Abstract
Taste receptor cells constitute a highly specialized cell type that perceives and conveys specific sensory information to the brain. The detailed molecular composition of these cells and the mechanisms that program their fate are, in general, poorly understood. We have generated serial analysis of gene expression (SAGE) libraries from two distinct populations of single, isolated sensory neuron classes, the gustatory neuron class ASE and the thermosensory neuron class AFD, from the nematode Caenorhabditis elegans. By comparing these two libraries, we have identified >1000 genes that define the ASE gustatory neuron class on a molecular level. This set of genes contains determinants of the differentiated state of the ASE neuron, such as a surprisingly complex repertoire of transcription factors (TFs), ion channels, neurotransmitters, and receptors, as well as seven-transmembrane receptor (7TMR)-type putative gustatory receptor genes. Through the in vivo dissection of the cis-regulatory regions of several ASE-expressed genes, we identified a small cis-regulatory motif, the “ASE motif,” that is required for the expression of many ASE-expressed genes. We demonstrate that the ASE motif is a binding site for the C2H2 zinc finger TF CHE-1, which is essential for the correct differentiation of the ASE gustatory neuron. Taken together, our results provide a unique view of the molecular landscape of a single neuron type and reveal an important aspect of the regulatory logic for gustatory neuron specification in C. elegans.
Keywords: Caenorhabditis elegans, development, gustatory neuron, transcription factor
Taste (“gustatory”) cells are defined by their ability to sense water-soluble cues and to transmit this information to the brain in order to evoke a specific behavioral response. In vertebrates, taste receptor cells are highly specialized cells of epithelial origin that express gustatory receptor proteins, generate action potentials, and communicate with neurons that innervate them (Northcutt 2004). In the invertebrate model systems Drosophila melanogaster and Caenorhabditis elegans, taste receptor cells are specialized sensory neurons embedded within specific support cells (Ward et al. 1975; de Bruyne and Warr 2006). The gustatory system of C. elegans has been particularly well studied both on the anatomical and functional level (Ward 1973; Dusenbery 1974; Ward et al. 1975; Bargmann and Horvitz 1991). Laser ablation studies have revealed the existence of five classes of gustatory neurons, each composed of a pair of bilaterally symmetric neurons (Fig. 1A; Bargmann and Horvitz 1991). The ASE neuron class is the most relevant of these five classes since it is the only neuron class whose laser ablation eliminates or significantly reduces the response to all known classes of gustatory cues, namely, salts, amino acids, and small metabolites (Bargmann and Horvitz 1991).
Figure 1.
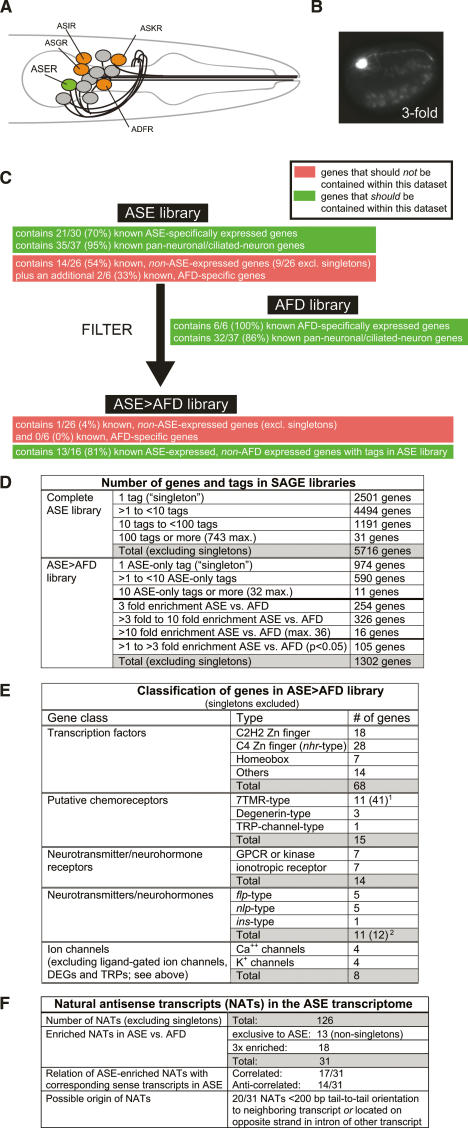
SAGE analysis of isolated ASER neurons. (A) Schematic representation of amphid chemosensory neurons. Gustatory neurons revealed by laser ablation studies (Bargmann and Horvitz 1991) are colored. (B) ASER-specific ntIs1 transgene (gcy-5prom∷gfp) used to isolate ASER from embryos. (C) Number of genes found in the raw and filtered ASE SAGE libraries. See Materials and Methods for more details and Supplementary Tables 1 and 3A–C for a list of genes. (D) Filtering of the ASE SAGE library by comparing it with an AFD SAGE library. The success of the filtering is assessed through the use of control data sets (Supplementary Table 2) that contain genes whose expression pattern was previously determined. It should be kept in mind that expression patterns in these control data sets are almost exclusively inferred from reporter gene analysis, which may not provide an accurate reflection of endogenous expression as they may be lacking relevant regulatory information and/or may not be sensitive enough to detect low-level, “leaky” expression. Previously known, ASE-expressed genes in the control data set include genes that at post-embryonic stages become restricted to ASEL (Supplementary Table 2); their representation in the ASE library confirms that the isolation of ASER from embryos indeed provides the gene expression profiles of both ASEL and ASER. (E) Summary of genes with specific predicted function in the ASE > AFD library. See Supplementary Tables 5–7 for more detail on the genes. [1An additional 30 singletons raises the total number of ASE-expressed 7TMRs to 41. 2A 10th neuropeptide/hormone, ins-1, is expressed in ASE (Kodama et al. 2006), but is not represented in any SAGE library, due to the absence of a restriction site for the tagging enzyme used to generate the SAGE library.] (F) NATs in the ASE transcriptome. See Supplementary Table 8 for a list of genes and more details. [1In eight of 20 cases, an overlap in transcript has been explicitly demonstrated by EST analysis (http://www.wormbase.org).]
Central aspects of the molecular biology of taste receptor cells, such as ASE, are incompletely understood in all organisms studied to date. First, the molecular composition of gustatory cells is poorly mapped. It is to be expected that these cells express a unique set of structural proteins and signaling proteins that determine their unique morphology and function. A crucial aspect of the molecular composition of taste receptor cells is the nature of taste receptor molecules, which are known for some but not all taste modalities. Sweet, bitter, and amino acid receptors have been described in vertebrates and flies, and all fall into the seven-transmembrane receptor (7TMR) family (Scott 2005). In contrast, the molecular identity of vertebrate and fly salt receptors is still under debate, although several promising candidates exist (Scott 2005). In nematodes, guanylyl cyclases have been proposed as candidate chemoreceptors (Yu et al. 1997; Ortiz et al. 2006), but no experimental evidence supports this notion yet. The identification of such receptors and their downstream signaling components may help to explain the unusual ability of the ASE gustatory neurons to process multiple distinct sensory cues and to either engage in an attractive or repulsive response, depending on the sensory input (Bargmann and Horvitz 1991; Sambongi et al. 1999).
Another poorly understood issue concerns the development of taste receptor cells. In all systems studied to date, the gene regulatory program that controls the terminal differentiation features of taste receptor cells remains mostly unknown. Homeobox and basic helix– loop–helix (bHLH) transcription factors (TFs) are known to be expressed in vertebrate taste cells, but their function in these cells has not been determined (Miura et al. 2003). Genetic analysis in Drosophila has revealed that the Paired box gene, PoxN, is involved in an early step of chemosensory neuron fate determination (Dambly-Chaudiere et al. 1992), but it is not known how PoxN affects chemosensory neuron differentiation.
Genetic analysis in C. elegans has revealed a phylogenetically conserved C2H2 zinc finger TF, encoded by the che-1 gene, that appears to be exclusively expressed in post-embryonic ASE gustatory neurons (B. Tursun and O. Hobert, unpubl.) and is essential for their development (Dusenbery et al. 1975; Chang et al. 2003; Uchida et al. 2003). Several molecular markers of the terminal fate of ASE are known, and in the absence of che-1, all these markers fail to be expressed and ASE completely loses its ability to sense water-soluble taste cues. However, ASE does maintain its pan-neuronal features and, according to dye-uptake assays and electron microscopical analysis, still remains a sensory neuron in the absence of che-1 (Ward et al. 1975; Uchida et al. 2003). che-1 is, therefore, a selector gene (Mann and Carroll 2002) that controls the identity of the ASE neuron and, thereby, distinguishes ASE from other sensory neurons. However, it is not clear how che-1 controls ASE development. Does it control downstream transcriptional regulatory programs or does it directly control the expression of terminal differentiation features of ASE? If so, what is the molecular identity of these terminal differentiation features?
In this study, we address these questions and thereby advance our understanding of the molecular architecture and development of gustatory neurons. Specifically, we first define the transcriptome of the ASE gustatory neurons using comparative serial analysis of gene expression (SAGE) of isolated sensory neurons. We then asked how the cell-type specificity of the ASE transcriptome is determined. Through the in vivo dissection of regulatory regions of selected members of the ASE transcriptome, we identify a single cis-regulatory motif present in many ASE-expressed genes and required for their expression in ASE. We show that the Zn finger TF CHE-1, which is both required and sufficient for the expression of ASE motif-containing genes, directly binds this regulatory motif. We have, therefore, revealed a strikingly simple regulatory architecture that determines a neuron-type-specific gene expression profile, composed of a simple regulatory motif and a single factor required for binding and activating this motif.
Results
Defining the ASE transcriptome by SAGE analysis
The ASE neuron class is one of 12 amphid sensory neuron classes located in the lateral head ganglia of C. elegans and is composed of a single pair of bilaterally symmetric cells, ASE left (ASEL) and ASE right (ASER) (Fig. 1A). Although these two neurons display several left/right asymmetric features in post-developmental stages (Yu et al. 1997; Ortiz et al. 2006), they appear largely equivalent after their birth (Johnston et al. 2005). To determine the molecular signature of both ASE neurons, we isolated ASER neurons from embryos, in which both ASEL and ASER still appear largely equivalent, using an ASER-specific gfp reporter transgene (Fig. 1B). Transgenic animals were gently disrupted and a >90% pure sample of ∼85,000 gfp-labeled ASER neurons was obtained by fluorescent activated cell sorting. We then used SAGE to identify the transcriptome of the ASE neurons. This technique has been used successfully to determine the transcriptome of groups of microdissected cell types (Blackshaw et al. 2001; Blacque et al. 2005; McGhee et al. 2006).
Five-thousand-seven-hundred-sixteen protein-coding genes were found to be represented in the ASE library with more than a single tag (Supplementary Table 1A; summarized in Fig. 1C). These genes fall into many broad functional categories, as illustrated by the association with a broad spectrum of Gene Ontology terms (data not shown). Most genes previously known to be expressed in ASE (including genes that later become restricted to ASEL or ASER) can be found in the ASE library, yet a substantial number of genes that one would not expect to be expressed in ASE can also be found in the library (Fig. 1C; Supplementary Table 2). The presence of such genes (e.g., the GABA transporter unc-47) indicates either an authentic, previously unappreciated expression of these genes at low levels in ASE (indeed, many of the “false-positive” hits are represented only as singleton tags) (Supplementary Table 2) or may be a reflection of some level of contamination of the isolated ASE cells. We sought to filter out these possible “false-positive” hits by the isolation and SAGE analysis of another individual sensory neuron type, the AFD thermosensory neurons. We reasoned that the comparison of the ASE SAGE library to a SAGE library prepared from AFD thermosensory neurons will not only filter out noise but will also filter out ubiquitously expressed generic cellular components and, thereby, provide a library of genes with ASE-specific functions.
After isolating individual, gcy-8∷gfp-labeled AFD neurons by FACS and establishing an AFD SAGE library, we composed what we call a comparative “ASE > AFD library” in which we assembled genes that fulfill at least one of the following three criteria: (1) represented exclusively in the ASE SAGE library (Supplementary Table 3A); (2) represented with at least three times as many tags in the ASE versus AFD SAGE library (Supplementary Table 3B; see Supplemental Material for a rationalization of the 3× cutoff value); and (3) enriched in ASE versus AFD by less than threefold but fulfilling the Audic-Claverie test of statistical significance (Supplementary Table 3C; this test can only be reliably applied for genes with high tag counts; see the Supplemental Material). The composite ASE > AFD library contains a total of 1302 genes that are represented with more than one tag (Fig. 1C). In this comparative library, possible false-positive hits are almost completely eliminated, yet the majority of known ASE-specifically expressed genes are still retained (Fig. 1D; Supplementary Table 2). In the reverse comparative library, in which we assembled genes enriched in AFD (either exclusive to AFD or at least 3× enriched) (Supplementary Table 4A–C), all previously known AFD-specifically expressed genes can be found (Supplementary Table 2), and a significant overlap exists with a previously published microarray analysis of isolated AFD neurons (Colosimo et al. 2004; Supplementary Table 4D). The approach of comparing SAGE libraries is, therefore, a powerful means to elucidate neuron-type-specific gene expression profiles. However, it needs to be kept in mind that ASE and AFD may both coexpress genes that define features specific to each of these two neurons but no other neuron types. Such a subtractive approach, therefore, likely underestimates the full battery of genes that determine cell-type-specific properties of each neuron class.
