All three components of the toluene dioxygenase system have been expressed, purified and crystallized.
Keywords: toluene 2,3-dioxygenase enzyme system
Abstract
Pseudomonas putida F1 can grow with toluene as its sole source of carbon and energy. The initial reaction of the degradation of toluene is catalyzed by a three-component toluene dioxygenase enzyme system consisting of a reductase (ReductaseTOL), a ferredoxin (FerredoxinTOL) and a Rieske non-heme iron dioxygenase (OxygenaseTOL). The three components and the apoenzyme of the dioxygenase (apo-OxygenaseTOL) were overexpressed, purified and crystallized. ReductaseTOL diffracts to 1.8 Å and belongs to space group P41212, with unit-cell parameters a = b = 77.1, c = 156.3 Å. FerredoxinTOL diffracts to 1.2 Å and belongs to space group P21, with unit-cell parameters a = 30.5, b = 52.0, c = 30.95 Å, β = 113.7°. Apo-OxygenaseTOL and OxygenaseTOL diffract to 3.2 Å and belong to space group P4332, with unit-cell parameters a = 235.9 Å and a = 234.5 Å, respectively.
1. Introduction
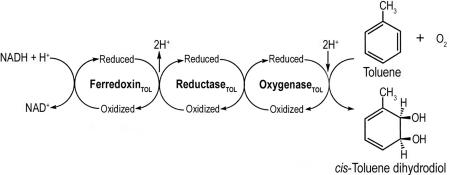
Toluene is used extensively as a gasoline component and as an industrial solvent (Greenberg, 1997 ▶). Unfortunately, this widespread use has resulted in the identification of toluene as a frequent pollutant of soils, groundwater and the urban atmosphere. Bacteria play a key role in the biodegradation of toluene. For example, Pseudomonas putida F1 can grow with toluene as its sole source of carbon and energy (Gibson et al., 1968 ▶). This organism initiates the catabolism of toluene by the enantioselective addition of dioxygen to the aromatic nucleus to form cis-(1R,2S)-dihydroxy-3-methylcyclohexa-3,5-diene (cis-toluene dihydrodiol; Gibson et al., 1970 ▶; Kobal et al., 1973 ▶). The reaction is catalyzed by the three-component toluene dioxygenase enzyme system organized as shown in Fig. 1 ▶. The reductase (ReductaseTOL) is a flavoprotein (Subramanian et al., 1981 ▶) which transfers electrons from NADH to a Rieske [2Fe–2S] ferredoxin (FerredoxinTOL; Subramanian et al., 1985 ▶). The latter shuttles electrons to the dioxygenase (OxygenaseTOL), which catalyzes the formation of cis-toluene dihydrodiol (Subramanian et al., 1979 ▶). OxygenaseTOL is a Rieske non-heme iron dioxygenase and a member of the toluene/biphenyl family (Gibson & Parales, 2000 ▶). Previous studies have suggested that OxygenaseTOL has an α2β2 subunit composition, with each α-subunit containing a Rieske [2Fe–2S] cluster and mononuclear iron at the active site (Jiang et al., 1996 ▶). More recent work on the structure of naphthalene (Kauppi et al., 1998 ▶), nitrobenzene (Friemann et al., 2005 ▶) and biphenyl dioxygenases (Furusawa et al., 2004 ▶) has shown that each dioxygenase component has an α3β3 subunit. The α-subunits from each protein contain a Rieske [2Fe–2S] cluster coordinated to two histidine and two cysteine residues and a His,His-carboxylate facial triad (Kauppi et al., 1998 ▶; Hegg & Que, 1997 ▶) which binds mononuclear iron at the active site. The genes encoding ReductaseTOL (todA), FerredoxinTOL (todB) and OxygenaseTOL (todC1 and todC2) have been cloned and their respective nucleotide sequences determined (Zylstra & Gibson, 1989 ▶; Huang, 1991 ▶). To date, more than 30 Rieske non-heme iron oxygenase systems have been identified by sequence analysis and/or protein purification. However, three-dimensional structures are only available for a few of them. In this paper, we present the overexpression, purification and crystallization of the three components of the OxygenaseTOL enzyme system, along with X-ray diffraction data.
Figure 1.
Dihydroxylation of toluene to cis-toluene dihydrodiol catalyzed by the three-component toluene dioxygenase enzyme system.
