Abstract
The armadillo-family protein, p120 catenin (p120), binds to the juxtamembrane domain of classical cadherins and increases cell–cell junction stability. Overexpression of p120 modulates the activity of Rho family GTPases and augments cell migratory ability. Here we show that down-regulation of p120 in epithelial MCF-7 cells by siRNA leads to a striking decrease in lamellipodial persistence and focal adhesion formation. Similar alterations in lamellipodial activity were observed in MCF-7 cells treated with siRNA to cortactin, an activator of Arp2/3-dependent actin polymerization. We found that, in many cell types, p120 is colocalized with cortactin-containing actin structures not only at cell–cell junctions, but also at extrajunctional sites including membrane ruffles and actin-rich halos around endocytotic vesicles. p120 depletion led to dramatic loss of cortactin and its partner, Arp3, from the cell leading edges. Cortactin and p120 are shown to directly interact with each other via the cortactin N-terminal region. We propose that the mechanism underlying p120 functions at the leading edge involves its cooperation with cortactin.
Keywords: adherens junctions, Arp2/3, endocytic vesicles, focal adhesions, leading edge
Catenin p120 (also known as p120ctn and p120cas) is an armadillo-family protein that binds to the juxtamembrane domain of classical cadherins (1). Knockout or knockdown of p120, as well as experimental interventions blocking its binding to cadherins, leads, in many cellular systems, to perturbation of adherens junction formation/maintenance and impeding cell spreading on the cadherin-coated substrates (2–10). These alterations in adhesion are often accompanied by a reduction in cellular cadherin levels (2, 3, 6, 8, 11) due to augmentation of its internalization and degradation (2, 11, 12). The degree of adhesion impairment depends on cellular context and varies from very subtle in the epidermis (8) to very severe in the salivary gland (3).
In addition to its role in cell–cell junctions, p120 appears to have other functions. It participates in transcription regulation via its interaction with transcriptional repressor Kaiso (13), controls inflammatory responses in the skin through regulation of NFκB (8), and regulates dendritic spine development (14). Possible involvement of p120 in the regulation of cell migration is especially intriguing because the increased migratory and invasive ability of several types of tumor cells correlates with increased cytoplasmic levels of p120 (15–18).
In culture, overexpression of p120 enhances the motility of fibroblasts (19, 20) and epithelial cells (21). These motility alterations are thought to be a result of changes in the activity of small Rho family GTPases. The activities of Rac and Cdc42 were shown to increase upon p120 overexpression (19, 20), whereas the activity of Rho can decrease (20, 22) or increase (21) depending on the cell type. p120 was reported to interact with Rho (22–24), the Rho family GTPases exchange factor Vav2 (20), and the Rho GTPase-activating protein p190RhoGAP (25). Thus, the mechanisms underlying the p120-mediated changes in small GTPase activities are complex and cell context-dependent.
To elucidate the mechanisms of p120 involvement in motility regulation, we characterize here the subcellular localization of p120 in cells and analyze its roles in different processes associated with cell motility and matrix adhesion by using RNAi-mediated gene silencing. We show that p120 is concentrated at major sites of actin polymerization such as membrane ruffles, lamellipodia, and actin halos associated with endocytotic vesicles. In all these sites, p120 colocalizes with the actin- and Arp2/3 complex-binding protein cortactin. We present evidence for direct interaction between cortactin and p120 and involvement of the latter in the proper localization of cortactin and Arp3 to the ruffles and lamellipodia. Down-regulation of p120 not only destabilizes adherens junctions, but also interferes with persistent lamellipodia extension and formation of focal adhesions. We propose that p120, in cooperation with cortactin, is involved in the regulation of cell leading edge dynamics.
Results
p120 Catenin Is Required for Lamellipodial Persistence.
We first produced p120-knockdown MCF-7 epithelial cells by using stable expression of a construct encoding p120 shRNA and confirmed that, similarly to other cultured epithelial cells lacking p120 (2, 3, 8), these cells demonstrate impaired cadherin-mediated cell–cell junctions [supporting information (SI) Fig. 4].
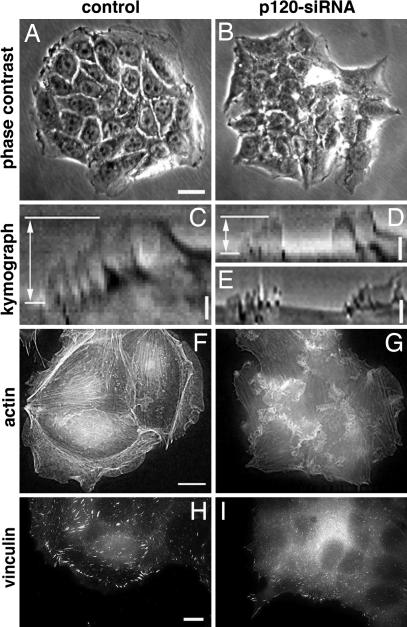
The lamellipodial activity of quiescent epithelial cells can be induced by a variety of motogenic factors, including neuregulin (heregulin) (26), the ligand for the Erb3 and Erb4 receptors (27). To test whether p120-knockdown in epithelial cells affects lamellipodial activity, we examined the response of serum-starved control and p120-shRNA-expressing MCF-7 cells to neuregulin stimulation. As early as 5–10 min after the addition of neuregulin, the majority of control cells exhibited intensive lamellipodial and ruffling activity at the periphery of the epithelial island (Fig. 1 A and F and SI Movie 1a). However, neuregulin-treated, p120-shRNA-expressing MCF-7 cells formed, at the periphery, only sporadic, discontinuous lamellipodial protrusions (Fig. 1 B and G and SI Movie 1b).
