Abstract
Transcription factors MITF and PU.1 collaborate to increase expression of target genes like cathepsin K (Ctsk) and acid phosphatase 5 (Acp5) during osteoclast differentiation. We show that these factors can also repress transcription of target genes in committed myeloid precursors capable of forming either macrophages or osteoclasts. The direct interaction of MITF and PU.1 with the zinc finger protein Eos, an Ikaros family member, was necessary for repression of Ctsk and Acp5. Eos formed a complex with MITF and PU.1 at target gene promoters and suppressed transcription through recruitment of corepressors CtBP (C-terminal binding protein) and Sin3A, but during osteoclast differentiation, Eos association with Ctsk and Acp5 promoters was significantly decreased. Subsequently, MITF and PU.1 recruited coactivators to these target genes, resulting in robust expression of target genes. Overexpression of Eos in bone marrow-derived precursors disrupted osteoclast differentiation and selectively repressed transcription of MITF/PU.1 targets, while small interfering RNA knockdown of Eos resulted in increased basal expression of Ctsk and Acp5. This work provides a mechanism to account for the modulation of MITF and PU.1 activity in committed myeloid progenitors prior to the initiation of osteoclast differentiation in response to the appropriate extracellular signals.
Osteoclasts are derived from committed myeloid progenitors that differentiate into highly specialized multinuclear cells capable of bone resorption in response to the action of colony-stimulating factor 1 (CSF-1) and receptor activator of NF-κB ligand (RANKL) (36). Osteoclasts and the bone-forming cells, osteoblasts, play reciprocal roles in the bone remodeling process necessary for maintaining bone integrity and function. It is now well established that many human bone disorders, including osteoporosis, Paget's disease, and rheumatoid arthritis result from the imbalance in the differentiation and function of these two cell types (1).
MITF (microphthalmia-associated transcription factor), a basic helix-loop-helix-leucine zipper protein, has been implicated in differentiation and survival of developmentally unrelated cell types, including osteoclasts, melanocytes, and pigmented retinal epithelial cells (7, 30, 40). Interactions between MITF and the ETS family transcription factor PU.1 at least partly account for the ability of MITF to selectively regulate target genes during osteoclast differentiation (22, 27, 39). However, MITF and PU.1 are expressed in macrophages and osteoclasts and in the common mononuclear precursor for both of these cell types (13, 38). This poses a general question concerning how gene regulation patterns are maintained in closely related cell lineages. In recent work, we demonstrated that in primary cells deprived of CSF-1, MITF was sequestered to the cell cytoplasm through interactions with 14-3-3 proteins, providing one potential mechanism that regulates MITF activity in myeloid precursor cells (2).
Ikaros is the founding member of a family of five related Kruppel zinc finger transcription factors that can act as repressors of gene expression (6, 9, 14, 29, 31). The results of both knockout and dominant-negative studies in mice suggest that Ikaros is one of the central regulators of lymphocyte differentiation (4, 41). Ikaros and other family members associate with two corepressors, the Sin3 and Mi-2 complexes (15, 17, 18). Ikaros and Eos can also interact with corepressor CtBP (C-terminal binding protein), which can repress gene expression in a histone deacetylase (HDAC)-independent manner (16, 33). Besides lymphocytes, the influence of the Ikaros family members on other hematopoietic cell types, including the myeloid lineage, is largely unknown. Eos has previously been reported to be expressed in most hematopoietic cell types but is present at its highest levels in monocytic cells (34).
Here we demonstrate that expression of the Ikaros family protein Eos is temporally regulated during osteoclast differentiation. Overexpression of Eos in bone marrow-derived precursors disrupted osteoclast differentiation and represses transcription of osteoclast marker genes, like cathepsin K (Ctsk) and acid phosphatase 5 (Acp5). We provide direct evidence that Eos interacts with both MITF and PU.1 to repress transcription from specific promoters through recruitment of corepressors Sin3A and CtBP. This work provides a mechanism by which MITF and PU.1 activity is regulated in myeloid precursors and macrophages to prevent inappropriate expression activation of osteoclast-specific genes.
MATERIALS AND METHODS
Antibodies.
The peptide SSGDSSLEKDSL (corresponding to amino acids [aa] 8 to 19 of mouse Eos) was used to make specific antibody against Eos in rabbits (QCB/Biosource, Hopkinton, MA). Antibodies against MITF and PU.1 were previously described (24). Anti-BRG1 antibody was described elsewhere (42). Anti-CBP antibody was a kind gift from Marc R Montminy, San Diego, CA. Other commercially available antibodies include: Flag M2 and hemagglutinin (HA) (Sigma-Aldrich, St. Louis, MO), His6, glutathione S-transferase (GST), mSin3A, histone deacetylase 1 (HDAC1), CtBP, and Mi-2 (Santa Cruz Biotechnology Inc., Santa Cruz, CA), and histone H3 (Upstate Cell Signaling, Charlottesville, VA).
Cell culture and transfections.
NIH 3T3 cells were transfected using the calcium phosphate procedure. Expression vector pCDNA3-Flag-Eos was a kind gift from Merlin Crossley, Sydney, Australia, and was described earlier (34). The luciferase reporter constructs and expression vector for MITF and PU.1 have been described previously (22). COS-7 cells were transfected using Lipofectamine reagent (Invitrogen Life Technologies, Carlsbad, CA). DNA constructs for Flag-tagged MITF and truncation mutants were recently described (2). HA-tagged PU.1 and deletions were constructed in pCGN vector. To generate pEBG-GST-Eos, full-length Eos cDNA or various truncations of Eos cDNA were PCR amplified from pCDNA3-Flag-Eos plasmid and subcloned into the mammalian GST expression vector (pEBG). All of the amplified sequences were verified by DNA sequencing.
Culture and analysis of osteoclasts.
C57B/6J mice were used to make bone marrow precursors for the in vitro osteoclast differentiation experiments. The Mitf-vga mice were obtained from Lynn Lamoreux (Texas A&M University). The conditional “floxed” Pu.1 allele and conditions for inducible Pu.1 deletion using poly(dI-C)induction of Mx1-Cre were previously described (12). Mice containing the Pu.1 “floxed” allele were a gift from Dan Tenen (Harvard Institutes of Medicine, Harvard Medical School). Use and care of mice in this study were approved by the Ohio State University Institutional Animal Care and Use Committee. Detailed procedures for osteoclast differentiation from bone marrow-derived macrophages (BMMs) prepared from mice have been previously described (25, 26). Briefly, bone marrow was flushed from femurs and cultured for 3 days in the presence of 50 ng/ml CSF-1 on non-tissue-culture dishes. At this point, the nonadherent cell population, containing the committed osteoclast progenitors, were either harvested (zero time point controls) or cultured with 50 ng/ml of CSF-1 (a gift from David Hume, University of Queensland) and 100 ng/ml RANKL (Roche Diagnostics, Indianapolis, IN) for the indicated time periods. Tartrate-resistant acid phosphatase (TRAP)/Acp5 staining was done using a leukocyte acid phosphatase kit (Sigma).
