Abstract
In neurons, intracellular calcium signals have crucial roles in activating neurotransmitter release and in triggering alterations in neuronal function. Calmodulin has been widely studied as a Ca2+ sensor that has several defined roles in neuronal Ca2+ signalling, but members of the neuronal calcium sensor protein family have begun to emerge as key components in a number of regulatory pathways and have increased the diversity of neuronal Ca2+ signalling pathways. The differing properties of these proteins allow them to have discrete, non-redundant functions.
Many different aspects of neuronal function are regulated by changes in the concentration of intracellular free Ca2+. An understanding of Ca2+ signalling pathways in neurons will not only increase our understanding of basic aspects of neuronal regulation but possibly also highlight abnormalities in signalling in disease states. There are two main conundrums: how changes in the concentration of a simple ion can modify neuronal function in a multitude of ways, and how the same ion can produce distinct outcomes over short, medium or long distances and timescales in the same types of neuron. It is well known that an increase in presynaptic Ca2+ concentration following influx though voltage-gated Ca2+ channels triggers neurotransmitter release. Ca2+ signals can lead to a range of alterations in neuronal function lasting from seconds to days through effects on ion channels and neurotransmission, and can also influence gene expression, neuronal growth, neuronal development, survival and death.
The diversity of events controlled by Ca2+ must partly be a consequence of the distinct types of Ca2+ signal that differ spatially, temporally and in magnitude1. The differing outcomes will also be a consequence of the actions of a range of Ca2+ sensor proteins that transduce the Ca2+ signals into specific changes in cellular function. The specificity of the outcomes will depend on factors such as the affinity of the different sensors for Ca2+, their localization in relation to the Ca2+ signal and their interactions with other proteins 2. One such Ca2+ sensor, calmodulin, has been very widely studied as it is a ubiquitous protein, and its role in synaptic plasticity, for example, has been defined in detail. Another Ca2+-binding protein, synaptotagmin, is now well established as the Ca2+ sensor for fast neurotransmission 3.
A number of other Ca2+-binding proteins related to calmodulin are enriched in or expressed only in the nervous system where they have distinct roles in the regulation of neuronal function. These include the neuronal calcium sensor (NCS) protein family, members of which have been implicated in a very wide range of Ca2+ signalling events in neurons and photoreceptors which have been reviewed previously 4, 5. These range from very specific single functions for particular NCS proteins in the retina to more broad ranging functions in neurotransmitter release, channel and receptor regulation, control of gene transcription, neuronal growth and survival. Members of the NCS family appeared early in evolution, and there has since been a progressive increase in the size of the family. Rather than discuss all of the functions of the NCS proteins, this review will concentrate on those that illustrate how they have diversified to have distinct properties and functions. The distinct properties of the NCS proteins means that they function not just as ‘other calmodulins’ but, instead, have very discrete and non-redundant roles that, in several cases, have been confirmed by genetic manipulations.
The significance of these Ca2+ sensors might not yet be fully appreciated, nor is there a general understanding of the distinct properties of the different family members that allow them to contribute to the specificity of Ca2+ signalling. This Review will outline general aspects of neuronal Ca2+ signalling, introduce the members of the NCS family and summarize their individual properties and physiological functions. I will highlight how the evolutionary expansion of this family, with the appearance of distinct variations from an original Ca2+-binding protein prototype, has generated an increased diversity in neuronal Ca2+ signalling.
Ca2+ signals in neurons
Changes in Ca2+ concentration
Neurons maintain a resting intracellular Ca2+ concentration in the range of 40-100 nM 6. The receipt of a neurotransmitter signal from another neuron, or some other form of stimulation, can lead to changes in Ca2+ concentration ranging from highly localized (see Fig. 1) and transient Ca2+ elevations to longer lasting and global changes throughout the neuron7. Ca2+ can enter cells through plasma membrane channels such as voltage-gated Ca2+ channels or NMDA (N-methyl-D-aspartate) receptors, or be released from intracellular stores in response to messengers such as inositol-1,4,5-trisphosphate 8. The existence of sites of local Ca2+ entry into the cytoplasm (from external or internal sources) has resulted in the concept of Ca2+ nanodomains and microdomains 7, in which the effective intracellular Ca2+ concentration could transiently reach up to hundreds of micromolar near the mouths of the Ca2+ channels.