Fig. 1.
Effect of p120-knockdown on the lamellipodial dynamics and focal adhesion formation in MCF-7 cells. (A and B) Phase contrast images of neuregulin (100 ng/ml)-treated colonies of control (A) and p120-shRNA-expressing (B) cells. (Scale bar: 20 μm.) Compare broad lamellipodia surrounding colony of control cells versus irregular protrusions in the poorly spread cells with down-regulated p120. See also SI Movie 1 a and b. (C–E) Typical kymographs illustrating dynamic behavior of lamellipodia in control (C), p120-shRNA-expressing (D), and cortactin shRNA-treated (E) cells. To generate the kymograph (28), phase contrast images of a 1-pixel-wide line drawn in the direction of protrusion were taken at 30-sec (C and D) or 40-sec (E) intervals and pasted side by side. The time axis is from left to right, and the duration of the sequence was 30 min (C and D) or 60 min (E). Note that protrusions in the control cell lasted longer than in the cell with down-regulated p120 or cortactin, where they were often interrupted by abrupt retractions. As a result, lamella extension (indicated by double-headed arrow in C and D) was higher in control cells than in the p120- or cortactin-depleted cells (see also Table 1). (Scale bars: 5 μm.) (F–I) Effect of 20-min neuregulin treatment on actin (F and G) and vinculin (H and I) distribution in control (F and H) and p120-shRNA-expressing (G and I) cells. (Scale bars: 10 μm.)
Kymograph analysis (28) revealed (Fig. 1 C and D and Table 1) that lamella extension rates were approximately the same in control MCF-7 cells and cells with p120-knockdown. However, the extension phase in control cells persisted longer than in the cells lacking p120, in which the development of protrusions was frequently interrupted by edge withdrawal (compare Fig. 1 C and D and Table 1). Consequently, maximal lamella extension (double-headed arrows in Fig. 1 C and D) and the net cell edge displacement per hour (Table 1) were significantly lower in the p120-deficient cells, compared with controls.
Table 1.
Kymograph analysis of lamellipodial activity in neuregulin-stimulated control, p120-knockdown, and cortactin-knockdown MCF-7 cells
| Control | p120-shRNA | Control | Cortactin-shRNA | |
|---|---|---|---|---|
| Lamella extension rate, μm/min | 1.1 ± 0.4 | 1.0 ± 0.3 | 1.9 ± 0.1 | 2.07 ± 0.13 |
| Average leading edge displacement, μm/h | 21.6 ± 2.6 | 6.8 ± 1.5 | 15.12 ± 0.79 | 5.73 ± 0.75 |
| Percent of cells not retracting lamella | 72.2 | 35.3 | 87 | 58 |
| Percent of cells not protruding lamella | 5.6 | 17.6 | 19 | 35 |
Cells (20 ± 34) from randomly selected colonies were used for each measurement. Values given represent the mean ± SEM. In experiments with p120-knockdown, period of observation was 30 min; in experiments with cortactin-knockdown, period of observation was 60 min.
Similar deficiency of lamellipodia persistence was described in cells after RNAi-induced down-regulation of cortactin, a component of ruffles and lamellipodia and a potent activator of Arp2/3-dependent actin polymerization (29). In MCF-7 cells, using kymograph analysis, we demonstrated that treatment with siRNA directed to cortactin, indeed, led to alteration of lamellipodial activity quantitatively similar to that described above for p120-depleated MCF-7 cells (Fig. 1E, SI Movie 1 c and d, and Table 1). It is noteworthy that, at the same time, overall morphology of cortactin-knockdown cells differed from that of p120-knockdown cells; in particular, cortactin-depleted cells, unlike p120-depleated cells, demonstrated augmented substrate spreading (SI Movie 1d) in agreement with a previous publication (30).
Visualization of actin cytoskeleton in control and p120-depleted MCF-7 cells treated with neuregulin revealed reduction of lamellipodia and stress fibers in cells lacking p120 (compare Fig. 1 F and G). At the same time, these cells still displayed nonpolarized actin-rich “internal protrusions” at the interface between neighboring cells (revealed by deconvolution microscopy; Fig. 1G) and, in some cases, dorsal ruffles (SI Movie 1b).
Although p120-knockdown significantly affects neuregulin-induced lamellipodial activity, the general mechanisms of neuregulin-dependent signal transduction were nevertheless preserved in the absence of p120. Indeed, both control and p120-deficient MCF-7 cells display similar activation of the MAP kinases Erk1/2 and PI3 kinase upon neuregulin treatment (SI Fig. 5 A and B).
Effect of p120-Knockdown on Focal Adhesion Formation and Cell Migration.
