Abstract
Over the past ten years, much progress has been made to understand the roles of the similar, yet distinct yeast SAGA and SLIK coactivator complexes involved in histone post-translational modification and gene regulation. Many different groups have elucidated functions of the SAGA complexes including identification of novel components, which have conferred additional distinct functions. Together, recent studies demonstrate unique attributes of the SAGA coactivator complexes in histone acetylation, methylation, phosphorylation, and deubiquitination. In addition to roles in transcriptional activation with the 19S proteasome regulatory particle, recent evidence also suggests functions for SAGA in elongation and mRNA export. The modular nature of SAGA allows this ∼1.8MDa complex to organize its functions and carry out multiple roles during transcription, particularly under conditions of cellular stress.
Keywords: SAGA, Acetyltransferase, Deubiquitinase, Transcription, Histone
INTRODUCTION
Access to nuclear DNA is critical for cells to accomplish all DNA-mediated events, however, the physiological nature of chromatin poses an accessibility problem with histone proteins tightly bound to the DNA all throughout the eukaryotic genome [1]. The nucleosome is the fundamental unit of chromatin and is composed of a core particle of DNA wrapped around a histone protein octamer and a linker region that joins adjacent core particles [2]. Histones are among the most highly conserved families of proteins, suggesting that the fundamental structure of chromatin has evolved from a common ancestor. The four core histone polypeptides are of low molecular weight and rich in the positively charged, basic amino acid lysine that is subject to a variety of post-translational modifications [3]. Each histone has an unstructured N-terminal ‘tail’ that ranges from about 16-44 residues in length. Protruding beyond the nucleosome core particle, the histone tails are known to be post-translationally modified at specific residues and have been shown to function in altering chromatin structure through direct interactions with elements of the nucleosome and components of nuclear machinery [4].
Besides covalent modifications to the histones, there are at least two other principal ways by which histones contribute to the dynamics of chromosome function. First, ATP-dependent chromatin remodeling complexes, such as SWI/SNF, can alter the positions of nucleosomes [5]. Second, incorporation of histone variants whose sequences are slightly different from the canonical histones can also have significant consequences when directly targeted to particular functional sites in the genome [6]. Chromatin remodeling and histone variant deposition both play essential roles in altering the structure and function of chromatin, however we are focusing this discussion on the functional implications of histone modifications coordinated by the SAGA family of complexes.
In the mid-1960's, the post-translational modifications acetylation and methylation were identified on histones, and the first evidence was presented that histones are hyper-acetylated on lysines at actively transcribed genes [7]. About a decade ago, the evolutionarily conserved transcriptional coactivator/adaptor protein Gcn5 was identified as a histone acetyltransferase (HAT) enzyme as part of the SAGA complex, directly linking histone acetylation to gene activation and establishing that histone acetylation is a targeted phenomenon [8,9]. Shortly after, a domain within Gcn5 and other nuclear proteins, called the bromodomain, was shown to directly interact with acetyllysines in the histone tails, providing the first evidence that histone modifications may function to recruit or stabilize machinery involved in DNA-mediated processes [10]. Besides targeted and functionally relevant histone acetylation, other studies have subsequently demonstrated that distinctive site-specific H3 methylation patterns and their associated complexes help maintain euchromatic and heterochromatic chromosomal domains [11]. Elucidation of sites of histone methylation has revealed that some methylation events confer transcriptional activation while others confer gene silencing, depending on the lysine residue modified [12]. Moreover, recent mass spectrometric analyses and characterization of full-length histones reveals that histone proteins are indeed modified throughout their entire sequences, in addition to the more studied histone tails [13].
One model to explain how charge-altering modifications might act to change histone-DNA or histone-protein interactions is by affecting chromatin structure through modification of the charge of a histone domain in a way that leads to regulated chromatin functions. For example, acetylation neutralizes the charge on a lysine, thereby potentially reducing interactions with the DNA phosphate backbone to make the DNA more accessible for active processes such as transcription [14]. An alternative, more complicated model accounting for recently identified functions and binding partners for multiple histone modifications has been termed the ‘histone code’. In this model, histone modifications acting at specific sites, either alone, in combination, or sequentially on one or more histones, could form a ‘histone code’ that specifies unique downstream chromatin functions [15-18]. Whether or not a literal ‘histone code’ exists is of substantial current debate in the field. There is no doubt that specific protein modules can directly bind distinct histone modifications to contribute to gene expression and DNA repair in important ways, but if a code exists, then, by definition, there must be a conversion or translation of modification inputs into a specific biological function or output. Further time and experiments are needed to distinguish between these two models, which very well may not be mutually exclusive.
Substrates and functions of the Gcn5 HAT enzyme
Mutation of multiple acetylation sites in H3 and/or H4 in budding yeast confers specific transcriptional and cell growth defects, underscoring their biological significance [19-21]. Mechanistically, histone tail acetylation was first shown to facilitate binding of transcriptional activators to nucleosomal DNA [22]. Acetylation of histones also affects higher order packing of chromatin and interactions of non-histone proteins with chromatin [23-25]. HAT enzymes transfer the acetyl group of acetyl coenzyme A (acetyl-CoA) to the ε-amino group of lysine residues within histone proteins. The removal of acetyl groups from histones by histone deacetylases (HDACs) play equally important roles to oppose HAT activities and, in general, function in transcriptional repression [26]. Several HDAC inhibitors are currently being developed and tested for their potency in cancer chemotherapy with the rationale of reactivating silenced tumor suppressor genes, underscoring the potential for epigenetics in health and disease [27].
The recombinant Gcn5 HAT enzyme displays in vitro activity on the N-terminal tails of free histones H3 and H4 [28]. The activity is specific for K14 of H3 and, to a lesser extent, K8 and K16 of H4 [28]. Mutations in GCN5 that eliminate HAT activity in vitro are defective in transcriptional activation in vivo [29-31], further indicating that histone acetylation is required for Gcn5-mediated activation events. To understand the in vivo specificity of Gcn5 and the importance of particular acetylated residues, a number of lysine mutations in H3 and H4 were constructed with and without GCN5. Simultaneous mutation of H3-K14 and H4-K8/16 to arginine, or deletion of either the H3 or H4 N-terminal tail, results in the death of gcn5 cells. Mutation of these same three sites to glutamine, mimicking acetylation, is not lethal, indicating that a critical level of histone acetylation involving Gcn5 must be maintained for cell viability, although GCN5 itself is not essential [32].
