Abstract
Treatment of HIV-infected individuals with antiretroviral agents selects for drug-resistant mutants, resulting in frequent treatment failures. Although the major antiretroviral resistance mutations are routinely characterized by DNA sequencing, treatment failures are still common, probably in part because undetected rare resistance mutations facilitate viral escape. Here we combined DNA bar coding and massively parallel pyrosequencing to quantify rare drug resistance mutations. Using DNA bar coding, we were able to analyze seven viral populations in parallel, overall characterizing 118 093 sequence reads of average length 103 bp. Analysis of a control HIV mixture showed that resistance mutations present as 5% of the population could be readily detected without false positive calls. In three samples of multidrug-resistant HIV populations from patients, all the drug-resistant mutations called by conventional analysis were identified, as well as four additional low abundance drug resistance mutations, some of which would be expected to influence the response to antiretroviral therapy. Methods for sensitive characterization of HIV resistance alleles have been reported, but only the pyrosequencing method allows all the positions at risk for drug resistance mutations to be interrogated deeply for many HIV populations in a single experiment.
INTRODUCTION
Antiretroviral resistance is a major threat to successful anti-HIV treatment. This problem is more frequent among individuals that started therapy in the 1990s, who were treated sequentially with single antiviral agents (1), allowing sequential development of resistance to each treatment. Therapy with combinations of antiretroviral drugs is more effective, but nevertheless up to 50% of individuals currently in care in the US harbor HIV-resistant viruses (2). These resistant strains can also be transmitted—more than 15% of recently infected individuals have acquired viruses that are resistant to at least one of the major antiretroviral classes (3–5). Current treatment guidelines in the United States recommend resistance testing before beginning or changing antiretroviral therapy.
In the developing world, where much of the burden of HIV infection is concentrated, combination therapy is increasingly available. A sharp increase in drug resistance is expected as patients become more treatment experienced. Unfortunately, resistance genotyping is generally unavailable in the developing world due to the prohibitive expense.
Genotypic and phenotypic methods are commonly used to detect antiviral resistance in clinical specimens. Genotypic methods use bulk sequencing of the protease (PR) and reverse transcriptase (RT) coding regions, which reports the sequence of the predominant circulating HIV variants. Resistance mutations conferring reduced sensitivity to the three most widely used drug classes (nucleoside and non-nucleoside RT inhibitors and PR inhibitors) are well characterized, allowing probable resistance patterns to be inferred from sequence information. Phenotypic methods rely on cloning the RT and PR-coding regions from patient samples into a standard HIV plasmid backbone, allowing generation of viral stocks and functional analysis of viral drug sensitivity in short-term culture (1).
Several studies have demonstrated that minor drug-resistant HIV populations that are not detectable in the standard assays can impair the response to therapy (6,7). This problem is particularly apparent in studies of pregnant women that received single doses of nevirapine, a non-nucleoside reverse transcriptase inhibitor (NNRTI), at the time of delivery to prevent vertical transmission of HIV. In these patients, the presence of minor populations with resistance to nevirapine—which were often undetectable by conventional sequencing—compromised the response to subsequent NNRTI therapy (8,9).
A variety of technologies have been devised to allow characterization of minor HIV drug-resistant populations (7,10,11). In one method, microarrays were designed to interrogate positions of drug resistance mutations (12). This technique allows analysis of many genomic positions in a single experiment, but the method has not been widely used, in part due to the high cost of each test. Another method involves allele-specific RT-PCR, which allows sensitive detection of single drug resistance mutations (10). A third approach takes advantage of the massively parallel polony sequencing method (11), and a fourth uses an early version of pyrosequencing to query single nucleotide positions for possible mutations (13). However, except for the microarray method, all of the above methods queried individual base pairs at a time (10). Given that there are over 60 amino acid positions just in PR and RT that can affect resistance to the three widely used drug classes, these methods are time consuming and difficult to use for comprehensive analysis. An ideal method for investigating drug resistance mutations would yield many complete sequences of HIV genomic regions at risk for mutations for each viral population, and allow analysis of many samples of HIV populations in a single experiment.