Besides destabilization of cell–cell junctions and decrease of lamellipodial persistence, adhesion to the extracellular matrix was also suppressed in p120-deficient cells. Significant reduction of the average projected area of p120-knockdown cells, as compared with their control counterparts, was observed in MCF-7 cells (SI Fig. 6 A Upper). Focal adhesions visualized by antivinculin antibody staining in p120-depleted MCF-7 cells (Fig. 1 H and I and SI Fig. 6 C and D) showed significant decrease in area and fluorescence intensity (SI Fig. 6B), as well as in total number per unit of cell area (SI Fig. 6A Lower), compared with focal adhesions in control cells. Moreover, observations of focal adhesion dynamics using cells transiently transfected with mCherry-vinculin demonstrated that the average rate of focal adhesion formation (number of newly formed adhesions per hour) was 32.7 (min 13 to max 59) in control cells and only 5.0 (min 2 to max 10) in p120-knockdown cells (P = 0.05, Wilcoxon's ranked-sum test). Taking into consideration the total number of focal adhesions in cells of each type, we calculated that the rate of focal adhesion turnover (fraction of focal adhesions, which appeared/disappeared per 1 h) was also somewhat higher in control cells than in p120-knockdown cells [0.32 ± 0.08 vs. 0.20 ± 0.08 (mean± SEM)].
The difference in focal adhesion formation between control and p120-depleted MCF-7 cells was especially spectacular in the situation of neuregulin-mediated stimulation, which caused a dramatic activation of focal adhesion formation in control cells (Fig. 1H). The effect of neuregulin on focal adhesions in p120-shRNA cells was manifested by the appearance of only a few mature focal adhesions at the cell peripheral processes (Fig. 1I).
To verify that the effects of RNAi described above are specific, we prepared a p120 construct bearing triple-point mutation in the 19-bp sequence that served as a target for shRNA-mediated knockdown in our experiments. This mutation conserved the amino acid sequence of the p120 protein, but rendered its expression insensitive to inhibition by the shRNA. Expression of this shRNA-insensitive p120 in the cells stably expressing the p120-shRNA, indeed, restored the ability of these cells to form typical adherens junctions (SI Fig. 7 A and B), as well as prominent actin-rich lamellipodia and focal adhesions (SI Fig. 7 C–F).
Requirement of p120 for cell spreading, migration, and focal adhesion formation was also validated in other cell types, including BALB/c 3T3 cells and B16 F1 melanoma cells expressing the p120-shRNA. We have shown that p120-knockdown inhibits migration speed and spreading efficiency in BALB/c 3T3 cells (SI Fig. 8) and reduces focal adhesion formation and spreading in B16 F1 cells (data not shown).
p120 Colocalizes and Directly Associates with Cortactin.
The involvement of p120 in lamellipodia and focal adhesion regulation suggests that it may interact with some proteins controlling formation of these structures. One of the apparent candidates for such interaction is cortactin, whose knockdown, as shown above, similarly to the p120-knockdown, suppresses lamellipodial persistence (see also ref. 29). To explore the relationships between p120 and cortactin, we investigated a possible interaction between these two proteins.
Immunofluorescence localization studies indicated that, in addition to cell–cell junctions, p120 was enriched in actin-containing ruffles and lamellipodia. Such localization, especially prominent in highly motile cells such as B16 F1 or MDA-MB 231 breast carcinoma (Fig. 2A, SI Movie 2a, and SI Fig. 9 A–D) was also observed in MCF-7 cells and BALB/c 3T3 fibroblasts (Fig. 2 B and C and SI Fig. 9 E and F). We used deconvolution-wide field microscopy and total interference reflection fluorescence microscopy to confirm that p120 is indeed enriched in ruffles and at the leading edge of the lamella close to the substrate (SI Fig. 9 A–D).
Fig. 2.
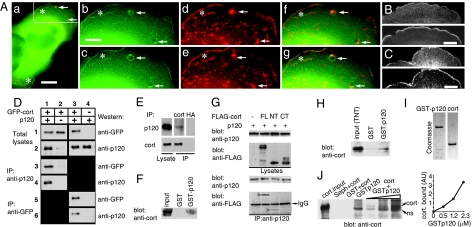
Association of p120 with cortactin. (A) Colocalization of p120 and cortactin at the cell leading edge, ruffles, and vesicle-associated halos in B16 F1 cells. Cells were cotransfected with GFP-p120 and RFP-cortactin and filmed using double color fluorescence microscopy (see SI Movie 2 a–c). A low-magnification merged image is shown in Aa, and enlarged images of the cell part shown in this photograph inside the rectangular frame are shown in A b–g. The time interval between images (b, d, and f) and (c, e, and g), respectively, is 30 sec. GFP-p120 fluorescence is shown in green (b and c), RFP-cortactin fluorescence is shown in red (d and e), and merged images are presented in f and g. Arrows indicate halos associated with moving vesicles. Asterisks indicate localization of the ruffles. (Scale bars: Aa, 10 μm; A b–g, 4.5 μm.) (B and C) Colocalization of endogenous p120 and cortactin at the lamellipodia of neuregulin-stimulated MCF-7 cells (B) and BALB/c-3T3 cells spreading on fibronectin (C). Staining with antibody against p120 (B and C Upper) and cortactin (B and C Lower). (Scale bars: 6 μm.) (D) Coimmunoprecipitation of exogenous cortactin and p120. CHO cells were cotransfected with plasmid-encoding p120 