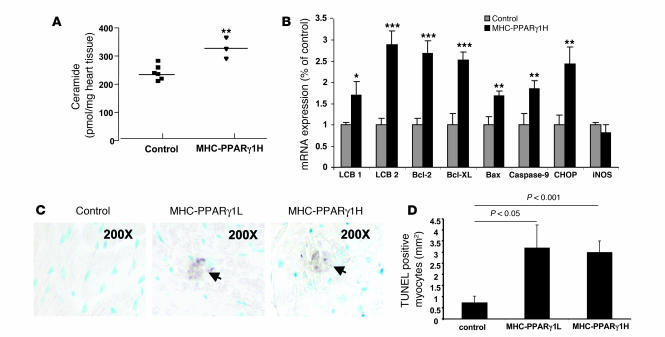
To explore potential mechanisms in the development of lipid-induced cardiomyopathy, we examined heart tissue for evidence of lipid accumulation triggering programmed cell death. Four-month-old MHC-PPARγ1H mice had a 40% increase in levels of cardiac ceramide, a lipid capable of inducing apoptosis (25), compared with littermate controls (Figure 7A). This was associated with increased expression of long chain base 1 (LCB1) and LCB2, the subunits of serine palmitoyl-CoA transferase. Expression of apoptosis-related genes — B cell leukemia/lymphoma 2 (Bcl2), Bcl-2-like protein (Bcl-XL), BCL2-associated X protein (Bax), and caspase-9 — also dramatically increased (Figure 7B). The CCAAT/enhancer-binding protein (C/EBP) homologous protein (CHOP), one of the regulators of ER stress–mediated cell death, was upregulated in the MHC-PPARγ1H mice. iNOS was unchanged. There was evidence of positive TUNEL staining of heart tissue in both MHC-PPARγ1L and MHC-PPARγ1H, but not control, hearts (Figure 7, C and D).
Figure 7. Lipoapoptosis in MHC-PPARγ1 hearts.
(A) Cardiac ceramide was measured in heart tissue from 4-month-old MHC-PPARγH male mice and their littermate controls using diacylglycerol kinase assay and normalized for tissue weight (n = 3–6). (B) Ceramide synthesis genes (LCB1 and LCB2), ER stress–induced apoptosis-related gene (CHOP), and apoptosis genes (Bax and caspase-9) were upregulated in MHC-PPARγ1H male mice hearts. Antiapoptosis genes Bcl2 and Bcl-XL were also upregulated (n = 8–9). (C) Cardiac ventricular tissues from control (left panel) and 8-month-old MHC-PPARγ1L (middle panel) and 4-month-old MHC-PPARγ1H male mice (right panel) were stained for DNA fragmentation by a TUNEL protocol (original magnification, ×200). Apoptotic cells containing fragmented nuclear chromatin exhibited a dark brown nuclear staining. (D) The TUNEL-positive myocytes were counted and expressed as the number of TUNEL-positive myocytes per millimeter squared tissue area. Data are shown as mean (± SD) and normalized to values for littermate controls (set as 1.0). *P < 0.05, **P < 0.01, and ***P < 0.001 versus littermate controls.
PPARγ1 expression in humans and streptozotocin-induced diabetic mouse hearts.
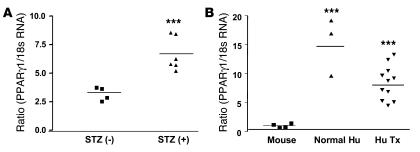
We then tested whether PPARγ expression could be induced in the hearts of diabetic mice. Hearts of diabetic mice had 2.1-fold higher expression of PPARγ than those of control mice (P = 0.003) (Figure 8A). We also assessed PPARγ expression in hearts from humans. Human hearts had 8.1- to 14.5-fold higher PPARγ mRNA levels than mouse hearts (Figure 8B). These data suggest that PPARγ is more functional in human hearts.
Figure 8. Upregulation of PPARγ1 expression in streptozotocin-induced diabetic mouse and humans hearts.
(A) Four-month-old wild-type C57BL/6 male mice were treated with streptozotocin (STZ), and 3 weeks later hearts from control and diabetic mice (glucose >300 mg/dl) were harvested and PPARγ1 mRNA assayed by qRT-PCR. (B) Heart tissue was obtained from a normal individual (Normal Hu) and from hearts of patients undergoing heart transplantation (Hu Tx). PPARγ1 and 18S rRNA were measured in human and wild type mouse mRNAs using primers that were common for human and mouse genes. ***P < 0.001 versus mice group.