Sequence analysis of genes revealed by SAGE
The ASE > AFD comparative library is expected to provide a broad view of the molecular apparatus that defines the ASE neuron and distinguishes it from other sensory neurons. We find that the library reveals an ample set of TFs, cell adhesion molecules, neurotransmitters and receptors, ion channels, and putative chemoreceptors that presumably shape the development and function of the ASE neurons (summarized in Fig. 1E; Supplementary Tables 5–7).
TFs
A large number of predicted DNA-binding TFs are present in the ASE > AFD library as nonsingleton hits (Supplementary Table 5; summarized in Fig. 1E). These include three TFs previously found to be expressed in ASE as well as 65 TFs that were not previously reported to be expressed or function in ASE. Two-thirds of these TFs are Zn-finger-type TFs, and of those, another two-thirds are nuclear hormone receptor-type C4 Zn finger TFs. Another five previously known ASE-expressed TFs are also contained within the ASE library but not specifically enriched compared with AFD (Supplementary Table 2).
Putative chemoreceptors
Forty-six putative chemoreceptors of the 7TMR family are present in the ASE library, of which 41 are enriched in the ASE > AFD library. None of these genes fall into specific subfamilies of 7TMRs genes. Eleven of these 41 genes are represented with more than one tag (Supplementary Table 6). Other than two predicted neuropeptide receptors (Uchida et al. 2003), no 7TMR has previously been found to be expressed in ASE (http://www.wormbase.org; Troemel et al. 1995). In comparison, the AFD thermosensory neuron contains substantially fewer 7TMRs (three nonsingletons in the AFD > ASE library compared with 11 in the ASE > AFD library).
While 7TMRs are the best characterized gustatory receptors to date, two other classes of proteins have been proposed to be involved in the gustatory response to salt, namely, TRP channels and ENaC/Degenerin channels (for review, see Scott 2005). One TRP channel and three ENaC/Degenerin channels are in the ASE > AFD library (singletons excluded) along with several additional TRP and degenerin channels represented with multiple tags in the complete ASE SAGE library (Supplementary Table 1A).
Channels and signaling genes
Other notable members of the ASE > AFD library are (1) neurotransmitter receptors of various kinds, including metabotropic and ionotropic receptors (14 genes); and (2) calcium channels and potassium channels, which are thought to propagate and shape the electrical property of neurons (eight genes) (Fig. 1E; Supplementary Table 7). The expression of these genes in the ASE neurons illustrates the potential of the ASE neurons to receive a multitude of distinct inputs and tune their membrane potentials by a variety of means. Additionally, the ASE neurons express a substantial number of putative peptidergic neurotransmitters, including FMRFamide-related (flp) genes, neuropeptide-like (nlp) genes, and insulin-related (ins) peptides (10 genes) (Fig. 1E; Supplementary Table 7). One member of the latter class of genes has recently been implicated in a form of behavioral plasticity mediated by the ASE neurons (Tomioka et al. 2006).
Antisense transcripts
Compared with microarray analysis, SAGE samples transcripts in a broader and more unbiased manner. We made use of this ability to specifically examine the ASE transcriptome for naturally occurring antisense transcripts (NATs). NATs constitute a significant fraction of animal transcriptomes and are thought to be involved in a complex variety of gene regulatory events (Lapidot and Pilpel 2006; Munroe and Zhu 2006). We found a total of 126 nonsingleton antisense transcripts in the ASE transcriptome, 31 of which are enriched in ASE versus AFD (Fig. 1F; Supplementary Table 8A,B). These 31 NATs are antisense to a variety of protein-coding genes, including TFs, signaling proteins, and putative chemoreceptors (Supplementary Table 8B). Less than half of the sense transcripts matching the NATs are represented by sense tags in the ASE transcriptome (“correlated expression”), while the remainder show anti-correlated expression. Examples for correlated and anti-correlated sense and antisense transcripts have been described before in the literature and have been proposed to be a reflection of distinct modes of function of antisense transcripts (Lapidot and Pilpel 2006).
Further experimental validation of the SAGE data
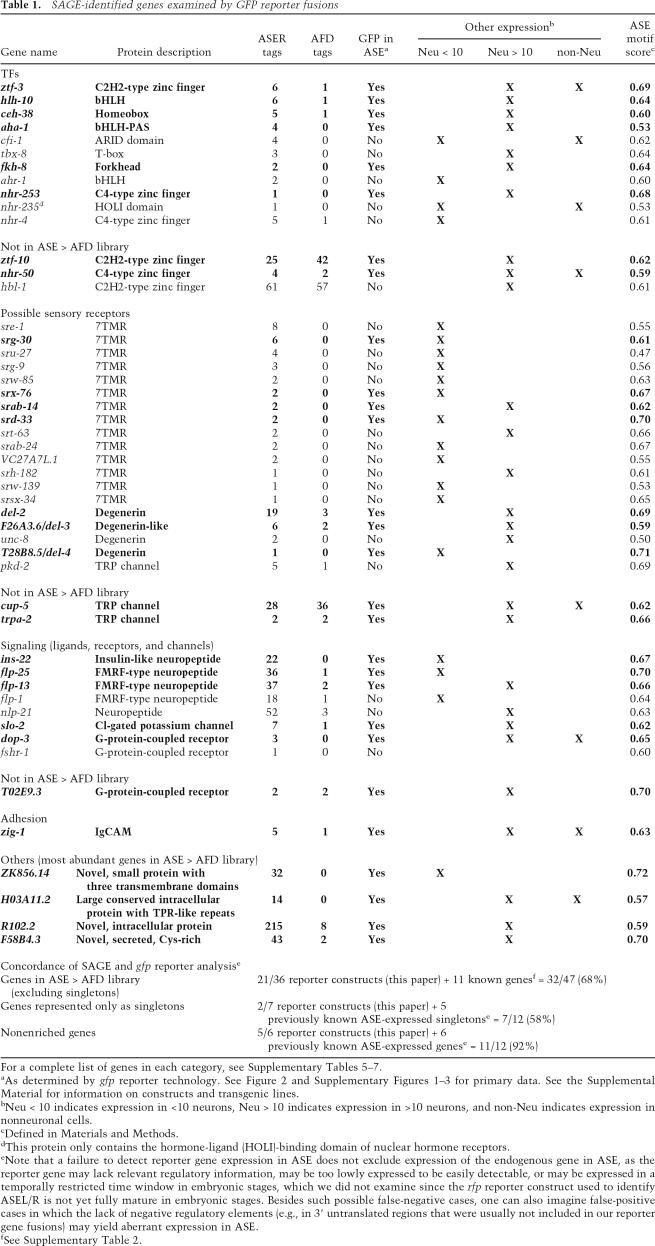
We validated selected aspects of the SAGE data using gfp reporter fusions expressed in transgenic animals. We focus in this study entirely on coding sense transcripts. Rather than exclusively focusing on genes with the most sequence tags, we broadly sampled genes with different tag numbers in the ASE SAGE library. We tested genes from the ASE > AFD comparative library but also genes not specifically enriched in the ASE library versus the AFD library. In total, we analyzed transgenic animals expressing reporter fusions to 49 genes, including 14 TFs, 14 candidate gustatory receptors of the 7TMR class, four candidate gustatory receptors of the ENaC/Degenerin class, three candidate gustatory receptors of the TRP channel class, 10 additional signaling genes (GPCRs and neuropeptides), and four novel genes that represent the most ASE-enriched genes in the ASE > AFD library. Almost 70% of all examined reporter genes are expressed in post-embryonic ASE neurons (Table 1; Fig. 2; Supplementary Figs. 1–3; Supplementary Tables 5–7). Technical reasons could account for the failure to detect ASE expression for the remaining reporter genes, as detailed in the legend to Table 1. Reporter gene expression ranges from relatively broad (but never ubiquitous) to very narrow (e.g., 7TMRs) (Fig. 2; Supplementary Fig. 2). We did not find any reporters to be exclusively expressed in the ASEL/R neurons or left/right asymmetrically in either ASEL or ASER only. The latter observation underscores that the two ASE neurons are largely bilaterally symmetric (White et al. 1986) and may diversify only in regard to some sensory functions (Pierce-Shimomura et al. 2001).
Table 1.
SAGE-identified genes examined by GFP reporter fusions
For a complete list of genes in each category, see Supplementary Tables 5–7.
aAs determined by gfp reporter technology. See Figure 2 and Supplementary Figures 1–3 for primary data. See the Supplemental Material for information on constructs and transgenic lines.
bNeu < 10 indicates expression in <10 neurons, Neu > 10 indicates expression in >10 neurons, and non-Neu indicates expression in nonneuronal cells.
cDefined in Materials and Methods.
dThis protein only contains the hormone-ligand (HOLI)-binding domain of nuclear hormone receptors.
eNote that a failure to detect reporter gene expression in ASE does not exclude expression of the endogenous gene in ASE, as the reporter gene may lack relevant regulatory information, may be too lowly expressed to be easily detectable, or may be expressed in a temporally restricted time window in embryonic stages, which we did not examine since the rfp reporter construct used to identify ASEL/R is not yet fully mature in embryonic stages. Besides such possible false-negative cases, one can also imagine false-positive cases in which the lack of negative regulatory elements (e.g., in 3′ untranslated regions that were usually not included in our reporter gene fusions) may yield aberrant expression in ASE.
fSee Supplementary Table 2.
Figure 2.
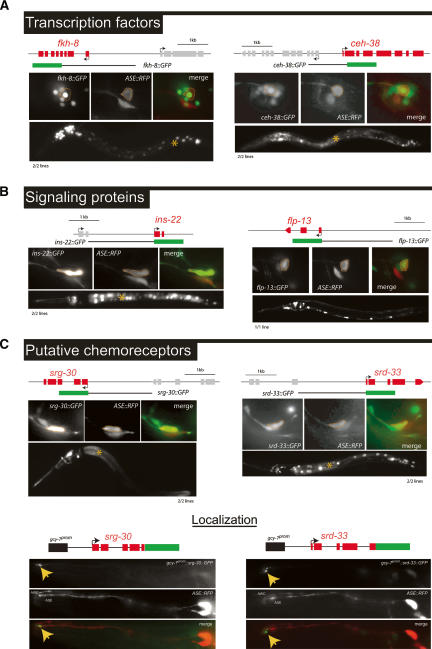
Expression patterns of genes from the SAGE library as assessed by gfp reporter gene technology. Only a selected and representative number of examples are shown in this figure. All analyzed reporter strains, including those shown here, are shown in Supplementary Figures 1–3. Green boxes indicate gfp coding sequences (rfp in the case of dop-3 and yfp in the case of hlh-10). If extrachromosomal arrays were generated, multiple lines were analyzed for each reporter construct, as indicated below each panel of micrographs. Each data set of micrographs shows expression of the reporter construct under investigation in a high-magnification (400×) image, the overlap with a red fluorescent marker for ASEL/ASER (otIs151 transgene; “ASE-RFP”), and a low-magnification (160×) overview of expression of the reporter construct under investigation throughout the whole worm. In the low-magnification images, the asterisk (*) indicates expression of the gut-specific injection marker elt-2∷gfp. (A) Expression patterns of two selected TFs from the SAGE data set. (B) Expression patterns of two selected peptidergic signaling proteins from the SAGE data set. (C) Expression patterns of two selected putative chemoreceptor proteins of the 7TMR family from the SAGE data set. The srg-30 7TMR-type chemoreceptor shows the most restricted expression pattern with strong expression in ASEL/R and weak expression in the chemosensory neurons ASIL/R and ADLL/R. Additional expression can be observed in a pharyngeal neuron. The srd-33 7TMR reporter gene fusion also shows a restricted expression pattern (ASEL/R, AWBL/R, ASHL/R, AVKL/R, PHAL/R, PHBL/R, and weakly in another unidentified tail neuron pair, possibly PHCL/R). The bottom panels in C demonstrate that 7TMR-type chemoreceptors localize to the dendritic ending of the ASE neurons. In order to boost expression in the ASE neurons, a 188-bp ASEL-specific regulatory element from the gcy-7 gene was appended to each locus (indicated as a black box). One transgenic line each was analyzed in detail. Yellow arrows point to the dendritic endings.
Furthermore, we tested whether putative chemoreceptors of the 7TMR class fulfill the chemoreceptor criterion of localizing to the exposed dendritic endings of the ASE sensory neurons. Using reporter genes in which the entire 7TMR coding region was fused to gfp, we found this to be indeed the case for both 7TMRs tested (srg-30 and srd-33) (Fig. 2C). We generated reporter gene fusions to other members of the srg or srd subfamily of 7TMRs (srg-25, srg-27, srg-29, srd-32) but found them to show no expression in the ASE neurons (data not shown).
In summary, our SAGE approach has successfully uncovered a large number of ASE-expressed genes, including a variety of gene regulatory factors, receptors, ion channels, and signaling genes, as well as novel genes that may eventually help to deconstruct the development and function of the ASE gustatory neuron on a molecular level.