and GFP-cortactin (lanes 1 and 3), with GFP-cortactin alone (lane 2), or with p120 alone (lane 4). Total-cell lysates were immunoblotted with anti-GFP (row 1) or anti-p120 (row 2) antibodies. Immunoprecipitation was performed with either anti-p120 (lanes 3 and 4) or anti-GFP (lanes 5 and 6) antibodies. Precipitates were immunoblotted with anti-GFP (rows 3 and 5) or anti-p120 (rows 4 and 6) antibodies. (E) Coimmunoprecipitation of endogenous cortactin and p120. Endogenous cortactin was immunoprecipitated from the lysate of MCF-7 cells with anticortactin antibody. Anti-HA antibody was used as a negative control. The presence of p120 in the precipitates was detected by Western blotting with anti-p120 antibody. MCF-7 lysate was loaded as a positive control (left lane). (F) Bacterially expressed GST-p120 pulls down endogenous cortactin from the lysates of HCT116 cells. (G) p120 interacts with the N-terminal domain of cortactin. Lysates of 293T cells transfected with p120 or cotransfected with p120 and FLAG-full-length (FL), FLAG-N terminus (NT), or FLAG-C terminus (CT) cortactin constructs were immunoprecipitated with anti-p120 antibody. Immune complexes were analyzed by Western blotting with anti-FLAG antibody. Only full-length and N terminus of cortactin (red arrowheads) coimmunoprecipitate with p120. (H) In vitro translated N-terminal domain of cortactin interacts with p120. FLAG-NT cortactin was transcribed and translated by using a TNT Promega kit and incubated with GST- or GST-p120 Sepharose beads. Bead-associated proteins were then analyzed by Western blotting with anticortactin antibody. TNT product (input) was loaded as a positive control. (I) Coomassie staining of purified GST-p120 and cortactin after SDS/PAGE. (J) Cortactin directly binds p120. Purified cortactin (120 nM) was incubated with 0.5, 1.2, and 2.3 μM GST-p120 bound to glutathione-Sepharose beads for 30 min. The bound cortactin was detected by Western blotting with anticortactin antibody and quantified by densitometry (Right). Cortactin input as well as negative controls (cortactin added to Sepharose beads only, cortactin added to Sepharose-bound GST, and Sepharose-bound GST-p120 without added cortactin) are shown in first four lanes of the blot (Left). ns, nonspecific band.
We next studied the colocalization of p120 and cortactin. Coexpression of GFP-p120 and RFP-cortactin in B16 F1 mouse melanoma cells revealed dynamic colocalization of the two in membrane ruffles at the leading edges (Fig. 2A, asterisks) and in the halos associated with macropinocytic vesicles (Fig. 2A, arrows; see also SI Movie 2 a–c). This observation is in agreement with previous data showing that cortactin is a component of such structures (31, 32). In addition, colocalization of endogenous p120 and cortactin in lamellipodia of neuregulin-stimulated MCF-7 (Fig. 2B) and BALB/c-3T3 cells spreading on fibronectin (Fig. 2C) was also observed.
To determine whether p120 and cortactin are associated with the same molecular complex, CHO cells were transiently cotransfected with p120 and GFP-cortactin, and the proteins were immunoprecipitated from cell lysate by using antibodies against GFP or p120. As shown in Fig. 2D, a complex containing both p120 and GFP-cortactin could be precipitated by using either antibody. To confirm these results with endogenous proteins, we also demonstrated that the endogenous p120 can be specifically immunoprecipitated from MCF-7 cell lysates with anticortactin antibody (Fig. 2E). In the reciprocal experiment, endogenous cortactin was also pulled down from cell extract by using bacterially expressed GST-p120 (Fig. 2F).
Using FLAG-fusion constructs encoding the N-terminal (amino acids 1–330) or C-terminal (amino acids 350–546) parts of cortactin, we demonstrated that GFP-p120 coimmunoprecipitates with the N-terminal portion of cortactin (Fig. 2G), which contains Arp2/3- and actin-binding sites (33–35), but not with the C-terminal region containing the SH3 domain. We next translated the N-terminal domain of cortactin in vitro by using the TNT kit (Promega, Madison, WI) and showed that the translation product interacts with bacterially expressed GST-p120, but not with GST alone (Fig. 2H), suggesting that the proteins directly interact with each other. This interaction was also observed in the presence of a high concentration of latrunculin B, indicating that the binding was not mediated through residual actin filaments (data not shown).
Finally, we expressed GST-fused p120 and cortactin in bacteria, purified both proteins by affinity chromatography on glutathione-Sepharose column and size-exclusion chromatography, and removed the GST tag from GST-cortactin by proteolysis. Then using these purified proteins (Fig. 2I), we assessed their interaction by incubation of increasing concentrations of GST-p120 with a constant amount of cortactin and pulling down the complex by using glutathione-Sepharose beads (Fig. 2J). Amounts of pulled-down cortactin were quantified densitometrically (Fig. 2J Right). Concentration-dependent pull down of cortactin by p120 shows that these proteins, indeed, directly interact with each other.
p120-Knockdown Leads to the Disappearance of Cortactin from the Leading Edge.