2. Materials and methods
2.1. Expression of the three components of the toluene dioxygenase enzyme system
Escherichia coli CGSC#7692 (pDTG601A) containing the cloned todC1C2BA genes that encode OxygenaseTOL, FerredoxinTOL and ReductaseTOL from P. putida F1 were expressed under the control of a IPTG-inducible tac promotor carried on the expression vector pKK223-3 (Pharmacia) (Zylstra & Gibson, 1991 ▶). E. coli CGSC#7692 is a tryptophanase (tnaA5) negative strain, which does not produce indigo on expression of the toluene dioxygenase enzyme system. The tryptophanase-negative strain was necessary in order to be able to grow the cells in rich media that contain tryptophan. Tryptophanase cleaves tryptophan and produces indole. Indole is a good substrate for the enzyme system and results in indigo production, which ends up being toxic to the cell and results in lower yields. Cells were grown at 291 K in a 8 l fermentor inoculated with a 500 ml seed culture to an OD600nm of 2.4 in a medium containing yeast extract (80 g), tryptone (80 g), NaCl (40 g), tryptophan (0.4 g), ampicillin (0.4 g) and ferrous ammonium sulfate (0.4 g). Protein expression was induced with IPTG (1 g) for 3 h. Cells were harvested by centrifugation at 14 300g at 277 K for 10 min, suspended in BTGD buffer (50 mM Bis-Tris pH 6.8, 5% glycerol, 1 mM dithiothreitol; Lee et al., 1997 ▶) and stored at 201 K until use.
2.2. Purification
All purification procedures were performed at 286 K using an automated FPLC system (Bio-Rad Laboratories, Hercules, CA, USA). Chromatography columns and column resins were from Pharmacia LKB.
Frozen IPTG-induced recombinant E. coli cells were thawed and DNase I was added to a final concentration of 1 µg ml−1. The cells were broken by passage through a chilled French pressure cell at 137 MPa. The crude cell-free extract was obtained from the supernatant following ultracentrifugation at 146 000g for 1 h at 277 K.
2.2.1. Separation of ReductaseTOL, FerredoxinTOL and OxygenaseTOL
Crude cell extract (6.6 g) was applied onto a XK50/30 chromatography column with Q-Sepharose FF (5 × 25 cm) pre-equilibrated with BTDG buffer. The column was washed with 400 ml BTGD buffer containing 0.2 M KCl at a flow rate of 2 ml min−1 to elute unbound proteins. Bound proteins were eluted with a 2 l linear KCl gradient (0.2–0.5 M) in BTGD buffer at a flow rate of 2 ml min−1. The proteins were eluted in the order ReductaseTOL, FerredoxinTOL and OxygenaseTOL.
2.2.2. Purification of ReductaseTOL
Fractions containing active ReductaseTOL were pooled, concentrated and desalted by ultracentrifugation over an Amicon YM-30 membrane. After centrifugation, the supernatant was applied onto a Blue Sepharose CL-6B column (2.6 × 28 cm) pre-equilibrated with BTGD buffer. The column was eluted with BTGD buffer at a flow rate of 0.5 ml min−1. Fractions containing ReductaseTOL were pooled, concentrated and desalted by ultrafiltration over an Amicon YM-30 membrane. After centrifugation, the supernatant was applied onto a DEAE cellulose DE52 column (2.6 × 12 cm) pre-equilibrated with BTGD buffer. The bound protein was eluted with a 450 ml linear KCl gradient (0–0.5 M) in BTGD buffer at a flow rate of 0.5 ml min−1. Fractions containing ReductaseTOL were pooled, desalted, concentrated by ultrafiltration and centrifugated. The supernatant protein solution in BTGD buffer was rapidly frozen in liquid nitrogen and stored at 345 K.
2.2.3. Purification of OxygenaseTOL
Fractions containing active OxygenaseTOL from the Q-Sepharose FF column were pooled and concentrated by ultrafiltration over an Amicon YM-100 membrane. Solid (NH4)2SO4 was slowly added under stirring to a final concentration of ∼1 M. After centrifugation, the supernatant was applied onto a butyl Sepharose 4B column (2.6 × 26 cm) equilibrated with BTGD buffer containing 1 M (NH4)2SO4. OxygenaseTOL was eluted with a 450 ml decreasing linear (NH4)2SO4 gradient (1–0 M) followed by 150 ml BTGD buffer at a flow rate of 0.5 ml min−1. Active fractions were pooled, desalted and concentrated by ultrafiltration over a YM100 membrane. The purified OxygenaseTOL in BTGD buffer was stored as described above.