Discussion
Of the 3 members of the PPAR family of transcription factors, PPARγ is expressed at the lowest level in the heart. However, recent studies of cardiac-specific PPARγ–knockout mice have shown a role for this transcription factor in maintaining normal heart function (4). PPARγ has several well-documented effects to alter glucose and lipid metabolism in tissues. In both the adipose tissue (26, 27) and liver (28), this transcription factor increases uptake of both glucose and lipid and increases storage of TG.
To study the pharmacologic actions of PPARs, these transcription factors, either alone or in the presence of their potent agonist VP16 (29), have been transgenically expressed within a tissue. Our objective was to study actions of PPARγ only in the heart and to determine whether exogenous ligands modulate its activity; this was necessary to differentiate local from systemic actions of PPAR agonists. We created several lines of PPARγ-transgenic mice to assess PPARγ function specifically in cardiomyocytes. In the low-expressing line, no downstream changes in cardiomyocyte genes were found in young animals. The effects of PPARγ activation were noted only in older MHC-PPARγ1L mice. In contrast, in the MHC-PPARγ1H line, increased expression of PPARγ downstream genes was seen in young mice. Expression of lipid oxidation and uptake genes was increased, glucose uptake was not reduced, and lipotoxicity with cardiac dilation and dysfunction ensued. Thus, cardiac glucolipotoxicity was created.
The 2 lines of PPARγ1-transgenic mice showed a similar phenotype, but the timing and severity differed. Young MHC-PPARγ1L mice had no obvious changes in PPARγ downstream genes such as CD36, ADRP, and CPT1, even though PPARγ was expressed in hearts at levels comparable to those in adipose tissue. This was quite surprising but in some ways echoed the lack of effect of cardiomyocyte PPARγ deficiency on expression of genes controlling lipid and glucose metabolism (4). We initially wondered whether a deficiency of agonist prevented the expected gene inductions. However, neither a high-fat diet nor rosiglitazone led to PPARγ activation in 4-month-old PPARγ1L mice (data not shown). We also noted that rosiglitazone treatment of wild-type mice reduced expression of PPARγ targets. We suspect that rosiglitazone redirected naturally occurring PPAR ligands to other tissues, especially adipose tissue. In another model of lipotoxicity, rosiglitazone decreased delivery of plasma lipids to the heart (13). In part, the lack of effect of the MHC-PPARγ1L transgene was due to the reduced heart expression of PPARα. It is of interest that tissues with the highest level of PPARγ, i.e., the adipose tissue, have relatively low levels of PPARα expression (30), whereas PPARα-rich tissues such as the heart have low levels of PPARγ. The reasons for this are not clear, but our data suggest that there is a reciprocal regulation of PPAR transcription factors.
With aging, the actions of the MHC-PPARγL transgene became apparent: expression of genes involved in both FA uptake and oxidation increased, and the hearts accumulated TG. The hearts were dilated, had decreased ejection fraction, and expressed higher levels of failure markers (BNP and ANF). Other models of cardiac dysfunction show greater pathology with aging. With aging, hearts shift to greater glucose and less FA use even in the absence of any cardiac pathology (31). Perhaps if a similar process occurs in mice, more PPARγ ligands that are normally oxidized might accumulate in older PPARγ1L hearts. Alternatively, the age-related decline in PPARα expression (19) might allow more ligands to bind to PPARγ and permit this transcription factor to associate with unoccupied PPAR response elements.
In contrast to the lack of effect in MHC-PPARγ1L young mice, the MHC-PPARγ1H line had gene changes and evidence of cardiac dysfunction by 2 months. The severe dilated cardiomyopathy in these animals led to 100% mortality by 5 months (Supplemental Figure 3E).
We performed several studies to quantify the in vivo uptake of substrates in the PPARγ1 mice. More FAs derived from VLDL-TG were found in PPARγ1 hearts. Cardiac glucose uptake and GLUT4 expression were either not reduced (PPARγ1L) or were increased (PPARγ1H). This was associated with greater storage of glycogen. These studies contrast with and parallel those found with cardiac overexpression of PPARα (8). MHC-PPARα transgenic mice also have dilated lipotoxic cardiomyopathy. Cardiomyocyte PPARα induced FA oxidation (e.g., AOX and CPT1) and FA uptake genes (CD36). This likely contributed to the increased cardiac lipid in the setting of greater oxidation. In contrast to MHC-PPARγ1 mice, MHC-PPARα mice had a marked reduction in GLUT4 expression and cardiac glucose uptake (8). Thus, PPARγ and PPARα increased the expression of genes for FA oxidation and uptake and caused TG accumulation but differed in their effects on glucose metabolism.