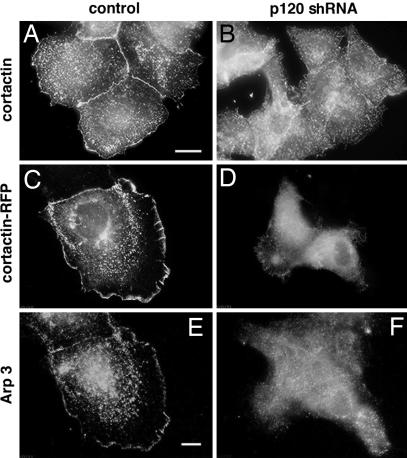
Down-regulation of p120 has a profound effect on cellular distribution of cortactin and its immediate partner, Arp3. In control MCF-7 cells, endogenous cortactin is enriched at the tip of the leading lamella and in cell–cell junctions (Fig. 3A). The levels of leading edge- and junction-associated cortactin were, however, dramatically reduced in cells with down-regulated p120 (Fig. 3B), although total cortactin levels in these cells were not altered, as estimated by Western blot analysis (SI Fig. 4). Exogenous RFP-cortactin in control cells is also recruited to the peripheral cell edge and cell–cell junctions (Fig. 3C), where it colocalizes with Arp3 (Fig. 3E). Cells expressing p120-shRNA failed to form a continuous RFP-cortactin- and Arp3-containing rim at the free cell edges and in cell–cell junctions (Fig. 3 D and F). Instead, both RFP-cortactin and Arp3 were abundant in the cytoplasm of these cells. Of note, the knockdown of cortactin does not abolish the localization of p120. Endogenous p120 was still prominent at the leading edges of cortactin-depleted MCF-7 and MDA-MB 231 cells (SI Fig. 10).
Fig. 3.
Effect of p120-knockdown on localization of cortactin and Arp3 in MCF-7 cells. (A and B) Immunofluorescence visualization of endogenous cortactin. (A) Cortactin enrichment at the cell–cell junction and peripheral lamellipodia in control cells. (B) Diffuse punctate cortactin staining in p120-knockdown cells. (Scale bar: 10 μm.) (C–F) Cells transfected with RFP-cortactin (C and D) and stained with anti-Arp3 antibody (E and F). Cells were fixed and stained 48 h after transfection and 20 min after neuregulin (100 ng/ml) addition. Note that cortactin and Arp3 colocalize at the cell–cell junctions and the cell edges in control cells (C and E), whereas in p120-shRNA-expressing cells (D and F), cortactin and Arp3 are diffusely distributed in the cytoplasm. (Scale bar: 5 μm.)
Because lamellipodial activity strongly depends on the function of small GTPase Rac and p120 overexpression was shown to augment Rac activity (19, 20), we checked whether knockdown of p120 affects Rac activity and localization. Pull-down assay (36) did not reveal a significant difference in Rac activity between control and p120-knockdown MCF-7 cells in conditions of serum starvation and neuregulin stimulation (SI Fig. 11 A and B). However, immunofluorescence staining with Rac antibody indicated that enrichment of Rac at the leading edges and cell–cell junctions typical for control MCF-7 cells (SI Fig. 11C) was not observed in the cells lacking p120.
Discussion
We have shown in this study that p120, in addition to its involvement in cell–cell junction stabilization, also plays an important role in the regulation of cell locomotion. In many cell types, we found p120 in extrajunctional sites such as ruffles, tips of advancing cell edges, and actin halos associated with macropinocytic vesicles. Furthermore, down-regulation of p120 by RNAi decreases leading edge extension persistence, cell spreading, and cell migration speed. These results are consistent with the recent observation of lamellipodial extensions' deficiency in p120-knockout primary keratinocytes (8). Interestingly, p120 is not found in focal adhesions, yet its down-regulation in MCF-7 cells impairs the development of these structures. The effects of p120-knockdown on protrusion stability, focal adhesion formation, and cell–cell junctions were all reversed by restoring p120 protein levels in the p120-shRNA-expressing cells.
One possible mechanism of p120 involvement in cell motility regulation could be based on the p120-dependent control of small GTPase activity. Neuregulin treatment was shown to activate Rac in MCF-7 cells (37). We were not able to detect a striking difference between control and p120-knockdown MCF-7 cells in the total Rac activity (basal and neuregulin-induced) by pull-down assay. However, we did see the difference in Rac localization. Although in control cells Rac was enriched at the leading edge and cell–cell junctions, it was not the case in the p120-knockdown cells. Thus, at the physiological expression level, p120 modulates Rac spatial distribution. Recent studies suggest that p120 is also involved in transient Rac-dependent translocation of p190RhoGAP to the plasma membrane and, hence, controls function of this major regulator of Rho activity (25).
In search of other mechanisms underlying p120 function in cell motility, we found that p120 interacts with the actin-binding protein cortactin, which controls Arp2/3-driven actin polymerization (33–35, 38). First, we showed that p120 and cortactin colocalize at the extrajunctional motility-related sites such as ruffles and macropinocytic vesicles. Next we showed that these two proteins coimmunoprecipitate, suggesting that they are associated with the same molecular complexes. We further showed that the N-terminal, actin-binding region of cortactin, but not its SH3 domain-containing C-terminal portion, interacts with p120. Finally, using bacterially expressed purified proteins, we demonstrated unequivocally that the interaction between p120 and cortactin is direct.