Co-IP, GST pull-down assays, and Western blotting.
Procedures for coimmunoprecipitation (Co-IP), GST pull-down assays, and Western blotting have all been recently described (2). Production of recombinant proteins in Escherichia coli and in vitro GST pull-down assays were performed as previously described (22).
EMSA.
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (21, 22). The sense strand oligonucleotides, representing the mouse Acp5 proximal sequences, was 5′-TTCTGGGGAAGTCCAGTGCTCACATGACCCA-3′. The consensus Eos binding site was mutated at two regions, M1 (GGAA to TTTT) and M2 (GTCC to CAAA).
Retrovirus production and transduction.
MSCV-FlagEos-IRES-GFP (where MSCV is murine stem cell virus, IRES is internal ribosome entry site, and GFP is green fluorescent protein) was constructed by inserting Flag-tagged Eos cDNA into XhoI-digested MSCV-IRES-GFP vector. Retrovirus packaging was performed using the Phoenix cell line. Bone marrow-derived osteoclast progenitors cultured for 2 days with 50 ng/ml CSF-1 were transferred into 12-well plates and transduced using retroviral supernatant as previously described (11). Twenty-four hours after transduction, cells were treated with the combination of CSF-1 and RANKL as described above.
siRNA knockdown of Eos.
Two separate small interfering RNA (siRNA) oligoribonucleotides (Eos siRNA1 [5′-CGGCCAACUUUCAUUGAUCtt3-′] and Eos siRNA2 [5′-CGGCCAACUUUCAUUGAUCtt3′]; lowercase letters distinguish the overhang in the siRNA design that does not have homology with the Eos sequence) directed against exons 6 and 7, respectively, were purchased from Ambion (Austin, TX), along with a control siRNA, which encodes a scrambled sequence with no particular homology to any known sequence. The combination of these two siRNAs at a concentration of 500 nM each in solution T (Amaxa Biosystems) was introduced into 5 × 106 myeloid precursors using program T-020 in a nucleofector (Amaxa Biosystems).
Cells were harvested 72 h posttransfection and analyzed for the Eos knockdown by real-time PCR and Western blot analysis. Effects of Eos knockdown were analyzed by relative expression analysis of Eos targets, such as Ctsk and Acp5.
Analysis of RNA expression.
RNA was extracted using TRIzol kit (Invitrogen) and reverse transcribed by Superscript III reverse transcriptase (Invitrogen). Real-time PCR was conducted, and the relative mRNA expression level was calculated as previously described (43). The primers used for real-time PCR were picked by Oligo v4.0 software, and the sequences used are available upon request.
ChIP and ReChIP.
Chromatin immunoprecipitation (ChIP) assays were performed by the method of Luo et al. (23). Briefly, bone marrow cells were plated at a density of 3 × 106 cells per 10-cm dishes and treated as indicated in the figure legends. Cells were cross-linked, and soluble chromatin with an average size of 200 to 1,000 bp was prepared by sonication. Precleared soluble chromatin was incubated with 5 μg of specific antibodies as indicated in figure legends. Immunoprecipitated DNA-protein complex was extensively washed, eluted, and decross-linked. Precipitated DNA sample was further purified by using a QIAGEN PCR purification kit according to the manufacturer's instructions (QIAGEN). For sequential ChIP (ReChIP) assays, the beads were washed after the first immunoprecipitation with Eos antibody, the immunoprecipitated chromatin was eluted in 10 mM dithiothreitol at 37°C for 30 min and diluted 50-fold in ChIP dilution buffer. This eluted immune complex was divided and immunoprecipitated with the second specific antibody indicated in the figures. Samples were analyzed by real-time PCR either by SYBR green super mix (Bio-Rad) for the Ctsk promoter or by the Roche universal probe library (Roche) probe using the Faststart TaqMan master kit (Roche) for the Acp5, Bcl-2, or c-Fms promoter. The threshold for the promoter being studied was adjusted by that of input values and represented as relative enrichment. All quantitative PCRs (qPCRs) were analyzed by melting curve analysis and agarose gels to confirm the presence of a single specific band.
RESULTS
Eos is downregulated during osteoclast differentiation and can repress Ctsk and Acp5 promoter activity.
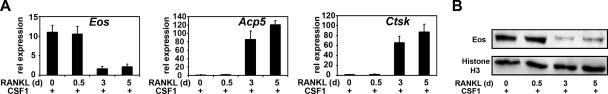
Gene expression profiles of osteoclast-like cells (OCL) derived in vitro were determined using DNA microarrays (data not shown). The Ikaros family member Eos was one gene downregulated during osteoclast differentiation initiated by CSF-1 and RANKL that was selected for further analysis. The kinetics of Eos mRNA expression at various stages of OCL differentiation was validated using quantitative reverse transcription-PCR (qRT-PCR). Bone marrow-derived precursors from wild-type mice were cultured for 3 days with CSF-1 only (BMMs) and subsequently stimulated with recombinant CSF-1 and RANKL to induce osteoclast differentiation in vitro (Fig. 1A). Consistent with the microarray data, Eos mRNA expression was highest in OCL precursors treated only with CSF-1 (day 0) or during the early stages of differentiation in cells (day 0.5). Eos mRNA expression was reduced more than fivefold after 3 or 5 days of cytokine stimulation at a time when osteoclast differentiation occurs and expression of MITF/PU.1 target genes, like cathepsin K (Ctsk) and acid phosphatase 5 (Acp5)/tartrate-resistant acid phosphatase (TRAP) are robust (Fig. 1A). Eos protein expression was evaluated using an antibody generated and characterized in our lab (see Materials and Methods), an analysis that demonstrated that Eos protein expression was also significantly reduced during CSF-1/RANKL-initiated osteoclast differentiation (Fig. 1B).
FIG. 1.
Eos expression is downregulated during osteoclast differentiation. (A) Relative (rel) expression of Eos, Acp5, and Ctsk mRNA was measured by qRT-PCR at the indicated times (in days [d]) and cytokine treatments. Results from three independent experiments are presented as means plus standard errors of the means (error bars). (B) Nuclear extracts from osteoclasts harvested at the indicated times (in days [d]) were analyzed by Western blotting using anti-Eos antibody. Histone H3 was used as a loading control.