Under normal physiologic conditions, the heart utilizes FAs as its chief energy substrate. Because there is limited capacity for TG storage in the cardiomyocyte, the uptake and oxidation of FAs are tightly coupled. Both increased FA delivery and impaired FA oxidation result in intramyocardial TG accumulation and lipotoxicity. The etiology of heart dysfunction in diabetes and severe obesity is thought to involve lipotoxicity (32).
Reduced cardiac function has been attributed to accumulation of toxic lipids (33), increased FA oxidation, and/or reduced glucose utilization. Reductions in glucose uptake and oxidation alone are associated with cardiac dysfunction; this is especially the case under ischemic conditions (32). GLUT4 mRNA levels are reduced in several models of cardiac lipotoxicity (8, 34) and with streptozotocin-induced diabetes (35, 36). However, the development of lipotoxic dilated cardiomyopathy in the MHC-PPARγ1 mice is not the result of defective glucose uptake. Thus, alterations in lipid metabolism, and not reduced glucose utilization, are likely to be the primary cause of heart dysfunction in the MHC-PPARγ1 mice and other lipotoxic models.
A number of factors, including MEF2 (20, 22) and PGC1β (23), regulate GLUT4 gene expression in skeletal muscle, adipose tissue, and heart. In one study, MEF2A, but not MEF2D, was associated with increased cardiac expression of GLUT4 (37). In our mice, cardiac expression of PPARγ was associated with upregulated MEF2C, MEF2D, and PGC-1β but not MEF2A. Others have suggested that PPARγ alone upregulates GLUT4 expression (38).
At first glance our results appear to conflict with the beneficial effects of PPARγ agonist treatment of animals and perfused hearts (16). However, the in vivo effects of these drugs on the heart are complicated by the changes in plasma and tissue lipid metabolism that accompany their use and non-PPAR effects of these agents (4). The actions of thiazolidinedione drugs on PPAR activation in the heart should be a balance between a reduction in the level of available ligands as a result of more plasma lipids being directed to adipose tissue (17), and a direct cardiac effect. Thus, the beneficial actions of troglitazone (12) and rosiglitazone (13) on lipotoxic cardiomyopathies may result, at least in part, from redirection of lipids away from the heart, rather than direct activation of cardiac PPARγ-regulated genes. Our data showing a reduction in cardiac gene expression in rosiglitazone-treated wild-type mouse hearts are consistent with this explanation. However, with high circulating levels of PPARγ agonists or with very potent agents, cardiac dysfunction similar to that found in our MHC-PPARγ1 mice could result.
The reasons for lipotoxicity are only partially understood. In our model, in addition to TG, ceramide content was increased in MHC-PPARγ1H, but not MCH-PPARγ1L, hearts. Heart ceramide levels are increased in other lipotoxic cardiomyopathies (11, 12). Recently, inhibition of ceramide synthesis has been shown to correct the lipotoxicity leading to skeletal muscle insulin resistance in mice fed a high-fat diet (39). Ceramide has been suggested to lead to apoptosis in some cells (40).
ER stress, which is associated with FA excess and was increased in both lines of MHC-PPARγ1 mice (33), is a cellular response triggered by disruption of the ER homeostasis and consequent accumulation of misfolded proteins in its lumen. To date, several proteins involved in ER stress–induced apoptosis have been identified, including CHOP and Bcl-2 family members (41). CHOP is a transcription factor of the C/EBP family. Its basal expression is low under nonstressed conditions, but it increases markedly in response to physiological and pharmacological stresses (42). Upregulation of CHOP expression indicates that the ER stress–mediated apoptosis pathway was involved in the MHC-PPARγ mouse hearts.
In our MHC-PPARγ mouse hearts, there was evidence of cellular apoptosis; some proapoptotic genes were induced (Bax and caspase-9), but expression of some antiapoptotic genes also increased (Bcl2 and Bcl-XL). It appears likely that cardiac overexpression of PPARγ leading to either increased ceramide or ER stress triggered the intrinsic apoptosis-signaling pathway mediated by caspase-9 but also induced survival signals.
PPARγ agonist treatment of humans is associated with peripheral edema and increased heart failure (43); this was seen in the recent PROactive (44) and Diabetes Reduction Assessment with Ramipril and Rosiglitazone Medication (DREAM) studies (45). In addition, the use of a potent PPARα/PPARγ dual agonist caused cardiomyopathy and more heart failure (46). Tissue-specific deletion of PPARγ in the kidney reduced sodium uptake (47). Thus, it has been postulated that a side effect of PPARγ agonists is salt and water retention, a process that could lead to edema and heart failure (48). However, as has been noted by others (49), salt retention usually leads to hypertension; PPARγ agonists are associated with reduced blood pressure (44). Thus, the reasons for increased heart failure in some patients treated with these drugs may be complex and involve toxic effects of the drugs on the myocardium, a process found only in a minority of patients.