Gcn5 has also been shown to interact genetically with other HATs in yeast. Double mutants of sas3 gcn5, for example, are synthetically lethal and arrest in the G2/M phase of the cell cycle, illustrating the importance of histone acetylation both in cell-cycle progression and gene expression [33]. Also, the central subunit of the general transcription factor TFIID, Taf1, contains HAT activity that is specific for H3 and H4 in vitro [34]. Recent evidence, however, suggests that Taf1 is not a major HAT in yeast because Taf1 is not functionally redundant with other HATs, including Gcn5, Elp3, Hat1, Hpa2, Sas3, and Esa1 [35]. However, other studies show functional redundance with taf1 gcn5 double mutations, indicating some functional interaction between Gcn5-containing complexes and TFIID [36,37]. Additionally, combining an elp3 mutation, the HAT subunit of the Elongator complex, with H3 or H4 tail mutations confers lethality, and gcn5 elp3 double mutants display a number of severe phenotypes [38].
Although Gcn5 displays HAT activity on free histones and genetics implicates its function with specific tail lysine residues, the recombinant enzyme fails to acetylate the more physiological nucleosomal histone substrate in vitro [39]. This led to the discovery of native nucleosomal HAT activities that were generally found to exist as multisubunit complexes [40]. Incorporation into complexes enables several HATs to recognize and acetylate specific histone tails within the native nucleosomal substrate. The first of these native activities isolated were the Gcn5-containing ADA and SAGA complexes that can acetylate nucleosomal histones H3 and H2B in vitro [9,41,42]. A number of nucleosomal HAT activities have been isolated from yeast to humans, some of which have been shown to contain a Gcn5-related acetyltransferase (GNAT) as the catalytic subunit. In yeast, these include the SAGA, SLIK, ADA, and HAT-A2 complexes, whereas in humans they have been termed STAGA, TFTC, and PCAF [43]. Different components of these complexes are major determinants in specifying HAT substrate preference and also gene-specific targeting. Other HAT activities have subsequently been identified although here we are limiting our discussion to advances in functional elucidation of the archetypal SAGA family of HAT complexes containing Gcn5, which include both SAGA and SLIK.
General functions and organization of the SAGA family of complexes
The yeast SAGA (Spt-Ada-Gcn5-acetyltransferase) and SLIK (SAGA-like)/SALSA (SAGA altered, Spt8 absent) HAT activities are two related high molecular weight (∼1.8MDa) protein coactivator complexes that have been shown to be required for the expression of a subset of Pol II-transcribed genes as well as being functionally redundant with the general transcription factor, TFIID [9,37,44-48]. Both complexes acetylate nucleosomal H3 and H2B, and share groups of proteins that were previously known be transcriptional regulators (Figure 1): the Spt group consisting of Spt7, Spt3, and Spt20/Ada5 interact with the TATA-binding protein (TBP); the Ada group consisting of Ada1, Ada2, Ada3, Ada4/Gcn5, and Ada5/Spt20 is functionally linked to the nucleosomal HAT activity; Tra1 is an ATM/PI-3 kinase-related protein that targets DNA-bound activators for recruitment to promoters; and the TBP-associated factor (TAF) proteins consisting of Taf5, Taf6, Taf9, Taf10, and Taf12 mediate nucleosomal HAT activity and are thought to help recruit the basal transcription machinery [9,45,49-52]. Mutations that disrupt the structural integrity of both complexes, such as ada1Δ, spt20Δ, or spt7Δ, do not display severe phenotypes in yeast grown in dextrose-containing media. However, the SAGA and SLIK HAT complexes are required for growth under stressful conditions to activate transcription of stress-responsive genes, indicating that these complexes play an auxiliary role in transcriptional activation [9,44,45,48].
Figure 1.
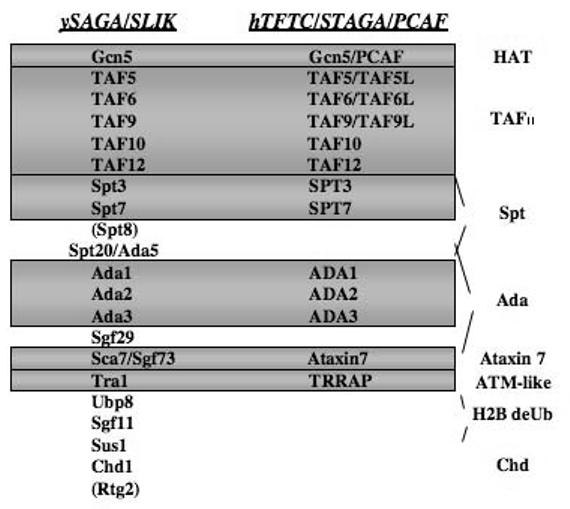
Composition of the SAGA family of HAT/coactivator complexes. Shown is a representation of the reported subunits of the SAGA and SLIK complexes and homologues in the mammalian SAGA family of complexes PCAF, STAGA and TFTC. Proteins are grouped according to their designation as histone acetyltransferase (HAT), TAF, Spt, Ada, Ataxin-7, Tra1 (ATM-like), Chd1 or proteins involved in histone H2B deubiquitination (H2B deUb). The yeast Spt8 and Rtg2 proteins are unique to the SAGA and SLIK complexes respectively. Shaded horizontal bars represent those yeast proteins with homologues identified in one or more of the mammalian complexes. Additional TFTC and STAGA associated proteins, such as SAP130 and DDB1, are not shown.
Amidst the similarities between these two complexes, SAGA and SLIK have been shown to have distinct and separate functions that are established through compositional differences. A SLIK-specific component, Rtg2, links SLIK function to the yeast retrograde response pathway that is important for gene expression changes during mitochondrial dysfunction [45]. Mutation of RTG2 cripples the integrity of the SLIK complex while leaving SAGA essentially intact [45]. Also, the Spt7 subunit is processed in SLIK, through an unknown mechanism, which dissociates the SAGA-specific Spt8 subunit. This processing of Spt7 appears to define the two complexes since a constitutive spt7 truncation allele dramatically alters complex levels in favor of SLIK by chromatography [45,46,53]. Despite their biochemically distinct activities and subunit compositions, double mutants disrupting both SAGA and SLIK function, suggest that the two complexes may have multiple redundant activities that play critical roles in transcription by Pol II [45,53].