2.2.4. Purification of FerredoxinTOL
Because the level of expression of FerredoxinTOL was not sufficient in E. coli CGSC#7692 (pDTG601A), we used E. coli JM109 (pDTG614) (Zylstra & Gibson, 1991 ▶) for purification of FerredoxinTOL. The conditions for production of the cell extract and running the first Q-Sepharose FF column with the cell extract of E. coli JM109 (pDTG614) were the same as described in §2.2.1.
Fractions containing active FerredoxinTOL from the Q-Sepharose FF column were pooled and concentrated by ultracentrifugation over an Amicon YM-10 membrane as described above. (NH4)2SO4 was added under slow stirring to a final concentration of 1.6 M. The precipitate was centrifugated and the supernatant was applied onto a Octyl Sepharose column (2.6 × 16.5 cm) equilibrated with BTGD buffer containing 1.6 M (NH4)2SO4. The column was eluted isoratically with a flow rate of 0.4 ml min−1. Fractions containing FerredoxinTOL were pooled, concentrated, washed with BTGD buffer by ultrafiltration and stored as described above.
2.3. Analytical procedures and enzyme assays
Samples of purified proteins were analyzed on 12.5% SDS–PAGE and by dynamic light scattering using a light-scattering apparatus from Protein Solutions. The fractions containing the active TDO components during purification were assayed with 14C toluene as the substrate in the presence of the other two components as described previously (Jiang et al., 1996 ▶). In addition, the active fractions containing FerredoxinTOL and OxygenaseTOL components could be identified and pooled from their brown colour. Protein concentrations were determined by the BCA assay (Pierce, Rockford, IL, USA) using bovine serum albumin as the standard. The activity of OxygenaseTOL was determined by measuring the rate of oxygen consumption with a Clark-type oxygen electrode (Rank Brothers, Cambridge, England) in a chamber containing 1 ml 50 mM MES buffer pH 6.8, 0.25 mM NADH, 0.61 µM ReductaseTOL, 2.5 µM FerredoxinTOL and 0.1 µM OxygenaseTOL by injection of 2 µl toluene (0.1 M dissolved in methanol) in the presence or absence of ferrous iron (0.125 mM) as described previously (Lee, 1999 ▶).
2.4. Crystallization
Crystallization was performed by the hanging-drop or sitting-drop vapor-diffusion method. In all the experiments except for OxygenaseTOL (see §3), 1.5–2 µl protein solution and 1.5–2 µl precipitant solution were equilibrated against 0.7–1 ml precipitant solution. The initial crystallization conditions were found using Hampton Crystal Screen (Hampton Research, USA) and Wizard I and II Screens (Emerald Biostructures, USA).
3. Results and discussion
The three components of the toluene dioxygenase enzyme system have been purified from wild-type P. putida strain F1 and characterized (Subramanian et al., 1979 ▶, 1981 ▶, 1985 ▶). In this study, we have further optimized the purifications from recombinant E. coli cells to obtain large quantities of proteins for crystallization. The procedures described in §2 gave 25 mg ReductaseTOL, 19.2 mg FerredoxinTOL and 380 mg OxygenaseTOL, with purification folds of 66, 103 and 8, respectively. The preparations were homogenous when analyzed by SDS–PAGE and gave the same absorption spectra as reported previously (Subramanian et al., 1979 ▶, 1981 ▶, 1985 ▶). The purified OxygenaseTOL yielded a specific activity of 2.15 µmol min−1 mg−1 (equivalent to a turnover of 2.7 s−1) in the presence of ferrous iron in the assay mixture. However, the activity was negligible in the absence of added ferrous iron, indicating that labile mononuclear iron was lost during purification, as OxygenaseTOL retained the visible spectrum arising from Rieske [2Fe–2S] clusters. Thus, we referred to the initially purified protein as OxygenaseTOL apoenzyme (apo-OxygenaseTOL).
3.1. Crystallization
Photographs of the crystals of ReductaseTOL, FerredoxinTOL and OxygenaseTOL are presented in Fig. 2 ▶.
Figure 2.
Photographs of crystals of ReductaseTOL (a), FerredoxinTOL (b) and OxygenaseTOL (c).
ReductaseTOL (25 mg ml−1 in half-strength BTGD buffer) initially crystallized in 0.1 M Tris pH 8.5, 1.5 M (NH4)2SO4 and 12% glycerol at 286 K. These conditions were optimized to 1.4 M (NH4)2SO4 and 0.1 M HEPES pH 7.7. Crystals (0.1 × 0.1 × 0.3 mm) were formed in two weeks.
FerredoxinTOL (6 mg ml−1 in half-strength BTGD buffer) crystallized at 286 K in 38% PEG 8000, 0.1 M MES pH 6.1 or 37% PEG 8000, 0.1 M MES pH 5.8. Crystals (0.1 × 0.2 × 0.3 mm) formed within 3 d.