The interaction between p120 and cortactin is especially interesting in view of the fact that cortactin knockdown affects lamellipodial activity in exactly the same way as p120-knockdown. Although the overall cell morphology was very different (p120-knockdown cells were less spread, whereas cortactin-knockdown cells were more spread than control) (see ref. 30), lamellipodial persistence was compromised in both cases. The impairment of lamellipodial persistence was also observed previously in cortactin-knockdown fibrosarcoma cells (29). A key observation, which explains this similarity, is the aberrant localization of cortactin in p120-deficient cells demonstrated in our study. p120-knockdown leads to the disappearance of cortactin from the residual cell–cell junctions and to significant reduction of its content at the cell leading edges. It is noteworthy that localization of Arp3, a cortactin-binding component of the Arp2/3 complex (35), is changing in the same way.
Another striking feature of the p120-knockdown phenotype is the decreased ability of these cells to form focal adhesions. Cortactin knockdown was also reported to reduce focal adhesion formation in fibrosarcoma cells (29). Thus, the function of p120 in the regulation of focal adhesions could be related to its interaction with cortactin. It is worth noting that in some cell types cortactin localizes to specialized microdomains at the focal adhesion/stress fiber interface and cooperates with p190RhoGAP in the control of focal adhesion turnover (39). As mentioned above, p190RhoGAP also was recently shown to be a partner of p120 (25).
How could p120 control the proper localization and function of cortactin? The simplest scenario is that p120 functions as a cortactin docking site, binding to corresponding locations before and independently of cortactin. This hypothesis might be true for the adherens junction localization because p120 is enriched there via its direct binding to cadherin (1, 9). Moreover, p120 can localize to the cell leading edge independently of cortactin because we observed that cortactin knockdown did not prevent localization of p120 to the leading edge. Thus, p120 controls cortactin localization, but not vice versa. It is possible that p120 not only controls the localization, but also affects the function of cortactin. In our recent experiments performed in collaboration with Dr. A. Bernheim-Groswasser (Ben Gurion University, Negev, Israel), we studied the formation of actin superstructures (40) in a cell-free solution containing the major proteins participating in the assembly of lamellipodia: actin-nucleating Arp2/3 complex and its activator, VCA (the constitutively active verprolin homology, cofilin homology, and acidic region-containing domain of the Wiskott–Aldrich syndrome protein). Our studies revealed that p120 can efficiently modulate the cortactin effect on the self-assembly of actin arrays in this in vitro system (A. Bernheim-Groswasser, S.B., and A.B., unpublished data).
It is conceivable that, similarly to other Arp2/3 activators, such as WAVE and N-WASP (41), cortactin functions as a core of a large multimolecular complex, which determines its proper localization and activation in response to various signals. p120 is a good candidate for being a component of such a complex because it can either attract to cortactin many potentially important regulatory proteins or protect cortactin from their action. In particular, p120 binds tyrosine kinases, Fer and Fyn (42, 43), which are known to bind, and phosphorylate cortactin (34). Binding of cortactin to p120 may also trigger cortactin interaction with its partners, actin, Arp2/3 complex, and N-WASP, and thus affect actin-branching polymerization. Finally, p120 was reported to interact with kinesin (44, 45) and therefore can promote microtubule-based cortactin delivery to the proper localization sites.
In conclusion, we demonstrated here that p120-knockdown hinders several cell motility-related processes, including lamellipodial extension and focal adhesion formation. p120 directly interacts with cortactin and is required for its proper localization. We hypothesize that p120 functions as a local regulator of cortactin-dependent actin assembly.
Materials and Methods
Cell Culture.
MCF-7, MDA-MB 231, BALB/c-3T3, B16 F1, and HCT 116 cells were routinely cultured in DMEM supplemented with antibiotics, glutamine, and 10% heat-inactivated FCS (Gibco, Gaithersburg, MD). Establishment of MCF-7 cells, stably expressing shRNA for p120, as well as a transient cell-transfection procedure are described in SI Materials and Methods. Recombinant human neuregulin (NRG1-beta1 EGF Domain, R&D Systems, Minneapolis, MN) was added to serum-starved MCF-7 cells at a final concentration of 100 ng/ml.
cDNA Constructs.
Mouse p120 cDNA in pRcCMV vector (46) was kindly provided by A. Reynolds (Vanderbilt University, Nashville, TN). The GFP-fused p120 construct in the pEGFP-C1 vector was described previously (19), and cloning of GST-p120 is described in SI Materials and Methods. GFP- and RFP-fused cortactin constructs (31) were kindly provided by M. Kaksonen (University of California, Berkeley, CA), and GST-cortactin construct was kindly provided by A. M. Weaver (Vanderbilt University, Nashville, TN). The full-length N- and C-terminal portions of cortactin cloned into pcDNA3 vector (47) were kindly provided by J. T. Parsons (University of Virginia, Charlottesville, VA). mCherry-C2-vinculin plasmid was kindly provided by J. V. Small (Institute of Molecular Biotechnology, Vienna, Austria).
