Abstract
microRNAs (miRNAs) serve as post-transcriptional regulators of gene expression, by guiding effector complexes (miRNPs) to target RNAs. Although considerable progress has been made in computational methods to identify miRNA targets, only a relatively limited assessment of their ability to function in vivo has been reported. Here we describe an alternative approach to miRNA target identification based on a biochemical method for purifying miRNP complexes with associated miRNAs and bound mRNA targets. Microarray analysis revealed a high degree of enrichment for miRNA complementary sites in the 3′UTRs of the miRNP-associated mRNAs. mRNAs specifically associated with an individual miRNA were identified by comparing the miRNP-associated mRNAs from wild-type flies and mutant flies lacking miR-1, and their regulation by the miRNA was validated. This approach provides a means to identify functional miRNA targets based on their physical interaction in vivo.
Keywords: Drosophila, RISC, Argonaute, AGO1
INTRODUCTION
microRNAs (miRNAs) have recently been recognized as important regulators of gene expression at the post-transcriptional level. miRNAs act as guide molecules that recruit miRNP effector complexes, including Argonaute, and a variety of other proteins to the target RNA (Hutvagner and Zamore 2002; Zamore and Haley 2005). miRNP association can lead to translational repression, destabilization of the mRNA, or mRNA cleavage (for review, see Bartel 2004; Jackson and Standart 2007; Wang et al. 2007). In plants, the miRNAs have extensive complementarity with their target mRNAs and mediate cleavage; while in animals, their complementarity is typically limited and translation of the mRNA is blocked (Du and Zamore 2005; Pillai et al. 2005; Schwab et al. 2005; Jones-Rhoades et al. 2006; Zhang et al. 2007). Despite the rarity of site-specific cleavage by animal miRNAs, miRNP binding can cause deadenlyation and destabilization of the target mRNA (Giraldez et al. 2006; Wu et al. 2006).
One approach to understanding the biological roles of miRNAs has been to identify their targets. Experimental analyses of miRNA target relationships have shown that only limited pairing is needed for miRNAs to confer regulation (Doench and Sharp 2004; Kiriakidou et al. 2004; Brennecke et al. 2005). A variety of computational approaches, some based on rules derived from experimentally validated miRNA target pairs, have been successful in predicting miRNA target relationships (Grun et al. 2005; Krek et al. 2005; Lewis et al. 2005; Robins et al. 2005; Stark et al. 2005; Xie et al. 2005; Lall et al. 2006). Yet much remains to be learned about factors that might influence target site function in vivo (for a recent review, see Rajewsky 2006).
Whereas siRNA-mediated target cleavage is compatible with a transient association of the Ago-containing protein complex with the target mRNA, miRNA-mediated translational inhibition can be expected to depend on stable physical association between the miRNP and the target mRNA. This opens the possibility of making use of this interaction to identify the miRNA–target interactions that occur in vivo. Here we present a method for identifying miRNA targets, based on their physical association with miRNAs in Argonaute protein–containing miRNPs, and show that this approach can be used to identify new miRNA targets.
RESULTS AND DISCUSSION
The two Drosophila Argonaute proteins have distinct functions, with Ago1 acting in miRNA-mediated translational repression and Ago2 specifically involved in mi/siRNA-mediated cleavage of target mRNAs (Okamura et al. 2004; Behm-Ansmant et al. 2006; Rehwinkel et al. 2006). We introduced a FLAG tag followed by an HA epitope tag into the N terminus of the Ago1 protein (Meister et al. 2005) and generated stably transformed Schneider SL2 (S2) cell lines expressing this protein and transgenic Drosophila strains. Functionality of the tagged Ago1 protein was confirmed by using this transgene to completely rescue the lethality of an ago1 mutant strain, thereby replacing the endogenous Ago1 protein with the epitope tagged version (data not shown).