In summary, we have studied the actions of PPARγ1 directly in cardiomyocytes. As is known to occur in adipocytes, PPARγ expression increased both lipid and glucose uptake, the former by an increase in CD36 expression and the latter, in part, by upregulation of GLUT4. The ensuing dilated cardiomyopathy is due to glucolipotoxicity, as both TG and glycogen are inappropriately stored. Rosiglitazone, a PPARγ agonist, increased this toxicity but did not increase cardiac PPARγ downstream gene expression in wild-type mice. In part, this might be due to a redirection of normal PPARγ ligands to other tissues. PPARγ in the mouse heart is increased with streptozotocin-induced diabetes. In addition, this transcription factor is expressed at higher levels in human heart. It is possible that cardiotoxic effects of PPARγ agonists in humans occur due to glucolipotoxocity. Fortunately, this is seen in only a minority of patients whose diabetes and perhaps genetic variation make them unusually sensitive to what is otherwise a useful form of therapy.
Methods
Generation and identification of the MHC-PPARγ mice.
Creation of the mice and all metabolic and genetic studies were reviewed and approved by the Columbia University Institutional Animal Care and Use Committee. A transgenic construct containing a 1.7-kb mouse PPARγ1 cDNA, which differs from PPARγ2 by a lack of 30 additional NH2-terminal amino acids, was cloned downstream of the α-MHC promoter (5.4 kb). Sac1 and Nae1 digestion of MHC-PPARγ1 produced a linear 7.8-kb fragment that was used for microinjection (Figure 1A). Transgenic mice were produced by microinjection of the MHC-PPARγ1 construct into fertilized 1-cell C57BL/6 × CBA F1 eggs. Founder mice and transgenic expression of PPARγ1 were identified by analysis of genomic DNA with primer A (5ι-AGCTGTGGTCCACATTCTTC-3ι; a sense primer specific to MHC promoter exon 2) and antisense primer (5ι-AGAATGGCATCTCTGTGTC-3ι) specific to PPARγ1 cDNA nucleotides 173–192. PPARγ1 mRNA expression (both transgene and endogenous) in the heart was confirmed by RT-PCR with PPARγ1-specific primer B, sense: 5ι-GAGTGTGACGACAAGATTTG-3ι, and antisense: 5ι-GGTGGGCCAGAATGGCATCT-3ι. To compare heart PPARγ expression with that of adipose tissue, we measured heart and adipose tissue PPARγ expression simultaneously using primers that were common to PPARγ1 and PPARγ2 (primer C, sense: 5ι-ATCTACACGATGCTGGC-3ι, and antisense: 5ι-GGATGTCCTCGATGGG-3ι) (Figure 1A).
Western blot analysis.
Fresh heart tissues from 4- and 8-month-old MHC-PPARγ1L and 4-month-old MHC-PPARγ1H mice and their littermate controls were prepared and nuclear proteins were isolated using a nuclear extraction kit according to the manufacturer’s instructions (Active Motif). Thirty micrograms of nuclear proteins were subjected to Western blot analysis with PPARγ antibody (sc-7196; 1:100 dilution; Santa Cruz Biotechnology Inc.). For control of protein loading, the blots were stripped and reacted with anti–lamin B1 antibody (gift from Howard Worman, Columbia University). Bands were quantified by densitometry using Molecular Analysis Software (Bio-Rad).
Heart and plasma lipids.
Blood from fasted (6 hours) mice was collected from retro-orbital plexus for the measurement of plasma total cholesterol (TC), TG, and FFAs. To measure tissue lipids, hearts were perfused with PBS and homogenized in 4°C in 1 M NaCl buffer containing lipase inhibitors to prevent TG hydrolysis. Lipids were extracted from heart tissues (50 mg) according to methods modified from that of Folch et al. (50). The dried lipids were solubilized in PBS containing 2% Triton X-100. Heart and plasma TC, TG, and FFA were measured enzymatically using an Infinity kit (Thermo Electron Corp.) and a NEFA C kit (Wako).
Tissue gene expression.