We have adapted pyrosequencing (14), combined with a DNA bar coding system (15,16), to characterize rare drug-resistant HIV variants in many samples in parallel. In a single experiment, we determined 118 093 sequences from ∼100 bp segments of the PR and RT-coding regions for seven samples of viral populations (Table 1). These data identified a variety of minor drug resistance alleles in patient samples of potential clinical significance, and demonstrate the feasibility of using pyrosequencing for efficient HIV genotyping. Multiplex analysis of many bar coded samples in a single sequencing experiment offers the potential to drive down the cost of each genotype determination.
Table 1.
Viral populations characterized in this study
| Sample | Amplification template | Description | Bar code | Number of sequence reads |
|---|---|---|---|---|
| 1 | Viral RNA | HIV LAI RNA from particles | d(ACTG) | 17434 |
| 2 | Plasmid DNA | HIV LAI cloned DNA | d(AGTC) | 13856 |
| 3 | Viral RNA | HIV NL4-3 mixed with drug resistant NL4-3 | d(CTGA) | 7398 |
| 4 | Viral RNA | Mixture of HIV particles from subtypes A-E | d(CGTA) | 28751 |
| 5 | Viral RNA | HIV quasispecies from patient 1 | d(GCTA) | 19079 |
| 6 | Viral RNA | HIV quasispecies from patient 3 | d(TAGC) | 10730 |
| 7 | Viral RNA | HIV quasispecies from patient 2 | d(TGAC) | 20845 |
MATERIALS AND METHODS
Nucleic acid purification
DNA primers for PCR amplification were obtained from Invitrogen (Carlsbad, CA, USA). The HIV LAI plasmid (pLAI) was obtained from Dr Michael Emerman (Fred Hutchinson Cancer Research Center). HIV LAI particles were made by transfection of the pLAI and pVSV-G into 293T cells using standard methods. Supernatants were collected 3 days after transfection, concentrated by centrifugation, then treated with DNAse I to remove possible DNA carried over from the initial transfection. HIV NL4-3 particles were made similarly but without adding the pVSV-G plasmid. The following viral stocks were obtained from the AIDS Reference Reagent Repository: HIV NL4-3 (L10R/M46I/L63P/V82T/I84V; Catalog #2840); HIV subtype A (Catalog #7683); HIV subtype B (Catalog #7686); HIV subtype C (Catalog #7694); HIV subtype D (Catalog #7699); HIV subtype E (Catalog #7701).
HIV RNA from patient plasma samples was purified in the high-throughput facility at the Children's Hospital of Philadelphia using the Magna-Pure System (Roche) by Dr R. Hodinka. Viral loads were patient 1: 736 000 c/ml; patient 2: 105 000 c/ml; patient 3: 105 000 c/ml. Other HIV RNA samples were purified using the Illustra RNAspin mini RNA Isolation Kit (GE Health Care, Piscataway, NJ, USA).
PCR amplification
For the primer design, representative sequences (341 total) from subtypes A–D and G were downloaded from the Los Alamos HIV Database and Compendia (http://www.hiv.lanl.gov). Subtype consensus sequences were formed by aligning sequences within each subtype, then ‘Pan-HIV’ primers were designed to anneal to sequences conserved among subtypes (Supplementary Table 1).
All samples were amplified using the OneStep RT-PCR Kit (QIAGEN, Valencia, CA, USA) following the manufacturer's instructions. RNasin RNAse inhibitor (Promega, Madison, WI, USA) was added to the reaction to a final concentration of 0.5 units/μl. Five thousand copies of nucleic acid templates per reaction were used for the LAI DNA, LAI RNA and NL4-3 RNA mixture. For the patient samples, 10 μl of eluted material from the nucleic acid extraction was used as template. The RT-PCR reaction was carried out using a GeneAmp® PCR System 9700 (Applied Biosystems, Foster City, CA, USA). The program used was: 1 cycle of 45°C for 45 min followed by 15 min at 95°C; 14 touch-down cycles with annealing temperature from 55°C to 48°C for 30 s followed by 26 cycles with annealing temperature of 48°C for 30 s. A final extension of 10 min at 72°C was used. The denaturing and extension temperatures were 94°C and 72°C, respectively, for all cycles. The estimated error rates for the amplification step using Taq polymerase is 2–3 misincorporation events in 100 000, and for the RT step <1 substitution in 30 000 (estimates provided by Qiagen, Valencia, CA, USA).
