Abstract
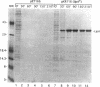
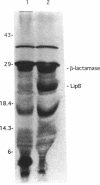
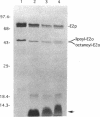
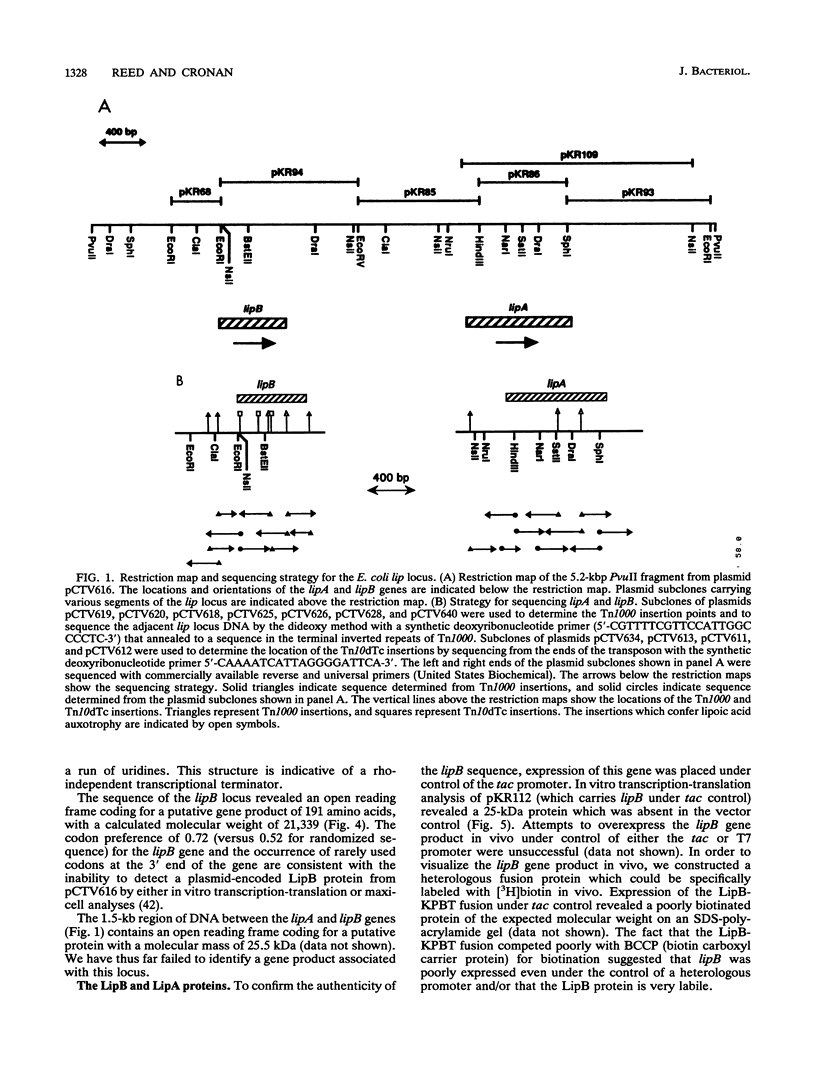
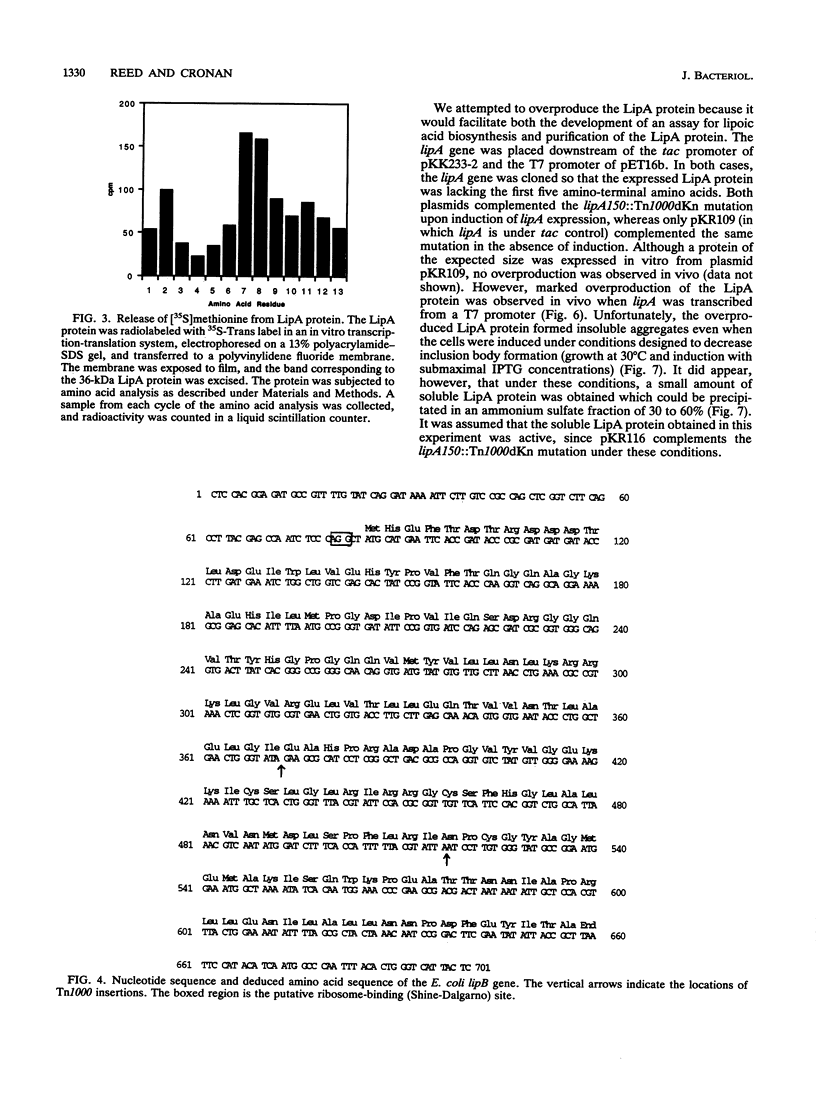
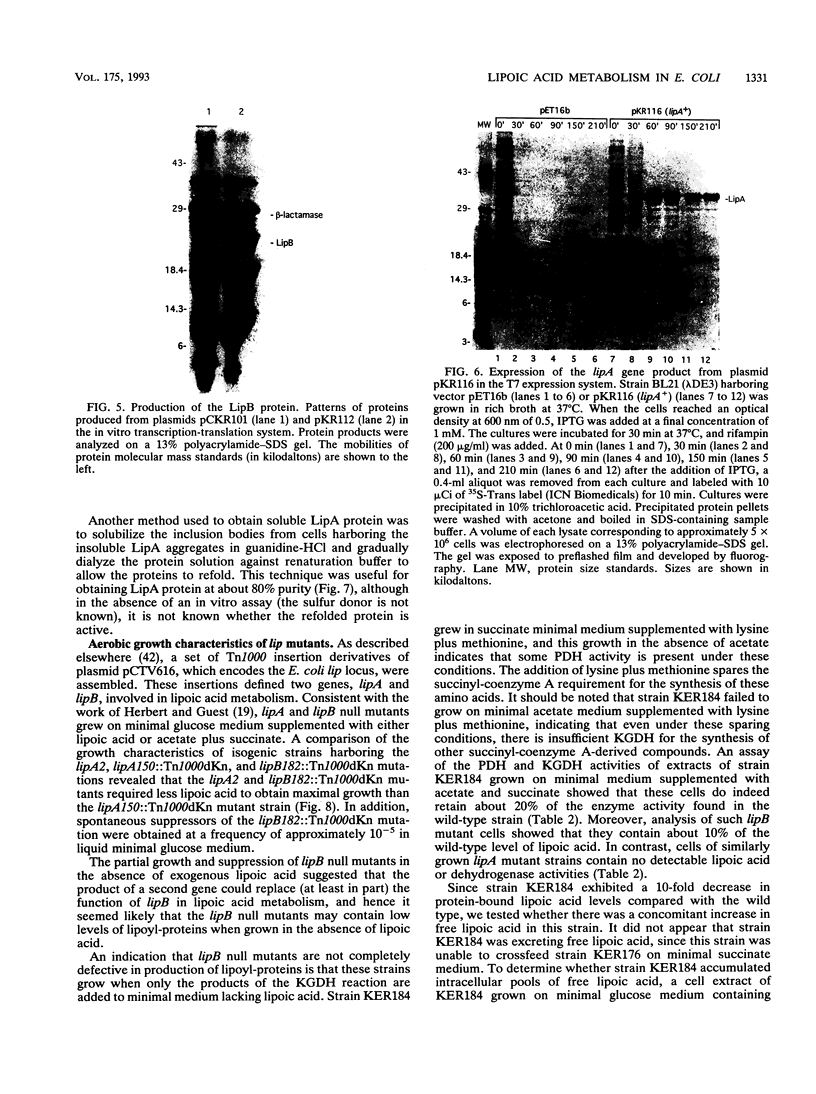
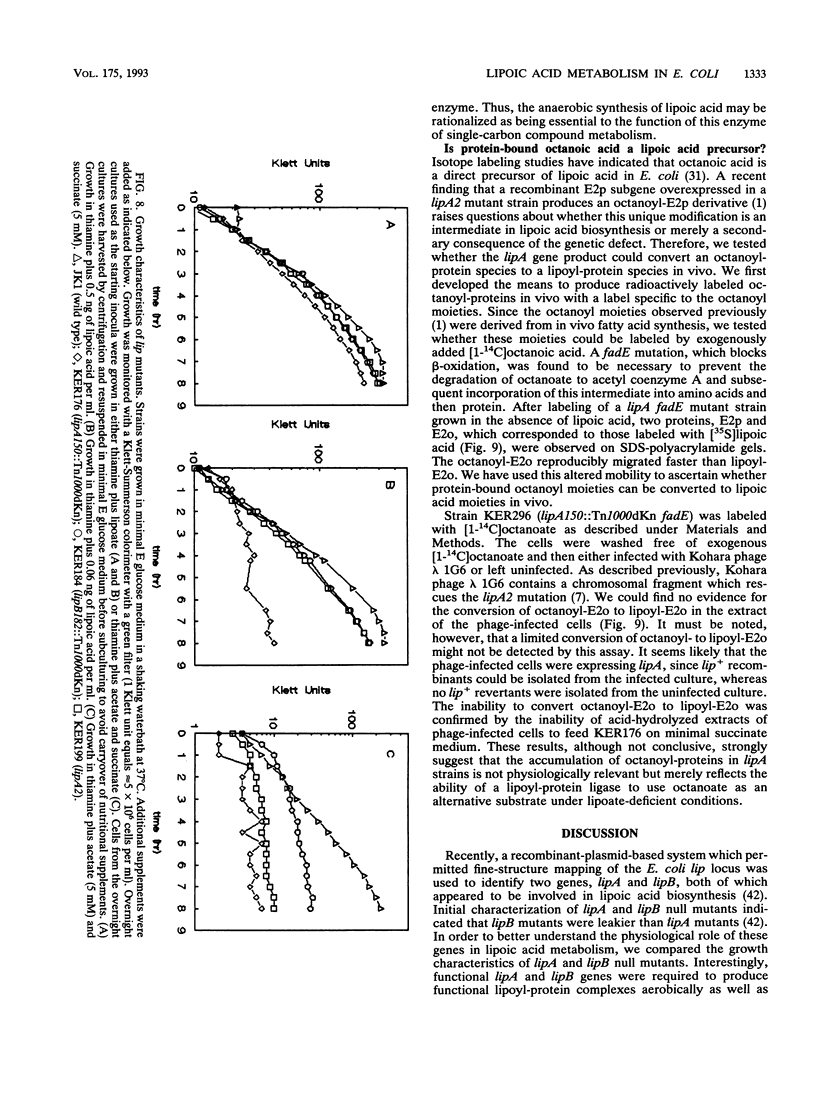
Two genes, lipA and lipB, involved in lipoic acid biosynthesis or metabolism were characterized by DNA sequence analysis. The translational initiation site of the lipA gene was established, and the lipB gene product was identified as a 25-kDa protein. Overproduction of LipA resulted in the formation of inclusion bodies, from which the protein was readily purified. Cells grown under strictly anaerobic conditions required the lipA and lipB gene products for the synthesis of a functional glycine cleavage system. Mutants carrying a null mutation in the lipB gene retained a partial ability to synthesize lipoic acid and produced low levels of pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase activities. The lipA gene product failed to convert protein-bound octanoic acid moieties to lipoic acid moieties in vivo; however, the growth of both lipA and lipB mutants was supported by either 6-thiooctanoic acid or 8-thiooctanoic acid in place of lipoic acid. These data suggest that LipA is required for the insertion of the first sulfur into the octanoic acid backbone. LipB functions downstream of LipA, but its role in lipoic acid metabolism remains unclear.