Total RNA was prepared using a Pure Link Micro-to-Midi Total Purification System kit (Invitrogen). One microgram of RNA was initially treated with DNase I (Invitrogen) for 15 minutes. The RNA samples were then reverse transcribed using the ThermoScript RT-PCR Kit (Invitrogen). Real-time quantitative RT-PCR (qRT-PCR) was performed using an ABI 7700 (Applied Biosystems). Amplification was performed using SYBR Green PCR Master Mix (Applied Biosystems). Primers used for PCR amplification are listed in Supplemental Tables 2 and 3. Analysis was performed using Sequence Detection Software (Applied Biosciences). Standard curves were generated using undiluted and diluted (1:10, 1:100, and 1:1,000) cDNA samples from heart tissue. Correlation coefficients were 0.98 or greater. Data were normalized with 18S rRNA.
Histological analysis.
Neutral lipids were assessed in hearts taken from 24-hour-fasted male and female mice perfused with PBS. The hearts were embedded in Tissue-Tek Optimal Cutting Temperature compound (Sakura). Midventricular sections of myocardium (6 μM in thickness) were stained with oil red O and counterstained with hematoxylin.
PAS and PAS-D staining were used to demonstrate heart glycogen. Hearts from 6-hour-fasted male and female mice were placed in 10% neutral buffered formalin. Midventricular sections were fixed in methyl alcohol for 10 minutes and stained with Schiff Reagent (Poly Scientific) and H&E. The specificity of glycogen staining was demonstrated by treating heart sections with diastase of malt (Sigma-Aldrich), which removes the glycogen.
TUNEL staining.
Cardiac ventricular tissues from 4- and 8-month-old MHC-PPARγ1L and 4-month-old MHC-PPARγ1H were fixed in formalin, embedded in paraffin, and sectioned. Tissues were stained for DNA fragmentation by a TUNEL protocol according to the manufacturer’s specifications (R&D Systems). The data were quantified by counting TUNEL-positive myocytes per square millimeter cellular area.
Echocardiographic analysis.
Two-dimensional echocardiography was performed in conscious mice using techniques described previously (Sonos 5500 system; Philips Medical Systems) (10). Two-dimensional echocardiographic images were obtained and recorded in a digital format. Images were than analyzed off-line by a researcher blinded to the murine genotype. Left ventricular end-diastolic dimension (LVDd) and left ventricular end-systolic dimension (LVDs) were measured. Percent fractional shortening was calculated as: % FS = ([LVDd — LVDs] / LVDd) × 100.
Electron microscopy.
Left ventricles from 8-month-old mice were fixed with 2.5% glutaraldehyde in 0.1 M Sorensen’s buffer (0.2 M monobasic phosphate/0.2 M dibasic phosphate 1:4 vol/vol; pH 7.2), postfixed in osmium tetroxide, and embedded in EPON 812 (Electron Microscopy Sciences). Ultrathin sections were stained with uranyl acetate and lead citrate and examined under a JEM-1200ExII electron microscope (JEOL).
Uptake of VLDL and glucose.
Human VLDL was isolated from normal subjects by sequential ultracentrifugation and was labeled with [carboxyl-14C]triolein (PerkinElmer) via cholesterol ester transfer protein (CETP) as previously described (51). Sixteen-hour-fasted MHC-PPARγ1 mice and littermates were first injected intravenously with 1 × 106 decays per minute (DPM) of 2-deoxy-d-[3H]glucose (PerkinElmer). Fifty-five minutes after 2-deoxy-d-[3H]glucose injection, 1 × 106 DPM of [14C-TG]VLDL was injected. Five minutes after VLDL injection, blood was collected, and the vasculature was thoroughly perfused with 10 ml of PBS via cardiac puncture. Tissues were then excised, and accumulated radioactivity for [3H]glucose and [14C-TG]VLDL was measured. Amounts of glucose and VLDL injected were adjusted by plasma radioactivity counts at 30 seconds after each injection and were compared with plasma counts at the end of the experiments. Tissue uptake for transgenic mice was compared with uptake of control littermate controls.
Ceramide content.
Cardiac ceramide levels were determined using the diacylglyceride kinase method as described previously (52).
Human tissues.
Normal human heart tissues were obtained from National Disease Research Interchange. Hearts from 11 orthotopic heart transplant recipients (50.3 ± 8.8 years old) with idiopathic dilated cardiomyopathy were obtained from the Department of Surgery at Columbia University Medical Center (CUMC). The Institutional Review Board at CUMC approved all study protocols. Normal human left ventricle total RNA was purchased from Ambion (no. 5856). Human cardiac PPARγ1 mRNA expression was measured by qRT-PCR and normalized with human 18S rRNA. Primer sequences used for qRT-PCR were shared by both mouse and human genes (Supplemental Table 3). BLAST (http://www.ncbi.nlm.nih.gov/blast/) analysis showed that these primers were 97% and 90% homologous between human and mouse PPARγ1 mRNA and 18S rRNA, respectively.