Immunopurification of Ago1-containing protein complexes was performed with anti-HA antibody (Fig. 1A). Immunoblot analysis of the protein content of the immunoprecipitates showed purification of Ago1, and depletion of proteins not expected to be in the miRNP complex, such as tubulin (Fig. 1B). Northern blot analysis revealed the presence of the mature forms of miR-2b and bantam miRNAs in the purified fractions from tagged Ago1-expressing cells, but not in samples purified from untransfected control cells (Fig. 1C). The corresponding pre-miRNAs were not recovered in the purified fractions. Thus the immunopurified Ago1 complexes contain mature miRNAs. To ask whether these complexes also contained miRNA-associated target RNAs, we performed quantitative RT-PCR to compare the levels of a known miRNA target from S2 cells, reaper (Stark et al. 2003), with an S2 cell–expressed mRNA, CG1969, that is not a predicted target of the miRNAs expressed in S2 cells. reaper mRNA was enriched in total RNA extracted from Ago1 immunoprecipitates compared with control immunoprecipitates, but CG1969 was not (Fig. 1D, levels normalized to rp49). These data suggest that mRNAs can be enriched by virtue of their association with miRNAs in the purified Ago1 complexes.
FIGURE 1.
miRNP complex purification. (A) Schematic representation of the purification protocol. F/H-dAgo1 indicates Drosophila Ago1 protein tagged at the N terminus with a FLAG epitope followed by an HA epitope. Although two epitope tags were incorporated in the Ago-1 construct, we found that the specific enrichment of Ago-containing complexes and miRNAs was not significantly improved using a two-step purification. (B) Immunoblot to visualize F/H-dAgo1 purification. Lanes labeled + contain samples from cells stably transfected to express F/H-dAgo1. The lane labeled − contains sample from untransfected S2 cells. The input and eluate samples were from the same gel, but intervening lanes are not shown. The blot was first probed with anti-HA and reprobed with anti-tubulin. (C) Northern blot probed to visualize bantam miRNA and miR-2b. Ago1 denotes eluate from F/H-dAgo1 transfected cells; cont. denotes eluate from untransfected cells. (D) Quantitative RT-PCR to detect reaper (rpr) and CG1969 mRNAs in the eluate fraction, normalized to rp49. “1” and “2” are independent purifications.
Total RNA copurified with Ago1 was then used to probe cDNA microarrays to assess the complexity of the miRNP-associated mRNA pool. RNA samples from three independent purifications from Ago1 transfected cells and from untransfected control cells were analyzed. Each was compared to a reference sample of total RNA from 0- to 24-h embryos. Eighty-nine RNAs showed reproducible enrichment in the Ago1 samples compared with the controls, indicating the method consistently purifies a subset of cellular mRNAs (Supplemental Table S1). Previous studies have suggested that sequences in the 5′ end of the miRNA are the primary determinants of target specificity (Lewis et al. 2003, 2005; Doench and Sharp 2004; Brennecke et al. 2005; Grun et al. 2005). To assess whether the observed enrichment of the 89 mRNAs can be attributed to association with miRNAs, we searched for sequences complementary to the seed regions of Drosophila miRNAs.
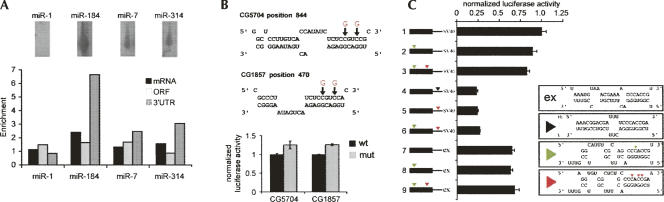
The number of miRNA seed matches per kilobase of 3′UTR sequence was compared for the immunopurified mRNA set and the UTRs of the whole transcriptome set. The ratio provides an enrichment value that corrects for overall UTR length and for the frequency of seed matches in the entire data set. A value of one means that a sequence is represented with the same frequency in the experimental data set as in the whole transcriptome. Enrichment values for the complete set of miRNAs are provided in Supplemental Figure S1A. We chose to focus on the small number of miRNAs with an enrichment of greater than two and that are expressed in S2 cells (Fig. 2A). miR-184 is highly expressed in S2 cells, and sequences complementary to its seed region were overrepresented in the enriched mRNAs compared with their occurrence in the whole Drosophila transcriptome. Although twofold enrichment for miR-184 seed matches was seen in the whole mRNA data set, the enrichment was much higher if the analysis was limited to 3′UTRs. A similar pattern of enrichment for UTR sites was observed for miR-7 and miR-314. miR-1 is not expressed in S2 cells and was used as a control. No enrichment was seen for miR-1 complementary sequences. In the analysis of the complete miRNA set, miR-124 stands out as it gives an enrichment signal despite not being expressed in S2 cells. In this case, the enrichment signal can be attributed to the co-occurrence of miR-124 sites in three of the five UTRs that have miR-184 sites (Supplemental Fig. S1B).