In order to carry out their functions as coactivators and critical regulators of transcription, the SAGA and SLIK complexes generally are targeted to promoters by DNA-bound transcriptional activators [54]. The SAGA complex interacts directly with multiple activators generally through its Tra1 subunit and stimulates in vitro transcription from chromatin templates in an acetyl-CoA-dependent manner [54]. SAGA-dependent promoters require different combinations of components of the complex for distinct functions, such as TBP recruitment, revealing a complex combinatorial network for transcription activation in vivo [55] (Figure 2). Some subunits have been implicated to have different functions when targeted to different locations. For example, SPT3 and SPT8 are required to promote TBP interaction at the ADH1, PHO84, and VTC3 genes [55], but inhibit TBP interaction at the HIS3, TRP3, and HO genes [56,57], even though expression of these genes is dependent on SAGA function. The localizations of some SAGA subunits within the complex have been identified through three-dimensional modeling of the complex generated by electron microscopy [58]. These studies show a high degree of structural conservation to human TFTC and illustrate the modular nature of the complex that helps organize all of its functions.
Figure 2.
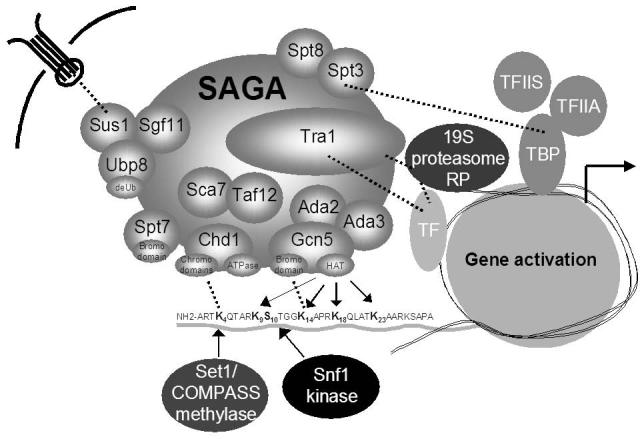
Multi-tasking on chromatin with the SAGA coactivator complexes. SAGA and SLIK subunits are organized into modules that carry out distinct functions. Shown is a representation of SAGA recruited to a promoter functioning in transcriptional activation. Tra1 associates with DNA-binding transcription factor/activator (TF) proteins and this interaction is strengthened by the 19S proteasome regulatory particle (RP). Set1-mediated H3-K4 methylation and Snf1-mediated H3-S10 phosphorylation promote the HAT activity of Gcn5. The bromo- and chromodomains within SAGA mediate additional chromatin interaction through association with modified histone lysines. The Ubp8/Sgf11/Sus1 module functions in H2B deubiquitination as well as mRNA export through interaction with pores within the nuclear membrane. Ada2 and Ada3, as well as Sca7, Taf12, and Chd1 function, in part, to regulate Gcn5-mediated HAT activity. The Spt3/Spt8 module interacts with the TATA-binding protein (TBP), and TBP and SAGA together interact strongly with the general transcription factor TFIIA and the elongation factor TFIIS. The histone H3 N-terminal tail is shown to illustrate interaction of SAGA with multiple resides, indicated by position number, within the peptide. Dotted lines indicate interactions, while arrows indicate enzymatic activities.
The Spt7 subunit of the complexes may be one of the most critical because disruption of this gene reduces the protein levels of both Spt20 and Ada1, providing evidence as to why ada1 and spt20 mutants have similar severe phenotypes to spt7 yeast [53]. Interestingly, message levels of SPT20 and ADA1 are not changed in the spt7 mutant, suggesting that at least some SAGA and SLIK subunits are mutually required for protein stability, but also potentially for other post-transcriptional events. One report, in fact, suggests that SAGA is directly involved in the unfolded protein response [59], which could also help explain the low protein levels in spt7 mutants.
Besides proteolytic processing of Spt7, which may define the two complexes, other post-translational modifications of SAGA and SLIK components also have been reported. The Spt7 subunit has been shown to be ubiquitinated in a TOM1-dependent manner; Tom1 is a HECT domain-containing E3 ubiquitin ligase that can physically associate with SAGA [60]. Set9, a human H3-K4 methyltransferase, methylates another SAGA/SLIK/TFIID subunit, Taf10, which was shown to potentiate pre-initiation complex formation and transcription at some genes [61]. Gcn5 is phosphorylated in human cells by the DNA-dependent protein kinase catalytic subunit involved in genome stability and was shown to inhibit its HAT activity [62]. There is also evidence that Taf12 is phosphorylated as well, although the biological significance is this finding is currently unknown (M. Torok, unpublished observation). A different report provides some evidence that H3 and H4 can physically associate with the SAGA complex under certain conditions [63]. Taken together, components of the SAGA complexes, as well as histones, are post-translationally modified, adding an additional level of regulation to this multi-subunit complex.
Transcriptional regulation by SAGA in a chromatin environment
Although the yeast SAGA, SLIK, and TFIID complexes regulate the expression of some distinct genes, these activities may have redundant roles in activation, although the extent of which is still controversial [35,37]. Besides their interaction with DNA-bound activators and HAT activities, a redundancy may be explained through the common Taf components of both complexes [64]. A recent study shows that only the Gcn5 and Esa1 HATs contribute substantially to gene expression genome wide, and, interestingly, histone acetylation at promoter regions throughout the genome does not require Taf1 or Pol II [35]. These results indicate that most acetylation is likely to precede transcription and not depend upon it [35]. Essential functional interactions have also been shown between SAGA and SLIK and both SWI/SNF and Mediator complexes, highlighting the redundancy between these large transcription complexes in vivo [65-67].
There is some evidence for SAGA interaction with general transcription factors other than TBP. For example, overexpression of TFIIA suppresses a gcn5 spt15 synthetic lethality, suggesting that Gcn5-mediated acetylation can stimulate transcription by promoting the formation of TBP-TFIIA-DNA complexes [57]. Also, TFIIA mutants are lethal in gcn5 mutant yeast strains [68]. Additionally, several SAGA mutants displayed a greater impact on promoter occupancy of Pol II versus TBP at Gcn4-dependent genes, suggesting the SAGA can promote Pol II binding independently of its stimulatory effects on TBP recruitment [69].
Although recruitment and functions of SAGA have been studied at a number of loci including the PHO genes [70] and those required for response to heat stress [71] and amino acid starvation [69], a number of groups studying SAGA have turned to the GAL genes in yeast as a system to study gene expression because of their galactose-induced, transcriptionally active, chromatin state that is completely dependent on SAGA [44,72,73]. During activation of the GAL genes in yeast, alleviation of Gal80 inhibition establishes a functional Gal4 activation domain that targets the Tra1 and TAF12 subunits of the SAGA complex [74-76]. Upon galactose induction, SAGA undeniably localizes to GAL1-10 gene upstream activating sequence (UAS) DNA through Gal4 and is critical for TBP recruitment and, thus, expression of the GAL genes [72,73,77]. The Mediator complex also plays an important role in GAL gene activation along with SAGA [78]. Several subunits of Mediator, including Gal11, are critical for growth on galactose and accumulating evidence suggests that Mediator is recruited subsequently to SAGA although its precise role is still unclear [75,79].