PCR products were separated by electrophoresis on TAE agarose gels, slices containing the bands of interest were excised, DNA was extracted using the Qiagen Gel Extraction Kit (Valencia, CA, USA), then samples were pooled (75 ng per amplicon). For sequencing, the concentrations of DNA were adjusted to ∼5–15 ng/μl.
Conventional analysis of HIV genotypes in patients
Patient 1 was treated with tenofovir, FTC, atazanvir and ritonavir; patient 2 is deceased and treatment history is unknown, and patient 3 was treated with retrovir (zidovudine), epivir (lamivudine; 3TC) and viracept (nelfinavir). The conventional clinical analysis of HIV genotypes for the three patients studied was carried out as follows. Plasma viral RNA was extracted, amplified by RT-PCR (Roche Amplicor v 1.0; Roche Diagnostic Systems, Inc., Branchburg, NJ, USA), and sequenced using the Viroseq HIV Genotyping System v 2.0 (Applied Biosystems, Foster City, CA, USA). The sequences were analyzed for the resistance mutations using the HIV Drug Resistance Database at Stanford University (http://hivdb.stanford.edu/) (17), then calls were edited to match the slightly different catalog from the International AIDS Society Drug Resistance Mutations in HIV (Fall 2006) (2).
Pyrosequencing
Pyrosequencing was carried out using the 454 Life Sciences Technology at the University of Florida. The pyrosequencing method, as implemented by 454 Life Sciences, involves the following steps. Genomic DNA samples of interest are amplified using primers that include 5′ extensions providing binding sites for the 454 A and B primers. DNA fragments are then mixed with beads that have bound on their surfaces oligonucleotides complementary to the primers. This step is carried out in dilute solution so that on average a single DNA strand binds to each bead. A dilute mixture of beads is then added to an oil–water emulsion, arranged so that on average each aqueous droplet contains a single bead with a single bound strand. PCR amplification is then carried out in the emulsion. Each DNA strand becomes amplified and then binds by sequence complementarity to the bead, thereby creating beads that are each conjugated to DNA strands of a single homogenous sequence. Beads with bound DNA are then distributed on a picotiter plate at a density of ∼150 000 beads per plate. A primer is then bound to each DNA, and a polymerase used to extend a DNA chain. The four nucleotide triphosphates are sequentially flowed over the plate. An enzyme system is present in the buffer, which directs incorporation of pyrophosphate liberated by nucleotide addition into ATP, which then activates purified luciferase enzyme in the buffer to produce light. A CCD camera records each flash from each well on the plate. Sequential application of the four nucleotides allows DNA sequences of ∼100 bp to be built up ∼150 000 at a time (14).
The initial sequence reaction, carried out on a single plate, yielded 135 528 sequence reads, of which 118 093 ultimately passed quality control. For the sequences to pass, we required that each have a perfect match to the bar code and primer region, and no more than one N in the determined sequence. A total of 5.1% of the initial sequence reads had bar codes that were not included in the original experiment. The pyrosequencing method is error-prone at homopolymers, and of the incorrect bar codes, 54% created homopolymeric sequences within the bar codes and were excluded on that basis. To suppress bar code ‘crossover’, the different viral samples were separated into separate quadrants of the sequencing plate as follows (numbers from Table 1): samples 1 and 2, quadrant 1; samples 3 and 5, quadrant 2; samples 6 and 7, quadrant 3; sample 4, quadrant 4. Inspection of drug resistance calls for samples within the same quadrant showed no obvious bar code crossover.
For scoring drug resistance mutations, we used the Sierra Webservice at the HIV Drug Resistance Database at Stanford University (http://hivdb.stanford.edu/). To query the Drug Resistance Database, the pyrosequence reads were embedded in ‘dummy’ HIV flanking sequences, and the resistance alleles identified over the sequenced region. To pass quality control at this step, we required that the full sequence be recognized as HIV by the HIV Drug Resistance database, and that all pyrosequence reads cover at least 60% of the genomic window interrogated. Resistance mutations called by the Stanford Database were filtered to match the International AIDS Society definitions.