FIGURE 2.
Enrichment of sequences complementary to miRNA seeds in Ago1 immunoprecipitates. (A, top) Northern blots probed to visualize miR-1, miR-184, miR-7, and miR-314 in total RNA from S2 cells. (Bottom) Relative enrichment of 7mer sequences complementary to positions 2–8 of the indicated miRNAs in the messenger RNAs immunopurified with Ago1. mRNA indicates the sequence matches found in the whole transcript. ORF indicates matches found in coding sequence. UTR indicates sequence matches found in 3′UTRs. Enrichment is normalized to the overall frequency of occurrence of the sequences in the mRNA, ORF, and UTR data sets. (B, top) Predicted miR-184 sites in the ORFs of CG5704 and CG1857 (top, strand mRNA; bottom, strand miR-184). Arrows indicate residues altered in the mutant constructs to disrupt pairing to miR-184, while retaining the ORF in the mRNA. The ORFs of the wild-type and mutant forms of CG5704 and CG1857 were cloned in frame with firefly luciferase. (Bottom) Luciferase assays comparing regulation by endogenous miR-184 in S2 cells transfected to express the ORF–luciferase fusion proteins. Levels were normalized to the wild-type construct. Elevated activity in the mutant constructs indicates alleviation of repression mediated by miR-184. (C) Luciferase assays for regulation of the reporter constructs depicted at left by miR-278. SV40 and expanded indicate the origin of the 3′UTR. Colored arrowheads indicate the introduced miR-278 sites. Boxes at right show these sites: “ex” shows the endogenous miR-278 site in the expanded 3′UTR; black triangle shows a synthetic miR-278; green and red triangles denote the alteration of the luciferase ORF to produce sites matching the miR-278 seed region.
We noted that some miRNAs known to be expressed in S2 cells failed to give a clear enrichment signal. Because the enrichment signal reflects the occurrence of mRNAs with a site in the purified set compared with the frequency in the whole transcriptome, it is very sensitive to loss of mRNAs. Loss of a target RNA, for example, due to unstable association with the miRNP might lead to loss of the signal, even if other targets were recovered. Low target RNA levels, perhaps due to low endogenous expression or destabilization of the target RNA as a consequence of miRNP association, could make their detection difficult, leading to loss of an enrichment signal even if they were present. These observations suggest that while the approach may not capture all miRNA target associations, it may be useful to capture a subset of the stable ones.
Although 3′UTR sites appear to contribute most of the observed enrichment, some of the miR-184–associated mRNAs had sites in the coding region. Previous studies have indicated the possibility of miRNAs being able to act on target sites in the coding region of an mRNA (Lewis et al. 2005; Nakamoto et al. 2005). To ask whether the potential miR-184 ORF sites can contribute to regulation, we selected two such mRNAs, which lack 3′UTR sites, for further analysis (Fig. 2B). The ORFs of the CG5704 and CG1857 mRNAs were cloned in-frame with firefly luciferase. Similar constructs were made in which two C residues in the putative target sites were changed to G, so that the ORF was maintained while disrupting complementarity to the miR-184 seed (Fig. 2B). These constructs were expressed in S2 cells together with a Renilla luciferase control, and normalized luciferase levels were determined. In both cases disruption of seed complementarity led to a modest, but reproducible, increase in luciferase activity. This indicates that sites within the coding sequence can contribute to miRNA-mediated regulation in vivo. However, the limited enrichment for ORF sites overall suggests that their contribution to the association of mRNAs with the miRNP is minor compared with the contribution of sites located in 3′UTRs.