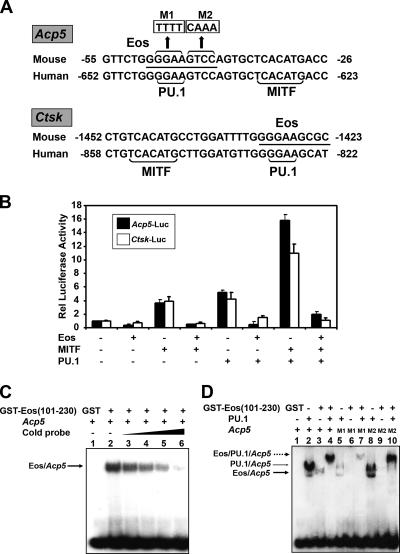
Eos was previously reported to recognize the consensus Ikaros binding site GGGAATRCC and related sequences (29, 34). A potential consensus Eos recognition site overlaps with the functional PU.1 sites present in the Acp5 and Ctsk promoter sequences (Fig. 2A) (22, 27). To examine whether Eos could regulate Acp5 and Ctsk genes through these recognition motifs, transient-transfection assays with luciferase reporters were performed (Fig. 2B). These assays demonstrated that Eos expression repressed both Acp5 and Ctsk reporter activation by MITF and PU.1, either singly or in combination. The c-Fms promoter, which is regulated by PU.1 but not MITF, is not repressed by Eos overexpression (data not shown).
FIG. 2.
Eos represses both Acp5 and Ctsk promoter activity. (A) Acp5 and Ctsk promoter sequences from mouse and human cells with conserved MITF and PU.1 binding sites (shown by brackets), and the putative Eos binding site (underlined). M1 and M2 show the sequence replacements within the Eos DNA consensus, as indicated. (B) Transient-transfection assays in NIH 3T3 cells. Either Acp5 luciferase reporter (Acp5-luc) or the Ctsk luciferase reporter (Ctsk-luc) was transfected alone or together with indicated combinations of expression vectors encoding MITF, PU.1, and Eos. Total DNA in each transfection was kept constant by adding empty expression vector. Relative (Rel) luciferase activity was represented as the difference from basal promoter activity (n-fold) (set at 1). Results from three independent experiments are presented as means plus standard errors of the means (error bars). (C) EMSAs using γ-32P end-labeled Acp5 oligonucleotide and recombinant GST or GST-Eos (101-230) protein. The formation of the DNA-Eos complex (arrow) was competed with increasing amounts of cold Acp5 probe. (D) EMSAs using γ-32P end-labeled wild-type or mutated Acp5 oligonucleotides in the presence of recombinant His6-PU.1 and GST-Eos (101-230) protein. The Eos-DNA complex (thick arrow), PU.1-DNA complex (thin arrow), and the supershifted band containing both Eos and PU.1 (broken arrow) are indicated.
EMSAs were performed with oligonucleotides representing the Acp5 promoter region and recombinant Eos protein containing the N-terminal DNA binding domain (34). The experiments demonstrated that Eos could directly bind to the Acp5 target sequence, and the binding was competed by the addition of unlabeled probe to the EMSA reaction mixture (Fig. 2C). Weaker binding of Eos to the Acp5 probe compared to recombinant PU.1 was observed (Fig. 2D, lane 3 versus lane 2). Unexpectedly, incubation of both PU.1 and Eos resulted in formation of a larger complex that contained both factors (Fig. 2D, lane 4). Mutation of either half of the Eos consensus sequence (M1 [GGAA mutated to TTTT] and M2 [GTCC mutated to CAAA]; Fig. 2A) resulted in loss of Eos binding (Fig. 2D, lanes 6 and 9). PU.1 binding to Acp5 mutation M1 was also significantly reduced (Fig. 2D, lane 5). However, the larger PU.1/Eos complex formed with either Acp5 mutation M1 or M2 (Fig. 2D, lanes 7 and 10). This result indicated that formation of the larger complex might not require Eos to bind directly to DNA.
Eos directly interacts with both PU.1 and MITF through two distinct domains.
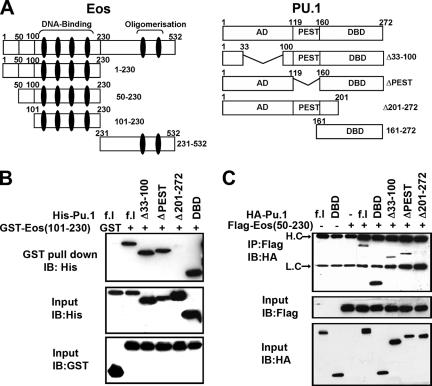
The EMSA results suggested that Eos and PU.1 might directly interact, a possibility that was tested by GST pull-down and Co-IP assays (Fig. 3). The various PU.1 and Eos constructs used for these experiments are graphically depicted in Fig. 3A. Eos shares two domains in common with other Ikaros family members: an N-terminal domain containing four zinc fingers crucial for sequence-specific DNA binding and a C-terminal domain with two zinc fingers that is involved in homo- or heterodimerization (29, 34). GST-Eos (101-230) (GST-Eos with aa 101 to 230) protein, containing zinc fingers 1 to 4, was used to pull-down various PU.1 peptides (Fig. 3B). These experiments mapped the Eos interaction region to the C-terminal DNA binding domain (DBD)/ETS domain of PU.1 (Fig. 3B). In a parallel analysis, the C-terminal domain (aa 231 to 532) of Eos fused to GST was unable to pull down either full-length HA-PU.1 or HA-DBD-PU.1 (see Fig. S1A in the supplemental material). The in vitro results were confirmed by Co-IP assays using Flag-tagged Eos (50-230) (Eos with aa 50 to 230) and HA-tagged PU.1 expression constructs expressed in COS-7 cells (Fig. 3C). Identical results were obtained when Flag-tagged full-length Eos protein was used in the Co-IP assays (data not shown).
FIG. 3.
Physical interaction of Eos with PU.1. (A) Schematic representation of Eos and PU.1 domains and the respective deletion mutations used in the present study. Zinc fingers in Eos are represented as a black vertical eclipse. AD, activation domain; DBD, DNA binding domain. (B) Recombinant GST-Eos (101-230) and His-tagged full-length (f.l) PU.1, as well as different deletions of PU.1 as indicated, were used for in vitro GST pull-down assays and analyzed by Western blotting with anti-His antibody. (C) Co-IP assays using extracts from COS-7 cells transfected with expression vector encoding Flag-Eos (50-230) and HA-PU.1, as indicated. Arrows indicate heavy chain (H.C) and light chain (L.C). In both panels B and C, input controls were analyzed by Western blotting, or immunoblotting (IB), using appropriate antibodies as indicated.