Defining a cis-regulatory element required for expression in ASE
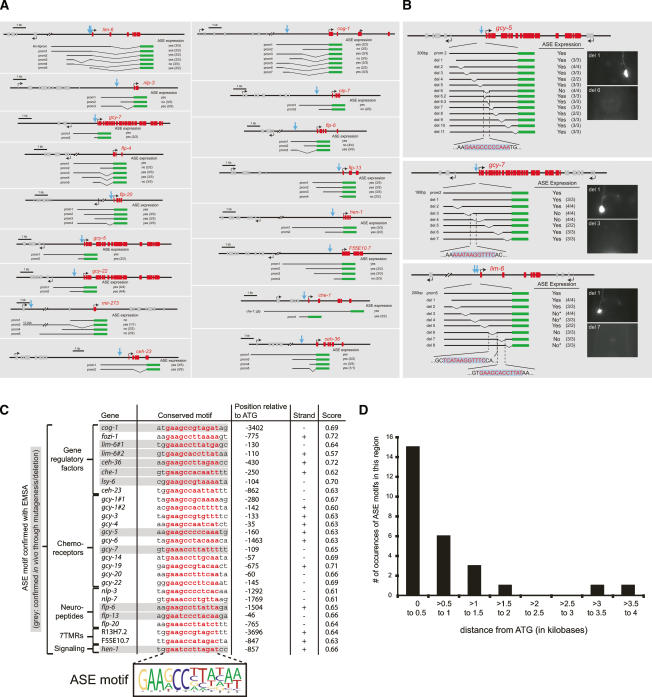
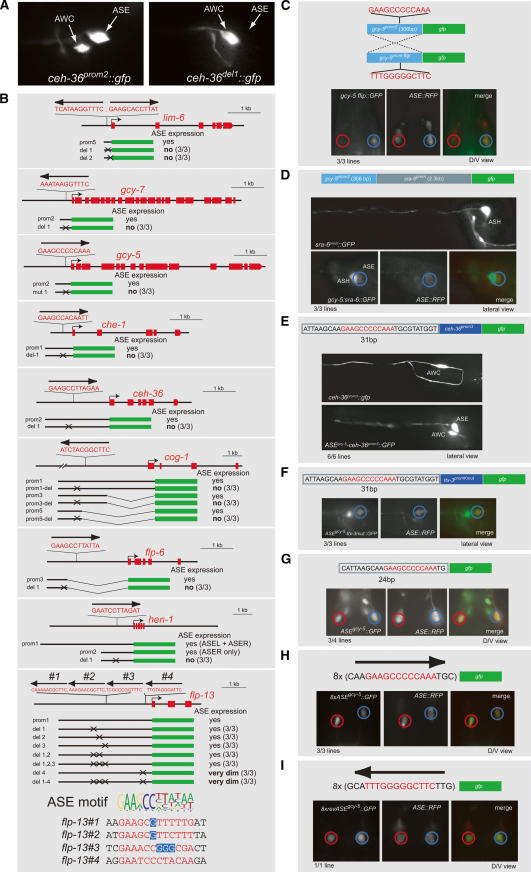
In combination with previously described ASE-expressed genes, the new SAGE data set provides an unprecedented view into the molecular makeup of a gustatory neuron and prompts the question as to how the expression of this gene battery is controlled. To elucidate the regulatory architecture of the ASE gene battery, we analyzed the cis-regulatory control regions of a set of ASE-expressed genes in detail using in vivo promoter dissection approaches. Since our goal was to analyze the cis-regulatory control mechanisms that are specific for the ASE neurons, we ignored genes that are expressed in ASE but also broadly expressed in a large number of other cells, such as ubiquitously expressed genes, pan-neuronally expressed genes, or genes expressed in most or all ciliated sensory neurons. We focused on a set of 17 genes previously reported to be expressed in a selected number of neurons, including ASE (either ASEL and ASER, ASEL-only, or ASER-only; for simplicity, we refer to all these genes as “ASE-expressed”) (Fig. 3; Supplementary Table 2). Sequence alignment of the promoters of these genes and motif-finding algorithms did not yield any obvious patches of common sequence elements, likely because the sequence space is too large. Therefore, we used two parallel approaches to dissect the cis-regulatory architecture of ASE-specific gene expression, both of which rely on the generation of a large set of transgenic C. elegans lines. First, we dissected gfp reporter constructs derived from 17 ASE-expressed genes, thereby defining small cis-regulatory elements within these constructs that are required and/or sufficient for ASE-specific expression (Fig. 3A). Second, we performed a systematic scanning deletion mutagenesis of the cis-regulatory elements from three ASE-expressed genes—gcy-5, gcy-7, and lim-6—yielding 25-base-pair (bp) elements required for ASE expression (Fig. 3B). Comparison of the sequences of minimal elements from these 17 genes identifies a conserved motif present in all but one cis-regulatory region, which we termed the “ASE motif” (Fig. 3C). The motif contains an essentially invariant, 6-bp core at its 5′ end [GAADCC] followed by an additional, more degenerate, A/T-rich 6-bp sequence.
Figure 3.
Dissection of the cis-regulatory architecture of ASE-expressed genes reveals a common motif, the “ASE motif.” (A) Determining minimal regulatory elements required for ASE expression. Blue arrows indicate the presence of a conserved site, the ASE motif (detailed in C). The number of transgenic lines scored is indicated in parentheses next to each construct. (B) Scanning mutational analysis of the regulatory elements of the ASE-expressed genes gcy-5 (ASER), gcy-7 (ASEL), and lim-6 (ASEL). The ASE motif is indicated in red. The number of transgenic lines scored is indicated in parentheses next to each construct. Only cases in which a loss of gfp expression (rather than ectopic expression) is observed are indicated here. In the case of the gcy-5 gene, the deletion of a single 25-bp region causes loss of expression in ASER (“del 6”). “del 6.2” and “del 6.3” are deletions that eliminate sequences directly adjacent to the ASE motif. In the case of the gcy-7 gene, deletions of two neighboring 25-bp regions affect the ASE motif and cause loss of expression in ASEL. In the case of the lim-6 gene, the deletion of four 25-bp regions each causes loss of expression in ASEL. Two of these regions each contain a single copy of the ASE motif (ASE motif score: 0.64 and 0.57). Deletion of another 25-bp element specifically causes a failure to maintain ASE expression. Since we previously showed that lim-6 genetically autoregulates its own expression (Johnston et al. 2005), we presume that these 25 bp contain a lim-6 autoregulatory element. The 25-bp region contains a GAATAAA motif that is conserved in three nematode species. When this motif is deleted alone, similar maintenance defects are observed (data not shown). The fourth 25-bp region that is required for ASEL expression also contains a phylogenetically conserved motif; however, its precise excision has no impact on ASE expression (data not shown). We note that the presence of multiple sites required for ASE expression is consistent with our previous genetic analysis, which points to several regulatory inputs into the lim-6 locus (see final model figure). (C) Alignment of a conserved motif present in ASE-expressed regulatory elements defined by reporter gene analysis. Shown here are only those ASE motifs whose binding to the CHE-1 TF have been confirmed by EMSA, as shown in Figure 6. Other previously known ASE-expressed genes also harbor ASE motifs (data not shown), whose relevance was not further tested. Gray shading indicates that the functional relevance of this motif was explicitly confirmed by deleting the motif in a reporter gene construct and observing a loss of gfp expression in ASE, as shown in Figure 4. The sequence alignment defines a position weight matrix (PWM) that is represented by a sequence logo; the quality of the match of each individual ASE motif with the sequence logo is assigned what we term an “ASE motif score” (see Materials and Methods). ASE motifs in genes defined by SAGE analysis are represented in the form of ASE motif scores in Table 1. (D) ASE motifs preferentially cluster within the first ∼1 kb upstream of the ATG start codon of a gene. This graph contains all genes shown in C.
We tested the functional relevance of the ASE motif by mutating or deleting it in the context of different cis-regulatory control regions from various ASE-expressed genes. In each case tested, the motif is required for expression of the reporter gene in ASE (Fig. 4A,B). Mutating the ASE motif generally causes a complete loss of ASE expression of the respective reporter gene, as shown in a representative example in Figure 4A. The only exception is the ASE motif in the flp-13 locus whose deletion largely reduces, but does not completely abolish, ASE expression. Further mutational analysis showed that the remaining low-level gfp expression is not due to the presence of three other motifs in the flp-13 promoter that share limited homology with the ASE motifs (Fig. 4B).
Figure 4.
Functional characterization of the ASE motif. (A,B) Requirement of the ASE motif for gene expression in ASE. A shows representative animals expressing a reporter construct for the ceh-36 gene with the normal ASE motif either present (left panel) or deleted (right panel). B shows a schematic representation of normal or ASE motif-mutated/deleted reporter constructs and their expression. “No ASE expression” indicates a complete loss of detectable gfp expression in ASE that we observed upon deletion of all motifs (representative example is shown in A), with the exception of flp-13, where some weak gfp expression persists in ASE after the ASE motif mutation. The number of transgenic lines analyzed is indicated in parenthesis. All lines for each construct show similar expression patterns. Note that besides its functional ASE motif (with an ASE motif score of 0.66; see text and Materials and Methods for explanation of how an ASE score is calculated), several low-scoring ASE motifs (ASE motif score: 0.54, 0.56, and 0.53) can be found in the flp-13 promoter. None of these low-scoring ASE motifs is required for expression in ASE (last panel). (C) Orientation independence of the ASE motif. A 306-bp regulatory element from the gcy-5 locus drives expression in ASER in either orientation. (D) Distance independence of the ASE motif. The 306-bp regulatory element from the gcy-5 gene is functional when artificially separated by several kilobases from the start of the gfp reporter. Separation was achieved through the addition of the sra-6 promoter, which is active in ASH, among other cells (Troemel et al. 1995). (E–I) Sufficiency of the ASE motif. ASE expression is observed upon insertion of 31 bp, containing the ASE motif, either upstream of an AWC-specific promoter fragment from the ceh-36 gene (E), or upstream of a fragment of the ttx-3 promoter, in which the AIY motif, normally required for expression of this motif in AIY, is deleted (Wenick and Hobert 2004) (F). (G) A 24-bp element (12 bp of ASE motif + 6 bp on either side) also drives ASE expression; additional gfp expression can be observed in a few other neurons. Eight multimerized copies of an 18-bp element that contains the gcy-5 ASE motif (plus 3 bp of flanking sequences on either side) direct gfp expression in only the ASE neurons, in either the forward (H) or reverse (I) orientation. In all panels, “ASE-RFP” indicates the presence of the otIs151 array, which labels the ASE neurons with dsRed2. The vector backbone for all constructs is pPD95.75. The number of transgenic lines analyzed is indicated.
In the majority of the experimentally examined cases, the ASE motif is located within ∼1 kb upstream of the ATG start codon of the regulated gene (Fig. 3D). Nevertheless, motifs located >1 kb away from the ATG are also functional. For example, the deletion of the ASE motif in the cog-1 locus, which is 3.4 kb upstream of the ATG, eliminates ASE expression (Fig. 4B).
The ASE motif usually occurs in a single copy. Only two ASE-expressed genes each contain two clear matches to the ASE motif (gcy-1 and lim-6) Fig. (3C). In the case of lim-6, we found that deletion of either motif disrupts ASE expression (Fig. 4B). The flp-13 locus contains, besides a clear match to the ASE motif, three poorly matching variants of the ASE motif (Fig. 4B). However, as mentioned above, we find these variant motifs to not be required for ASE expression, when deleted either alone or in combination (Fig. 4B). The lim-6 gene is also the only gene for which we found, through promoter dissection, a motif with no similarity to the ASE motif that is required for ASE expression of lim-6. However, such an additional element could not be identified in other ASE-expressed genes, either experimentally (in the case of the gcy-5 or gcy-7 scanning mutagenesis) or by sequence gazing. We suspect that the additional element required for ASE expression in the lim-6 locus relates to the gene’s left/right asymmetric expression in ASEL versus ASER (see final model figure).
We further analyzed the orientation and distance dependence of the ASE motif through various modes of experimental manipulation. The occurrence of functional ASE motifs in both sense and antisense orientations upstream of ASE-expressed genes strongly suggests orientation independence. We confirmed this notion by inverting the orientation of a 306-bp fragment from the gcy-5 promoter that contains the ASE motif. The inverted gcy-5 construct shows unaltered ASE expression (Fig. 4C). Although the large majority of ASE motifs are found within the first 1 kb upstream of the predicted start codon of ASE-expressed genes (Fig. 3C), the ASE motif can also work at larger distances. As mentioned above, this is exemplified by the cog-1 gene, whose ASE motif, located 3402 bp upstream of the start codon, is required for ASE expression, as determined by deletion analysis (Fig. 4B). Moreover, adding the minimal ASER-specific gcy-5 promoter of 306 bp upstream of the ∼2.4-kb sra-6 promoter, normally expressed in ASH and PVQ, yielded additional expression in ASER (Fig. 4D).