To further explore the potential for sites in coding sequence to function, we constructed a series of luciferase reporters, based on the experimentally validated miR-278 target expanded (Teleman et al. 2006). Single sites for miR-278 were generated in the ORF of luciferase, by altering wobble positions while retaining the coding sequence of luciferase. Two such sites were possible, and introducing one (construct 2) or both (construct 3) marginally reduced luciferase activity in the presence of miR-278 (Fig. 2C). These sites were active when placed within the context of the 3′UTR (constructs 5 and 6), yielding repression similar in magnitude to an optimal miR-278 site (construct 4). Likewise, for a luciferase construct containing the expanded 3′UTR, with an endogenous miR-278 site, adding sites to the luciferase ORF had no impact (constructs 7–9). These observations suggest that miRNA sites in the coding region can confer regulation but that their effect is marginal compared with the regulation conferred by sites in 3′UTRs. All our further analysis therefore focused on 3′UTR sites.
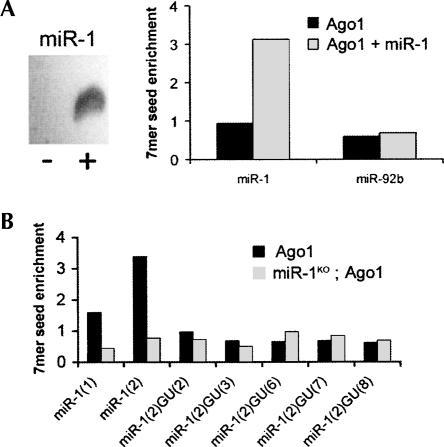
We next assessed the effects of expressing a new miRNA on the profile of target RNAs associated with Ago1 in S2 cells. miR-1 was expressed by transient transfection of an expression vector into S2 cells that had been selected for stable expression of F/H-Ago1 (Fig. 3A, left). Ago1 purification followed by microarray analysis revealed a threefold enrichment of sequences complementary to miR-1 in the miR-1 transfected F/H-Ago1 cells compared with untransfected Ago1-expressing cells (Fig. 3A, right), while the overall enrichment profile of the S2 cells was relatively unchanged (Supplemental Fig. S2). For example, no enrichment was seen for sequences complementary to another miRNA, miR-92b, which is not expressed in S2 cells. This suggests that miR-1 was incorporated into Ago1-containing complexes when expressed in S2 cells and was able to enrich for potential miR-1 target RNAs by immunopurification.
FIGURE 3.
Alterations in the profile of Ago1-associated RNAs by adding or removing miR-1. (A, left) Northern blot comparing miR-1 expression in untransfected (−) and transfected (+) S2 cells. (Right) Enrichment of 7mer sequences complementary to positions 2–8 of miR-1 or miR-92b in RNA copurified with Ago1. (B) Enrichment of 7mer sequences complementary to miR-1 in total RNA copurified with Ago1 from control or miR-1 mutant embryos. miR-1(1) indicates complementarity to positions 1–7; mir-1(2) to positions 2–8. GU(#) indicates sequence matches with a GU base pair at the indicated position.
To examine the effects that depleting a miRNA in vivo would have on the profile of Ago1-associated mRNAs, we made use of transgenic flies expressing the epitope tagged Ago1 transgene, and introduced into the miR-1 mutant background (Sokol and Ambros 2005). mRNAs purified by association with Ago1 from 18- to 24-h control and miR-1 mutant embryos were compared. Microarray analysis revealed an enrichment for the 6mer and 7mer sequences complementary to the miR-1 seed in the 3′UTRs of the immunopurified mRNAs from Ago1-expressing control embryos, but not from the miR-1 mutant embryos (Fig. 3B; Supplemental Table S2). In a total of 108 mRNAs that were differentially recovered in the immunopurifed samples, 32 had 6mer seed matches to positions 2–7 or 3–8 of miR-1, consistent with previous evidence that positions 2–8 are the primary determinant of miRNA specificity (Brennecke et al. 2005). This represents a significant overrepresentation of potential miR-1 targets among the immunopurified RNAs compared with the occurrence of these seed matches in the total mRNA pool (P = 8 × 10−5) (for analysis, see Supplemental Table S2). Figure 3B shows that the strongest enrichment for 7mer seed matches corresponded to positions 2–8 in miR-1 mutants. No overrepresentation was seen for sequence matches that require a G:U base-pair at any of the possible positions complementary to the miR-1 seed, consistent with the previous finding that GU base pairs, although compatible with regulation in a miRNA overexpression scenario, are always detrimental (Doench and Sharp 2004; Brennecke et al. 2005).