Apo-OxygenaseTOL (50 mg ml−1 in BTGD buffer) crystallized at 286 K in 40%(w/v) polyethylene glycol 600, 0.1 M sodium citrate pH 5.8 at 286 K. Cubic crystals (0.1 × 0.1 × 0.1 to 1.0 × 1.0 × 1.0 mm) were obtained within one week.
Co-crystallization experiments with Fe(NH4)2(SO4)2 were performed under strict anaerobic conditions as described for apo-OxygenaseTOL. Cubic crystals (0.1 × 0.1 × 0.1 to 1.0 × 1.0 × 1.0 mm) were obtained within 24 h in 40%(w/v) polyethylene glycol 600, 0.1 M sodium citrate pH 6.1, 20–50 mM Fe(NH4)2(SO4)2 at 296 K.
3.2. X-ray diffraction study
Diffraction data were collected under liquid-nitrogen cryoconditions at 100 K. Data-collection statistics are summarized in Table 1 ▶.
Table 1. X-ray data-collection statistics.
Values in parentheses indicate the statistics for the last resolution shell.
| ReductaseTOL | FerredoxinTOL | Apo-OxygenaseTOL | OxygenaseTOL | |
|---|---|---|---|---|
| Space group | P41212 | P21 | P4332 | P4332 |
| Unit-cell parameters (Å, °) | a = b = 77.13, c = 156.38 | a = 30.54, b = 52.09, c = 30.95, β = 113.67 | a = b = c = 235.87 | a = b = c = 234.53 |
| Beamline | X26A, BNL | ID14-1, ESRF | 17-ID, APS | ID14-2, ESRF |
| Resolution (Å) | 19.9–1.8 (1.86–1.8) | 28.3–1.2 (1.26–1.20) | 48.0–3.2 (3.37–3.2) | 30.0–3.2 (3.37–3.2) |
| No. of observations | 409869 | 91725 | 366056 | 886708 |
| Unique reflections | 44352 | 25558 | 37544 | 36887 |
| Completeness (%) | 99.0 (96.4) | 98.2 (96.8) | 100 (100) | 99.9 (100) |
| Rmerge† | 7.6 (38.6) | 8.8 (39.9) | 12.3 (67.2) | 11.8 (38.9) |
| I/σ(I) | 14.6 (4.0) | 3.7 (2.1) | 5.6 (1.2)‡ | 6.0 (2.0) |
| Multiplicity | 9.2 (7.0) | 3.4 (3.0) | 9.7 (8.5) | 24.0 (24.7) |
R
merge = 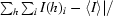
 , where I(h) is the intensity of a reflection h,
, where I(h) is the intensity of a reflection h,  is the sum over all reflections and
is the sum over all reflections and  is the sum over I measurements of reflection h.
is the sum over I measurements of reflection h.
I/σ(I) is 2.00 for the apo-Oxygenase at 3.5 Å resolution.
ReductaseTOL crystals were cryoprotected using reservoir solution supplemented with 10%(v/v) glycerol and flash-cooled by rapidly moving them into a cold nitrogen stream. X-ray diffraction data were collected at beamline X26A at Brookhaven National Laboratories (BNL), Upton, NY, USA using a ADSC Q4 CCD detector. The crystals diffracted to 1.8 Å and belong to space group P41212, with unit-cell parameters a = b = 77.13, c = 156.38 Å. The asymmetric unit contain one molecule of ReductaseTOL, corresponding to a solvent content of 54%. The data were indexed, integrated, scaled and merged using d*TREK (Pflugrath, 1999 ▶).
FerredoxinTOL crystals were soaked in a cryoprotection solution consisting of the reservoir solution with the addition of 25%(v/v) polyethylene glycol 400 for 10–30 s upon flash-cooling in liquid nitrogen. X-ray diffraction data were collected at beamline ID14-1 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France using an ADSC Q4 CCD detector. The crystals belongs to space group P21, with unit-cell parameters a = 30.54, b = 52.09, c = 30.95 Å, β = 113.67°, and diffracted to 1.2 Å. These crystals have a solvent content of 35%. The data were indexed, integrated, scaled and merged using MOSFLM (Powell, 1999 ▶) and SCALA (Collaborative Computational Project, Number 4, 1994 ▶).