We next tested whether the ASE motif is sufficient to drive expression in ASE. As a first step, we fused 31 nucleotides (nt), containing the 12-bp ASE motif from the gcy-5 gene, to a 368-bp promoter fragment of the ceh-36 gene that is only expressed in AWC. This synthetic construct shows expression in ASE (Fig. 4E). Similarly, when the same 31 nt are fused to a 243-bp promoter fragment of the ttx-3 promoter (Wenick and Hobert 2004), expression is observed in ASE (Fig. 4F). Moreover, we find single or multimerized ASE motifs with very little flanking sequence (24-bp or 18-bp elements) from the gcy-5 promoter to be sufficient to yield reporter expression in ASE when fused to the bare pPD95.75 gfp expression vector (Fig. 4G–I) (by itself, pPD95.75 has no transcriptional activity) (data not shown). This activity is independent of the orientation of the element. The few base pairs that flank the ASE motif in these minimal constructs are unlikely to play a role since deletions in the gcy-5 promoter that are directly adjacent to the ASE motif (Fig. 3B) show no effect on expression in ASE.
The ASE motif is a binding site for the Zn finger TF CHE-1
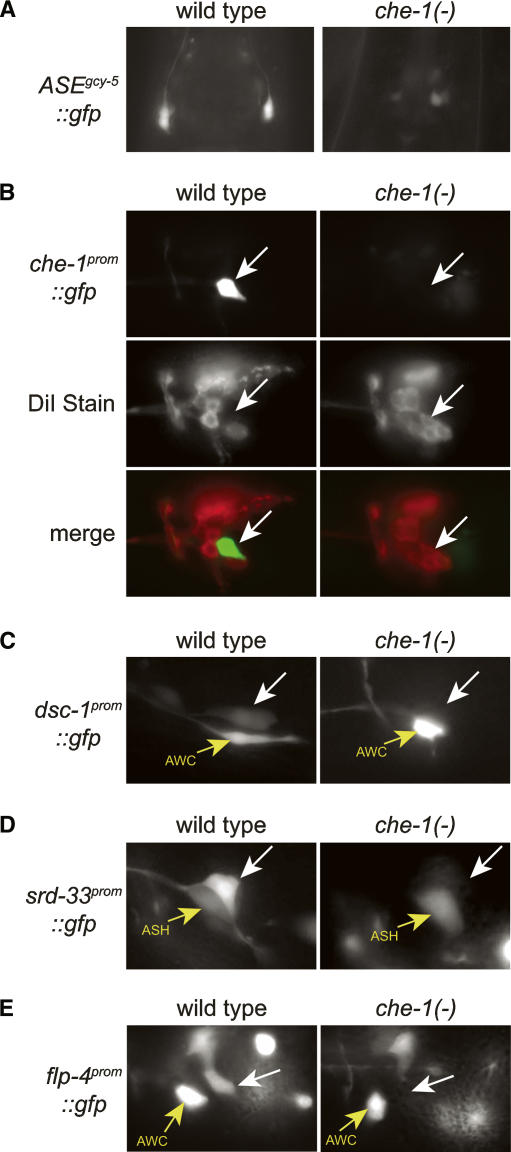
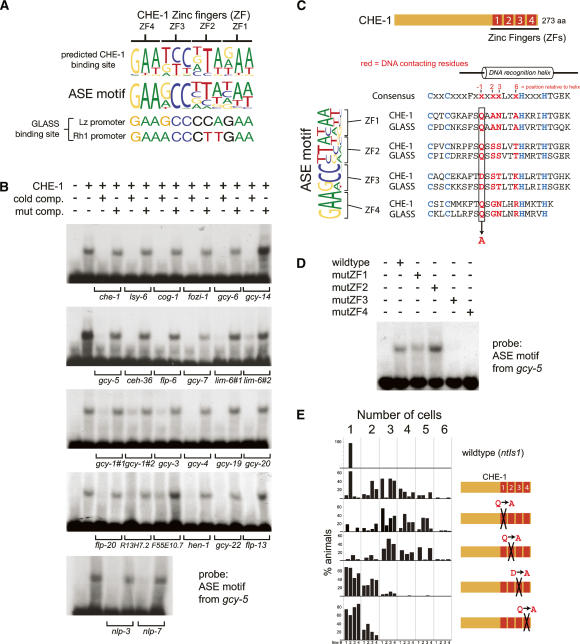
What is the nature of the trans-acting factor(s) that recognizes the ASE motif? Four lines of evidence point to the ASE-expressed C2H2 zinc finger TF CHE-1. (1) In che-1 mutants, the expression of all tested ASE-expressed genes is lost exclusively in the ASE neurons, but not in other cells that express these genes (Fig. 5; Chang et al. 2003; Uchida et al. 2003). We find that expression of the “ASE motif-alone” reporter construct, which contains 24 bp encompassing the gcy-5 ASE motif, is lost in che-1 mutant animals as well (Fig. 5A). (2) Ectopic expression of che-1 in several sensory neurons causes ectopic activation of the ASE motif-containing gcy-5 promoter (see below) (Uchida et al. 2003). (3) To further support the notion that CHE-1 directly controls the ASE motif, we used a previously described probabilistic recognition code (Benos et al. 2002) that predicts the DNA-binding site of C2H2 Zn finger TFs. We find that the predicted CHE-1-binding site shares striking similarity with the ASE motif (Fig. 6A). (4) The Drosophila ortholog of CHE-1, called GLASS, shares 100% identity with CHE-1 in the residues within the Zn fingers that are predicted to contact DNA (Fig. 6A) and has been found to bind directly to two regulatory elements in fly promoters (Moses and Rubin 1991; Yan et al. 2003), each of which contain a site similar to the ASE motif (Fig. 6A).
Figure 5.
che-1 controls the expression of various reporter constructs in vivo. che-1 regulates a variety of ASE-expressed genes, as inferred by crossing reporter gene arrays from a wild-type background into a che-1(ot27) mutant background. All white arrows point to the location of the ASE neuron. (A) Control of the “ASE motif-only” construct from Figure 4G by che-1. (B) Control of the ASE motif-containing che-1 promoter by che-1, demonstrating that che-1 autoregulates its expression. Note that in che-1 mutants, the ASE neuron is aberrantly taking up the dye DiI, thereby demonstrating that even in the absence of che-1, ASE remains a sensory neuron (Uchida et al. 2003). The analysis of several cell fate markers has, however, failed to show any specific identity to which the aberrant ASE neuron may have switched in che-1 mutants (Uchida et al. 2003). In contrast to Uchida et al. (2003), we were able to demonstrate that che-1 positively regulates its own expression. We have no explanation for these differences in results. (C) Regulation of an ASE motif-containing promoter of a TF from the SAGE data set. (D) Regulation of an ASE motif-containing promoter of a 7TMR gene from the SAGE data set. (E) che-1 controls the expression of flp-4, which does not contain a functional ASE motif (see below). Apart from the genes shown here, che-1 had been shown previously to regulate several additional genes (tax-2, gcy-5, gcy-6, gcy-7, ceh-23, flp-6, F55E10.7, R13H7.2, ceh-36, and cog-1) (Chang et al. 2003; Uchida et al. 2003).
Figure 6.
The ASE motif is a binding site for the CHE-1 Zn finger TF. (A) Comparison of the ASE motif consensus motif (represented as a sequence logo) with the predicted binding site of CHE-1 determined by C2H2-enoLOGOS and with the experimentally determined binding site of the Drosophila CHE-1 ortholog GLASS in the Lozenge and Rhodopsin promoters. (B) EMSA of bacterially produced CHE-1 and the ASE motif from the gcy-5 locus. In order to compare binding affinities and to avoid potential problems with probe-labeling efficiencies, binding to ASE motifs was determined by competition assays in which binding to the radiolabeled gcy-5 probe was competed with unlabeled ASE motifs from other genes. As specificity controls, the same unlabeled oligos were used in which the core GAANCC motif is mutated; those probes are not able to compete for binding. Supplementary Table 9 contains sequences of probes. (C) Alignment of the Zn fingers of CHE-1 and the Drosophila ortholog GLASS with predicted DNA-contacting residues. Predictions are according to Pavletich and Pabo (1991). The DNA-contacting residues mutated in the constructs used in D and E are indicated. (D) EMSA with wild-type CHE-1 and point-mutated CHE-1, as indicated. (E) Ectopic expression of CHE-1 in other sensory neurons under control of the gpa-10 promoter results in ectopic activation of the ASE motif-containing gcy-5 promoter, as previously reported (Uchida et al. 2003) and as assayed by counting of ectopically gfp-expressing cells, using the ntIs1 Is[gcy-5∷gfp] transgene. This activity largely depends on the last two Zn fingers (#3 and #4) of CHE-1. For each experiment, four transgenic lines were scored, except for mutated Zn finger 2, in which three transgenic lines were scored. For the wild-type control, ntIs1 Is[gcy-5∷gfp] was scored in an otherwise wild-type background.
We tested whether CHE-1 binds to the ASE motif in vitro using electrophoretic mobility shift assays (EMSAs). We find that bacterially expressed and purified full-length CHE-1 protein is, indeed, able to bind the ASE motif present in all tested genes (Fig. 6B). The binding is sequence specific since binding of CHE-1 to a given ASE motif can only be competed with other ASE motifs but not with point-mutated ASE motifs.
Similar to many TFs (e.g., see Way and Chalfie 1989; Hobert et al. 1997; Satterlee et al. 2001), che-1 regulates its own expression. A che-1 promoter gfp fusion, which is normally expressed in ASEL/R only, fails to be expressed in che-1 mutants (Fig. 5B). The che-1 promoter contains a single ASE motif to which CHE-1 exhibits selective binding in vitro. The autoregulation of che-1 provides an explanation for the persistent che-1 expression throughout the life of the animal (data not shown) and suggests that che-1 not only induces the expression of terminal differentiation features but also actively maintains them.
CHE-1 activates the ASE motif via two of its four Zn fingers
Based on crystallographic studies, it is known that each Zn finger of a canonical C2H2 Zn finger TF contacts 3 nt (Pavletich and Pabo 1991). The predicted CHE-1 site is, therefore, 12 bp long (Fig. 6A,C). Since only the first half of the 12-bp ASE motif is strongly conserved (Fig. 6A), the site-specific affinity of CHE-1 for the ASE motif may be primarily dictated by only two of its four Zn fingers, a situation that would be analogous to the selective usage of a subset of Zn fingers in other multi-Zn finger domain TFs (Evans-Galea et al. 2003). The “anti-parallel” binding of Zn fingers to their recognition sites (Pavletich and Pabo 1991) furthermore predicts that Zn fingers #3 and #4 bind to the conserved [GAADCC] 5′ end of the ASE motif. To test this prediction, we used EMSA and bacterially expressed/purified mutant variants of CHE-1 in which individual Zn fingers carry a single amino acid substitution. Specifically, we mutated the residue at position −1 relative to the DNA-binding helix (Fig. 6C). Based on available crystal structures, this residue is predicted to be in direct contact with DNA (Pavletich and Pabo 1991). We found that mutations in the third and fourth Zn fingers but not in the first or second Zn fingers significantly disrupt binding to the ASE motif (Fig. 6D).
che-1 is not only genetically required to activate ASE motif-containing genes, but is also sufficient to do so upon ectopic expression in distinct neuron types (Fig. 6E; Uchida et al. 2003). The ability of che-1 to activate the ASE motif in vivo shows the same domain requirements that we have defined in vitro for ASE motif binding. Ectopically expressed che-1, in which either of the first two Zn fingers is mutated, does not affect che-1’s ability to induce ASE motif-dependent reporter gene expression in other neuron types. However, mutations in either the third or fourth Zn finger do affect this ability (Fig. 6E).
The ASE motif is not a sufficient predictor for expression in ASE
Loss of che-1 precisely phenocopies the behavioral consequences observed upon laser ablation of ASE (Dusenbery et al. 1975; Bargmann and Horvitz 1991; Uchida et al. 2003). Moreover, every gene tested that is expressed in both ASE and a restricted set of additional cells (i.e., not pan-neuronal, not pan-ciliated) fails to be expressed specifically in the ASE neurons of che-1 mutants (15/15 genes tested) (Fig. 5; Chang et al. 2003; Uchida et al. 2003). This includes several genes newly identified in the SAGE analysis (Fig. 5). We, therefore, presume that che-1 is a selector gene that controls all cell-type-specific properties of the ASE neuron, including the ASE-specific gene battery that we have defined by our SAGE analysis. One can envision two models of how che-1 controls ASE fate specification: In one model that is analogous to the control of intestinal cell fate specification by the GATA factor elt-2 (McGhee et al. 2006), the expression of most or all members of the ASE gene battery is directly controlled by the ASE motif. In an alternative model, only a subset of ASE-expressed genes may contain an ASE motif; those containing an ASE motif are directly controlled by che-1, and those not containing an ASE motif may be controlled by TFs that che-1 controls via an ASE motif.