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ali S. T., Moir A. J., Ashton P. R., Engel P. C., Guest J. R. Octanoylation of the lipoyl domains of the pyruvate dehydrogenase complex in a lipoyl-deficient strain of Escherichia coli. Mol Microbiol. 1990 Jun;4(6):943–950. doi: 10.1111/j.1365-2958.1990.tb00667.x. [DOI] [PubMed] [Google Scholar]
- Brookfield D. E., Green J., Ali S. T., Machado R. S., Guest J. R. Evidence for two protein-lipoylation activities in Escherichia coli. FEBS Lett. 1991 Dec 16;295(1-3):13–16. doi: 10.1016/0014-5793(91)81373-g. [DOI] [PubMed] [Google Scholar]
- Chambers S. P., Prior S. E., Barstow D. A., Minton N. P. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988 Aug 15;68(1):139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- Chang Y. Y., Cronan J. E., Jr, Li S. J., Reed K., Vanden Boom T., Wang A. Y. Locations of the lip, poxB, and ilvBN genes on the physical map of Escherichia coli. J Bacteriol. 1991 Sep;173(17):5258–5259. doi: 10.1128/jb.173.17.5258-5259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan J. E., Jr Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J Biol Chem. 1990 Jun 25;265(18):10327–10333. [PubMed] [Google Scholar]
- Cronan J. E., Jr, Li W. B., Coleman R., Narasimhan M., de Mendoza D., Schwab J. M. Derived amino acid sequence and identification of active site residues of Escherichia coli beta-hydroxydecanoyl thioester dehydrase. J Biol Chem. 1988 Apr 5;263(10):4641–4646. [PubMed] [Google Scholar]
- Davis T. N., Muller E. D., Cronan J. E., Jr The virion of the lipid-containing bacteriophage PR4. Virology. 1982 Jul 30;120(2):287–306. doi: 10.1016/0042-6822(82)90031-9. [DOI] [PubMed] [Google Scholar]