The OxygenaseTOL apoenzyme crystals were flashed-cooled in liquid nitrogen after a 10–30 s soak in a cryoprotection solution consisting of the reservoir solution with the addition of 25%(v/v) glycerol. Crystals of OxygenaseTOL co-crystallized with Fe(NH4)2(SO4)2 were flashed-cooled in liquid nitrogen inside an anaerobic box after a 10–30 s soak in a cryoprotection solution consisting of the reservoir solution with the addition of 25%(v/v) glycerol. X-ray diffraction data were collected on beamline ID14-2 at the ESRF, Grenoble, France using an ADSC Q4 CCD detector and on beamline 17-ID (IMCA-CAT) at the Advanced Photon Source (APS), Argonne, IL, USA using an ADSC Q4 CCD detector. The apoenzyme and OxygenaseTOL co-crystallized with Fe(NH4)2(SO4)2 belong to space group P4332. The asymmetric unit contain one αβ heterodimer, with a solvent content of 84% as estimated by the program TRUNCATE (Collaborative Computational Project, Number 4, 1994 ▶). The data were indexed, integrated, scaled and merged using MOSFLM (Powell, 1999 ▶) and SCALA (Collaborative Computational Project, Number 4, 1994 ▶).
Acknowledgments
We thank Karin Valegård and Janos Hajdu for help with the anaerobic box. The work was supported by US Public Health Service grant GM62904 to SR.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Friemann, R., Ivkovic Jensen, M. M., Lessner, D. J., Yu, C.-L., Gibson, D. T., Parales, R. E., Eklund, H. & Ramaswamy, S. (2005). J. Mol. Biol.348, 1139–1151. [DOI] [PubMed] [Google Scholar]
- Furusawa, Y., Nagarajan, V., Tanokura, M., Masai, E., Fukuda, M. & Senda, T. (2004). J. Mol. Biol.342, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Gibson, D. T., Hensley, M., Yoshioka, H. & Mabry, T. J. (1970). Biochemistry, 9, 1626–1630. [DOI] [PubMed] [Google Scholar]
- Gibson, D. T., Koch, J. R. & Kallio, R. E. (1968). Biochemistry, 7, 2653–2662. [DOI] [PubMed] [Google Scholar]
- Gibson, D. T. & Parales, R. E. (2000). Curr. Opin. Biotechnol.11, 236–243. [DOI] [PubMed] [Google Scholar]
- Greenberg, M. M. (1997). Environ. Res.72, 1–7. [DOI] [PubMed] [Google Scholar]
- Hegg, E. L. & Que, L. Jr (1997). Eur. J. Biochem.250, 625–629. [DOI] [PubMed] [Google Scholar]
- Huang, S.-L. (1991). PhD thesis. University of Iowa, USA.
- Jiang, H., Parales, R. E., Lynch, N. A. & Gibson, D. T. (1996). J. Bacteriol.178, 3133–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi, B., Lee, K., Carredano, E., Parales, R. E., Gibson, D. T., Eklund, H. & Ramaswamy, S. (1998). Structure, 6, 571–586. [DOI] [PubMed] [Google Scholar]
- Kobal, V. M., Gibson, D. T., Davis, R. E. & Garza, A. (1973). J. Am. Chem. Soc.95, 4420–4421. [DOI] [PubMed] [Google Scholar]
- Lee, K. (1999). J. Bacteriol.181, 2719–2725. [Google Scholar]
- Lee, K., Kauppi, B., Parales, R. E., Gibson, D. T. & Ramaswamy, S. (1997). Biochem. Biophys. Res. Commun.241, 553–557. [DOI] [PubMed] [Google Scholar]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- Powell, H. R. (1999). Acta Cryst. D55, 1690–1695. [DOI] [PubMed] [Google Scholar]
- Subramanian, V., Liu, T. N., Yeh, W. K. & Gibson, D. T. (1979). Biochem. Biophys. Res. Commun.91, 1131–1139. [DOI] [PubMed] [Google Scholar]
- Subramanian, V., Liu, T. N., Yeh, W. K., Narro, M. & Gibson, D. T. (1981). J. Biol. Chem.256, 2723–2730. [PubMed] [Google Scholar]
- Subramanian, V., Liu, T. N., Yeh, W. K., Serdar, C. M., Wackett, L. P. & Gibson, D. T. (1985). J. Biol. Chem.260, 2355–2363. [PubMed] [Google Scholar]
- Zylstra, G. J. & Gibson, D. T. (1989). J. Biol. Chem.264, 14940–14946. [PubMed] [Google Scholar]
- Zylstra, G. J. & Gibson, D. T. (1991). Genet. Eng. (N. Y.), 13, 183–203. [DOI] [PubMed] [Google Scholar]