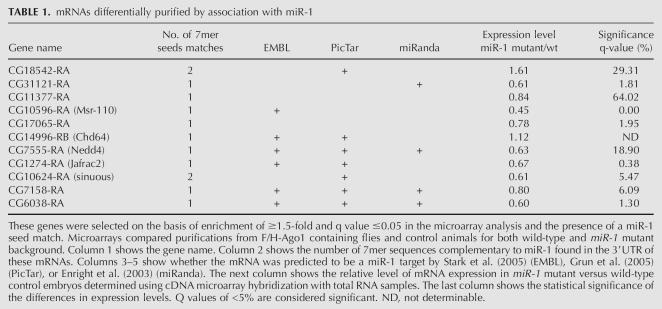
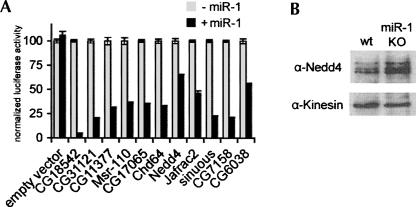
Thirty-two genes with potential miR-1 target sites were identified on the basis of their differential immunopurification with miRNP from wild-type animals compared with miR-1 mutants (Table S2). For further analysis, we selected the 11 genes with 7mer seed matches to positions 2–8 (Table 1). To test if these mRNAs can be regulated by miR-1, luciferase reporters were prepared containing the 3′UTRs of the 11 genes and expressed in S2 cells with or without coexpression of miR-1. Luciferase activity was reduced for all 11 constructs when miR-1 was coexpressed, although to varying extents (Fig. 4A). The combination of their association with miR-1 containing miRNP and their potential to be post-transcriptionally regulated by miR-1 in S2 cells indicate that these 11 mRNAs are likely to be miR-1 targets in vivo. For one of these genes, an antibody to the encoded protein was available, and immunoblot analysis of embryo lysates revealed an increase in the level of Nedd4 protein in the miR-1 mutant sample compared with wild-type embryos (Fig. 4B). We note that only five of the 11 newly validated miR-1 target RNAs had been previously predicted by more than one method and that two had not been predicted by any method. More extensive analysis of the miRNA–target relationships that exist in vivo for multiple miRNAs may provide new tools with which to improve the specificity and sensitivity of target prediction.
TABLE 1.
mRNAs differentially purified by association with miR-1
FIGURE 4.
mRNAs differentially enriched in control and miR-1 mutant embryos. (A) Histogram showing regulation by miR-1 of luciferase reporters containing the 3′UTRs of the 11 mRNAs that contributed to the enrichment signal from panel B. All were down-regulated in cells transfected to express miR-1 compared with control cells. A Renilla luciferase construct with the sv40 3′UTR was cotransfected to provide a normalization control. (B) Immunoblot of proteins samples from 18- to 24-h control embryos (wt) and miR-1 mutant embryos (miR-1KO), probed with antibody to Nedd4 and then reprobed with anti-kinesin as a loading control.
In this analysis we have assumed that the miRNA–target associations that we detect reflect the existence of a complex in the miRNA expressing cells that is stable enough to survive the immunopurification protocol. A priori we cannot exclude the possibility that some equilibration occurs in the lysate during the immunopurification, so that a target RNA that is not normally coexpressed with the miRNA could bind during the purification. Differential recovery of mRNAs in control and mutant samples nonetheless allows us to conclude that the recovery is due to association with the specific miRNA, even if some of the association were to occur post-lysis. The differentially recovered mRNAs are therefore good candidates to be targets, if coexpressed with the miRNA. For example, the increase in Nedd4 protein level in the miR-1 mutant embryos is most easily explained if Nedd4 is a genuine target in miR-1 expressing cells.