SAGA genetically and biochemically interacts with TBP through its Spt3 subunit and this interaction is critical for GAL activation [44,72]. In spt20 and spt3 mutants, the expression of numerous GAL genes is abolished [73,77], while GAL expression is only slightly impaired in gcn5 mutants [77]. These data provide further evidence that SAGA functions beyond HAT activity to activate the GAL genes [77]. Nevertheless, numerous reports have provided evidence that the H3 tail is acetylated upon galactose induction in a Gcn5-dependent manner [80-83]. Therefore, even though Gcn5 mediates histone acetylation at the GAL UAS, it is not required for activation in this case. However, earlier studies have made it clear that the H3 and H4 tails do in fact play a role in GAL gene regulation [78]. In the shift from the galactose-induced active chromatin state to one with glucose present, Gal80 mediates the deposition of nucleosomes to repress transcription, at least in part by, displacing SAGA from Gal4 [78,84].
Although there have been no direct roles for SAGA functioning in transcriptional elongation, there is some evidence for this notion, mostly from genetic studies. A large number of SAGA mutants display strong synthetic genetic interactions with components of the elongation machinery that include Rpb9, Rpb4, TFIIS, and Elongator [85-88]. Additionally, TFIIS has been shown to physically interact with SAGA in cell-free extracts [86]. However, thus far SAGA has only been shown to localize specifically to promoter regions [89,90], and so further work is needed to understand if there are any direct mechanistic functions for SAGA in elongation.
Transcription is also coupled with the concomitant assembly of RNA-binding proteins to nascent mRNAs to generate a stable and export-competent mRNP (messenger ribonucleoprotein) particle [91]. Recent data suggest a direct connection between SAGA and the mRNA export machinery through the Sus1 protein (Figure 2). Sus1 was identified as both a stable component of SAGA and the Sac3-Thp1 complex, which functions in mRNA export with specific nuclear pore proteins [92]. Also, sites of active transcription, including some SAGA-dependent genes, have been found to be located at nuclear pore complexes (NPCs) and this localization and subsequent expression is dependent on SUS1 [93-95]. Using live-cell imaging at the GAL genes, components of the SAGA complex were shown to confine sub-diffusion of transcribed genes to the nuclear envelope [94]. This may help ensure efficient mRNP export of SAGA-dependent gene expression during stressful cellular conditions. In sum, a continuum of all stages of transcription from activation to RNA export may be linked by SAGA for at least some genes, although it remains to be determined how universal this connectivity is.
Regulation of histone acetylation by SAGA
Along with conferring nucleosomal HAT activity of Gcn5, the SAGA and SLIK complexes also expand the lysine specificity of this enzyme. Within the context of the complexes, Gcn5 acetylates nucleosomal H3 at K9, 18, and 23, in addition to K14 in vitro [39]. This expansion of lysine specificity can also be reconstituted in vitro minimally with an Ada2/Ada3/Gcn5 trimer complex, biochemically demonstrating the role of these two Ada proteins in histone acetylation [52]. Evidence suggests that Ada2 potentiates the catalytic HAT activity while Ada3 facilitates nucleosomal acetylation and expands the lysine specificity of Gcn5 within the complex [52]. The SAGA and SLIK complexes preferentially acetylate K14>K18>K9=K23 in vitro on synthetic H3 peptides substrates and on nucleosomal H3 [39,45]. Importantly, these in vitro results are corroborated with genetic studies discussed above demonstrating that Gcn5 is required for H3-K9 and K18 acetylation in vivoand that mutation of H3-K14 or deletion of the H3 tail causes a synthetic growth defect or lethality, respectively, when combined with gcn5 [32]. The Taf12 and Sca7 components of SAGA and SLIK are also important for their nucleosomal HAT activities [50,96].
The Taf12 subunit of SAGA and SLIK is required for the nucleosomal HAT activities of the complexes in vitro and at a subset of SAGA-dependent promoters in vivo [M. Torok, unpublished observations and Ref. 50]. A domain outside of its histone-fold is required for this function of Taf12. Deletion of this Taf12 domain does not alter biochemical interaction with nucleosomes or recruitment of the complexes to target promoters (M. Torok, unpublished observations), suggesting that one function of Taf12 may be to present the H3 tail to Gcn5 for acetylation. Another subunit of SAGA and SLIK that regulates its HAT acitivity is Sca7 [96]. The SCA7 gene is the budding yeast homologue of human ataxin-7, which in its polyglutamine-expanded protein form causes the neurodegenerative disease, spinocerebellar ataxia type 7 (SCA7) [96-98]. Polyglutamine-expanded human ataxin-7 can be incorporated into both yeast and human SAGA complexes and inactivates its HAT function in vitro and in vivo through depletion of critical proteins that regulate the ability of SAGA to acetylate nucleosomes including Ada2 and Taf12 [96,97]. Based upon numerous histological studies reporting that expanded ataxin-7 aggregates within nuclear inclusions [99], one model for understanding the pathology of the human SCA7 disease is that the expanded glutamine tract ataxin-7 protein assembles a dysfunctional and putatively dominant-negative SAGA complex that may be recruited to specific promoters but be inactive for transcription-linked nucleosomal acetylation by Gcn5. Additional research on the ataxin-7 subunit of SAGA may provide clues for treatment of this trinucleotide repeat disease and the possibility of using HDAC inhibitors to counter loss of SAGA-dependent histone acetylation.
Acetyllysine recognition by SAGA
One way to transduce an acetylation signal is through binding to bromodomains. The bromodomain protein module recognizes specific acetyllysines and regulates protein-protein interactions in numerous cellular processes including chromatin remodeling and transcriptional activation [100]. The yeast and human Gcn5 bromodomains were found to associate specifically with an H4-K16 acetyl peptide [101-103], and it was predicted that H3-K14 acetylation would also interact strongly with this bromodomain based on histone contacts observed in the crystal structure [102].