A 19-base sequence (655gatggttatccaggtggca673) common to all known isoforms of human and mouse p120, but absent in other p120 family members, was selected for the design of p120-shRNA. Details of retroviral plasmid construction are described in SI Materials and Methods. Mutant pEGFP-p120 preserving the same amino acid sequence, but containing three point mutations in the target nucleotide sequence (655gatggttaCccaggCggGa673) and therefore resistant to p120-shRNA, was produced as described in SI Materials and Methods. For cortactin-knockdown, we used anticortactin SMARTpool siRNA obtained from Dharmacon (Lafayette, CO).
Immunochemical and Biochemical Procedures.
Details of immunoprecipitation and Western blot procedures are described in SI Materials and Methods. Activation of PI3-kinase/PKB and MAPK pathways upon neuregulin treatment was analyzed by using monoclonal anti-active MAPK antibodies (DP ERK) and rabbit polyclonal anti-phospho-Ser 473 Akt/PKB antibodies purchased from Sigma–Aldrich Israel (Rehovot, Israel). Total ERK and PKB proteins were detected by using anti-MAPK and anti-Akt/PKB Pan (Sigma–Aldrich Israel), respectively. The Rac-GTP pull-down assay was performed by using a Rac activation assay kit (Cytoskeleton, Denver, CO). For in vitro expression of cortactin, coupled transcription–translation reactions were performed by using rabbit reticulocyte lysates (Promega, Madison, WI) as specified by the manufacturer (Promega). Purification of bacterially expressed GST-p120 and cortactin and in vitro binding assay between these two proteins are described in SI Materials and Methods.
Microscopy, Time Lapses, and Quantification.
Immunofluorescence staining was performed as previously described (19). Antibodies and reagents used are characterized in SI Materials and Methods. For live cell imaging, cells were plated on glass-bottom microwell dishes (MatTek, Ashland, MA). Movies were taken using a DeltaVision microscopy system (Applied Precision, Issaquah, WA). Details of focal adhesion measurements, cell motility quantification, and kymograph analysis of lamellipodial activity (28) are given in SI Materials and Methods. The Olympus (Tokyo, Japan) total interference reflection fluorescence (48) was used to determine the localization of endogenous p120 at the ventral cell surface (see details in SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Drs. A. Reynolds and A. M. Weaver (Vanderbilt University, Nashville, TN), T. Parsons (University of Virginia, Charlottesville, VA), and M. Kaksonen (University of California, Berkeley, CA) for providing plasmids; Dr. Bernheim-Groswasser (Ben Gurion University, Negev, Israel) for permission to mention unpublished results of our collaborative work; Drs. S. Albeck and T. Unger (Weizmann Institute, Rehovot, Isreal) for help in protein purification; and M. Lev Ran for excellent technical assistance. This work was supported, in part, by grants from the Israel Sciences Foundation, Minerva Foundation, and Pasteur-Weizmann Council (to A.B.). B.G. is the incumbent of the E. Neter Chair in Cell and Tumor Biology. A.B. holds the Joseph Moss Professorial Chair in Biomedical Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702731104/DC1.
References
- 1.Reynolds AB. Biochim Biophys Acta. 2007;1773:2–7. doi: 10.1016/j.bbamcr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis MA, Ireton RC, Reynolds AB. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis MA, Reynolds AB. Dev Cell. 2006;10:21–31. doi: 10.1016/j.devcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Gavard J, Lambert M, Grosheva I, Marthiens V, Irinopoulou T, Riou JF, Bershadsky A, Mege RM. J Cell Sci. 2004;117:257–270. doi: 10.1242/jcs.00857. [DOI] [PubMed] [Google Scholar]
- 5.Goodwin M, Kovacs EM, Thoreson MA, Reynolds AB, Yap AS. J Biol Chem. 2003;278:20533–20539. doi: 10.1074/jbc.M213171200. [DOI] [PubMed] [Google Scholar]
- 6.Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, Anastasiadis PZ, Matrisian L, Bundy LM, Sealy L, Gilbert B, van Roy F, Reynolds AB, et al. J Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer S, Ferreri DM, DeCocco NC, Minnear FL, Vincent PA. Am J Physiol Lung Cell Mol Physiol. 2004;286:L1143–L1153. doi: 10.1152/ajplung.00305.2003. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E. Cell. 2006;124:631–644. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoreson MA, Anastasiadis PZ, Daniel JM, Ireton RC, Wheelock MJ, Johnson KR, Hummingbird DK, Reynolds AB. J Cell Biol. 2000;148:189–202. doi: 10.1083/jcb.148.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yap AS, Niessen CM, Gumbiner BM. J Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, Kowalczyk AP. J Cell Biol. 2003;163:535–545. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao K, Oas RG, Chiasson CM, Kowalczyk AP. Biochim Biophys Acta. 2007;1773:8–16. doi: 10.1016/j.bbamcr.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Daniel JM. Biochim Biophys Acta. 2007;1773:59–68. doi: 10.1016/j.bbamcr.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 14.Elia LP, Yamamoto M, Zang K, Reichardt LF. Neuron. 2006;51:43–56. doi: 10.1016/j.neuron.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellovin DI, Bates RC, Muzikansky A, Rimm DL, Mercurio AM. Cancer Res. 2005;65:10938–10945. doi: 10.1158/0008-5472.CAN-05-1947. [DOI] [PubMed] [Google Scholar]