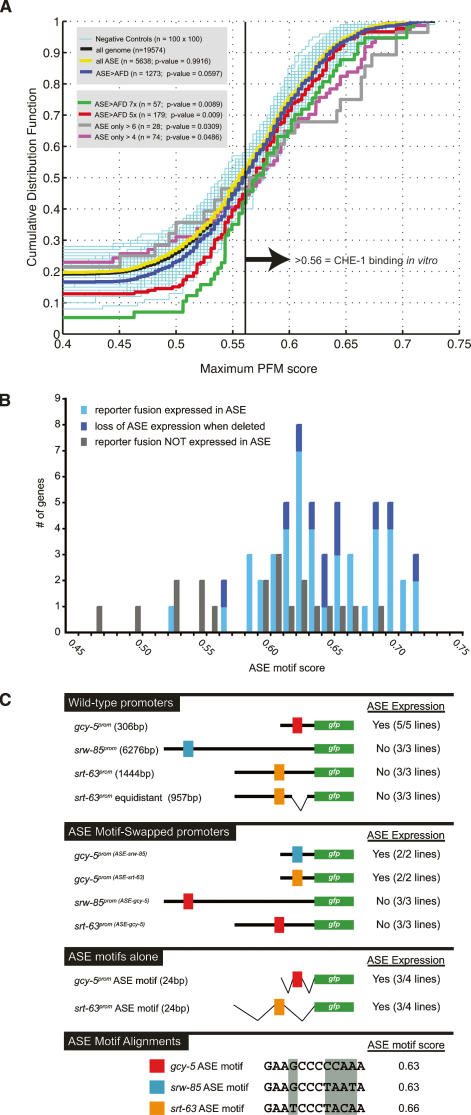
To test these models, we first examined the ASE > AFD SAGE data set for the occurrence of high-affinity che-1-binding sites in the form of an ASE motif. To this end, we developed a metric with which we could predict the presence of a high-affinity che-1-binding site in genomic input sequence. Using a position frequency matrix (PFM) of experimentally confirmed ASE motifs (Fig. 6; see the Supplemental Material), we assigned any given 12-bp motif a score that reflects its similarity to the ASE motif. A score of 0.73 as the best possible score. Gel shift analysis of variations of the ASE motif reveals that ASE motifs with a score of 0.56 or lower are not able to bind CHE-1 (Supplementary Fig. 4). We find that approximately half of the genes in the ASE > AFD library contain an ASE motif score of 0.57 or better within a 1.5-kb upstream region (see the Supplemental Material; note that 1.5 kb was chosen based on the occurrence of experimentally validated ASE motifs) (Fig. 3D). These genes are candidates to be directly regulated by CHE-1.
The other half of genes in the ASE > AFD data set with low-scoring ASE motifs may be regulated by che-1 indirectly. One example is the flp-4 gene, a FMRFamide-encoding gene that is expressed in ASE (Johnston et al. 2005). Its minimal promoter region sufficient to drive expression in ASE contains only low-scoring ASE motifs (<0.56), which all fail to bind CHE-1 in vitro (Supplementary Fig. 3A). We find that flp-4∷gfp expression is nevertheless entirely dependent on che-1 (Fig. 5). We conclude that che-1 regulates flp-4 in an ASE motif-independent manner by intermediary TFs. A good candidate for such a regulator is the LIM homeobox gene lim-6, an ASE motif-containing direct target of CHE-1, which we previously found to partially affect flp-4 expression (Johnston et al. 2005). Any one of the >60 ASE-expressed TFs that we described here is a candidate to regulate other non-ASE motif-containing genes in the ASE neurons.
The flp-4 analysis as well as the large number of non-ASE motif-containing genes in the ASE > AFD library suggest that the absence of an ASE motif is not a reliable predictor for absence of expression in ASE. Conversely, is the presence of a high-scoring ASE motif a reliable predictor for expression of a gene in ASE? We addressed this issue by examining the genome-wide occurrence of the ASE motif. To this end, we identified the best-scoring ASE motifs in the 1.5-kb upstream regions of all predicted genes in the genome and plotted the frequency of specific motif scores as a cumulative distribution function (Fig. 7A). We find that the distribution of ASE motifs in the ASE > AFD SAGE library is slightly enriched relative to the distribution of the ASE motif in all genes in the genome (P-value = 0.0597). When we sequentially narrow down the ASE > AFD data set to genes with increasingly larger enrichment factors in ASE versus AFD and/or larger tag numbers, we detect a strong increase in statistically significant enrichment of high-scoring ASE motifs and a statistically significant depletion of low-scoring ASE motifs in these data sets (Fig. 7A).
Figure 7.
Genome-wide distribution and context dependency of the ASE motif. (A) Distribution of the ASE motif in the genome and various SAGE data sets. The occurrence of ASE motifs in various data sets are shown as a cumulative distribution function, in which the highest-scoring match to the ASE motif in a particular promoter (X-axis) is plotted against the fraction of all promoters (in this particular set) that have site scores up to and including this particular score (Y-axis). The promoter size was chosen as 1.5 kb since most experimentally verified ASE motifs locate within this region (Fig. 3D). The distribution of ASE motif scores in the ASE > AFD library (blue line) is slightly different (P < 0.0597) from the distribution of the ASE motifs in all genes in the genome (black line). If one considers genes with progressive stringency in the ASE > AFD data set (5×, 7× enriched tag number in ASE vs. AFD, or >4 or >6 tags only in ASE, not in AFD) one observes a further increase in the statistical significance of enrichment of high-scoring ASE motifs and a depletion of low-scoring ASE motifs when compared with the genome-wide occurrence of ASE motifs. One-hundred sets of 100 genes were drawn randomly from the “all genes” set to form a graphical negative control in thin cyan lines. Kolmogorov-Smirnov statistics were calculated for all sets to determine the probability that each set was drawn from the same distribution of scores as that of all genes in the genome. See the Supplemental Material for more details. (B) The presence of an ASE motif is not a sufficient predictor of ASE expression. ASE motif scores from genes analyzed in the course of the SAGE data validation are shown. The difference between light and dark blue is that the relevance of the ASE motifs in the latter category was explicitly confirmed through deletion of the motifs. See Supplementary Tables 5–7 for reporter gene fusions, expression in ASE, and ASE motif score. (C) Manipulations of the context of ASE motifs demonstrate the context dependency of the ASE motif. Colored boxes represent ASE motifs as shown in the bottom panel. The swaps were conducted by mutating the dark-gray-shaded residues from one ASE motif to that of another ASE motif. The primary data for the “ASE motif-alone” construct from the gcy-5 promoter is shown in Figure 4G. The “equidistant” srt-63 ASE motif is a construct in which the ASE motif of srt-63 was positioned at exactly the same distance from the gfp start site as the ASE motif in the gcy-5 construct. “ASE expression” indicates reporter gene expression in ASE neurons.
In spite of the enrichment of the ASE motif in these comparative ASE versus AFD data sets, it is clear that high-scoring ASE motifs occur broadly enough in the genome that they are unlikely to be sufficient predictors for gene expression in ASE. We tested this prediction experimentally by examining the cis-regulatory sequences of genes not expressed in the ASE neurons for the presence of ASE motifs. We found that several reporter genes we generated to validate SAGE expression data and did not find to be expressed in ASE, indeed, contain high-scoring ASE motifs (genes are listed in Supplementary Tables 5–7, and their ASE motifs are graphically summarized in Fig. 7B). This confirms the notion that the presence of an ASE motif (i.e., a specific CHE-1-binding site) is not a sufficient predictor for ASE-specific gene expression.
As our dissection of several ASE-expressed promoters failed to uncover sequences other than the ASE motif required for expression in ASE (Fig. 3; note that the lim-6 gene is a special case due to its left/right asymmetric expression), we considered the possibility that a less sequence-specific context in a given promoter region determines whether an ASE motif results in ASE expression or not. To this end, we conducted ASE motif swaps using the promoters of three genes that all contain high-scoring ASE motifs. These genes include the ASE-expressed gcy-5 gene and two 7TMR-encoding genes, srt-63 and srw-85, whose promoters do not drive expression in ASE (Fig. 7C). When we replaced the ASE motif in the gcy-5 reporter construct (ASE motif score of 0.63) with the ASE motif from the srt-63 locus (score 0.66), we observe expression in ASE (Fig. 7C). The same holds for the srw-85 gene. An srw-85 promoter fusion is not expressed in ASE, but its ASE motif (score 0.63) drives ASE expression if transplanted into the context of the gcy-5 promoter (Fig. 7C). Conversely, when we replace the ASE motif in the non-ASE-expressed srt-63 or srw-85 promoters with the ASE motif of the gcy-5 locus, no expression in ASE is observed (Fig. 7C), thereby corroborating the notion of a context-dependent activity of the ASE motif. Distance from the transcriptional start site cannot account for the inability of the ASE motifs of srt-63 and srw-85 to function since the introduction of a deletion in the srt-63 reporter construct that moves its ASE motif to the same distance from the gfp start site as in the gcy-5 promoter still yields no expression in ASE (Fig. 7C). Lastly, when we test the ASE motif from the non-ASE-expressed srt-63 gene in isolation (12 bp of ASE motif + 6 bp of flanking sequence on either side), we observed gfp expression in ASE, in analogy to what we observed with the isolated ASE motif from the gcy-5 gene (Figs. 7C, 4G). We conclude that the ASE motif alone is, in principle, sufficient to determine ASE expression, but that the ASE motif is active only in specific genomic contexts.
Discussion
The molecular composition of a single gustatory neuron class
Our analysis provides an unprecedented view into the molecular composition of a taste receptor cell. Five-thousand-seven-hundred-sixteen protein-coding genes (i.e., more than one-quarter of all protein-coding genes in the C. elegans genome) are represented by multiple tags in the ASE SAGE library. This number is in concordance with the number of genes expressed in other single cell types as determined by SAGE or microarray analysis in both worms and vertebrates (e.g., see Inoue et al. 1999; McGhee et al. 2006). As expected, the transcriptome of the ASE neuron is composed of generic genes involved in cellular viability and structure, pan-neuronal features, sensory neuron-specific features, such as genes that instruct the ciliated morphology of ASE (and other ciliated neurons), and genes that define neuron-type-specific properties (e.g., chemoreceptors, neurotransmitter receptors, and ion channels). Very few genes of the latter category are absolutely exclusive to ASE. It is rather a unique combination of only relatively restrictively expressed genes that defines the molecular signature of ASE. This notion is well in accordance with the study of the molecular signature of other individual neuron types in C. elegans (Zhang et al. 2002; Colosimo et al. 2004; Wenick and Hobert 2004; Cinar et al. 2005; Fox et al. 2005).
Several features of the cell-type-specific molecular signature of ASE are of particular note. First, we note the presence of several distinct classes of putative chemoreceptors, including 7TMR-type putative chemoreceptors, TRP channels, and degenerins. Together with the previously reported expression of putative chemoreceptors of the receptor guanyl cyclase class in ASE (Yu et al. 1997; Ortiz et al. 2006), these proteins form a diverse array of putative chemoreceptors, which may endow the ASE neuron with its reported responsiveness to a diverse set of chemosensory cues, including salts, amino acids, and small metabolites (Bargmann and Horvitz 1991; Sambongi et al. 1999).
Second, ASE apparently expresses a substantial number of putative metabotropic and ionotropic neurotransmitter receptors of various types. These receptor systems may provide the molecular correlate for the diverse synaptic inputs that the ASE neurons receive from several distinct neuron types, according to the electron micrographic reconstruction of the C. elegans nervous system (approximately six sensory neurons and approximately four interneurons) (White et al. 1986). The interneuronal synaptic partners may provide feedback control, as has been recently shown explicitly for the AIA interneurons, which release an insulin ligand after activation by ASE, to affect the association of a gustatory stimulus with a starvation stimulus (Tomioka et al. 2006). Presynaptic input from other sensory neurons to ASE, as deduced by synaptic connectivity and possibly mediated by the neurotransmitter receptors described here, may enable distinct sensory modalities to tune the responsiveness of the worm to other sensory modalities.
The ASE neuron class not only appears to perceive a variety of distinct molecular inputs via a substantial array of receptor proteins, but it may also be able to transmit a variety of discrete signals via a diverse set of neuropeptides and neurohormones, including several FMRFamides, NLPs, and insulin-related peptides. Since ASE does not use a panel of classic neurotransmitters (GABA, acetylcholine, serotonin, dopamine, octopamine, glutamate) (Rand and Nonet 1997), these ASE-expressed neuropeptides are prime candidates to be the primary neurotransmitters of ASE. The multitude of putative neurotransmitters may indicate that activation of ASE by distinct means (e.g., activation by presynaptic neuron or by distinct gustatory cues) may be conveyed by the use of distinct signaling systems. Neurotransmitters used in the gustatory system of other organisms are not known, and it will be interesting to see whether those systems also largely rely on nonclassical neurotransmitters.
A notable number of calcium and potassium channels are expressed in the ASE neurons, suggesting that they may be capable of finely tuning their electrical properties, a feature of C. elegans neurons previously proposed (Salkoff et al. 2001). The potential ability of ASE to modulate its electrochemical membrane properties in a variety of distinct ways may possibly relate to the potential ability of ASE to independently process multiple distinct types of sensory cues.
Besides its repertoire of receptors and signaling molecules, the identity of a neuron is defined by the combinatorial expression of regulatory factors that trigger the expression of neuron-type-specific gene batteries. Our analysis reveals a striking richness in the repertoire of transcriptional regulators in ASE. Together with previously identified TFs, the comparative SAGE analysis reveals the expression of a total of >60 TFs that are enriched in ASE versus AFD.