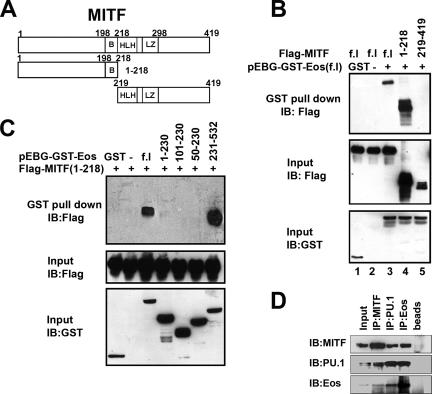
Whether Eos also directly interacted with MITF was studied using the MITF constructs depicted in Fig. 4A. GST pull-down assays using COS-7 cell extracts cotransfected with GST-Eos and Flag-MITF expression constructs demonstrated that MITF was pulled down with full-length GST-Eos (Fig. 4B, lane 3). Further, the N-terminal region of MITF (1-218) (MITF with aa 1 to 218), but not the C-terminal basic helix-loop-helix-leucine zipper region (aa 219 to 419), formed a complex with GST-Eos (Fig. 4B, lane 4 and 5, respectively). A reciprocal analysis demonstrated that Flag-MITF (1-218) formed a complex with C-terminal Eos (aa 231 to 532, containing zinc fingers 5 and 6) but not N-terminal Eos fragments (Fig. 4C). In vitro GST pull-down assays using recombinant GST-tagged full-length MITF and His-tagged C-terminal Eos (aa 231 to 532) demonstrated that the interaction between these two proteins was direct (see Fig. S1B in the supplemental material). GST-MITF and His-Eos (231-532) could also form a complex in the presence of the Acp5 target DNA sequence as determined by EMSA (see Fig. S1C, lane 3, in the supplemental material), while His-Eos (231-532) did not form a complex with the Acp5 sequence (see Fig. S1C, lane 1, in the supplemental material).
FIG. 4.
Physical interaction between Eos and MITF. (A) Schematic representation of MITF constructs used in the present study. B, basic; HLH, helix-loop-helix; LZ, leucine zipper. (B) GST pull-down assays using COS-7 cells expressing full-length (f.l) GST-Eos and Flag-tagged full-length, N-terminal (aa 1 to 218) and C-terminal (aa 219 to 419) fragments of MITF. (C) GST pull-down assays using COS-7 cells cotransfected with Flag-tagged N-terminal MITF (1-218) and full-length (f.l) GST-Eos, or Eos deletion mutations as indicated. Input controls were analyzed by Western blotting, or immunoblotting (IB), using appropriate antibodies as indicated. (D) Coimmunoprecipitation of endogenous Eos, MITF, and PU.1 proteins with specific antibodies from nuclear extracts of bone marrow progenitors grown with CSF-1 alone, as indicated. The rightmost lane (beads only) is a nonspecific rabbit immunoglobulin G control.
The association of endogenous MITF, PU.1, and Eos was studied in primary bone marrow-derived progenitors by coimmunoprecipitation analysis (Fig. 4D). The analysis demonstrated that Eos, MITF, and PU.1 could be found in a complex in the primary cells (Fig. 4D). Taken together, the results demonstrated that Eos can associate with PU.1 and MITF via distinct N-terminal (aa 101 to 230) and C-terminal (aa 231 to 532) domains, respectively.
Eos/MITF/PU.1 complexes are selectively enriched at target genes in committed osteoclast progenitors.
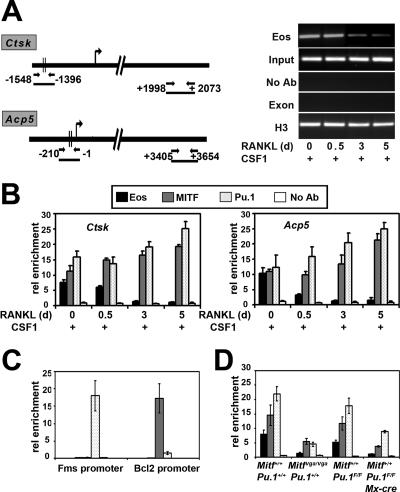
ChIP was used to determine whether endogenous Eos is present in complexes with MITF and PU.1 at osteoclast target promoters. The regions of Ctsk and Acp5 genes that were analyzed by qPCR following the ChIP procedures are depicted in Fig. 5A. PCR primers were designed to amplify Ctsk (−1548 to −1396) and Acp5 (−210 to −1) promoter regions that have been previously shown to contain functional MITF and PU.1 binding sites (21, 32). In addition to the 5′ regions of the Ctsk and Acp5 genes, a 3′ region corresponding to internal exon/intron regions was used as a negative control (Fig. 5A, left panel). An additional negative control was a reaction in which no primary antibody was added to ChIP reaction mixtures. A representative control experiment is shown for the Ctsk gene with Eos antibody over the time course studied (Fig. 5A, right panel). Following 40 cycles of PCR, no product could be detected in the negative controls. Histone H3 was routinely used as a positive control. The same set of controls was used for all subsequent ChIP experiments presented for both Ctsk and Acp5 genes (data not shown).
FIG. 5.
Recruitment of Eos, MITF, and PU.1 to the Acp5 and Ctsk promoters during osteoclast differentiation. (A) (Left) Graphic representation of regions analyzed for Ctsk and Acp5 genes in ChIP assays. (Right) Representative gel pictures for Eos ChIPs on Ctsk gene, following 40 cycles of PCR. Input indicates the total DNA in each assay before antibody was added. Negative controls included no antibody (No Ab) and 3′ exon/intron region (Exon). Anti-histone H3 was used as a positive control. d, days. (B) ChIP assays to study the association of Eos, MITF, and PU.1 with Ctsk and Acp5 promoters in cells treated with CSF-1 alone (day 0) or subsequently with CSF-1 and RANKL for 0.5, 3, and 5 days. rel enrichment, relative enrichment. (C) Analysis of enrichment of MITF, PU.1, and Eos at Bcl2 and c-Fms promoters in committed osteoclast precursors. (D) Analysis of enrichment of MITF, PU.1, and Eos at Acp5 promoter in cells treated with CSF-1 alone derived from hypomorphic MITFvga/vga mice (left) or Pu.1 conditional knockout cells (right). Results from three independent experiments are represented as means ± standard errors of the means (error bars) for each experiment in panels B to D.
The ChIP experiments demonstrated that Eos was enriched at both Ctsk and Acp5 target promoters in osteoclast progenitors grown in the presence of CSF-1 alone for 3 days (Fig. 5B). In contrast, levels of Eos rapidly decreased by 7- to 10-fold following treatment of this same population with both CSF-1 and RANKL for an additional 3 to 5 days (Fig. 5B). The amounts of MITF and PU.1 at Ctsk and Acp5 promoters remained largely unchanged after addition of CSF-1/RANKL, increasing less than twofold after 3 to 5 days of treatment with both cytokines.
Whether Eos was bound to promoters that contained only binding sites for PU.1 or MITF, but not both factors, was next addressed. The proximal c-Fms promoter contains PU.1 binding sites but is devoid of MITF binding sites (10), whereas Bcl2 is an established MITF target that lacks PU.1 sites (28). As predicted, ChIP demonstrated that in cells treated with CSF-1, only PU.1 was enriched at the proximal c-Fms promoter, but neither MITF or Eos could be detected (Fig. 5C). Similarly, MITF was enriched at the Bcl2 promoter, but neither PU.1 nor Eos was detected (Fig. 5C).