- 16.Sarrio D, Perez-Mies B, Hardisson D, Moreno-Bueno G, Suarez A, Cano A, Martin-Perez J, Gamallo C, Palacios J. Oncogene. 2004;23:3272–3283. doi: 10.1038/sj.onc.1207439. [DOI] [PubMed] [Google Scholar]
- 17.Shibata T, Kokubu A, Sekine S, Kanai Y, Hirohashi S. Am J Pathol. 2004;164:2269–2278. doi: 10.1016/S0002-9440(10)63783-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanagisawa M, Anastasiadis PZ. J Cell Biol. 2006;174:1087–1096. doi: 10.1083/jcb.200605022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grosheva I, Shtutman M, Elbaum M, Bershadsky AD. J Cell Sci. 2001;114:695–707. doi: 10.1242/jcs.114.4.695. [DOI] [PubMed] [Google Scholar]
- 20.Noren NK, Liu BP, Burridge K, Kreft B. J Cell Biol. 2000;150:567–580. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cozzolino M, Stagni V, Spinardi L, Campioni N, Fiorentini C, Salvati E, Alema S, Salvatore AM. Mol Biol Cell. 2003;14:1964–1977. doi: 10.1091/mbc.E02-08-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford HC, Zheng Y, Reynolds AB. Nat Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- 23.Castano J, Solanas G, Casagolda D, Raurell I, Villagrasa P, Bustelo XR, Garcia de Herreros A, Dunach M. Mol Cell Biol. 2007;27:1745–1757. doi: 10.1128/MCB.01974-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magie CR, Pinto-Santini D, Parkhurst SM. Development (Cambridge, UK) 2002;129:3771–3782. doi: 10.1242/dev.129.16.3771. [DOI] [PubMed] [Google Scholar]
- 25.Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, Reynolds AB. Cell. 2006;127:1027–1039. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
- 26.Chausovsky A, Waterman H, Elbaum M, Yarden Y, Geiger B, Bershadsky AD. Oncogene. 2000;19:878–888. doi: 10.1038/sj.onc.1203410. [DOI] [PubMed] [Google Scholar]
- 27.Yarden Y, Sliwkowski MX. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 28.Hinz B, Alt W, Johnen C, Herzog V, Kaiser HW. Exp Cell Res. 1999;251:234–243. doi: 10.1006/excr.1999.4541. [DOI] [PubMed] [Google Scholar]
- 29.Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Curr Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 30.van Rossum AG, Moolenaar WH, Schuuring E. Exp Cell Res. 2006;312:1658–1670. doi: 10.1016/j.yexcr.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 31.Kaksonen M, Peng HB, Rauvala H. J Cell Sci. 2000;113:4421–4426. doi: 10.1242/jcs.113.24.4421. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, Parsons JT. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daly RJ. Biochem J. 2004;382:13–25. doi: 10.1042/BJ20040737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lua BL, Low BC. FEBS Lett. 2005;579:577–585. doi: 10.1016/j.febslet.2004.12.055. [DOI] [PubMed] [Google Scholar]
- 35.Weaver AM, Heuser JE, Karginov AV, Lee WL, Parsons JT, Cooper JA. Curr Biol. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- 36.Benard V, Bohl BP, Bokoch GM. J Biol Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 37.Yang C, Liu Y, Lemmon MA, Kazanietz MG. Mol Cell Biol. 2006;26:831–842. doi: 10.1128/MCB.26.3.831-842.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weed SA, Parsons JT. Oncogene. 2001;20:6418–6434. doi: 10.1038/sj.onc.1204783. [DOI] [PubMed] [Google Scholar]
- 39.Burgstaller G, Gimona M. J Cell Sci. 2004;117:223–231. doi: 10.1242/jcs.00839. [DOI] [PubMed] [Google Scholar]
- 40.Haviv L, Brill-Karniely Y, Mahaffy R, Backouche F, Ben-Shaul A, Pollard TD, Bernheim-Groswasser A. Proc Natl Acad Sci USA. 2006;103:4906–4911. doi: 10.1073/pnas.0508269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stradal TE, Scita G. Curr Opin Cell Biol. 2006;18:4–10. doi: 10.1016/j.ceb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Lee SH. Mol Cells. 2005;20:256–262. [PubMed] [Google Scholar]
- 43.Piedra J, Miravet S, Castano J, Palmer HG, Heisterkamp N, Garcia de Herreros A, Dunach M. Mol Cell Biol. 2003;23:2287–2297. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Kojima S, Borisy GG, Green KJ. J Cell Biol. 2003;163:547–557. doi: 10.1083/jcb.200305137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanagisawa M, Kaverina IN, Wang A, Fujita Y, Reynolds AB, Anastasiadis PZ. J Biol Chem. 2004;279:9512–9521. doi: 10.1074/jbc.M310895200. [DOI] [PubMed] [Google Scholar]
- 46.Reynolds AB, Herbert L, Cleveland JL, Berg ST, Gaut JR. Oncogene. 1992;7:2439–2445. [PubMed] [Google Scholar]
- 47.Weed SA, Karginov AV, Schafer DA, Weaver AM, Kinley AW, Cooper JA, Parsons JT. J Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Axelrod D, Thompson NL, Burghardt TP. J Microsc. 1983;129:19–28. doi: 10.1111/j.1365-2818.1983.tb04158.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.