An additional level of regulatory complexity in ASE can be inferred from the existence of several NATs in the ASE transcriptome. NATs can be found for up to almost 30% of genes in animal genomes (Lapidot and Pilpel 2006), and we find >100 in the ASE transcriptome, one-quarter of which are enriched in ASE versus AFD. These NATs match a variety of functionally distinct genes. Many of these genes are very closely positioned in a tail-to-tail orientation to neighboring genes, thereby suggesting that NATs derive from partial overlaps of two coding transcripts. It is also possible, however, that tags of NATs derive from overlapping noncoding antisense transcripts. To distinguish between these possibilities, ASE-enriched NATs have to be more precisely mapped. The sense counterparts of about half of the ASE-enriched NATs are coexpressed in ASE (“correlated expression”). Double-stranded sense–antisense pairs are not necessarily degraded by the RNA interference (RNAi) machinery (Lu et al. 2005) but have rather been proposed to be involved in other regulatory phenomena such as the masking of cis-regulatory sequences on the sense mRNA (Lapidot and Pilpel 2006; Munroe and Zhu 2006). In contrast, the anti-correlated expression of the other half of ASE-enriched NATs with their matching sense counterparts may indicate that the NATs negatively regulate their sense counterparts by some sort of transcriptional interference (Lapidot and Pilpel 2006; Munroe and Zhu 2006).
In summary, our analysis has revealed several striking features of the molecular signature of the ASE gustatory neurons. It remains to be seen whether some of these features, such as the abundance of TFs, neurotransmitter signaling systems, and ion channels, reveal a complexity in gustatory neuron makeup that is common to gustatory neurons across phylogeny. In any case, the newly identified ASE gene battery provides a plethora of entry points into a detailed, molecular understanding of ASE development and function.
Insights into the regulatory architecture of ASE cell fate specification
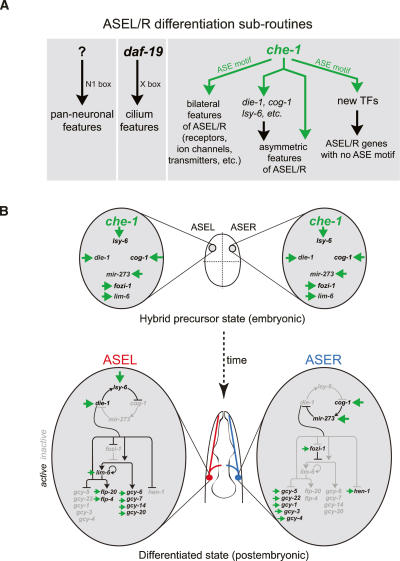
ASE differentiation appears to depend on several genetically separable regulatory programs (Fig. 8A). ASE expresses pan-neuronal features as well as ciliated features independent of the che-1 Zn finger TF (Uchida et al. 2003). Pan-neuronal features of ASE may be controlled, at least in part, by a previously described cis-regulatory motif found in many pan-neuronally expressed genes, activated by an as-yet-unknown trans-acting factor (Ruvinsky et al. 2006). Ciliated features of ASE may be controlled by daf-19, an RFX-box TF controlling generic features of all ciliated neurons through an X-box motif (Swoboda et al. 2000). The cell-type-specific signature of ASE that distinguishes it from other sensory neurons appears to be under complete control of che-1. In contrast to pan-neuronal and pan-ciliated features, all tested genes expressed in both ASE and other limited cell types fail to be expressed specifically in the ASE neurons of che-1 mutants. Moreover, ASE is completely dysfunctional in che-1 mutants. che-1 is not only absolutely required for the ASE-specific differentiation program, but ectopic misexpression studies have shown that it also appears sufficient to trigger ASE-specific features (Uchida et al. 2003). The specificity of che-1 function is brought about by the ASE-exclusive expression of che-1 (B. Tursun and O. Hobert, unpubl.). Upstream regulators of che-1 are currently unknown.
Figure 8.
CHE-1 in the context of ASE development and gene regulatory networks. All interactions involving the ASE motif and CHE-1 are shown in green. (A) Neuronal gene expression programs are defined by parallel gene regulatory pathways that determine general and cell-type-specific features and can be classified as “differentiation subroutines.” The N1 motif is a functionally relevant motif found in many pan-neuronal genes (Ruvinsky et al. 2006). We described here that che-1 regulates three aspects of ASE neuron differentiation: It regulates scores of bilaterally symmetric, terminal differentiation markers, such as ion channels, chemoreceptors, neurotransmitters, etc.; it triggers the expression of components of a bistable feedback loop that eventually controls left/right asymmetric terminal differentiation features of the ASE neurons, such as the gcy genes (in whose regulation che-1 also directly participates; see B); and it controls the expression of newly identified TFs that control as-yet-unknown aspects of ASE development and/or function. (B) che-1 appears to be directly responsible for inducing the previously described hybrid precursor state in the embryo, characterized by the bilateral expression of genes that later in embryogenesis and larval development become restricted to either ASEL or ASER. This restriction is mediated by a bistable feedback loop composed of TFs and miRNAs, each of which contain a che-1-responsive ASE motif. The left/right differential activity of the bistable feedback loop appears to be programmed into the system by as-yet-unknown means at an early embryonic stage (Poole and Hobert 2006).
Members of the che-1-controlled ASE-specific gene battery, revealed through our SAGE analysis, fall into several classes (Fig. 8A). About half of the genes contain an ASE motif and are likely to be under direct control of CHE-1, and the other half do not contain a high-affinity CHE-1-binding site and, therefore, may be regulated by che-1 in an indirect manner. This indirect regulation may be conferred by one of the several TFs that are directly regulated by che-1 via ASE motifs in their promoters. An ongoing functional analysis of these TFs reveals that several of them may, indeed, be involved in regulating specific subaspects of ASE function, such as the responsiveness to some but not all chemosensory cues sensed by ASE (J.F. Etchberger and O. Hobert, unpubl.).
CHE-1 controls the expression of terminally differentiated features of ASE such as neuropeptide genes (flp genes) and putative chemoreceptors (gcy genes) directly via ASE motifs, yet CHE-1 also triggers downstream regulatory events via ASE motif-containing TFs. Other TFs that act like che-1 as selector genes of specific cell or tissue types also both trigger transcriptional regulatory cascades and control terminal differentiation genes directly. For example, the Eyeless/Pax-6 gene controls a complex transcriptional regulatory network that induces eye development (Chen et al. 1997), but also directly binds to and activates the promoters of terminal eye differentiation genes, such as rhodopsins (Sheng et al. 1997). Other examples include the elt-2 GATA factor (McGhee et al. 2006), the unc-86 homeobox gene (Duggan et al. 1998), and the pha-4 forkhead gene (Gaudet and Mango 2002). This regulatory architecture may be a reflection of an evolutionary co-option of a TF from one function (regulator of terminal features) into additional functions (upstream regulator of other regulatory factors) that served to further diversify cellular fates.
Since several of the che-1-regulated TFs are relatively broadly expressed, it is possible that they fulfill functions that are common to many neuron types and that che-1 serves to “plug” these functions into ASE. The regulation of relatively broadly expressed regulatory factors by highly cell-type-specific TFs is not uncommon. For example, the ttx-3 homeobox gene, which is expressed in only four head neuron classes, appears to directly regulate the expression of the pan-neuronal unc-13 gene (Wenick and Hobert 2004).
The ASE motif is distributed broadly across the genome, and we experimentally demonstrated that not all genes with high-scoring ASE motifs—that is, predicted high-affinity binding sites for CHE-1—are expressed in ASE; at least not at the levels sufficient to observe by reporter gene expression. What determines whether an ASE motif is functional or not? In principle, two scenarios can be envisioned: In one scenario, regulatory regions of genes that are expressed in ASE contain in addition to an ASE motif other discrete sequence motif(s) required for ASE expression. However, our promoter dissection approaches, which included deletions distal but also directly adjacent to the ASE motif (Fig. 3; data not shown), have not identified regions other than the ASE motif required for ASE expression. Moreover, appending the ASE motif to heterologous promoters or even the ASE motif in isolation appears sufficient to drive reporter gene expression in ASE. We therefore favor an alternative scenario to explain what determines the activity of the ASE motif. In this scenario, regulatory regions of genes with high-scoring ASE motifs that are not expressed in ASE may be antagonized by the genomic context in which the ASE motif is located. This genomic context may harbor discrete regulatory element(s) that bind to transcriptional repressor proteins that antagonize CHE-1 function. Alternatively, the genomic context may dictate the positioning of nucleosomes that may in turn control ASE motif accessibility. This would not be unprecedented, since, for example, in yeast the presence of potential binding sites for specific TFs only partially correlates with in vivo occupancy of the site by its cognate TF, and this effect has been ascribed to nucleosome positioning (Lieb et al. 2001; Yuan et al. 2005). In vivo nucleosome positions are partly determined by nucleosome positioning signals encoded in the genomic sequence (Ioshikhes et al. 2006; Segal et al. 2006), but these are difficult to predict at present in C. elegans. Further dissections of regulatory regions of genes with inactive ASE motifs will help to clarify the nature of the contextual information that determines the functionality of the ASE motif.
We have previously described a simple cis-regulatory element, termed the AIY motif, that controls expression of most if not all cell-type-specific characteristics of the AIY interneuron class (Wenick and Hobert 2004). The ASE and AIY motifs are similar in several regards. Both motifs appear to be able to act in isolation, but in both cases, the activity of the motif is lost in some genomic contexts. One difference between the motifs is that the AIY motif is larger (16 bp) than the ASE motif (12 bp) and synergistically activated by two homeodomain proteins, while the ASE motif is smaller, being activated apparently only by a single TF. Owing to its smaller size, the ASE motif occurs more frequently in genomes, and to provide specificity, genomic context of whatever type may have a greater impact on ASE motif function. Other neuronal differentiation events, such as touch neuron differentiation (Zhang et al. 2002) or aspects of ciliated neuron differentiation (Swoboda et al. 2000), also appear to be under control of simple cis-regulatory motifs. Such simplicity in cis-regulatory control mechanisms may facilitate the diversification of neuronal expression programs in the course of evolution (Wenick and Hobert 2004).
The small size of the ASE motif and its apparent context dependence complicates the identification of functional ASE motifs in different species. Conservation is easier to assess on the trans-acting level. Genome sequence searches reveal that the CHE-1 protein, in particular the DNA-contacting residues, is highly conserved from protostomes to deuterostomes (worms, flies, sea urchins). The fly che-1 ortholog, glass, has been characterized for its function in eye development (Moses et al. 1989), but it is also expressed in several other neurons whose identity has not been determined (Moses and Rubin 1991). It will be interesting to examine the expression and possible function in gustatory neurons not only of fly glass, but also of a newly identified C2H2 Zn finger protein in vertebrates, ZNF500, whose Zn fingers are phylogenetically distantly related to che-1/glass.
che-1 and left/right asymmetric diversification of ASE cell fate
Neuron class specification by CHE-1 is a prerequisite for another ensuing neuronal diversification event. The ASE neuron class consists of two subclasses, the ASE left (ASEL) and the ASE right (ASER) neurons. While sharing most differentiated features, these two subclasses can be distinguished by the selective expression of a small number of terminal differentiation markers, namely, gcy genes and neurotransmitter-encoding genes. The left/right asymmetric expression of these terminal differentiation markers is controlled by the left/right asymmetric expression of a set of gene regulatory factors, including TFs and miRNAs (Chang et al. 2003, 2004; Johnston and Hobert 2003, 2005; Johnston et al. 2005, 2006). Immediately following the birth of the ASE neurons, these factors are initially expressed in both ASEL and ASER, but, through the activity of a bistable feedback loop, become restricted to either ASEL or ASER (Fig. 8B; for review, see Hobert 2006). Notably, all the eventually left/right asymmetric gene regulatory factors, as well as the left/right asymmetrically expressed terminal differentiation genes, appear to be directly controlled by the bilaterally expressed che-1 gene (Fig. 8B). That is, all these genes are genetically downstream from che-1, they harbor ASE motifs, and their ASE motifs are required for expression in ASEL and/or ASER. The restriction of the regulatory factors that constitute the bistable feedback loop to ASEL and ASER may then induce negative regulatory factors that overcome activation of several of the che-1-activated terminal differentiation genes and may cause their repression in either ASEL or ASER. The model of che-1 being a “permissive” activator of ASEL and ASER fate, which is then subsequently modified by repressive mechanisms, may represent a paradigm for an emerging theme in other regulatory systems as well (Muhr et al. 2001; Lee et al. 2004).
Materials and methods
Strains and transgenes
The strains used in this study were N2 wild-type, che-1(ot27), ntIs1: Is[gcy-5prom∷gfp; lin-15(+)], otIs151: Is[ceh-36prom∷ DsRed2; rol-6(d)], otIs125 Is[flp-6∷gfp], otIs178 Is[flp-4∷gfp] (Chang et al. 2003; Johnston et al. 2005), and oyIs17: Is[gcy-8prom∷ gfp; lin-15(+)] (kindly provided by P. Sengupta). In addition, we received several transgenic strains expressing gfp constructs of previously described genes, whose expression in ASEL/R had not previously been determined. These strains were crossed with the otIs151 transgene to identify expression in ASE and are listed in the Supplemental Material.
Transgenic strains constructed during this study are listed in the Supplemental Material. All transgenic strains were generated in an otIs151 background in which the ASEL/R cells are labeled with the ceh-36∷DsRed2 transgene to facilitate the identification of expression of gfp-based reporter genes in ASEL/R. Due to its slow maturation, DsRed2 fluorescence becomes visible post-embryonically. We therefore focused our expression pattern analysis on post-embryonic ASE neurons. Hence, temporally restricted, earlier expression of a gfp reporter in the embryo, which is often difficult to score due to the absence of clear landmarks and is often confounded by the presence of other gfp-expressing cells, has not been examined.