Rosiglitazone treatment.
Four- and 8-month-old mice were fed rosiglitazone-containing chow for 15 days. The chow was produced by Research Diets and contained 200 mg/kg of drug; this is equivalent to a dose of 10 mg/kg/d in an adult mouse that consumes on average 4.5 g/d of chow. These doses were similar to those used by other investigators (53).
Statistics.
We analyzed data using the Prism software package (GraphPad Software). Comparisons between 2 groups were performed using unpaired 2-tailed Student’s t tests. All values are presented as mean ± SD. Differences between groups were considered statistically significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank T. Seo and Y. Hu for help with the kinetic studies and animal husbandry. This work was funded by grants HL73029, HL77113 Specialized Centers of Clinically Oriented Research (SCCOR), and HL62583 from the National Heart, Lung, and Blood Institute. M. Yokoyama and H. Yamashita were supported by a Mentor-Based Postdoctoral Fellowship from the American Diabetes Association.
Footnotes
Nonstandard abbreviations used: ADRP, adipose differentiation–related protein; AOX, acyl-CoA oxidase; CD36, FA translocase; CHOP, CCAAT/enhancer-binding protein (C/EBP) homologous protein; CPT1, carnitine palmitoyl transferase–1; FA, fatty acid; FAS, FA synthase; GLUT4, glucose transporter 4; MEF, myocyte enhancer factor; α-MHC, α–myosin heavy chain; PGC1, PPARγ coactivator 1; qRT-PCR, real-time quantitative RT-PCR; TG, triglyceride.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2791–2801 (2007). doi:10.1172/JCI30335
References
- 1.Lee C.H., Olson P., Evans R.M. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- 2.Staels B., Fruchart J.C. Therapeutic roles of peroxisome proliferator-activated receptor agonists. Diabetes. 2005;54:2460–2470. doi: 10.2337/diabetes.54.8.2460. [DOI] [PubMed] [Google Scholar]
- 3.Barger P.M., Kelly D.P. PPAR signaling in the control of cardiac energy metabolism. Trends Cardiovasc. Med. 2000;10:238–245. doi: 10.1016/s1050-1738(00)00077-3. [DOI] [PubMed] [Google Scholar]
- 4.Duan S.Z., Ivashchenko C.Y., Russell M.W., Milstone D.S., Mortensen R.M. Cardiomyocyte-specific knockout and agonist of peroxisome proliferator-activated receptor-gamma both induce cardiac hypertrophy in mice. Circ. Res. 2005;97:372–379. doi: 10.1161/01.RES.0000179226.34112.6d. [DOI] [PubMed] [Google Scholar]
- 5.Yu S., et al. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J. Biol. Chem. 2003;278:498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 6.Metzger D., et al. Functional role of RXRs and PPARgamma in mature adipocytes. Prostaglandins Leukot. Essent. Fatty Acids. 2005;73:51–58. doi: 10.1016/j.plefa.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Schaffer J.E. Lipotoxicity: when tissues overeat. Curr. Opin. Lipidol. 2003;14:281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Finck B.N., et al. The cardiac phenotype induced by PPARα overexpression mimics that caused by diabetes mellitus. J. Clin. Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barouch L.A., Berkowitz D.E., Harrison R.W., O’Donnell C.P., Hare J.M. Disruption of leptin signaling contributes to cardiac hypertrophy independently of body weight in mice. Circulation. 2003;108:754–759. doi: 10.1161/01.CIR.0000083716.82622.FD. [DOI] [PubMed] [Google Scholar]
- 10.Yagyu H., et al. Lipoprotein lipase (LpL) on the surface of cardiomyocytes increases lipid uptake and produces a cardiomyopathy. J. Clin. Invest. 2003;111:419–426. doi: 10.1172/JCI16751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu H.C., et al. A novel mouse model of lipotoxic cardiomyopathy. J. Clin. Invest. 2001;107:813–822. doi: 10.1172/JCI10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y.T., et al. Lipotoxic heart disease in obese rats: implications for human obesity. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1784–1789. doi: 10.1073/pnas.97.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vikramadithyan R.K., et al. Peroxisome proliferator-activated receptor agonists modulate heart function in transgenic mice with lipotoxic cardiomyopathy. J. Pharmacol. Exp. Ther. 2005;313:586–593. doi: 10.1124/jpet.104.080259. [DOI] [PubMed] [Google Scholar]