Previous reports have indicated that miRNAs can reduce the level of some of their target transcripts (Lim et al. 2005; Giraldez et al. 2006; Rehwinkel et al. 2006; Teleman et al. 2006; for review, see Jackson and Standart 2007). Transcript levels for the 11 miR-1–associated target genes changed only modestly in wild-type versus miR-1 mutant embryos, in most cases less than twofold up or down, and few of these differences were statistically significant (Table 1, last two columns). miRNA-mediated regulation of these genes does not appear to involve significant destabilization of their mRNAs. A priori it seems likely that mRNAs identified by virtue of stable association with the Ago1 complex would be unlikely to include those strongly affected at the RNA level by miRNAs. Such targets are more likely to reflect mRNAs abundantly expressed in the miRNA expressing cells, and for which the miRNA has a role in limiting or buffering expression levels perhaps “tuning” target activity levels (Bartel and Chen 2004), rather than reducing them to an inconsequential level.
MATERIALS AND METHODS
Molecular cloning
The FLAG/HA epitope tag was constructed by annealing two oligonucleotides 5′ CATGGACTACAAGGACGACGATGACAAGCTCGATGGAGGATACCCCTACGACGTGCCCGACTACGCCGGAGGC and 5′ GGCCGCCTCCGGCGTAGTCGGGCACGTCGTAGGGGTATCCTCCATCGAGCTTGTCATCGTCGTCCTTGTAGTCCATGGTAC. The resulting duplex was digested with KpnI and NotI and ligated into KpnI/NotI sites of a P-element vector containing the tubulin promoter (Lecuit et al. 1996). Ago1 sequences were from EST clone LD09501 (Kataoka et al. 2001). A 300-bp fragment of Ago1 was amplified by PCR using the primers 5′ AAGGAAAAAAGCGGCCGCATGTATCCAGTTGGACAACAGTC and 5′ GTTGCGGTGGCAATTGCTGG to introduce a NotI site, and a NotI/MfeI fragment was cloned together with a MfeI/XhoI fragment comprising the remainder of the Ago1 cDNA, into NotI/XhoI digested pCaSpEr4-Tub>FLAG/HA such that the Ago1 cDNA lies downstream of the HA and in frame. This plasmid was used to generate transgenic flies. For cell culture assays, the Tub>FLAG/HA-Ago1 fragment was recloned into modified pRmHa vector (engineered to contain a Puromycin resistance gene).
Luciferase 3′UTR reporter constructs and miRNA plasmids were expressed under the Tubulin promoter as in Teleman et al. (2006). Details of all constructs provided on request.
Cell culture
Stable cell lines expressing F/H-Ago1 were generated by transfection of Schneider S2 cells in six-well plates with 1 μg of the plasmid and Cellfectin (Invitrogen) according to the manufacturer's protocol. Selection for the plasmid was applied using 10 μg/mL of Puromycin, beginning 48 h after transfection. The Puromycin-resistant cell population was tested for expression of F/H-Ago1 by immunoblotting with anti-HA antibody. The stable lines were amplified to ∼1010 cells, corresponding to 1 L of suspension culture for extract preparation.
Immunoblotting
The samples for immunoblots were separated by SDS-PAGE and transferred using the semi-dry technique to a nitrocellulose membrane. Primary antibodies used were anti-HA (Roche), anti-tubulin (Sigma), anti-kinesin (Cytoskeleton), and anti-Nedd4 (Sakata et al. 2004). Peroxidase-conjugated secondary antibodies were used, and the bound antibodies were visualized by the ECL System according to the manufacturer's protocol (Amersham-Biosciences).