SAGE analysis and bioinformatics
We used the recently established LongSAGE technique, which uses the enzyme MmeI to generate 21-bp tags (Saha et al. 2002). Cell isolation was done similarly to what has been described earlier (McGhee et al. 2006) and is detailed together with the SAGE data analysis in the Supplemental Material.
DNA constructs
A list of all constructs, as well as primers used to generate the constructs, can be found in the Supplemental Material and in Supplementary Table 9. In brief, gfp-based reporter constructs were all made using the pPD95.75 vector as a backbone. Most constructs were generated by subcloning into the multiple cloning site of pPD95.75. Some constructs were created by PCR fusion (Hobert 2002). Mutagenesis reactions were performed using the QuickChangeII XL Site-Directed Mutagenesis Kit (Stratagene). Some gfp reporter constructs were kindly provided by members of the worm community; these include ceh-38∷gfp (Cassata et al. 1998), hlh-10∷gfp (Portman and Emmons 2004), fkh-8∷gfp (Hope et al. 2003), dsc-1∷gfp (Branicky and Hekimi 2005), and nhr-50∷gfp (Miyabayashi et al. 1999). All reporter constructs were injected or crossed into otIs151 to allow for easy identification of ASEL/R. Gut-specific elt-2∷gfp was commonly used as an injection marker, which we needed to use since otIs151 contains rol-6(d) on the array. The gpa-10∷che-1 misexpression construct was previously described (Uchida et al. 2003).
EMSA
Wild-type and point-mutated che-1 cDNAs were cloned into the bacterial expression vector pET14b and expressed in BL21-DE3 pLysS bacteria to produce N-terminal 6xHis-tagged CHE-1 protein. Protein was purified using Ni-Agarose beads, and EMSA was performed essentially as previously described (Wenick and Hobert 2004). A detailed description in provided in the Supplemental Material.
Acknowledgments
We thank Q. Chen for expert injection assistance; D. Baillie, G. Cassata, S. Hekimi, I. Hope, M. Koelle, M. Koga, C. Li, D. Miller, D. Portman, J. Powell-Coffman, L. Salkoff, and S. Shaham for DNA constructs; L. Schroeder for generating several of the DEG and TRP gfp fusion constructs; A. Zhang for help with the flp-6 promoter analysis; and P. Sengupta, I. Greenwald, R. Mann, and Hobert laboratory members for discussion and comments on the manuscript. This work was funded by NIH R01 NS050266-01 and 2R01NS039996-05 to O.H. This research was also supported by a grant from Genome Canada and Genome British Columbia to D.G.M., S.J.J., and M.A.M. M.A.M. and S.J.J. are scholars of the Michael Smith Foundation for Health Research. M.C.S. is a trainee of the Michael Smith Foundation for Health Research. O.H. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1560107
References
- Bargmann C.I., Horvitz H.R., Horvitz H.R. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- Benos P.V., Lapedes A.S., Stormo G.D., Lapedes A.S., Stormo G.D., Stormo G.D. Probabilistic code for DNA recognition by proteins of the EGR family. J. Mol. Biol. 2002;323:701–727. doi: 10.1016/s0022-2836(02)00917-8. [DOI] [PubMed] [Google Scholar]
- Blackshaw S., Fraioli R.E., Furukawa T., Cepko C.L., Fraioli R.E., Furukawa T., Cepko C.L., Furukawa T., Cepko C.L., Cepko C.L. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- Blacque O.E., Perens E.A., Boroevich K.A., Inglis P.N., Li C., Warner A., Khattra J., Holt R.A., Ou G., Mah A.K., Perens E.A., Boroevich K.A., Inglis P.N., Li C., Warner A., Khattra J., Holt R.A., Ou G., Mah A.K., Boroevich K.A., Inglis P.N., Li C., Warner A., Khattra J., Holt R.A., Ou G., Mah A.K., Inglis P.N., Li C., Warner A., Khattra J., Holt R.A., Ou G., Mah A.K., Li C., Warner A., Khattra J., Holt R.A., Ou G., Mah A.K., Warner A., Khattra J., Holt R.A., Ou G., Mah A.K., Khattra J., Holt R.A., Ou G., Mah A.K., Holt R.A., Ou G., Mah A.K., Ou G., Mah A.K., Mah A.K., et al. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Branicky R., Hekimi S., Hekimi S. Specification of muscle neurotransmitter sensitivity by a Paired-like homeodomain protein in Caenorhabditis elegans. Development. 2005;132:4999–5009. doi: 10.1242/dev.02069. [DOI] [PubMed] [Google Scholar]
- Cassata G., Kagoshima H., Pretot R.F., Aspock G., Niklaus G., Burglin T.R., Kagoshima H., Pretot R.F., Aspock G., Niklaus G., Burglin T.R., Pretot R.F., Aspock G., Niklaus G., Burglin T.R., Aspock G., Niklaus G., Burglin T.R., Niklaus G., Burglin T.R., Burglin T.R. Rapid expression screening of Caenorhabditis elegans homeobox open reading frames using a two-step polymerase chain reaction promoter–gfp reporter construction technique. Gene. 1998;212:127–135. doi: 10.1016/s0378-1119(98)00137-1. [DOI] [PubMed] [Google Scholar]
- Chang S., Johnston R.J., Hobert O., Johnston R.J., Hobert O., Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes & Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston R.J., Frokjaer-Jensen C., Lockery S., Hobert O., Johnston R.J., Frokjaer-Jensen C., Lockery S., Hobert O., Frokjaer-Jensen C., Lockery S., Hobert O., Lockery S., Hobert O., Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Chen R., Amoui M., Zhang Z., Mardon G., Amoui M., Zhang Z., Mardon G., Zhang Z., Mardon G., Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Cinar H., Keles S., Jin Y., Keles S., Jin Y., Jin Y. Expression profiling of GABAergic motor neurons in Caenorhabditis elegans. Curr. Biol. 2005;15:340–346. doi: 10.1016/j.cub.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Colosimo M.E., Brown A., Mukhopadhyay S., Gabel C., Lanjuin A.E., Samuel A.D., Sengupta P., Brown A., Mukhopadhyay S., Gabel C., Lanjuin A.E., Samuel A.D., Sengupta P., Mukhopadhyay S., Gabel C., Lanjuin A.E., Samuel A.D., Sengupta P., Gabel C., Lanjuin A.E., Samuel A.D., Sengupta P., Lanjuin A.E., Samuel A.D., Sengupta P., Samuel A.D., Sengupta P., Sengupta P. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr. Biol. 2004;14:2245–2251. doi: 10.1016/j.cub.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Dambly-Chaudiere C., Jamet E., Burri M., Bopp D., Basler K., Hafen E., Dumont N., Spielmann P., Ghysen A., Noll M., Jamet E., Burri M., Bopp D., Basler K., Hafen E., Dumont N., Spielmann P., Ghysen A., Noll M., Burri M., Bopp D., Basler K., Hafen E., Dumont N., Spielmann P., Ghysen A., Noll M., Bopp D., Basler K., Hafen E., Dumont N., Spielmann P., Ghysen A., Noll M., Basler K., Hafen E., Dumont N., Spielmann P., Ghysen A., Noll M., Hafen E., Dumont N., Spielmann P., Ghysen A., Noll M., Dumont N., Spielmann P., Ghysen A., Noll M., Spielmann P., Ghysen A., Noll M., Ghysen A., Noll M., Noll M. The paired box gene pox neuro: A determinant of poly-innervated sense organs in Drosophila. Cell. 1992;69:159–172. doi: 10.1016/0092-8674(92)90127-x. [DOI] [PubMed] [Google Scholar]
- de Bruyne M., Warr C.G., Warr C.G. Molecular and cellular organization of insect chemosensory neurons. Bioessays. 2006;28:23–34. doi: 10.1002/bies.20338. [DOI] [PubMed] [Google Scholar]
- Duggan A., Ma C., Chalfie M., Ma C., Chalfie M., Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- Dusenbery D.B. Analysis of chemotaxis in the nematode Caenorhabditis elegans by countercurrent separation. J. Exp. Zool. 1974;188:41–47. doi: 10.1002/jez.1401880105. [DOI] [PubMed] [Google Scholar]
- Dusenbery D.B., Sheridan R.E., Russell R.L., Sheridan R.E., Russell R.L., Russell R.L. Chemotaxis-defective mutants of the nematode Caenorhabditis elegans. Genetics. 1975;80:297–309. doi: 10.1093/genetics/80.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans-Galea M.V., Blankman E., Myszka D.G., Bird A.J., Eide D.J., Winge D.R., Blankman E., Myszka D.G., Bird A.J., Eide D.J., Winge D.R., Myszka D.G., Bird A.J., Eide D.J., Winge D.R., Bird A.J., Eide D.J., Winge D.R., Eide D.J., Winge D.R., Winge D.R. Two of the five zinc fingers in the Zap1 transcription factor DNA binding domain dominate site-specific DNA binding. Biochemistry. 2003;42:1053–1061. doi: 10.1021/bi0263199. [DOI] [PubMed] [Google Scholar]
- Fox R.M., Von Stetina S.E., Barlow S.J., Shaffer C., Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., Von Stetina S.E., Barlow S.J., Shaffer C., Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., Barlow S.J., Shaffer C., Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., Shaffer C., Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., Olszewski K.L., Moore J.H., Dupuy D., Vidal M., Miller D.M., Moore J.H., Dupuy D., Vidal M., Miller D.M., Dupuy D., Vidal M., Miller D.M., Vidal M., Miller D.M., Miller D.M. A gene expression fingerprint of C. elegans embryonic motor neurons. BMC Genomics. 2005;6:42. doi: 10.1186/1471-2164-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J., Mango S.E., Mango S.E. Regulation of organogenesis by the Caenorhabditis elegansFoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hobert O. Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb. Symp. Quant. Biol. 2006;71:181–188. doi: 10.1101/sqb.2006.71.006. [DOI] [PubMed] [Google Scholar]
- Hobert O., Mori I., Yamashita Y., Honda H., Ohshima Y., Liu Y., Ruvkun G., Mori I., Yamashita Y., Honda H., Ohshima Y., Liu Y., Ruvkun G., Yamashita Y., Honda H., Ohshima Y., Liu Y., Ruvkun G., Honda H., Ohshima Y., Liu Y., Ruvkun G., Ohshima Y., Liu Y., Ruvkun G., Liu Y., Ruvkun G., Ruvkun G. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- Hope I.A., Mounsey A., Bauer P., Aslam S., Mounsey A., Bauer P., Aslam S., Bauer P., Aslam S., Aslam S. The forkhead gene family of Caenorhabditis elegans. Gene. 2003;304:43–55. doi: 10.1016/s0378-1119(02)01175-7. [DOI] [PubMed] [Google Scholar]
- Inoue H., Sawada M., Ryo A., Tanahashi H., Wakatsuki T., Hada A., Kondoh N., Nakagaki K., Takahashi K., Suzumura A., Sawada M., Ryo A., Tanahashi H., Wakatsuki T., Hada A., Kondoh N., Nakagaki K., Takahashi K., Suzumura A., Ryo A., Tanahashi H., Wakatsuki T., Hada A., Kondoh N., Nakagaki K., Takahashi K., Suzumura A., Tanahashi H., Wakatsuki T., Hada A., Kondoh N., Nakagaki K., Takahashi K., Suzumura A., Wakatsuki T., Hada A., Kondoh N., Nakagaki K., Takahashi K., Suzumura A., Hada A., Kondoh N., Nakagaki K., Takahashi K., Suzumura A., Kondoh N., Nakagaki K., Takahashi K., Suzumura A., Nakagaki K., Takahashi K., Suzumura A., Takahashi K., Suzumura A., Suzumura A., et al. Serial analysis of gene expression in a microglial cell line. Glia. 1999;28:265–271. doi: 10.1002/(sici)1098-1136(199912)28:3<265::aid-glia10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Ioshikhes I.P., Albert I., Zanton S.J., Pugh B.F., Albert I., Zanton S.J., Pugh B.F., Zanton S.J., Pugh B.F., Pugh B.F. Nucleosome positions predicted through comparative genomics. Nat. Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Hobert O., Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Hobert O., Hobert O. A novel C. elegans zinc finger transcription factor, lsy-2, required for the cell type-specific expression of the lsy-6 microRNA. Development. 2005;132:5451–5460. doi: 10.1242/dev.02163. [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Chang S., Etchberger J.F., Ortiz C.O., Hobert O., Chang S., Etchberger J.F., Ortiz C.O., Hobert O., Etchberger J.F., Ortiz C.O., Hobert O., Ortiz C.O., Hobert O., Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Copeland J.W., Fasnacht M., Etchberger J.F., Liu J., Honig B., Hobert O., Copeland J.W., Fasnacht M., Etchberger J.F., Liu J., Honig B., Hobert O., Fasnacht M., Etchberger J.F., Liu J., Honig B., Hobert O., Etchberger J.F., Liu J., Honig B., Hobert O., Liu J., Honig B., Hobert O., Honig B., Hobert O., Hobert O. An unusual Zn-finger/FH2 domain protein controls a left/right asymmetric neuronal fate decision in C. elegans. Development. 2006;133:3317–3328. doi: 10.1242/dev.02494. [DOI] [PubMed] [Google Scholar]