- Dietrich J., Henning U. Regulation of pyruvate dehydrogenase complex synthesis in Escherichia coli K 12. Identification of the inducing metabolite. Eur J Biochem. 1970 Jun;14(2):258–269. doi: 10.1111/j.1432-1033.1970.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Fujiwara K., Okamura-Ikeda K., Motokawa Y. Chicken liver H-protein, a component of the glycine cleavage system. Amino acid sequence and identification of the N epsilon-lipoyllysine residue. J Biol Chem. 1986 Jul 5;261(19):8836–8841. [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M. A., Huang I., Bussiere D. E., Ashley G. W. The biosynthesis of lipoic acid. Cloning of lip, a lipoate biosynthetic locus of Escherichia coli. J Biol Chem. 1992 May 15;267(14):9512–9515. [PubMed] [Google Scholar]
- Herbert A. A., Guest J. R. Biochemical and genetic studies with lysine+methionine mutants of Escherichia coli: lipoic acid and alpha-ketoglutarate dehydrogenase-less mutants. J Gen Microbiol. 1968 Oct;53(3):363–381. doi: 10.1099/00221287-53-3-363. [DOI] [PubMed] [Google Scholar]
- Herbert A. A., Guest J. R. Lipoic acid content of Escherichia coli and other microorganisms. Arch Microbiol. 1975 Dec 31;106(3):259–266. doi: 10.1007/BF00446532. [DOI] [PubMed] [Google Scholar]
- ITZHAKI R. F., GILL D. M. A MICRO-BIURET METHOD FOR ESTIMATING PROTEINS. Anal Biochem. 1964 Dec;9:401–410. doi: 10.1016/0003-2697(64)90200-3. [DOI] [PubMed] [Google Scholar]