Cell extracts and embryo lysates
S2 cell extracts were prepared as described (Dignam et al. 1983; Rigaut et al. 1999; Puig et al. 2001; Meister et al. 2004) with certain modifications; 0.5–1 × 1010 cells were collected by centrifugation (200g, 15 min). The cell pellet washed twice with PBS, resuspended in 3 volumes of hypotonic buffer (20 mM HEPES at pH 7.9, 2 mM MgCl2, 10 mM KCl, 2% complete protease inhibitor cocktail [Sigma], 100 U/mL RNasin [Promega]) and incubated on ice for 20 min. The cells were lysed using a Dounce homogenizer. Nuclei were removed by centrifugation at 10,000g for 20 min. The supernatant was adjusted to 150 mM KCL, 20% glycerol at a total protein concentration of 5 mg/mL. For lysates from Drosophila embryos, large-scale embryo collection was set up with wild-type, Tub>F/H-Ago1, and miR-1/CyOKr-GFP;tub>f/h-ago1 flies. Embryos were collected for 6 h and aged for 17 h. Homozygous miR-1 mutant embryos were selected by separating GFP-expressing embryos (carrying the CyOKr-GFP balancer chromosome) from GFP-negative miR-1 mutants using the Copas Select embryo sorter (Union Biometrica). Embryos were washed and crushed in liquid nitrogen before resuspending them in the hypotonic buffer. The rest of the procedure was same as for cell extracts.
Immunopurification
Ten microliters of monoclonal anti-HA conjugated agarose beads (Sigma) were washed once with 0.1 M glycine-HCl (pH 2.5) and equilibrated with 0.2 M Tris-HCl (pH 8). The beads were then blocked in wash buffer (20 mM HEPES at pH 7.9, 150 mM KCl, 20% glycerol, 0.5% Tween-20) containing 0.2 mg/mL heparin for 2 h at 4°C with rotation. The lysates were cleared by centrifugation at 10,000g for 10 min and then incubated with the beads for 1 h at 4°C. The beads were collected and washed sequentially for 10 min each with buffer containing 150 mM, 200 mM, and 75 mM and then 200 mM KCl. The bound complexes were eluted by shaking the beads vigorously, with 50 μL of wash buffer containing 50 μg/mL of HA peptide (Sigma) for 30 min at room temperature. The elution step was repeated and eluates pooled.
RNA extraction and microarray analysis
The eluate was Proteinase K–digested and RNA extracted using TRIzol reagent (Invitrogen). Ten micrograms of yeast tRNA was added to serve as a carrier for ethanol precipitation, and the RNA pellet was resuspended in 10 μL of water. Microarray experiments were performed as in Teleman et al. (2005) with arrays containing the INDAC oligomers representing the Flybase, version 4.0, transcripts of the genome. The cell culture experiments were used to probe arrays containing DGC1 and DGC2 gene sets. Cy5-labeled total RNA from 0- to 24-h embryos was used as a reference sample. RNA extracted from the immunopurifications was Cy3 labeled. Three biological replicates, representing independent immunopurifications, were measured in each case. A median log2 ratio of >0.6 and a q-value of <0.05 (<5% false positive) were considered as reliable enrichment. 3′UTR and coding sequence databases were extracted from http://flybase.bio.indiana.edu/, and the miRNA sequences of Drosophila were obtained from http://microrna.sanger.ac.uk/.
RNA analysis
Northern blots for miRNAs were prepared as in Brennecke et al. (2003). For Q-PCR, total RNA extracted from the immunopurified samples was used to generate first-strand cDNA primed with oligo(dT) using StrataScript RT (Stratagene). Quantitative PCR was done using SYBR Green (ABI) using the ABI Prism 7000 or 7500 Sequence Detection System. All measurements were done in duplicate and normalized to rp49.
Reporter assays
We transfected 0.1 μg of a firefly luciferase reporter plasmid, 0.1 μg of a Renilla luciferase expressing plasmid (transfection control), and 1 μg of the miRNA-expression plasmid or empty vector into S2 cells. Transfections were performed in triplicate. Dual-luciferase assay system (Promega) was used 3 d post-transfection, according to the manufacturer's instructions.