- 14.Asakawa M., et al. Peroxisome proliferator-activated receptor gamma plays a critical role in inhibition of cardiac hypertrophy in vitro and in vivo. Circulation. 2002;105:1240–1246. doi: 10.1161/hc1002.105225. [DOI] [PubMed] [Google Scholar]
- 15.Abdelrahman M., Sivarajah A., Thiemermann C. Beneficial effects of PPAR-gamma ligands in ischemia-reperfusion injury, inflammation and shock. Cardiovasc. Res. 2005;65:772–781. doi: 10.1016/j.cardiores.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Golfman L.S., et al. Activation of PPARgamma enhances myocardial glucose oxidation and improves contractile function in isolated working hearts of ZDF rats. Am. J. Physiol. Endocrinol. Metab. 2005;289:E328–E336. doi: 10.1152/ajpendo.00055.2005. [DOI] [PubMed] [Google Scholar]
- 17.Rangwala S.M., Lazar M.A. Peroxisome proliferator-activated receptor gamma in diabetes and metabolism. Trends Pharmacol. Sci. 2004;25:331–336. doi: 10.1016/j.tips.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Chawla A., et al. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 19.Iemitsu M., et al. Aging-induced decrease in the PPAR-alpha level in hearts is improved by exercise training. Am. J. Physiol. Heart Circ. Physiol. 2002;283:H1750–H1760. doi: 10.1152/ajpheart.01051.2001. [DOI] [PubMed] [Google Scholar]
- 20.Thai M.V., Guruswamy S., Cao K.T., Pessin J.E., Olson A.L. Myocyte enhancer factor 2 (MEF2)-binding site is required for GLUT4 gene expression in transgenic mice. Regulation of MEF2 DNA binding activity in insulin-deficient diabetes. J. Biol. Chem. 1998;273:14285–14292. doi: 10.1074/jbc.273.23.14285. [DOI] [PubMed] [Google Scholar]
- 21.Knight J.B., Eyster C.A., Griesel B.A., Olson A.L. Regulation of the human GLUT4 gene promoter: interaction between a transcriptional activator and myocyte enhancer factor 2A. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14725–14730. doi: 10.1073/pnas.2432756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mora S., Pessin J.E. The MEF2A isoform is required for striated muscle-specific expression of the insulin-responsive GLUT4 glucose transporter. J. Biol. Chem. 2000;275:16323–16328. doi: 10.1074/jbc.M910259199. [DOI] [PubMed] [Google Scholar]
- 23.Michael L.F., et al. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M.Y., Unger R.H. Role of PP2C in cardiac lipid accumulation in obese rodents and its prevention by troglitazone. Am. J. Physiol. Endocrinol. Metab. 2005;288:E216–E221. doi: 10.1152/ajpendo.00004.2004. [DOI] [PubMed] [Google Scholar]
- 25.Unger R.H. Lipotoxic diseases. Annu. Rev. Med. 2002;53:319–336. doi: 10.1146/annurev.med.53.082901.104057. [DOI] [PubMed] [Google Scholar]
- 26.Jones J.R., et al. Deletion of PPARgamma in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6207–6212. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J., et al. Selective disruption of PPARgamma 2 impairs the development of adipose tissue and insulin sensitivity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10703–10708. doi: 10.1073/pnas.0403652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gavrilova O., et al. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J. Biol. Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y.X., et al. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 2003;113:159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 30.Braissant O., Foufelle F., Scotto C., Dauca M., Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 31.Soto P.F., et al. Impact of aging on myocardial metabolic response to dobutamine. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H2158–H2164. doi: 10.1152/ajpheart.00086.2003. [DOI] [PubMed] [Google Scholar]
- 32.Taegtmeyer H., McNulty P., Young M.E. Adaptation and maladaptation of the heart in diabetes: Part I: general concepts. Circulation. 2002;105:1727–1733. doi: 10.1161/01.cir.0000012466.50373.e8. [DOI] [PubMed] [Google Scholar]
- 33.Listenberger L.L., Schaffer J.E. Mechanisms of lipoapoptosis: implications for human heart disease. Trends Cardiovasc. Med. 2002;12:134–138. doi: 10.1016/s1050-1738(02)00152-4. [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama M., et al. Apolipoprotein B production reduces lipotoxic cardiomyopathy: studies in heart-specific lipoprotein lipase transgenic mouse. J. Biol. Chem. 2004;279:4204–4211. doi: 10.1074/jbc.M311995200. [DOI] [PubMed] [Google Scholar]