- Langley D., Guest J. R. Biochemical genetics of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: genetic characterization and regulatory properties of deletion mutants. J Gen Microbiol. 1978 May;106(1):103–117. doi: 10.1099/00221287-106-1-103. [DOI] [PubMed] [Google Scholar]
- Lee E. C., MacWilliams M. P., Gumport R. I., Gardner J. F. Genetic analysis of Escherichia coli integration host factor interactions with its bacteriophage lambda H' recognition site. J Bacteriol. 1991 Jan;173(2):609–617. doi: 10.1128/jb.173.2.609-617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meedel T. H., Pizer L. I. Regulation of one-carbon biosynthesis and utilization in Escherichia coli. J Bacteriol. 1974 Jun;118(3):905–910. doi: 10.1128/jb.118.3.905-910.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa I., Speck D., Kisou T., Hayakawa K., Zinsius M., Gloeckler R., Lemoine Y., Kamogawa K. Cloning of the biotin synthetase gene from Bacillus sphaericus and expression in Escherichia coli and Bacilli. Gene. 1989 Aug 1;80(1):39–48. doi: 10.1016/0378-1119(89)90248-5. [DOI] [PubMed] [Google Scholar]
- Otsuka A. J., Buoncristiani M. R., Howard P. K., Flamm J., Johnson C., Yamamoto R., Uchida K., Cook C., Ruppert J., Matsuzaki J. The Escherichia coli biotin biosynthetic enzyme sequences predicted from the nucleotide sequence of the bio operon. J Biol Chem. 1988 Dec 25;263(36):19577–19585. [PubMed] [Google Scholar]
- Plamann M. D., Rapp W. D., Stauffer G. V. Escherichia coli K12 mutants defective in the glycine cleavage enzyme system. Mol Gen Genet. 1983;192(1-2):15–20. doi: 10.1007/BF00327641. [DOI] [PubMed] [Google Scholar]