SUPPLEMENTAL DATA
Supplemental data are available at http://www.tll.org.sg/stephen.asp.
ACKNOWLEDGMENTS
We thank Ann-Mari Voie for preparing transgenic flies. This work was supported in part by EU FP6 grant LSHG-CT-2006-037900; www.sirocco-project.eu.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.563707.
REFERENCES
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P., Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant, I., Rehwinkel, J., Doerks, T., Stark, A., Bork, P., Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Stark, A., Russell, R.B., Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., Roeder, R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Sharp, P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, T., Zamore, P.D. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., John, B., Gaul, U., Tuschl, T., Sander, C., Marks, D.S. MicroRNA targets in Drosophila . Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez, A.J., Mishima, Y., Rihel, J., Grocock, R.J., Van Dongen, S., Inoue, K., Enright, A.J., Schier, A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Grun, D., Wang, Y.L., Langenberger, D., Gunsalus, K.C., Rajewsky, N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G., Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Jackson, R.J., Standart, N. How do microRNAs regulate gene expression? Sci. STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., Bartel, D.P., Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kataoka, Y., Takeichi, M., Uemura, T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells. 2001;6:313–325. doi: 10.1046/j.1365-2443.2001.00427.x. [DOI] [PubMed] [Google Scholar]
- Kiriakidou, M., Nelson, P.T., Kouranov, A., Fitziev, P., Bouyioukos, C., Mourelatos, Z., Hatzigeorgiou, A. A combined computational-experimental approach predicts human microRNA targets. Genes & Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, A., Grun, D., Poy, M.N., Wolf, R., Rosenberg, L., Epstein, E.J., MacMenamin, P., da Piedade, I., Gunsalus, K.C., Stoffel, M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lall, S., Grun, D., Krek, A., Chen, K., Wang, Y.L., Dewey, C.N., Sood, P., Colombo, T., Bray, N., Macmenamin, P., et al. A genome-wide map of conserved microRNA targets in C. elegans . Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Lecuit, T., Brook, W.J., Ng, M., Calleja, M., Sun, H., Cohen, S.M. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., Burge, C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Peters, L., Chen, P.Y., Urlaub, H., Luhrmann, R., Tuschl, T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Nakamoto, M., Jin, P., O'Donnell, W.T., Warren, S.T. Physiological identification of human transcripts translationally regulated by a specific microRNA. Hum. Mol. Genet. 2005;14:3813–3821. doi: 10.1093/hmg/ddi397. [DOI] [PubMed] [Google Scholar]
- Okamura, K., Ishizuka, A., Siomi, H., Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Artus, C.G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., Filipowicz, W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., Seraphin, B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rajewsky, N. microRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Rehwinkel, J., Natalin, P., Stark, A., Brennecke, J., Cohen, S.M., Izaurralde, E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster . Mol. Cell. Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., Seraphin, B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Robins, H., Li, Y., Padgett, R.W. Incorporating structure to predict microRNA targets. Proc. Natl. Acad. Sci. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata, T., Sakaguchi, H., Tsuda, L., Higashitani, A., Aigaki, T., Matsuno, K., Hayashi, S. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr. Biol. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Sokol, N.S., Ambros, V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Russell, R.B., Cohen, S.M. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:e60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Bushati, N., Russell, R.B., Cohen, S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Teleman, A.A., Chen, Y.W., Cohen, S.M. Drosophila Melted modulates FOXO and TOR activity. Dev. Cell. 2005;9:271–281. doi: 10.1016/j.devcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Teleman, A.A., Maitra, S., Cohen, S.M. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes & Dev. 2006;20:417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Stricker, H.M., Gou, D., Liu, L. MicroRNA: Past and present. Front. Biosci. 2007;12:2316–2329. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- Wu, L., Fan, J., Belasco, J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X., Lu, J., Kulbokas, E.J., Golub, T.R., Mootha, V., Lindblad-Toh, K., Lander, E.S., Kellis, M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Haley, B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang, B., Wang, Q., Pan, X. MicroRNAs and their regulatory roles in animals and plants. J. Cell. Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]