Statistical analysis
Inspection of the raw counts of drug resistance calls in controls (Supplementary Table 2) showed that different positions showed differing error rates. For this reason, a statistical model was used that took into account the error rate measured at each position. The proportion of drug-resistant mutant calls in the combined HIV LAI DNA and HIV LAI RNA data sets were taken as the background error for each position. We took advantage of the Fisher's exact test to investigate whether drug resistance mutations were significantly enriched in the patient samples compared to controls. To control for multiple comparisons, a Bonferroni correction was applied. The P-values were multiplied by the number of comparisons within each individual (62 comparisons corresponding to 62 codons queried for drug resistance). Drug resistance calls with corrected P-values <0.05 were judged to be significantly enriched. Statistical analysis was carried out in the R environment (http://www.r-project.org/).
RESULTS
DNA bar coding and pyrosequencing
Primers were designed for amplifying the regions of HIV pol that are known from previous work to be sites of substitutions that result in resistance to PR and RT inhibitors. Because the most useful design would allow amplification of HIV sequences from any of the viral subtypes, we designed ‘Pan-HIV’ primers that would amplify subtypes A, B, C, D and 01_AE, the major subtypes circulating world-wide.
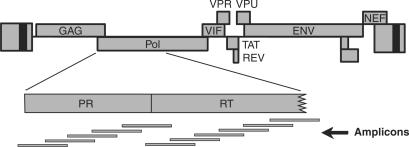
Typical read lengths for the pyrosequencing method at the time of this experiment were ∼100 bp. For the sequencing procedure, it is desirable to use fragments that are somewhat longer than this, to allow electrophoretic separation of the PCR products away from short contaminating sequences such as primer dimers. For this reason, we designed 11 overlapping amplicons, as shown in Figure 1, to allow analysis of all known positions of PR and RT drug resistance mutations [cataloged by the International AIDS Society (Fall 2006 Revision) (18)] while allowing purification of fragments in the 200–400 bp size range.
Figure 1.
Amplicons querying positions of HIV drug resistance mutations in the PR and RT-coding regions. The HIV genetic map is shown at the top. Below is shown an expanded version of the segment of pol encoding PR and part of RT. At the bottom is shown the location of the amplicons used to interrogate positions of potential drug resistance mutations.
Viral populations studied
A key consideration in analyzing our results was distinguishing authentic drug-resistant mutations from erroneous calls. After isolation of HIV RNA from particles, reverse transcription followed by PCR (RT-PCR) has the potential to introduce mutations. The pyrosequencing procedure, as implemented at 454 Life Sciences (14), also requires a PCR amplification step, and the pyrosequencing method is more error prone than the Sanger method.
Several controls were therefore included in the experiment to allow estimation of the background error rate (Table 1). Plasmid DNA from the HIV isolate LAI was used as template for PCR amplification by the 11 pan-HIV primers and the products analyzed. Because the HIV LAI DNA sequence is known, this serves as a control for error in the RT-PCR and pyrosequencing steps. RNA from HIV LAI viral particles was included as another control. This provided a second measure of error and also allowed us to assess the misincorporation rate due to transcription and reverse transcription of the HIV RNA, which proved to be undetectable using the methods described later.
As a test of the detection sensitivity, we studied RNA from particles of another strain, HIV NL4-3, which were ‘spiked’ with one-twentieth the amount of a drug-resistant HIV NL4-3 derivative containing five substitutions in the PR-coding region (encoding L10R/M46I/L63P/V82T/I84V). Viral stocks were normalized by measuring the amount of the p24 capsid protein in each stock by ELISA.
A fourth sample contained a pool of five HIV subtypes (A, B, C, D and 01AE) (19), and was included to illustrate the ability of the pan-HIV primers to amplify sequences from all five, and the downstream bioinformatic methods to distinguish sequence data from each.
We also studied three samples of plasma from patients with drug-resistant HIV. For each of the three, we obtained standard diagnostic sequence data generated previously using the Viroseq HIV Genotyping System v 2.0 (Applied Biosystems, Foster City, CA, USA) and used in managing each patients’ treatment. Analysis of these samples allowed us to compare the sensitivity of the pyrosequencing method to a standard method.