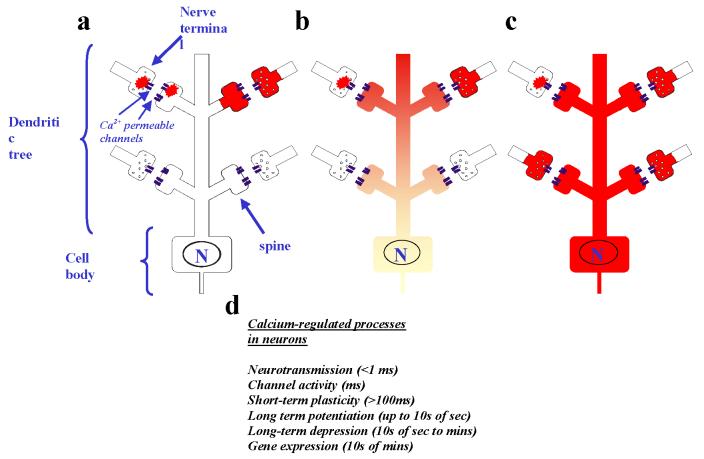
Figure 1.
Diversity of Ca2+ signals in neurons. A wide variety of different Ca2+ signals that range from very local pre- or post-presynaptic signals to an increase in Ca2+ concentration throughout the neuron can be generated in mature neurons. Three types of Ca2+ signal are shown schematically. a Local Ca2+ signals that are generated and remain in the pre-synaptic nerve terminal or the post-synaptic dendritic spine as very local Ca2+ elevations near to Ca2+ channels (nano- or micro-domains) are shown on the left of the figure; more diffuse Ca2+ elevations that fill the terminals or spines are shown on the right. b A Ca2+ signal that has propagated into part of the dendritic tree close to the site of synaptic inputs but that dissipates before reaching the cell body is shown. c A global Ca2+ signal is shown that would occur following more extensive stimulation owing to multiple active synaptic inputs along the dendrite with the generation of a propagating Ca2+ wave throughout the neuron (including the nucleus - labelled N). d Different Ca2+-regulated processes occur in neurons over a wide range of timescales and their selective activation will depend on the temporal nature of the Ca2+ signal.
In presynaptic nerve terminals, local elevations in Ca2+ levels are involved, for example, in triggering neurotransmitter release through exocytosis of synaptic vesicles less than 100 μsec after Ca2+ entry9. Post-synaptic Ca2+ entry following neurotransmission can subsequently generate localized Ca2+ elevations in dendritic spines. Ca2+ concentration can be increased in dendritic spines without changes in the adjacent dendrite due to limited diffusion of Ca2+ through the narrow necks of the spines 6 . More extensive or sustained synaptic input to a neuron can result in Ca2+ waves in the dendrites or global Ca2+ elevations throughout the neuron (Fig. 1).
Different signals, diverse outcomes
The significance of the differing types of Ca2+ signals in neurons is that they result in a wide range of different physiological outcomes. Indeed, even quite similar Ca2+ signals can have distinct or even opposite results. Examples of this are the opposing phenomena of long-term potentiation (LTP) and long-term depression (LTD), either of which can be brought about in the same neurons by transient Ca2+ signals depending on their exact timing and magnitude. LTP is triggered by larger (micromolar) and shorter-lived Ca2+ elevations (a few seconds) and LTD by a Ca2+ elevation to only a few hundred nM but for a longer duration 10 . Another example is the ability of Ca2+ signals of differing magnitude in neuronal growth cones to stimulate either attraction or repulsion 11 . In addition to these effects are long-term changes that can lead to potentially permanent changes in gene expression 12 that, for example, underlie learning and memory. Also, global Ca2+ signals can lead to neuronal cell death 13.
Ca2+ sensor proteins: calmodulin
Changes in intracellular Ca2+ concentration can be detected by Ca2+-binding proteins that act as Ca2+ buffers or that act as Ca2+ sensors. The latter are characterized by their ability to bind and release Ca2+ over the physiological range of Ca2+ concentrations, to undergo a significant conformational change on Ca2+ binding and to consequently bind and regulate specific target proteins to modify their function. In some cases they can also regulate target proteins in their Ca2+-free form. The best studied of these Ca2+ sensors is the ubiquitous Ca2+ sensor calmodulin, which contains four EF-hand Ca2+-binding motifs (BOX 1). One key pathway in which calmodulin functions involvse activation of Ca2+/calmodulin-dependent protein kinases (CaMKs), and CaMKII has an established important role in synaptic plasticity 14 . Another calmodulin target, the protein phosphatase calcineurin, has a key role in dephosphorylation, and thereby activation, of the transcription factor nuclear factor of activated T cells (NFAT) 15.
Box 1. The EF-hand motif.