A number of resulting studies have gone on to demonstrate that the recruitment and stabilization of bromodomain-containing complexes to promoter chromatin is crucial for efficient transcriptional activation of many genes. By using immobilized template pull-down assays, it was shown that retention of the SWI/SNF and SAGA complexes to promoter nucleosomes requires either the continued binding of activator or acetylated histones. Without the activator, this stable occupancy by SWI/SNF and SAGA on acetylated chromatin was shown to require the bromodomains of the Swi2/Snf2 and Gcn5 subunits, respectively [104-106]. These reconstituted chromatin-transcription results indicate that histone acetylation may in fact be ‘read’ as a code during the establishment of a transcriptionally competent chromatin structure. Also, these experiments and others support the model in which histone acetylation occurs prior to ATP-dependent chromatin remodeling [107-109], although other evidence indicate the opposite [110], leading us to suggest that order of recruitment of these complexes appears to be locus-dependent.
Methyllysine recognition by SAGA
Chromodomains are another chromatin interaction motif and has been shown to directly associate with methyllysine residues [111]. The chromodomain of HP1, for example, associates with H3-K9 methylation to establish and maintain heterochromatin [112]. Chd1 was recently shown to be a component of SAGA and SLIK as well as associate with H3-K4 methylation through its chromodomains [113,114]. In our studies, the second of two chromodomains (CD2) from yeast Chd1 was shown to be able to associate with di- and trimethylated H3-K4 in pull-down assays with perhaps somewhat higher affinity for a trimethylated peptide [113]. Also, the SLIK complex displayed a preference to acetylate H3-K4 methylated peptides over unmodified peptides and this preference was completely dependent on a functional Cd2 of Chd1 [113]. Our studies point to a biological significance of this H3-K4 methyl/Chd1 interaction in the context of SAGA, which is to stabilize and potentiate Gcn5-mediated histone acetylation that contributes to the establishment or maintenance of active chromatin [113] (Figure 2). Recognition of methylated H3-K4 by SAGA and SLIK may play a role in recruitment and/or stabilization of the complex onto chromatin and/or have a direct allosteric role in regulation of the HAT activity of Gcn5 [113]. Yeast Chd1 also physically associates with Spt16, Pob3, casein kinase II, and the Paf elongation complex [115,116], demonstrating that other complexes of proteins besides SAGA and SLIK may be recruited to chromatin through Chd1 and may function in processes other than transcriptional activation. The extent to which H3-K4 methyl recognition by Chd1 plays in recruitment/stabilization of SAGA, SLIK, and possibly other complexes to chromatin is still unclear.
A structure of the tandem chromodomains of a human CHD isoform have recently been solved in isolation and in complex with H3-K4 methylated tail peptides, demonstrating that the interaction with H3-K4 di- and trimethylation may be evolutionarily conserved [114,117]. The crystal structure of the human CHD1 tandem chromodomains in complex with methylated H3-K4 peptides suggests that both chromodomains are required to stabilize this interaction and that methylammonium recognition occurs through only two aromatic residues within the first chromodomain [114]. While these results argue against the biophysical interaction between the yeast chromodomain polypeptide fragment and K4-methylated H3 tail peptide, it does in no way preclude the possibility of a physiological interaction in the context of a native complex and nucleosomes. Taken together, these results suggest that the binding specificity of Chd1 may be highly context-dependent, indicative of a higher-level regulatory mechanism. An important question that still remains is what regulates Chd1 in vivo to allow for interaction with H3-K4 methylation within the context of SAGA. One model is that Chd1 harbors a structure within SAGA that confers more favorable methyllysine interaction. This model is supported by data in which the chromodomains of Chd1 are required for Chd1 to stably associate with the SLIK complex [113]. A different model, that is not mutually exclusive, is that post-translational modification of Chd1 itself affects binding specificity. Further experiments, both in vitro and more importantly in vivo, will determine the resolution of this discrepancy.
Consistent with yeast Chd1 as an effector protein for Set1-mediated H3-K4 methylation, a recent genetic study showed that disruption of CHD1 strongly suppresses growth defects of a set1Δ mutant on media lacking uracil or containing 6-azauracil [118]. Testing the specificity of this compelling set1Δ chd1Δ genetic interaction through further CHD1 and H3-K4 mutagenesis would more elegantly demonstrate a biologically relevant role for yeast Chd1 through interaction with H3-K4 methyl marks.
Regulation of SAGA by the Snf1 kinase and H3-S10 phoshphorylation
The recombinant Gcn5 protein displays a preference to acetylate H3 tail peptides that are phosphorylated at S10 over unmodified peptides, suggesting that H3 phosphorylation can also affect the efficiency of subsequent acetylation [119]. Histone H3-S10 phosphorylation functions in cell division with the Ipl1 kinase (Aurora B in mammals) and in transcriptional activation with the Snf1 kinase [120]. SNF1 is required for H3-S10 phosphorylation and recruitment of SAGA to a subset of genes to facilitate Gcn5-mediated H3-K14 acetylation [36,121,122] (Figure 2). Although GCN5 and H3-S10 genetically and biochemically interact with SNF1 [121-124], H3-S10E and S10D mutations that mimic phosphorylation severely impair global H3-K9 acetylation levels [124]. Together, data thus far suggest that Snf1-mediated H3-S10 phosphorylation directly causes site-specific changes in acetylation levels at K9 and K14 within H3 and highlight the importance of histone phosphorylation in subsequent SAGA function.
SAGA as a histone deubiquitinase
In addition to H3 and H2B acetylation, the SAGA family of complexes also modifies histones through Ubp8-mediated H2B-K123 deubiquitination [125,126] (Figure 2). Ubiquitination is a reversible process that is catalyzed by deubiquitinating enzymes that hydrolyze the isopeptide bond at glycine 76 of ubiquitin and the substrate lysine to liberate free ubiquitin and unmodified target protein as products [127]. This allows another level of regulation for ubiquitin-mediated events. Rad6/Bre1-mediated H2B-K123 ubiquitination is known to have roles in transcription, gene silencing, and mitotic and meiotic cell growth [128]. There are likely multiple reasons for removal of ubiquitin from H2B, though evidence suggests that its function is to facilitate transcription, partially through modulation of histone methylation [125,126]. Transcription of the GAL genes in yeast are only moderately impaired in the ubp8Δ mutant compared with the severe spt20Δ mutant, suggesting that other components of SAGA have redundant functions with Ubp8 in transcriptional activation [125]. Indeed, recent data show that double ubp8 gcn5 mutants display synthetic growth and transcriptional defects compared with each single mutant [126,129]. There is also evidence that prior H2B acetylation by SAGA slightly inhibits its in vitro H2B deubiquitinase activity, suggesting that these two histone modifying activities may be coupled [129]. Additionally, two novel components of SAGA and SLIK called Sgf11 and Sus1 cooperate with Ubp8 for their association within the complexes, and results suggest that the functions of Ubp8, Sgf11, and Sus1 are related and separable from those of other components of SAGA [63,87,129,130]. All together, these results indicate that a balance of cellular H2B ubiquitination regulates transcription. Moreover, they support the notion that SAGA is organized into modules that carry out specific functions [58] and that double mutants crippling two different functionalities within the complex display more severe phenotypes [131].