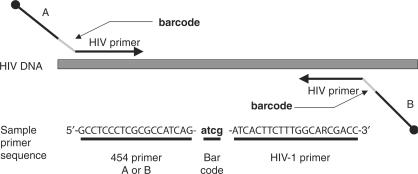
The DNA bar coding strategy
Since one of our goals was developing methods for testing many samples in single sequencing experiments, we developed a DNA bar coding strategy to allow sample multiplexing (Figure 2). Each primer consisted of the 454 A and B sequences at the 5′ ends, required for the emulsion PCR, and the HIV-complementary regions at the 3′ end. To distinguish different amplicons in a pool, we added a 4 bp DNA bar code sequence between the 454 A or B sequence and the HIV-complementary region. Because the 454 sequencing method has an increased error rate at positions of adjacent nucleotides of the same type (homopolymers), we avoided using bar codes containing adjacent identical nucleotides.
Figure 2.
The DNA bar coding system. Primers are shown diagrammatically, illustrating the placement of the DNA bar code between the 454 primers (A and B) and the HIV-complementary sequences. An example of a primer sequence is shown at the bottom. DNA 5′ ends in the primers are shown as the black balls.
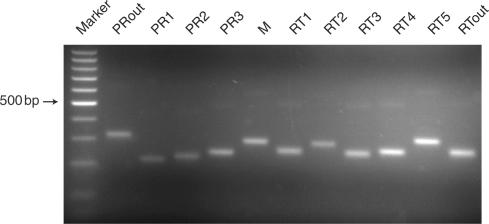
Sample amplification and initial sequence analysis
PCR amplification was carried out on the seven HIV nucleic acid templates listed in Table 1 using the pan-HIV primers. All amplification reactions yielded DNA chains of the expected length when analyzed by agarose gel electrophoresis and ethidium bromide staining. Figure 3 shows a sample amplification of subtype C viral RNA. The PCR amplicons were then pooled and subjected to pyrosequencing using the method implemented by 454 Life Sciences (14). Samples were attached to beads and amplified using emulsion PCR, the beads with bound DNA were applied to a picoliter plate, and sequences on beads were determined using the pyrosequencing method. A total of 135 528 sequence reads were returned, of average length 103 bp. After quality control, 118 093 sequences were available for analysis, corresponding to ∼12 million base pairs of DNA sequence. Of the seven bar codes used, the number of sequences returned per bar code after quality control ranged from 28 751 (pooled subtypes) to 7398 (NL4-3 with doped drug resistance allele). More of the PCR products from the subtype pool was used for sequencing because we expected a relatively higher level of diversity from this sample due to the diversity of the templates. Overall, 121 amplification products were pooled and analyzed, and sequences from all were recovered in good yield.
Figure 3.
Amplification of HIV subtype C using the pan-HIV primer set. The figure shows products of RT-PCR reactions analyzed by agarose gel electrophoresis and ethidium bromide staining. The amplicons extend from the 5′ side of the PR-coding region (left) to an internal region of the RT-coding region (right), thereby covering all the known positions of drug resistance mutations in the PR and RT-coding regions (primer sequences are in Supplementary Table 1). The marker ladder has ‘steps’ of 100 bp. The position of the 500 bp fragment is marked.
We next sought to identify mutations within the HIV sequences expected to confer drug resistance. Sequences passing quality control were analyzed at the HIV Drug Resistance Database at Stanford University (http://hivdb.stanford.edu/) (17), and the results tabulated. For our initial analysis, we restricted our attention to the best documented drug resistance mutations as summarized by the International AIDS Society Drug Resistance Mutations in HIV (Fall 2006 Revision) (2). Overall, 62 positions of drug resistance were queried, and the frequency of drug resistance calls at each codon tabulated (raw data in Supplementary Table 2).
The pool of HIV subtypes (Table 1, sample 4) was separated into subtype-specific groups by aligning the sequences against subtype-specific consensus sequences. Sequences were recovered for all five subtypes in good yield. Supplementary Table 3 catalogs the sequences identified and the drug resistance mutations detected in each subtype, and Supplementary Table 4 presents the rate of polymorphism at sites not involved in drug resistance.