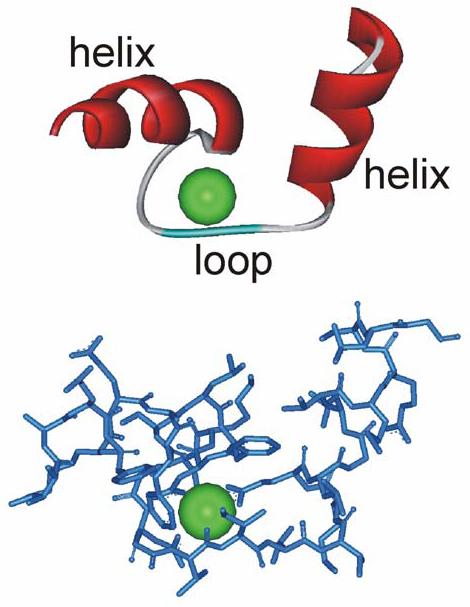
Two representations of the same view of the helix-loop-helix structure of the first EF hand of calmodulin with a bound Ca2+ ion.
The EF-hand as a Ca2+-binding motif was first characterized for the protein parvalbumin and is known to be one of the most common domains encoded by the human genome, being present in 122 predicted proteins. This motif has a helix-loop-helix structure, consisting of 29 amino acids, that binds a single Ca2+ ion. There are six residues involved in Ca2+ binding at positions 1, 3, 5, 7, 9 and 12, and these residues are denoted X, Y, Z, -Y, -X and -Z. Despite their related structure, there is considerable variability in the affinities of different EF-hands for Ca2+ and the extent to which they undergo a conformational change on Ca2+ binding. EF-hand-containing proteins can have very fast on-rates, with Ca2+ binding limited only by the rate of Ca2+ diffusion. The equilibrium dissociation constant can be variable (over a 1000-fold range), however, owing to differences in the Ca2+ off-rate between different EF-hands, which depends on the nature of the amino acid at the position 9 of the EF hand 111 . The slowest off-rates are found in proteins that act as Ca2+ buffers. Variability in off-rate is seen for the EF hands of calmodulin, in which the off-rate for the EF-hands in the N-terminal lobe is over 100 times faster than for the two in the C-terminal lobe 106 , which allows independent regulation by each lobe of calmodulin. The equilibrium dissociation constant for Ca2+-calmodulin based on in vitro Ca2+ binding is ∼10 μM but the off-rates are substantially modified by the interaction of calmodulin with its target proteins, leading to slowing of Ca2+ dissociation and an increase in the apparent affinity for Ca2+ 106, 107.
Ca2+ sensor proteins: NCS proteins
Members of the NCS protein family are among the Ca2+-binding proteins expressed in neurons (Table 1). Although these proteins possess four EF-hand motifs they have limited similarity to calmodulin (no more than 20% identity). NCS-1 was discovered originally as frequenin 16 in Drosophila melanogaster and was designated NCS-1 as it was thought to be expressed only in neuronal cell types 17 . However, this is not the case, and it is the most widely expressed of the NCS proteins 18-20 . Indeed, an orthologue of NCS-1 exists in Saccharomyces cerevisiae (60% identical to the human protein) and is essential for survival in this organism 21.
Table 1.
Summary of the members of NCS protein family represented in mammalian genomes
| Sub-group | First appearance in evolution | Mammalian protein | Expressed human splice variants | Proposed functions |
|---|---|---|---|---|
| A | Yeast | NCS-1 | 1 | Regulation of neurotransmission16, stimulation of constitutive79 and regulated exocytosis18, learning 77, short-term synaptic plasticity 128, Ca2+ channel 129, 130 and Kv4 K+ channel125, 126 regulation, phosphoinositide metabolism 21, 78, 79, 131, dopamine D2 receptor endocytosis 73, GDNF signalling 132, neuronal growth127 and survival133 |
| B | Nematodes | Hippocalcin | 1 | Anti-apoptotic134, AMPA receptor recycling in LTD135, MAPK signalling 136, 137, learning138 |
| Neurocalcin δ | 1 | Guanylyl cyclase activation 139 | ||
| VILIP-1 | 1 | Guanylyl cyclase activation and recycling 140, traffic of nicotinic receptors141, cAMP levels and secretion142 | ||
| VILIP-2 | 1 | Regulation of P/Q-type Ca2+ channels143 | ||
| VILIP-3 | 1 | Unnown | ||
| C | Fish | Recoverin | 1 | Light adaptation via inhibition of rhodopsin kinase 43 |
| D | Fish | GCAP1 | 1 | Regulation of retinal guanylyl cyclases23 |
| GCAP2 | 1 | Regulation of retinal guanylyl cyclases23 | ||
| GCAP3 | 1 | Regulation of retinal guanylyl cyclases23 | ||
| E | Insects | KChIP1 | 3 | Regulation of Kv4 61 and Kv 1.5 144 K+ channels, repression of transcription69 |
| KChIP2 | 5 | Regulation of Kv4 61 and Kv 1.5 144 K+ channels, repression of transcription69 | ||
| KChIP3 | 2 | Regulation of Kv461 K+ channels, presenilin- processing62, APP processing 63, repression of transcription 66, pro-apoptotic145, regulates ER Ca2+ 146 | ||
| KChIP4 | 6 | Regulation of Kv4 K+ channels100, presenilin- processing65, repression of transcription69 |
AMPA, a-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid; APP, amyloid precursor protein; cAMP, cyclic AMP; ER, endoplasmic reticulum; GDNF, glial-cell-derived neurotrophic factor; KChIP, Kv channel-interacting protein; LTD, long-term depression; MAPK, mitogen-activated protein kinase; NCS, neuronal Ca2+ sensor.