To more directly address whether the presence of both MITF and PU.1 were required for enrichment of Eos at target promoters, primary cells from genetic models with reduced levels of MITF and PU.1 proteins were examined by ChIP. Bone marrow cells derived from mice homozygous for the hypomorphic Mitf-vga9 produce 10 to 20% of wild-type MITF protein (see Fig. S2A in the supplemental material). ChIP assays performed on progenitors cultured for 3 days in the presence of CSF-1 demonstrated only three- to five-fold reduction in the levels of MITF, PU.1, and Eos at the Acp5 promoter in theses cells compared to those in the control cells (Fig. 5D). Bone marrow precursors were also obtained for mice homozygous for a previously described PU.1 conditional allele that also contained the interferon-inducible Mx1-cre transgene (12) (see Materials and Methods). PU.1 protein levels were reduced to 10 to 20% of the levels in the control in the bone marrow progenitors cultured for 3 days in the presence of CSF-1 from the PU.1f/f;Mx1-cre+ mice (f for floxed) treated with poly(dI-dC) (see Fig. S2B in the supplemental material). In this genetic model, Eos levels at the Acp5 promoter were reduced sevenfold (Fig. 5D). Similar results were obtained when the Ctsk promoter was studied (data not shown). These results are consistent with the hypothesis that Eos requires the MITF/PU.1 complex for its selective association with target genes.
Eos/MITF/PU.1 complexes recruit corepressors to target promoters in the absence of RANKL signaling.
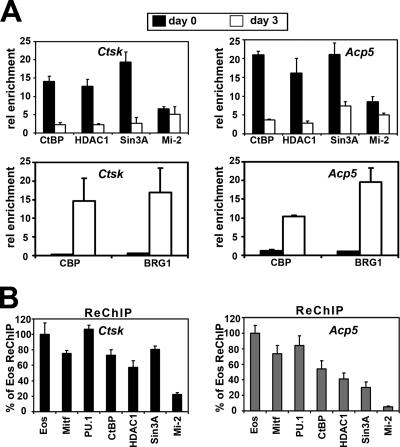
Eos and other Ikaros family members have been reported to interact with several corepressors, including Sin3, Mi-2, and CtBP (15-18, 33). ChIP assays were used to determine whether these corepressors were enriched at Ctsk and Acp5 promoters in bone marrow progenitors (Fig. 6A, top panels). For this analysis, cells grown in CSF-1 alone for 3 days were compared to cells grown for an additional 3 days with CSF-1 and RANKL, a time when visible differentiation of OCLs is first apparent and robust expression of target genes occurs (25, 26). These experiments demonstrated that corepressors CtBP, HDAC1, and Sin3A were all enriched at Ctsk and Acp5 promoters in cells grown with CSF-1 only, but the levels of their association with these target promoters were significantly reduced following 3 days of CSF-1/RANKL treatment. The Mi-2 protein was also detected at Ctsk and Acp5 promoters, but its association with these promoters upon CSF-1/RANKL stimulation was not significantly different.
FIG. 6.
Association of Eos, MITF, and PU.1 with corepressors on Ctsk and Acp5 promoters in osteoclast precursors. (A) Analysis of association of corepressors and coactivators with Ctsk and Acp5 promoters in osteoclast precursors grown in CSF-1 alone (day 0) and osteoclast-like cells after 3 days of CSF-1 and RANKL treatment (day 3) by ChIP assays. rel enrichment; relative enrichment. (B) ReChIP to analyze the simultaneous presence of Eos, MITF, PU.1, and corepressors in osteoclast precursors. Results from three independent experiments are represented as means plus standard errors of the means (error bars).
The recruitment of the coactivators CBP/p300 and BRG1 was also studied, since these coactivators were previously reported to interact with both MITF and PU.1 (3, 35, 37, 45). This analysis indicated that CBP and BRG1 were not detected at Ctsk and Acp5 promoters in cells grown with CSF-1 alone. However, both of the cofactors were enriched at these target promoters in cells grown for an additional 3 days with CSF-1 and RANKL (Fig. 6A, bottom panels), concurrent with robust expression of these target genes at day 3 (Fig. 1A).
ReChIP assays were used to determine whether MITF, PU.1, Eos, and corepressors Sin3A, CtBP, HDAC1, and Mi-2 were present at the target promoters within the same cells. The first round of immunoprecipitation was carried out with an Eos antibody, and the immunoprecipitated cross-linked DNA-protein complexes were then isolated and disassociated from the beads. Aliquots of this sample were subjected to reimmunoprecipitation using antibodies against Eos, MITF, PU.1, CtBP, HDAC1, Sin3A, and Mi-2. The amount of DNA present in the ReChIP samples using these antibodies was determined by real-time qPCR and compared to the amount of DNA present in the Eos ReChIP sample (Fig. 6B). This analysis indicated that more than 80% of the recovered Eos complex also contained MITF and PU.1 at both Ctsk and Acp5 promoters. In addition, more than 60% of the Eos complex contained CtBP, HDAC1, and Sin3A at the Ctsk promoter. Similar levels of CtBP were seen at the Acp5 promoter, but the levels of both HDAC1 and Sin3A measured at the Acp5 promoter were slightly lower, approximately 40% of complexes (Fig. 6B). The level of Mi-2 complex was much lower than the other corepressor complexes, indicating that Mi-2 may not be strongly associated with the Eos/MITF/PU.1 complex.
Eos overexpression or siRNA knockdown disrupts MITF/PU.1 target gene regulation.
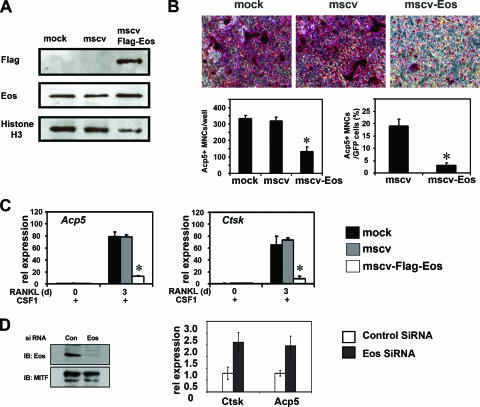
To further study the role of Eos as a negative regulator of MITF/PU.1 target genes, Eos was overexpressed in osteoclast progenitors via retroviral transduction. The bicistronic retroviral construct (MSCV-FlagEos-IRES-GFP) used to express Flag-tagged Eos also expressed the GFP under control of an IRES, allowing individual infected cells to be identified. Viral stocks produced from packaging cells mock transfected, transfected with MSCV-GFP alone (vector control), or transfected with Flag-tagged Eos/GFP vector were used to transduce precursors derived from wild-type mice following 2 days of growth with CSF-1. The efficiency of transduction 24 h postinfection as determined by expression of GFP was 70 to 80% for both vector control and MSCV-Flag-Eos (data not shown). Western blotting was performed to verify expression of exogenous Flag-Eos (Fig. 7A). Retroviral transduction resulted in a modest twofold increase in total Eos protein production as determined by Western blotting and densitometry (Fig. 7A).