Statistical analysis
A statistical procedure was devised to allow detection of significant enrichment of drug resistance mutations above the error rate. Inspection of the control data (HIV LAI DNA and RNA templates) revealed that, of the 124 codons queried (62 codons in each of the two controls), 57/124 codons showed zero calls for drug resistance (Supplementary Table 2). All 62 control codons showed <1% resistance calls. However, 4/124 positions showed erroneous calls for drug resistance between 0.5% and 1% of sequences. For this reason, a statistical procedure was devised to assess the possible presence of drug resistance mutations in the experimental samples in light of the position-specific error rate.
The proportion of drug resistance calls at each position in the controls (Table 1, samples 1 and 2) was assessed and taken as the position-specific background. The frequency of resistance calls in each sample was then tested against the position-specific background using Fisher's exact test. Inspection of the initial collection of P-values showed that there were more positions of significant enrichment than expected in the controls (Table 1, samples 1–3), which was not surprising because the large number of comparisons carried out (62 for each patient) increased the likelihood of obtaining significant enrichment by chance. To control for the inflation of error due to multiple comparisons, we applied a Bonferroni correction, in which the P-value from Fisher's exact test was multiplied by the number of comparisons in each patient (62). Thirty-six positions achieved significance after correction (P < 0.05), and are shown as black tiles in Figure 4.
Figure 4.
Summary of the drug resistance alleles detected. Results are shown for PR (top two grids) and RT (bottom two grids). The seven HIV samples studied are labeled to the left of each grid. The amino acid positions of drug-resistant mutations are marked at the top of the grid, labeled DRM for drug resistance mutation. Black tiles in the grid indicate positions that were significantly enriched for drug resistance mutations in the pyrosequencing data after correction for multiple comparisons. Raw counts are available in Supplementary Table 2. Black tiles with asterisks indicate positions that were enriched in drug resistance mutations in the pyrosequencing data that were not identified by a conventional genotyping method (Viroseq HIV Genotyping System v 2.0). The ‘i’ at RT position 69 denotes the insertions at this position that are known to confer drug resistance.
Proper performance of this procedure could be verified by analyzing the results for the HIV NL4-3 sample spiked with 5% virions encoding the L10R/M46I/L63P/V82T/I84V resistance substitutions. All five doped drug resistance alleles were found to be significantly enriched after the correction for multiple comparisons, and no additional mutations were called erroneously (Figure 4). The measured frequencies of the drug resistance alleles, initially added at 5%, were L10: 4.6%, M46: 1.3%, L63: 4.8%, V82: 2.9%, I84: 3.1%. We conclude that the pyrosequencing method together with the above statistical procedure is sensitive and accurate enough to selectively detect drug resistance substitutions present as 5% of the viral population.
Drug resistance alleles detected in patient samples
Drug resistance mutations were identified using pyrosequencing in the three patient samples (Table 1, samples 5–7), and the calls compared to those called by the Viroseq HIV Genotyping System (analyzed using the HIV Drug Resistance Database at Stanford University). The pyrosequencing study identified all 15 of the drug-resistant mutations that were called using the Viroseq genotyping pipeline (Figure 4, black bars without asterisks). The median proportion of mutations called by the Viroseq method was 88% drug resistance calls as measured in the pyrophosphate sequencing data (range: 28–99%). Four additional lower abundance drug resistance mutations were called in the pyrosequencing data that were not called by the Viroseq pipeline (Figure 4, black bars with white asterisks), one for patient 1, one for patient 2 and two for patient 3 (Figure 4, black bars with asterisks). The frequency of resistance mutations called by pyrosequencing only ranged from 11.6 to 0.65% of the total. A few positions were represented by many sequences, and appear to be just below the level of detection of the conventional genotyping assay. For example, the RT drug resistance substitution K70R was represented by 40 calls out of 305, and the RT mutation M184V was represented by 46 calls out of 726 (both from patient 3). At the other extreme, significant detection could be achieved by as few as nine drug resistance calls for codons in cases where the background was low—for the M46 position in patient 1, drug resistance for 9 out of 1377 calls achieved significance in the presence of a background of 1 out of 2395 calls (P = 0.0014, Fisher's exact test, two-sided comparison). The potential clinical significance of the minor drug-resistant alleles is discussed later.