Yeast cells with an spt20Δ mutation also accumulate uH2B, although not to the extent observed in the ubp8Δ mutant [126], suggesting that Ubp8 functions in H2B deubiquitination primarily as a component of SAGA and SLIK but that there still exists some residual Ubp8 activity present in these SAGA-deficient cells. H2B ubiquitination functions both in transcription and gene silencing, and the mechanisms for how this occurs are beginning to be investigated. Along with Ubp8, the ubiquitin protease Ubp10, which regulates gene silencing, has also recently been shown to deubiquitinate H2B [132,133]. These reports suggest a function for H2B deubiquitination that is distinct from the role of Ubp8 in gene activation with the finding that, different from ubp10 mutants, telomeric silencing is not affected in ubp8 mutants. However, ubp10 ubp8 double mutants result in a synergistic increase in the steady-state levels of uH2B and in the number of genes with altered expression [133], indicating that Ubp10 and Ubp8 likely overlap in some of their target chromatin regions, but generally have separate functions in silencing and SAGA-mediated transcription.
One way in which Ubp8-mediated H2B deubiquitination may regulate transcription is through modulation of H3-K4 and K36 methylation. Our results suggest that Ubp8 functions to promote global H3-K4 trimethylation [125], a mark highly correlated with transcriptional capacity [134]. Furthermore, UBP8 is required for efficient H3-K4 trimethylation at the GAL1-10 UAS upon transcriptional activation [125]. A different study provides evidence that, upon galactose induction and transcription activation, Ubp8 functions to inhibit this modification while promoting H3-K36 dimethylation at the GAL1 TATA element [126]. UBP8 has also been shown to inhibit H3-K4 di- and trimethylation at the PHO84 core promoter [130]. Thus far, Ubp8 appears to differentially regulate H3 methylation at some SAGA-dependent gene promoters and UAS elements, but not others.
On a different note, the ubp8Δ mutant confers global accumulation of uH2B and displays a modest GAL phenotype; paradoxically, the H2B-K123R ubiquitination site mutant has no detectable uH2B yet displays a similar phenotype [125,126]. This is an example of two mutants with opposite histone modification profiles, yet the wild-type alleles appear to have the same apparent biological function of promoting transcription. A similar result has recently been observed for activation of the ARG1 gene as well [129], and can be explained by a number of reasons, which are not mutually exclusive. First, the two mutants may have unexpected pleiotropic effects. For example, the H2B-K123 site may be post-translationally modified by a moiety other than ubiquitin. Evidence in support of this idea stem from one mass spectrometric analysis reporting that the conserved H2B ubiquitination site, K120, is acetylated in mammals [135]. Mutation of H2B-K123 could cause an unexpected phenotype if a different modification, such as lysine acetylation or methylation, is also functionally important. A different explanation could also be that both ubiquitination and deubiquitination are important for stable transcript levels. This idea is supported by recent studies suggesting that monoubiquitinated-H2B levels increase early during transcriptional activation, then decrease coincident with significant mRNA accumulation [126,136,137]. If this is the case, then ubiquitination of H2B may act as a type of licensing mechanism to mark chromatin through a signal that modulates subsequent histone methylation patterns and helps a cell gauge recent transcriptional activities; a phenomenon some have termed ‘transcriptional memory’ [138-140]. In the absence of Ubp8, the accumulated uH2B mark could then act as a recruiting platform for ubiquitin-binding proteins or alter nucleosome structure in a way that confers a repressive chromatin function.
Also, recent reports suggest that the Rad6/Bre1 complex and uH2B are not only localized to promoter regions but also throughout coding regions, implicating potential roles in elongation [126,136]. Along the same lines, human uH2B has recently been shown to stimulate elongation by Pol II with the FACT complex using a reconstituted chromatin transcription system [128]. In yeast, the Rad6 and uH2B association with chromatin is transient and can be explained at promoter regions by Ubp8-mediated H2B deubiquitination. Perhaps Rad6 leaves the promoter region upon Ubp8-mediated deubiquitination and then moves with Pol II along coding regions. Whether Ubp8 functions downstream within coding regions or whether the uH2B/H2A dimers are displaced from the core nucleosomes by FACT in vivo is currently unclear. Increasing evidence suggests that Ubp8 and SAGA localize specifically to UAS/promoter regions [89,90], possibly favoring the histone displacement model for uH2B within coding regions [48,89]. One model then could be that Ubp8 removes ubiquitin from H2B in promoter region chromatin while FACT ejects uH2B/H2A dimers in the coding region.
SAGA interaction with the 19S proteasome regulatory particle
The recent finding that the 19S proteasome regulatory particle (RP) alters SAGA and SLIK to enhance their interactions with transcriptional activators greatly advances our knowledge of 19S RP function in histone modification at promoters [141]. The 19S RP stimulates SAGA nucleosomal HAT and DNA binding activities in the absence of activators as well, suggesting that its ATP-dependent activity also affects SAGA functions subsequent to promoter loading. Also, two ATPases within the 19S RP of the proteasome were recently shown to be recruited in RAD6- and H2B-K123-dependent manners to promoters and coding regions of active genes and promote global H3-K4 trimethylation [142]. Therefore, accumulating evidence argues very strongly for roles of SAGA in this cascade of histone modifications. There is strong evidence that the 19S RP also associates with coding regions and functions in transcription elongation by Pol II [143]. Evidence that 19S RP recruitment requires H2B ubiquitination [142] and that SAGA contains the ubiquitin protease required to remove this modification [125,126] indicates that both complexes function subsequent to H2B ubiquitination. Ubp8-mediated deubiquitination could play a role in the progression of Rad6 and/or 19S RP from promoters into coding regions, similar to a suggested function of the PAF complex for Rad6 [137]. This model is consistent with the observation that Rad6 recruitment precedes that of SAGA to the GAL1 promoter region and is independent of Gcn5 [136].