FIG. 7.
Overexpression of Eos in BMMs disrupts osteoclast differentiation. Primary osteoclast precursors were either mock infected or retrovirally transduced with empty MSCV-IRES-GFP (mscv) and MSCV-Flag-Eos-IRES-GFP (mscv-Flag-Eos). (A) Expression of exogenous Eos analyzed by Western blotting using nuclear extracts 1 day postinfection. (B) Representative Acp5 (TRAP) staining for infected BMMs after 3 days of CSF-1 and RANKL treatment (top panels). Acp5-positive multinuclear cells (MNCs, three or more nuclei) were counted for each well (left bar graph) and among GFP-positive cells (right bar graph). (C) Relative (rel) mRNA expression of Acp5 and Ctsk genes measured by qRT-PCR at the indicated time points (in days [d]). Results in both panels B and C were from three independent experiments and presented as means plus standard errors of the means (error bars). Statistical analysis was conducted using two-sample t test (*, P < 0.01). (D) Eos knockdown by siRNA. (Left) Western blot analysis of Eos protein levels in scrambled siRNA-transfected (control [Con]) or Eos-specific siRNA-transfected (Eos) cells. MITF levels were measured as loading control. (Right) Levels of Ctsk and Acp5 mRNA determined by qPCR in Eos knockdown cells compared with control cells.
Twenty-four hours after viral transduction, cells were treated with both CSF-1 and RANKL for an additional 3 days. Control virus-infected bone marrow precursors were able to differentiate into multinucleated (containing three or more nuclei), TRAP/Acp5-positive osteoclasts. In contrast, multinuclear, TRAP/Acp5-positive cells were rarely detected in cells infected with retrovirus encoding Eos (Fig. 7B). Osteoclast precursors infected with the control retrovirus formed approximately the same number of OCLs as mock-infected cells, while cells infected with Eos virus produced about threefold-fewer OCLs, a reduction that was statistically significant (Fig. 7B, left bar graph, P < 0.01). When only the GFP-positive OCLs were considered, a sixfold reduction was observed in control versus Eos-infected cells (Fig. 7B, right bar graph, P < 0.01).
Supporting these results, qRT-PCR demonstrated that the levels of Ctsk and Acp5 mRNA were reduced six- to eightfold in cells expressing Flag-Eos versus control cells (Fig. 7C). In contrast, Eos overexpression did not affect the expression of two known PU.1 targets, c-Fms and RANK (see Fig. S3A, top panel, in the supplemental material), the receptors for CSF-1 and RANKL, respectively (10, 19). In addition, Bcl2 expression, a target of MITF, was not affected by Eos-overexpressed cells (see Fig. S3A, bottom panel, right, in the supplemental material). Expression of the calcitonin receptor (CTR), another marker gene for osteoclast differentiation, was not affected in cells overexpressing Eos compared with controls (see Fig. S3A, bottom panel, left, in the supplemental material).
In a complementary approach, siRNA was used to knock down Eos expression in primary osteoclast progenitors. When wild-type, bone marrow-derived precursors grown in the presence of CSF-1 alone were transfected with the combination of two Eos siRNAs (see Materials and Methods), both Eos mRNA levels (see Fig. S3B in the supplemental material) and protein levels were significantly reduced 72 h posttransfection (Fig. 7D). This resulted in a 2.5- to 3-fold increase in Acp5 and Ctsk mRNA expression of both Acp5 and Ctsk genes compared to scrambled siRNA-transfected cells (Fig. 7D). Expression of Bcl2 was not affected by Eos siRNA treatment (see Fig. S3B in the supplemental material). In the RAW264.7 cell culture model, cotransfection of the individual Eos siRNAs along with an MITF expression vector resulted in a fourfold increase in expression of basal levels of target genes, even though Eos protein levels were decreased only about 50% by the single siRNAs (see Fig. S3C in the supplemental material).
DISCUSSION
Both MITF and PU.1 are known transcriptional activators in osteoclasts, but results presented here demonstrate that they can also act as components of repressor complexes that suppress target gene expression in the absence of the appropriate extracellular signals necessary for osteoclast differentiation. Interaction of these factors with the zinc finger protein Eos appears to be critical for recruiting corepressors to the target genes, as the ability of Eos to interact directly with these corepressors is well documented (17, 33). Eos itself binds only weakly to the Acp5 target sequences in vitro, and interactions through distinct zinc finger domains with both MITF and PU.1 likely provide additional specificity for target gene repression. These results provide a mechanism for suppressing osteoclast target genes in committed myeloid precursors that have the potential to differentiate either to monocytes/macrophages, antigen-presenting dendritic cells, or osteoclasts (Fig. 8).
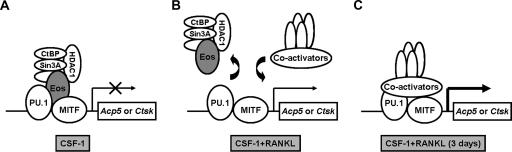
FIG. 8.
Schematic model of Acp5 and Ctsk gene regulation by transcription factors and their cofactors during osteoclast differentiation. (A) In the presence of CSF-1 only, MITF, PU.1, and Eos complexes recruit with corepressors, thus inhibiting Acp5 or Ctsk expression. (B) Combined CSF-1 and RANKL stimulation triggers dissociation of Eos and corepressor complexes and recruitment of coactivators. (C) Continued CSF-1 and RANKL treatment subsequently leading to robust induction of Acp5 and Ctsk expression.
Our results indicate that CSF-1/RANKL signaling can activate expression of osteoclast target genes by two mechanisms. The first mechanism is downregulation of Eos expression, both at the level of mRNA and protein, leading to dissociation of the corepressors from target genes (Fig. 8). The second mechanism is direct phosphorylation and activation of MITF by both Erk and p38 mitogen-activated protein kinase pathways, leading to recruitment of coactivators, like CBP/p300 and BRG1 (8, 25, 44). Release of negative regulation must accompany recruitment of coactivators before target genes can be fully activated as demonstrated by the Eos overexpression and knockdown studies. Our results do not rule out the possibility that direct signaling-dependent interactions between MITF and PU.1 and corepressor CtBP or Sin3 might be involved in this regulation, but the well-documented interaction of Eos with these corepressors provides a simpler explanation consistent with the results presented here. Whether CSF-1/RANKL signaling directly targets the posttranslational modification of Eos, affecting either its stability or activity, remains to be determined. For example, SUMO modification of Ikaros is reported to affect interaction with corepressors during lymphoid development (5), and Eos can also be modified by SUMO (R. Hu and M. C. Ostrowski, unpublished observation).