Eight resistance mutations were also identified in the collection of sequences from different HIV subtypes (Table 1, sample 4, data summarized in Supplementary Table 3). The HIV samples studied were isolated from patients worldwide in 1991 (19). The subtype B sample was isolated in the United States, subtypes A, C and D in Uganda, and subtype 01_AE in Thailand. None of the patients were on antiretroviral therapy at the time of collection. Eight drug resistance mutations were identified in the PR-coding region, and none in RT. In some cases the resistance mutations were present as substantial fractions of the populations, such as the M36I substitution in subtype D, which comprises more than 78%. Others were less common, comprising ∼20% of the population. It is uncertain how many of these mutations, which appear to be pre-existing polymorphisms, would influence sensitivity to protease inhibitors (20,21). Taken together, these findings demonstrate the ability of the pyrosequencing method to identify minor drug resistance mutations in individuals infected with diverse HIV subtypes.
DISCUSSION
Here we describe the use of DNA bar coding and pyrosequencing in detecting minor drug resistance mutations in HIV populations. We used pyrosequencing to generate 118 093 sequence reads from the pol region of seven samples of viral populations or controls. The seven samples were analyzed in a single sequencing experiment, made possible by use of DNA bar coding to distinguish the different samples. As controls we analyzed a DNA plasmid encoding HIV LAI and HIV LAI RNA from viral particles, allowing an empirical estimation of error at each codon at risk for mutation to drug resistance. To test the assay sensitivity, we analyzed a mixture containing HIV NL4-3 wild-type virions mixed with 5% of mutant virions containing five drug resistance mutations. All five mutations were called as present in the mixture without false positives, demonstrating the accuracy of the method. Analysis of viral populations from three patients harboring drug-resistant HIV revealed all the mutations called by the conventional genotyping method (Viroseq), plus four additional less abundant drug resistance mutations comprising from 11.6% to 0.65% of the population.
Previously several methods have been reported for sensitive detection of rare drug-resistant mutations, some with impressive sensitivity (10,11). However, only the combination of DNA bar coding and pyrosequencing offers the opportunity to determine the full sequence of genomic regions with drug-resistant mutations from many samples in a single sequencing experiment. Using this method, sequence polymorphisms that were not specifically targeted for analysis can be identified and analyzed for possible correlations with drug resistance, which is not possible with most of the alternative methods. Here we analyzed seven viral populations in a single picoliter sequencing plate, but there is no reason that the number could not be much larger. In another study, we have successfully sequenced 42 different DNA bar codes in a single plate (unpublished data), indicating the potential.
Several of the low abundance drug resistance mutations detected here are of potential clinical significance, since they may confer reduced sensitivity to drugs that otherwise might seem attractive for therapy. For example, patient 3 was found to harbor low level resistance alleles in RT at K70 and M184, which would be likely to impair therapy with several of the NRTIs, though not with all NRTIs. Patient 3 had been treated with NRTIs known to elicit these mutations, probably explaining their presence. Because alternative NRTIs are available for which all of the patient 3 viruses would remain sensitive, knowledge of the minor alleles could have improved the ability to choose effective therapy in this case. Looking forward, it will be important to test more fully the importance of minor HIV drug-resistant populations on antiretroviral responses and the impact of such information on treatment outcomes.
Lastly, the methods described here may be useful in implementing drug resistance genotyping in resource-limited settings. Using the DNA bar coding strategy, it should be possible to multiplex large numbers of patient samples in single sequencing runs, thereby driving down costs. Though many logistical obstacles would need to be overcome, the combination of DNA bar coding and pyrosequencing offers a prototype technology for affordable HIV genotyping.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Sridhar Hannenhalli and Rob Knight for helpful discussions, Rick Hodinka for high-throughput isolation of viral RNA, Heather Marshall for the purified HIV LAI particles and Tommy Lu and the creators of the Stanford HIV Database for allowing us to use their resource. This work was supported by the University of Pennsylvania. We also thank the AIDS Reagent Repository for HIV stocks. Funding to pay the Open Access publication charges for this article was provided by The University of Pennsylvania.
Conflict of interest statement. None declared.