- REED L. J., LEACH F. R., KOIKE M. Studies on a lipoic acid-activating system. J Biol Chem. 1958 May;232(1):123–142. [PubMed] [Google Scholar]
- Reed K. E., Cronan J. E., Jr Escherichia coli exports previously folded and biotinated protein domains. J Biol Chem. 1991 Jun 25;266(18):11425–11428. [PubMed] [Google Scholar]
- Reed L. J., Hackert M. L. Structure-function relationships in dihydrolipoamide acyltransferases. J Biol Chem. 1990 Jun 5;265(16):8971–8974. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. W., Neidhardt F. C. 2-Oxoacid dehydrogenase complexes of Escherichia coli: cellular amounts and patterns of synthesis. J Bacteriol. 1983 Oct;156(1):81–88. doi: 10.1128/jb.156.1.81-88.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M. E., Darlison M. G., Stephens P. E., Duckenfield I. K., Guest J. R. Nucleotide sequence of the sucB gene encoding the dihydrolipoamide succinyltransferase of Escherichia coli K12 and homology with the corresponding acetyltransferase. Eur J Biochem. 1984 Jun 1;141(2):361–374. doi: 10.1111/j.1432-1033.1984.tb08200.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem. 1983 Jul 1;133(3):481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Vanden Boom T. J., Reed K. E., Cronan J. E., Jr Lipoic acid metabolism in Escherichia coli: isolation of null mutants defective in lipoic acid biosynthesis, molecular cloning and characterization of the E. coli lip locus, and identification of the lipoylated protein of the glycine cleavage system. J Bacteriol. 1991 Oct;173(20):6411–6420. doi: 10.1128/jb.173.20.6411-6420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]