For lineage-determining factors, like PU.1, mechanisms for repressing potential target genes in progenitors are critical for maintaining the appropriate control of gene expression patterns. At an earlier stage of myeloid commitment, one mechanism by which PU.1 accomplishes this is to directly regulate expression of transcription factors, like Erg1 and Erg2, that can in turn repress neutrophil-specific genes while activating macrophage-specific genes (20). At the same time, the zinc finger factor Gfi-1 antagonizes macrophage differentiation by inhibiting expression of these PU.1 target genes (20). Our work provides an additional mechanism by which PU.1 activity is regulated in a more committed myeloid precursor. In this case, the zinc finger factor Eos directly modulates PU.1 activity at target promoters. An important distinction is that the effect of Eos is specific for the MITF/PU.1 complex, as other PU.1 target genes in both macrophages and osteoclasts, for example, c-Fms and RANK, were not affected when Eos is overexpressed.
Whether Eos actually functions as a lineage-decision factor in myeloid cells in a manner analogous to Gfi-1, or to Ikaros in lymphoid cells, seems unlikely based on current evidence. For example, the CTR gene, another definitive marker of osteoclast differentiation, is still upregulated in cells that overexpress Eos. Thus, Eos does not regulate the entire osteoclast gene expression program but selectively modulates the activity of the MITF/PU.1 complex and a subset of genes necessary for full activity of the differentiated osteoclast. At the same time, the presence of MITF and PU.1 at target promoters allows committed precursors the flexibility to respond rapidly to the bone microenvironment to reprogram gene expression in response to CSF-1/RANKL signaling. This model to explain how MITF/PU.1 activity is modulated in osteoclast progenitors may be generally applicable to other cell types in which committed progenitors can give rise to closely related cell types.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIAMS grant R01-AR-0447129 (M.C.O.).
Footnotes
Published ahead of print on 2 April 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Boyle, W. J., W. S. Simonet, and D. L. Lacey. 2003. Osteoclast differentiation and activation. Nature 423:337-342. [DOI] [PubMed] [Google Scholar]
- 2.Bronisz, A., S. M. Sharma, R. Hu, J. Godlewski, G. Tzivion, K. C. Mansky, and M. C. Ostrowski. 2006. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Mol. Biol. Cell 17:3897-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Serna, I. L., Y. Ohkawa, C. Higashi, C. Dutta, J. Osias, N. Kommajosyula, T. Tachibana, and A. N. Imbalzano. 2006. The microphthalmia-associated transcription factor requires SWI/SNF enzymes to activate melanocyte-specific genes. J. Biol. Chem. 281:20233-20241. [DOI] [PubMed] [Google Scholar]
- 4.Georgopoulos, K., M. Bigby, J. H. Wang, A. Molnar, P. Wu, S. Winandy, and A. Sharpe. 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79:143-156. [DOI] [PubMed] [Google Scholar]
- 5.Gómez-del Arco, P., J. Koipally, and K. Georgopoulos. 2005. Ikaros SUMOylation: switching out of repression. Mol. Cell. Biol. 25:2688-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahm, K., B. S. Cobb, A. S. McCarty, K. E. Brown, C. A. Klug, R. Lee, K. Akashi, I. L. Weissman, A. G. Fisher, and S. T. Smale. 1998. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 12:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallsson, J. H., J. Favor, C. Hodgkinson, T. Glaser, M. L. Lamoreux, R. Magnusdottir, G. J. Gunnarsson, H. O. Sweet, N. G. Copeland, N. A. Jenkins, and E. Steingrimsson. 2000. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics 155:291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershey, C. L., and D. E. Fisher. 2004. Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone 34:689-696. [DOI] [PubMed] [Google Scholar]
- 9.Honma, Y., H. Kiyosawa, T. Mori, A. Oguri, T. Nikaido, K. Kanazawa, M. Tojo, J. Takeda, Y. Tanno, S. Yokoya, I. Kawabata, H. Ikeda, and A. Wanaka. 1999. Eos: a novel member of the Ikaros gene family expressed predominantly in the developing nervous system. FEBS Lett. 447:76-80. [DOI] [PubMed] [Google Scholar]
- 10.Hume, D. A., X. Yue, I. L. Ross, P. Favot, A. Lichanska, and M. C. Ostrowski. 1997. Regulation of CSF-1 receptor expression. Mol. Reprod. Dev. 46:46-53. [DOI] [PubMed] [Google Scholar]
- 11.Humphrey, M. B., K. Ogasawara, W. Yao, S. C. Spusta, M. R. Daws, N. E. Lane, L. L. Lanier, and M. C. Nakamura. 2004. The signaling adapter protein DAP12 regulates multinucleation during osteoclast development. J. Bone Miner. Res. 19:224-234. [DOI] [PubMed] [Google Scholar]
- 12.Iwasaki, H., C. Somoza, H. Shigematsu, E. A. Duprez, J. Iwasaki-Arai, S. Mizuno, Y. Arinobu, K. Geary, P. Zhang, T. Dayaram, M. L. Fenyus, S. Elf, S. Chan, P. Kastner, C. S. Huettner, R. Murray, D. G. Tenen, and K. Akashi. 2005. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106:1590-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi, N., and M. Noda. 2000. Mitf is expressed in osteoclast progenitors in vitro. Exp. Cell Res. 260:284-291. [DOI] [PubMed] [Google Scholar]
- 14.Kelley, C. M., T. Ikeda, J. Koipally, N. Avitahl, L. Wu, K. Georgopoulos, and B. A. Morgan. 1998. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr. Biol. 8:508-515. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J., S. Sif, B. Jones, A. Jackson, J. Koipally, E. Heller, S. Winandy, A. Viel, A. Sawyer, T. Ikeda, R. Kingston, and K. Georgopoulos. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10:345-355. [DOI] [PubMed] [Google Scholar]
- 16.Koipally, J., and K. Georgopoulos. 2000. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 275:19594-19602. [DOI] [PubMed] [Google Scholar]
- 17.Koipally, J., and K. Georgopoulos. 2002. A molecular dissection of the repression circuitry of Ikaros. J. Biol. Chem. 277:27697-27705. [DOI] [PubMed] [Google Scholar]
- 18.Koipally, J., A. Renold, J. Kim, and K. Georgopoulos. 1999. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 18:3090-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon, O. H., C. K. Lee, Y. I. Lee, S. G. Paik, and H. J. Lee. 2005. The hematopoietic transcription factor PU.1 regulates RANK gene expression in myeloid progenitors. Biochem. Biophys. Res. Commun. 335:437-446. [DOI] [PubMed] [Google Scholar]