Concluding remarks
The nature of SAGA recruitment to promoters throughout the genome still remains largely elusive. Although SAGA directly interacts with some transcriptional activators through its Tra1 and Taf12 subunits, it is still unclear whether or not every SAGA-dependent gene bears a DNA-binding activator that recruits the complex. Also, there is evidence at Gcn4-dependent genes that SAGA recruitment is dependent on SPT3 as well subunits required for complex integrity [144]. These results indicate that while Tra1 can bind directly to Gcn4 in vitro, it requires other SAGA subunits for efficient recruitment in vivo [144]. Therefore, it is still possible that other chromatin interaction motifs, including the Chd1 chromodomains or bromodomains of Gcn5 or Spt7 to help recruit and retain the complex to promoters besides or along with Tra1. Genetic experiments investigating the requirement for these different chromatin-binding functions are necessary to further understand the in vivo role of these components.
Crystallographic data of yeast nucleosome core particles suggest that acetylation by the SAGA or SLIK complex may occur on a nucleosome different from the one undergoing deubiquitination. In a single nucleosome, the H3-K9/14/18/23 acetylation sites in the unstructured N-terminal tails are polar-positioned to the H2B-K123 ubiquitination site, which lies within the C-terminal helix involved in internucleosomal interactions [145]. However, because of the molecular masses of these multi-enzymatic complexes being ten fold that of the nucleosome, different components could conceivably interact with multiple nucleosomes and/or multiple sites within a single particle.
Evidence from ChIP on CHIP experiments in fly and human cells demonstrate that H3-K4 trimethylation and H3-K9/14 acetylation co-localize at transcription start sites and correlate with levels of transcription, whereas inactive genes contain hypoacetylation and hypomethylation at these sites [146-148]. Specifically, one study demonstrated that, on human chromosomes 21 and 22, H3-K4 trimethylation and H3-K9/14 acetylation sites coincide almost 100%, and 66% of those sites are near transcription starts [148]. Taking these data into consideration, recognition of H3-K4 methylation by the SAGA family of complexes may help coordinate the near binary pattern of covalent histone modifications found at transcriptionally active genes.
Acknowledgements
Research in the laboratory of PAG is funded from National Institutes of Health R01 grants DK58646 and NS049065.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 3.Luger K. Structure and dynamic behavior of nucleosomes. Curr Opin Genet Dev. 2003;13:127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 4.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 5.Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker PB, Brehm A. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. Embo J. 2002;21:2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 7.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 9.Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 10.Winston F, Allis CD. The bromodomain: a chromatin-targeting module? Nat Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 11.Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 13.Freitas MA, Sklenar AR, Parthun MR. Application of mass spectrometry to the identification and quantification of histone post-translational modifications. J Cell Biochem. 2004;92:691–700. doi: 10.1002/jcb.20106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wade PA, Pruss D, Wolffe AP. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 15.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 16.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 17.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 18.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 19.Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- 20.Smith MM. Histone structure and function. Curr Opin Cell Biol. 1991;3:429–437. doi: 10.1016/0955-0674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- 21.Roth SY. Chromatin-mediated transcriptional repression in yeast. Curr Opin Genet Dev. 1995;5:168–173. doi: 10.1016/0959-437x(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 22.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 23.Fletcher TM, Hansen JC. The nucleosomal array: structure/function relationships. Crit Rev Eukaryot Gene Expr. 1996;6:149–188. doi: 10.1615/critreveukargeneexpr.v6.i2-3.40. [DOI] [PubMed] [Google Scholar]
- 24.Edmondson DG, Roth SY. Chromatin and transcription. Faseb J. 1996;10:1173–1182. doi: 10.1096/fasebj.10.10.8751719. [DOI] [PubMed] [Google Scholar]
- 25.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 26.Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 27.Marks PA, Miller T, Richon VM. Histone deacetylases. Curr Opin Pharmacol. 2003;3:344–351. doi: 10.1016/s1471-4892(03)00084-5. [DOI] [PubMed] [Google Scholar]
- 28.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 29.Candau R, Zhou JX, Allis CD, Berger SL. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. Embo J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis CD, Berger SL. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. Embo J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, Nakatani Y, Allis CD. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 35.Durant M, Pugh BF. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:2791–2802. doi: 10.1128/MCB.26.7.2791-2802.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Oevelen CJ, van Teeffelen HA, Timmers HT. Differential requirement of SAGA subunits for Mot1p and Taf1p recruitment in gene activation. Mol Cell Biol. 2005;25:4863–4872. doi: 10.1128/MCB.25.12.4863-4872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 38.Svejstrup JQ. Chromatin elongation factors. Curr Opin Genet Dev. 2002;12:156–161. doi: 10.1016/s0959-437x(02)00281-2. [DOI] [PubMed] [Google Scholar]
- 39.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 40.Grant PA, Berger SL. Histone acetyltransferase complexes. Semin Cell Dev Biol. 1999;10:169–177. doi: 10.1006/scdb.1999.0298. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Garcia AB, Sendra R, Pamblanco M, Tordera V. Gcn5p is involved in the acetylation of histone H3 in nucleosomes. FEBS Lett. 1997;403:186–190. doi: 10.1016/s0014-5793(97)00049-5. [DOI] [PubMed] [Google Scholar]
- 42.Pollard KJ, Peterson CL. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torok MS, Grant PA. Histone acetyltransferase proteins contribute to transcriptional processes at multiple levels. Adv Protein Chem. 2004;67:181–199. doi: 10.1016/S0065-3233(04)67007-0. [DOI] [PubMed] [Google Scholar]
- 44.Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, 3rd, Grant PA. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sterner DE, Belotserkovskaya R, Berger SL. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc Natl Acad Sci U S A. 2002;99:11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 48.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 49.Winston F, Sudarsanam P. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harb Symp Quant Biol. 1998;63:553–561. doi: 10.1101/sqb.1998.63.553. [DOI] [PubMed] [Google Scholar]
- 50.Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 51.Grant PA, Schieltz D, Pray-Grant MG, Yates JR, 3rd, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- 52.Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 53.Wu PY, Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 55.Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu Y, Eriksson P, Bhoite LT, Stillman DJ. Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein. Mol Cell Biol. 2003;23:1910–1921. doi: 10.1128/MCB.23.6.1910-1921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 59.Welihinda AA, Tirasophon W, Kaufman RJ. The transcriptional co-activator ADA5 is required for HAC1 mRNA processing in vivo. J Biol Chem. 2000;275:3377–3381. doi: 10.1074/jbc.275.5.3377. [DOI] [PubMed] [Google Scholar]
- 60.Saleh A, Collart M, Martens JA, Genereaux J, Allard S, Cote J, Brandl CJ. TOM1p, a yeast hect-domain protein which mediates transcriptional regulation through the ADA/SAGA coactivator complexes. J Mol Biol. 1998;282:933–946. doi: 10.1006/jmbi.1998.2036. [DOI] [PubMed] [Google Scholar]