- 35.Garvey W.T., Hardin D., Juhaszova M., Dominguez J.H. Effects of diabetes on myocardial glucose transport system in rats: implications for diabetic cardiomyopathy. Am. J. Physiol. 1993;264:H837–H844. doi: 10.1152/ajpheart.1993.264.3.H837. [DOI] [PubMed] [Google Scholar]
- 36.Depre C., et al. Streptozotocin-induced changes in cardiac gene expression in the absence of severe contractile dysfunction. J. Mol. Cell. Cardiol. 2000;32:985–996. doi: 10.1006/jmcc.2000.1139. [DOI] [PubMed] [Google Scholar]
- 37.Mora S., Yang C., Ryder J.W., Boeglin D., Pessin J.E. The MEF2A and MEF2D isoforms are differentially regulated in muscle and adipose tissue during states of insulin deficiency. Endocrinology. 2001;142:1999–2004. doi: 10.1210/endo.142.5.8160. [DOI] [PubMed] [Google Scholar]
- 38.Al-Khalili L., et al. Enhanced insulin-stimulated glycogen synthesis in response to insulin, metformin or rosiglitazone is associated with increased mRNA expression of GLUT4 and peroxisomal proliferator activator receptor gamma co-activator 1. Diabetologia. 2005;48:1173–1179. doi: 10.1007/s00125-005-1741-3. [DOI] [PubMed] [Google Scholar]
- 39.Holland W.L., et al. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 2007;5:167–179. doi: 10.1016/j.cmet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Shimabukuro M., Zhou Y.T., Levi M., Unger R.H. Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2498–2502. doi: 10.1073/pnas.95.5.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J., Kaufman R.J. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 42.Oyadomari S., Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 43.Nesto R.W., et al. Thiazolidinedione use, fluid retention, and congestive heart failure: a consensus statement from the American Heart Association and American Diabetes Association. Circulation. 2003;108:2941–2948. doi: 10.1161/01.CIR.0000103683.99399.7E. [DOI] [PubMed] [Google Scholar]
- 44.Dormandy J.A., et al. Secondary prevention of macrovascular events in patients with type 2 diabetes in the PROactive Study (PROspective pioglitAzone Clinical Trial In macroVascular Events): a randomised controlled trial. Lancet. 2005;366:1279–1289. doi: 10.1016/S0140-6736(05)67528-9. [DOI] [PubMed] [Google Scholar]
- 45.Gerstein H.C., et al. Effect of rosiglitazone on the frequency of diabetes in patients with impaired glucose tolerance or impaired fasting glucose: a randomised controlled trial. Lancet. 2006;368:1096–1105. doi: 10.1016/S0140-6736(06)69420-8. [DOI] [PubMed] [Google Scholar]
- 46.Nissen S.E., Wolski K., Topol E.J. Effect of muraglitazar on death and major adverse cardiovascular events in patients with type 2 diabetes mellitus. JAMA. 2005;294:2581–2586. doi: 10.1001/jama.294.20.joc50147. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H., et al. Collecting duct-specific deletion of peroxisome proliferator-activated receptor gamma blocks thiazolidinedione-induced fluid retention. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9406–9411. doi: 10.1073/pnas.0501744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan Y., et al. Thiazolidinediones expand body fluid volume through PPARgamma stimulation of ENaC-mediated renal salt absorption. Nat. Med. 2005;11:861–866. doi: 10.1038/nm1278. [DOI] [PubMed] [Google Scholar]
- 49.Staels B. Fluid retention mediated by renal PPARgamma. Cell Metab. 2005;2:77–78. doi: 10.1016/j.cmet.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 51.Seo T., et al. Lipoprotein lipase-mediated selective uptake from low density lipoprotein requires cell surface proteoglycans and is independent of scavenger receptor class B type 1. J. Biol. Chem. 2000;275:30355–30362. doi: 10.1074/jbc.M910327199. [DOI] [PubMed] [Google Scholar]
- 52.Perry D.K., Bielawska A., Hannun Y.A. Quantitative determination of ceramide using diglyceride kinase. Methods Enzymol. 2000;312:22–31. doi: 10.1016/s0076-6879(00)12897-6. [DOI] [PubMed] [Google Scholar]
- 53.Watkins S.M., Reifsnyder P.R., Pan H.J., German J.B., Leiter E.H. Lipid metabolome-wide effects of the PPARgamma agonist rosiglitazone. J. Lipid Res. 2002;43:1809–1817. doi: 10.1194/jlr.m200169-jlr200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.