- Kodama E., Kuhara A., Mohri-Shiomi A., Kimura K.D., Okumura M., Tomioka M., Iino Y., Mori I., Kuhara A., Mohri-Shiomi A., Kimura K.D., Okumura M., Tomioka M., Iino Y., Mori I., Mohri-Shiomi A., Kimura K.D., Okumura M., Tomioka M., Iino Y., Mori I., Kimura K.D., Okumura M., Tomioka M., Iino Y., Mori I., Okumura M., Tomioka M., Iino Y., Mori I., Tomioka M., Iino Y., Mori I., Iino Y., Mori I., Mori I. Insulin-like signaling and the neural circuit for integrative behavior in C. elegans. Genes & Dev. 2006;20:2955–2960. doi: 10.1101/gad.1479906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M., Pilpel Y., Pilpel Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Jurata L.W., Funahashi J., Ruiz E.C., Pfaff S.L., Jurata L.W., Funahashi J., Ruiz E.C., Pfaff S.L., Funahashi J., Ruiz E.C., Pfaff S.L., Ruiz E.C., Pfaff S.L., Pfaff S.L. Analysis of embryonic motoneuron gene regulation: Derepression of general activators function in concert with enhancer factors. Development. 2004;131:3295–3306. doi: 10.1242/dev.01179. [DOI] [PubMed] [Google Scholar]
- Lieb J.D., Liu X., Botstein D., Brown P.O., Liu X., Botstein D., Brown P.O., Botstein D., Brown P.O., Brown P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Lu C., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Haudenschild C.D., Meyers B.C., Green P.J., Meyers B.C., Green P.J., Green P.J. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Mann R.S., Carroll S.B., Carroll S.B. Molecular mechanisms of selector gene function and evolution. Curr. Opin. Genet. Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- McGhee J.D., Sleumer M.C., Bilenky M., Wong K., McKay S.J., Goszczynski B., Tian H., Krich N.D., Khattra J., Holt R.A., Sleumer M.C., Bilenky M., Wong K., McKay S.J., Goszczynski B., Tian H., Krich N.D., Khattra J., Holt R.A., Bilenky M., Wong K., McKay S.J., Goszczynski B., Tian H., Krich N.D., Khattra J., Holt R.A., Wong K., McKay S.J., Goszczynski B., Tian H., Krich N.D., Khattra J., Holt R.A., McKay S.J., Goszczynski B., Tian H., Krich N.D., Khattra J., Holt R.A., Goszczynski B., Tian H., Krich N.D., Khattra J., Holt R.A., Tian H., Krich N.D., Khattra J., Holt R.A., Krich N.D., Khattra J., Holt R.A., Khattra J., Holt R.A., Holt R.A., et al. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 2006;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- Miura H., Kusakabe Y., Kato H., Miura-Ohnuma J., Tagami M., Ninomiya Y., Hino A., Kusakabe Y., Kato H., Miura-Ohnuma J., Tagami M., Ninomiya Y., Hino A., Kato H., Miura-Ohnuma J., Tagami M., Ninomiya Y., Hino A., Miura-Ohnuma J., Tagami M., Ninomiya Y., Hino A., Tagami M., Ninomiya Y., Hino A., Ninomiya Y., Hino A., Hino A. Co-expression pattern of Shh with Prox1 and that of Nkx2.2 with Mash1 in mouse taste bud. Brain Res. Gene Expr. Patterns. 2003;3:427–430. doi: 10.1016/s1567-133x(03)00081-4. [DOI] [PubMed] [Google Scholar]
- Miyabayashi T., Palfreyman M.T., Sluder A.E., Slack F., Sengupta P., Palfreyman M.T., Sluder A.E., Slack F., Sengupta P., Sluder A.E., Slack F., Sengupta P., Slack F., Sengupta P., Sengupta P. Expression and function of members of a divergent nuclear receptor family in Caenorhabditis elegans. Dev. Biol. 1999;215:314–331. doi: 10.1006/dbio.1999.9470. [DOI] [PubMed] [Google Scholar]
- Moses K., Rubin G.M., Rubin G.M. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes & Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- Moses K., Ellis M.C., Rubin G.M., Ellis M.C., Rubin G.M., Rubin G.M. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- Muhr J., Andersson E., Persson M., Jessell T.M., Ericson J., Andersson E., Persson M., Jessell T.M., Ericson J., Persson M., Jessell T.M., Ericson J., Jessell T.M., Ericson J., Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Munroe S.H., Zhu J., Zhu J. Overlapping transcripts, double-stranded RNA and antisense regulation: A genomic perspective. Cell. Mol. Life Sci. 2006;63:2102–2118. doi: 10.1007/s00018-006-6070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt R.G. Taste buds: Development and evolution. Brain Behav. Evol. 2004;64:198–206. doi: 10.1159/000079747. [DOI] [PubMed] [Google Scholar]
- Ortiz C.O., Etchberger J.F., Posy S.L., Frokjaer-Jensen C., Lockery S., Honig B., Hobert O., Etchberger J.F., Posy S.L., Frokjaer-Jensen C., Lockery S., Honig B., Hobert O., Posy S.L., Frokjaer-Jensen C., Lockery S., Honig B., Hobert O., Frokjaer-Jensen C., Lockery S., Honig B., Hobert O., Lockery S., Honig B., Hobert O., Honig B., Hobert O., Hobert O. Searching for neuronal left/right asymmetry: Genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics. 2006;173:131–149. doi: 10.1534/genetics.106.055749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich N.P., Pabo C.O., Pabo C.O. Zinc finger–DNA recognition: Crystal structure of a Zif268–DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura J.T., Faumont S., Gaston M.R., Pearson B.J., Lockery S.R., Faumont S., Gaston M.R., Pearson B.J., Lockery S.R., Gaston M.R., Pearson B.J., Lockery S.R., Pearson B.J., Lockery S.R., Lockery S.R. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature. 2001;410:694–698. doi: 10.1038/35070575. [DOI] [PubMed] [Google Scholar]
- Poole R.J., Hobert O., Hobert O. Early embryonic programming of neuronal left/right asymmetry in C. elegans. Curr. Biol. 2006;16:2279–2292. doi: 10.1016/j.cub.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Portman D.S., Emmons S.W., Emmons S.W. Identification of C. elegans sensory ray genes using whole-genome expression profiling. Dev. Biol. 2004;270:499–512. doi: 10.1016/j.ydbio.2004.02.020. [DOI] [PubMed] [Google Scholar]
- Rand J.B., Nonet M.L., Nonet M.L. Neurotransmitter assignments for specific neurons. In: Riddle D.L., et al., editors. Celegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 1049–1052. [Google Scholar]
- Ruvinsky I., Ohler U., Burge C.B., Ruvkun G., Ohler U., Burge C.B., Ruvkun G., Burge C.B., Ruvkun G., Ruvkun G. Detection of broadly expressed neuronal genes in C. elegans. Dev Biol. 2006;302:617–626. doi: 10.1016/j.ydbio.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Saha S., Sparks A.B., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Sparks A.B., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Vogelstein B., Kinzler K.W., Velculescu V.E., Kinzler K.W., Velculescu V.E., Velculescu V.E. Using the transcriptome to annotate the genome. Nat. Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- Salkoff L., Butler A., Fawcett G., Kunkel M., McArdle C., Paz-y-Mino G., Nonet M., Walton N., Wang Z.W., Yuan A., Butler A., Fawcett G., Kunkel M., McArdle C., Paz-y-Mino G., Nonet M., Walton N., Wang Z.W., Yuan A., Fawcett G., Kunkel M., McArdle C., Paz-y-Mino G., Nonet M., Walton N., Wang Z.W., Yuan A., Kunkel M., McArdle C., Paz-y-Mino G., Nonet M., Walton N., Wang Z.W., Yuan A., McArdle C., Paz-y-Mino G., Nonet M., Walton N., Wang Z.W., Yuan A., Paz-y-Mino G., Nonet M., Walton N., Wang Z.W., Yuan A., Nonet M., Walton N., Wang Z.W., Yuan A., Walton N., Wang Z.W., Yuan A., Wang Z.W., Yuan A., Yuan A., et al. Evolution tunes the excitability of individual neurons. Neuroscience. 2001;103:853–859. doi: 10.1016/s0306-4522(01)00079-3. [DOI] [PubMed] [Google Scholar]
- Sambongi Y., Nagae T., Liu Y., Yoshimizu T., Takeda K., Wada Y., Futai M., Nagae T., Liu Y., Yoshimizu T., Takeda K., Wada Y., Futai M., Liu Y., Yoshimizu T., Takeda K., Wada Y., Futai M., Yoshimizu T., Takeda K., Wada Y., Futai M., Takeda K., Wada Y., Futai M., Wada Y., Futai M., Futai M. Sensing of cadmium and copper ions by externally exposed ADL, ASE, and ASH neurons elicits avoidance response in Caenorhabditis elegans. Neuroreport. 1999;10:753–757. doi: 10.1097/00001756-199903170-00017. [DOI] [PubMed] [Google Scholar]
- Satterlee J.S., Sasakura H., Kuhara A., Berkeley M., Mori I., Sengupta P., Sasakura H., Kuhara A., Berkeley M., Mori I., Sengupta P., Kuhara A., Berkeley M., Mori I., Sengupta P., Berkeley M., Mori I., Sengupta P., Mori I., Sengupta P., Sengupta P. Specification of thermosensory neuron fate in C. elegans requires ttx-1, a homolog of otd/Otx. Neuron. 2001;31:943–956. doi: 10.1016/s0896-6273(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Scott K. Taste recognition: Food for thought. Neuron. 2005;48:455–464. doi: 10.1016/j.neuron.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J., Chen L., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J., Field Y., Moore I.K., Wang J.P., Widom J., Moore I.K., Wang J.P., Widom J., Wang J.P., Widom J., Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G., Thouvenot E., Schmucker D., Wilson D.S., Desplan C., Thouvenot E., Schmucker D., Wilson D.S., Desplan C., Schmucker D., Wilson D.S., Desplan C., Wilson D.S., Desplan C., Desplan C. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: Evidence for a conserved function in photoreceptors. Genes & Dev. 1997;11:1122–1131. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- Swoboda P., Adler H.T., Thomas J.H., Adler H.T., Thomas J.H., Thomas J.H. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Tomioka M., Adachi T., Suzuki H., Kunitomo H., Schafer W.R., Iino Y., Adachi T., Suzuki H., Kunitomo H., Schafer W.R., Iino Y., Suzuki H., Kunitomo H., Schafer W.R., Iino Y., Kunitomo H., Schafer W.R., Iino Y., Schafer W.R., Iino Y., Iino Y. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 2006;51:613–625. doi: 10.1016/j.neuron.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Troemel E.R., Chou J.H., Dwyer N.D., Colbert H.A., Bargmann C.I., Chou J.H., Dwyer N.D., Colbert H.A., Bargmann C.I., Dwyer N.D., Colbert H.A., Bargmann C.I., Colbert H.A., Bargmann C.I., Bargmann C.I. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Uchida O., Nakano H., Koga M., Ohshima Y., Nakano H., Koga M., Ohshima Y., Koga M., Ohshima Y., Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–1224. doi: 10.1242/dev.00341. [DOI] [PubMed] [Google Scholar]
- Ward S. Chemotaxis by the nematode Caenorhabditis elegans: Identification of attractants and analysis of the response by use of mutants. Proc. Natl. Acad. Sci. 1973;70:817–821. doi: 10.1073/pnas.70.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Thomson N., White J.G., Brenner S., Thomson N., White J.G., Brenner S., White J.G., Brenner S., Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J. Comp. Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Way J.C., Chalfie M., Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes & Dev. 1989;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- Wenick A.S., Hobert O., Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- White J.G., Southgate E., Thomson J.N., Brenner S., Southgate E., Thomson J.N., Brenner S., Thomson J.N., Brenner S., Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Yan H., Canon J., Banerjee U., Canon J., Banerjee U., Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev. Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Yu S., Avery L., Baude E., Garbers D.L., Avery L., Baude E., Garbers D.L., Baude E., Garbers D.L., Garbers D.L. Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc. Natl. Acad. Sci. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J., Wu L.F., Altschuler S.J., Rando O.J., Altschuler S.J., Rando O.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ma C., Delohery T., Nasipak B., Foat B.C., Bounoutas A., Bussemaker H.J., Kim S.K., Chalfie M., Ma C., Delohery T., Nasipak B., Foat B.C., Bounoutas A., Bussemaker H.J., Kim S.K., Chalfie M., Delohery T., Nasipak B., Foat B.C., Bounoutas A., Bussemaker H.J., Kim S.K., Chalfie M., Nasipak B., Foat B.C., Bounoutas A., Bussemaker H.J., Kim S.K., Chalfie M., Foat B.C., Bounoutas A., Bussemaker H.J., Kim S.K., Chalfie M., Bounoutas A., Bussemaker H.J., Kim S.K., Chalfie M., Bussemaker H.J., Kim S.K., Chalfie M., Kim S.K., Chalfie M., Chalfie M. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]