- 20.Laslo, P., C. J. Spooner, A. Warmflash, D. W. Lancki, H. J. Lee, R. Sciammas, B. N. Gantner, A. R. Dinner, and H. Singh. 2006. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126:755-766. [DOI] [PubMed] [Google Scholar]
- 21.Luchin, A., G. Purdom, K. Murphy, M. Y. Clark, N. Angel, A. I. Cassady, D. A. Hume, and M. C. Ostrowski. 2000. The microphthalmia transcription factor regulates expression of the tartrate-resistant acid phosphatase gene during terminal differentiation of osteoclasts. J. Bone Miner. Res. 15:451-460. [DOI] [PubMed] [Google Scholar]
- 22.Luchin, A., S. Suchting, T. Merson, T. J. Rosol, D. A. Hume, A. I. Cassady, and M. C. Ostrowski. 2001. Genetic and physical interactions between microphthalmia transcription factor and PU.1 are necessary for osteoclast gene expression and differentiation. J. Biol. Chem. 276:36703-36710. [DOI] [PubMed] [Google Scholar]
- 23.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 24.Mansky, K. C., K. Marfatia, G. H. Purdom, A. Luchin, D. A. Hume, and M. C. Ostrowski. 2002. The microphthalmia transcription factor (MITF) contains two N-terminal domains required for transactivation of osteoclast target promoters and rescue of mi mutant osteoclasts. J. Leukoc. Biol. 71:295-303. [PubMed] [Google Scholar]
- 25.Mansky, K. C., U. Sankar, J. Han, and M. C. Ostrowski. 2002. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J. Biol. Chem. 277:11077-11083. [DOI] [PubMed] [Google Scholar]
- 26.Mansky, K. C., S. Sulzbacher, G. Purdom, L. Nelsen, D. A. Hume, M. Rehli, and M. C. Ostrowski. 2002. The microphthalmia transcription factor and the related helix-loop-helix zipper factors TFE-3 and TFE-C collaborate to activate the tartrate-resistant acid phosphatase promoter. J. Leukoc. Biol. 71:304-310. [PubMed] [Google Scholar]
- 27.Matsumoto, M., M. Kogawa, S. Wada, H. Takayanagi, M. Tsujimoto, S. Katayama, K. Hisatake, and Y. Nogi. 2004. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J. Biol. Chem. 279:45969-45979. [DOI] [PubMed] [Google Scholar]
- 28.McGill, G. G., M. Horstmann, H. R. Widlund, J. Du, G. Motyckova, E. K. Nishimura, Y. L. Lin, S. Ramaswamy, W. Avery, H. F. Ding, S. A. Jordan, I. J. Jackson, S. J. Korsmeyer, T. R. Golub, and D. E. Fisher. 2002. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 109:707-718. [DOI] [PubMed] [Google Scholar]
- 29.Molnár, A., and K. Georgopoulos. 1994. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol. 14:8292-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore, K. J. 1995. Insight into the microphthalmia gene. Trends Genet. 11:442-448. [DOI] [PubMed] [Google Scholar]
- 31.Morgan, B., L. Sun, N. Avitahl, K. Andrikopoulos, T. Ikeda, E. Gonzales, P. Wu, S. Neben, and K. Georgopoulos. 1997. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 16:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motyckova, G., K. N. Weilbaecher, M. Horstmann, D. J. Rieman, D. Z. Fisher, and D. E. Fisher. 2001. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc. Natl. Acad. Sci. USA 98:5798-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perdomo, J., and M. Crossley. 2002. The Ikaros family protein Eos associates with C-terminal-binding protein corepressors. Eur. J. Biochem. 269:5885-5892. [DOI] [PubMed] [Google Scholar]
- 34.Perdomo, J., M. Holmes, B. Chong, and M. Crossley. 2000. Eos and pegasus, two members of the Ikaros family of proteins with distinct DNA binding activities. J. Biol. Chem. 275:38347-38354. [DOI] [PubMed] [Google Scholar]
- 35.Price, E. R., H. F. Ding, T. Badalian, S. Bhattacharya, C. Takemoto, T. P. Yao, T. J. Hemesath, and D. E. Fisher. 1998. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J. Biol. Chem. 273:17983-17986. [DOI] [PubMed] [Google Scholar]
- 36.Roodman, G. D. 1999. Cell biology of the osteoclast. Exp. Hematol. 27:1229-1241. [DOI] [PubMed] [Google Scholar]
- 37.Sato, S., K. Roberts, G. Gambino, A. Cook, T. Kouzarides, and C. R. Goding. 1997. CBP/p300 as a co-factor for the microphthalmia transcription factor. Oncogene 14:3083-3092. [DOI] [PubMed] [Google Scholar]
- 38.Scott, E. W., M. C. Simon, J. Anastasi, and H. Singh. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265:1573-1577. [DOI] [PubMed] [Google Scholar]
- 39.So, H., J. Rho, D. Jeong, R. Park, D. E. Fisher, M. C. Ostrowski, Y. Choi, and N. Kim. 2003. Microphthalmia transcription factor and PU.1 synergistically induce the leukocyte receptor osteoclast-associated receptor gene expression. J. Biol. Chem. 278:24209-24216. [DOI] [PubMed] [Google Scholar]
- 40.Steingrimsson, E., K. J. Moore, M. L. Lamoreux, A. R. Ferre-D'Amare, S. K. Burley, D. C. Zimring, L. C. Skow, C. A. Hodgkinson, H. Arnheiter, N. G. Copeland, et al. 1994. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 8:256-263. [DOI] [PubMed] [Google Scholar]
- 41.Wang, J. H., A. Nichogiannopoulou, L. Wu, L. Sun, A. H. Sharpe, M. Bigby, and K. Georgopoulos. 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5:537-549. [DOI] [PubMed] [Google Scholar]
- 42.Wang, L., R. A. Baiocchi, S. Pal, G. Mosialos, M. Caligiuri, and S. Sif. 2005. The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol. Cell. Biol. 25:7953-7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei, G., J. Guo, A. I. Doseff, D. F. Kusewitt, A. K. Man, R. G. Oshima, and M. C. Ostrowski. 2004. Activated Ets2 is required for persistent inflammatory responses in the motheaten viable model. J. Immunol. 173:1374-1379. [DOI] [PubMed] [Google Scholar]
- 44.Weilbaecher, K. N., G. Motyckova, W. E. Huber, C. M. Takemoto, T. J. Hemesath, Y. Xu, C. L. Hershey, N. R. Dowland, A. G. Wells, and D. E. Fisher. 2001. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol. Cell 8:749-758. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, H., F. Kihara-Negishi, T. Yamada, Y. Hashimoto, and T. Oikawa. 1999. Physical and functional interactions between the transcription factor PU.1 and the coactivator CBP. Oncogene 18:1495-1501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.