REFERENCES
- 1.Clavel F, Hance AJ. HIV drug resistance. See comment. N. Engl. J. Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 2.Richman DD, Morton SC, Wrin T, Hellmann N, Berry S, Shapiro MF, Bozzette SA. The prevalence of antiretroviral drug resistance in the United States. AIDS. 2004;18:1393–1401. doi: 10.1097/01.aids.0000131310.52526.c7. [DOI] [PubMed] [Google Scholar]
- 3.Little SJ, Holte S, Routy JP, Daar ES, Markowitz M, Collier AC, Koup RA, Mellors JW, Connick E, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 4.Smith D, Moini N, Pesano R, Cachay E, Aiem H, Lie Y, Richman D, Little S. Clinical utility of HIV standard genotyping among antiretroviral-naive individuals with unknown duration of infection. Clin. Infect. Dis. 2007;44:456–458. doi: 10.1086/510748. [DOI] [PubMed] [Google Scholar]
- 5.Cane P, Chrystie I, Dunn D, Evans B, Geretti AM, Green H, Phillips A, Pillay D, Porter K, et al. Time trends in primary resistance to HIV drugs in the United Kingdom: multicentre observational study. See comment. Br. Med. J. 2005;331:1368. doi: 10.1136/bmj.38665.534595.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellors J, Palmer S, Nissley D, Kearny M, Halvas E, Bixby C, Demeter L, Eshleman S, Bennett K, Hart S, Vaida F, Wantman M, Coffin J, Hammer S. Low-frequency NNRTI-resistant variants contribute to failure of efavirenz-containing regimen.. Program and Abstracts of the 11th Conference on Retroviruses and Opportunistic Infections Abstract 39.2004. [Google Scholar]
- 7.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey R.T., Jr, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jourdain G, Ngo-Giang-Huong N, Le Coeur S, Bowonwatanuwong C, Kantipong P, Leechanachai P, Ariyadej S, Leenasirimakul P, Hammer S, et al. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. See comment. N. Engl. J. Med. 2004;351:229–240. doi: 10.1056/NEJMoa041305. [DOI] [PubMed] [Google Scholar]
- 9.Lockman S, Shapiro RL, Smeaton LM, Wester C, Thior I, Stevens L, Chand F, Makhema J, Moffat C, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N. Engl. J. Med. 2007;356:135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- 10.Halvas EK, Aldrovandi GM, Balfe P, Beck IA, Boltz VF, Coffin JM, Frenkel LM, Hazelwood JD, Johnson VA, et al. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J. Clin. Microbiol. 2006;44:2612–2614. doi: 10.1128/JCM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai F, Chen H, Hicks CB, Bartlett JA, Zhu J, Gao F. Detection of minor drug-resistant populations by parallel allele-specific sequencing. Nat. Methods. 2007;4:123–125. doi: 10.1038/nmeth995. [DOI] [PubMed] [Google Scholar]
- 12.Kozal MJ, Shah N, Shen N, Yang R, Fucini R, Merigan TC, Richman DD, Morris D, Hubbell E, et al. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 13.O’Meara D, Wilbe K, Leitner T, Hejdeman B, Albert J, Lundeberg J. Monitoring resistance to human immunodeficiency virus type 1 protease inhibitors by pyrosequencing. J. Clin. Microbiol. 2001;39:464–473. doi: 10.1128/JCM.39.2.464-473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 17.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson VA, Brun-Vezinet F, Clotet B, Conway B, Kuritzkes DR, Pillay D, Schapiro JM, Telenti A, Richman DD. Update of the drug resistance mutations in HIV-1: Fall 2005. Top. HIV Med. 2005;13:125–131. [PubMed] [Google Scholar]
- 19.Jagodzinski LL, Wiggins DL, McManis JL, Emery S, Overbaugh J, Robb M, Bodrug S, Michael NL. Use of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 to assess the performance of viral RNA quantitation tests. J. Clin. Microbiol. 2000;38:1247–1249. doi: 10.1128/jcm.38.3.1247-1249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kantor R, Katzenstein D. Drug resistance in non-subtype B HIV-1. J. Clin. Virol. 2004;29:152–159. doi: 10.1016/S1386-6532(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 21.Holguin A, Paxinos E, Hertogs K, Womac C, Soriano V. Impact of frequent natural polymorphisms at the protease gene on the in vitro susceptibility to protease inhibitors in HIV-1 non-B subtypes. J. Clin. Virol. 2004;31:215–220. doi: 10.1016/j.jcv.2004.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.