- 61.Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 62.Barlev NA, Poltoratsky V, Owen-Hughes T, Ying C, Liu L, Workman JL, Berger SL. Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol Cell Biol. 1998;18:1349–1358. doi: 10.1128/mcb.18.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA Export Factor Sus1 Is Involved in SAGA-mediated H2B Deubiquitinylation through Its Interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006 doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grant PA, Workman JL. Transcription. A lesson in sharing? Nature. 1998;396:410–411. doi: 10.1038/24723. [DOI] [PubMed] [Google Scholar]
- 65.Roberts SM, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sudarsanam P, Cao Y, Wu L, Laurent BC, Winston F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. Embo J. 1999;18:3101–3106. doi: 10.1093/emboj/18.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biggar SR, Crabtree GR. Continuous and widespread roles for the Swi-Snf complex in transcription. Embo J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Biswas D, Imbalzano AN, Eriksson P, Yu Y, Stillman DJ. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex. Mol Cell Biol. 2004;24:8312–8321. doi: 10.1128/MCB.24.18.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu H, Hu C, Yoon S, Natarajan K, Swanson MJ, Hinnebusch AG. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol Cell Biol. 2004;24:4104–4117. doi: 10.1128/MCB.24.10.4104-4117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbaric S, Reinke H, Horz W. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol Cell Biol. 2003;23:3468–3476. doi: 10.1128/MCB.23.10.3468-3476.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zanton SJ, Pugh BF. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc Natl Acad Sci U S A. 2004;101:16843–16848. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 75.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reeves WM, Hahn S. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol. 2005;25:9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bash R, Lohr D. Yeast chromatin structure and regulation of GAL gene expression. Prog Nucleic Acid Res Mol Biol. 2001;65:197–259. doi: 10.1016/s0079-6603(00)65006-7. [DOI] [PubMed] [Google Scholar]
- 79.Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 80.Papamichos-Chronakis M, Petrakis T, Ktistaki E, Topalidou I, Tzamarias D. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol Cell. 2002;9:1297–1305. doi: 10.1016/s1097-2765(02)00545-2. [DOI] [PubMed] [Google Scholar]
- 81.Krebs JE, Fry CJ, Samuels ML, Peterson CL. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell. 2000;102:587–598. doi: 10.1016/s0092-8674(00)00081-7. [DOI] [PubMed] [Google Scholar]
- 82.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. Embo J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shukla A, Stanojevic N, Duan Z, Shadle T, Bhaumik SR. Functional analysis of H2B-K123 ubiquitination in regulation of H3-K4 methylation and recruitment of RNA polymerase II at the coding sequences of several active genes in vivo. J Biol Chem. 2006 doi: 10.1074/jbc.M513533200. [DOI] [PubMed] [Google Scholar]
- 84.Carrozza MJ, John S, Sil AK, Hopper JE, Workman JL. Gal80 confers specificity on HAT complex interactions with activators. J Biol Chem. 2002;277:24648–24652. doi: 10.1074/jbc.M201965200. [DOI] [PubMed] [Google Scholar]
- 85.Van Mullem V, Wery M, Werner M, Vandenhaute J, Thuriaux P. The Rpb9 subunit of RNA polymerase II binds transcription factor TFIIE and interferes with the SAGA and elongator histone acetyltransferases. J Biol Chem. 2002;277:10220–10225. doi: 10.1074/jbc.M107207200. [DOI] [PubMed] [Google Scholar]
- 86.Wery M, Shematorova E, Van Driessche B, Vandenhaute J, Thuriaux P, Van Mullem V. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. Embo J. 2004;23:4232–4242. doi: 10.1038/sj.emboj.7600326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Milgrom E, West RW, Jr., Gao C, Shen WC. TFIID and Spt-Ada-Gcn5-acetyltransferase functions probed by genome-wide synthetic genetic array analysis using a Saccharomyces cerevisiae taf9-ts allele. Genetics. 2005;171:959–973. doi: 10.1534/genetics.105.046557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 91.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 92.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 93.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 94.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 95.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 96.McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, 3rd, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci U S A. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 99.Kaytor MD, Duvick LA, Skinner PJ, Koob MD, Ranum LP, Orr HT. Nuclear localization of the spinocerebellar ataxia type 7 protein, ataxin-7. Hum Mol Genet. 1999;8:1657–1664. doi: 10.1093/hmg/8.9.1657. [DOI] [PubMed] [Google Scholar]
- 100.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 101.Ornaghi P, Ballario P, Lena AM, Gonzalez A, Filetici P. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J Mol Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 102.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. Embo J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J Mol Biol. 2000;304:355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- 104.Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- 105.Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 106.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 107.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 108.Yoon S, Qiu H, Swanson MJ, Hinnebusch AG. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol Cell Biol. 2003;23:8829–8845. doi: 10.1128/MCB.23.23.8829-9945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol Cell Biol. 2006;26:4095–4110. doi: 10.1128/MCB.01849-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fry CJ, Peterson CL. Chromatin remodeling enzymes: who's on first? Curr Biol. 2001;11:R185–197. doi: 10.1016/s0960-9822(01)00090-2. [DOI] [PubMed] [Google Scholar]
- 111.Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 112.Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 113.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 114.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 115.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 116.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. Embo J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang L, Schroeder S, Fong N, Bentley DL. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. Embo J. 2005;24:2379–2390. doi: 10.1038/sj.emboj.7600711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 120.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 121.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1--a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 122.Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. Embo J. 2005;24:997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu Y, Xu X, Singh-Rodriguez S, Zhao Y, Kuo MH. Histone H3 Ser10 phosphorylation-independent function of Snf1 and Reg1 proteins rescues a gcn5-mutant in HIS3 expression. Mol Cell Biol. 2005;25:10566–10579. doi: 10.1128/MCB.25.23.10566-10579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Edmondson DG, Davie JK, Zhou J, Mirnikjoo B, Tatchell K, Dent SY. Site-specific loss of acetylation upon phosphorylation of histone H3. J Biol Chem. 2002;277:29496–29502. doi: 10.1074/jbc.M200651200. [DOI] [PubMed] [Google Scholar]
- 125.Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- 126.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 128.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 129.Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 132.Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, Berger SL. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 133.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 135.Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- 136.Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 139.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 140.Gerber M, Shilatifard A. Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem. 2003;278:26303–26306. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- 141.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 142.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 143.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol Cell. 2001;7:981–991. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 144.Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol. 2005;25:3461–3474. doi: 10.1128/MCB.25.9.3461-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. Embo J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, Jones PA. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]