During evolution there has been a progressive increase in the complexity of the NCS family, such that there are now 5 classes of NCS proteins (A-E) that have been defined based on their amino acid sequences. Three NCS proteins are expressed in Caenorhabditis elegans, whereas D. melanogaster has four NCS proteins — two very similar frequenins, a neurocalcin and a single Kv channel-interacting protein (KChIP). Zebrafish have two NCS-1 orthologues 22 , up to 8 class B proteins, a recoverin, at least 8 guanylyl cyclase-activating proteins (GCAPs)23 and 5 KChIPs. Mammals have a highly conserved set of 14 NCS genes that encode only a single NCS-1, 5 class B proteins, one recoverin, three GCAPs and four KChIPs (Table 1). There are also several alternatively spliced versions of the mammalian KChIPs 24 . In all species, recoverin and GCAPs are expressed in the retina whereas the other NCS proteins are expressed to varying extents in neurons — in some cases, with very specific patterns of expression 25 . Hippocalcin, for example, is expressed at high levels in hippocampal pyramidal neurons and only moderately in certain other neuronal cell types 25, 26.
Ca2+ binding and membrane localization
A common feature of the NCS proteins is that their first EF-hand cannot bind Ca2+ owing to inactivating substitutions in the EF-hand loop in all the family In structural studies, human and yeast NCS-1 27, 28 , bovine neurocalcin δ 29 bovine GCAP2 30 and human GCAP3 31 were found to have three bound Ca2+ ions. By contrast, recoverin 32 and KChIP1 33, 34 were isolated with only two bound Ca2+ ions as a result of an inactivating substitution of residues required for Ca2+ coordination in another one of their EF hands.
The NCS proteins all have very similar structures which, in contrast to the dumb-bell shape of Ca2+-loaded calmodulin, are more compact and globular even when Ca2+ is bound (Fig.2). All mammalian NCS proteins of classes A-D, as well as KChIP1, are N-terminally myristoylated 4 . Some KChIP2 and 3 isoforms instead have putative palmitoylation sites35 . Both myristoylation and palmitoylation would be predicted to allow membrane association of these NCS proteins. In fact, as discussed below, some NCS proteins such as recoverin and the class B NCS proteins only expose their myristoyl group when Ca2+ is bound to them 32, 36, 37 . These NCS proteins are cytosolic at resting Ca2+ and reversibly associate with the plasma membrane and the Golgi complex when Ca2+is elevated. By contrast, others such NCS-1 are always associated with the plasma membrane and Golgi complex 38 and KChIP1 with distinct vesicular structures 39, 40 . The distinct localisations of these myristoylated proteins are in part determined by interactions of their N-terminal amino acids with specific phosphoinositides 39, 41.
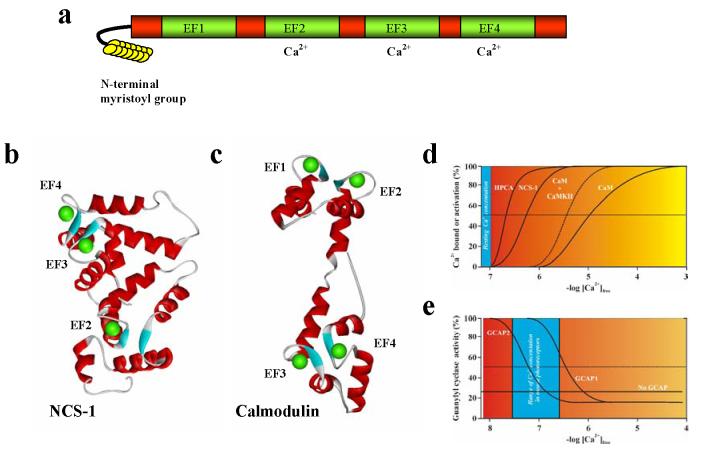
Figure 2.
The structure of NCS proteins . The structure and Ca2+-binding properties of NCS proteins, in comparison to calmodulin, are shown. a A schematic representation of the general domain structure of NCS proteins. These proteins contain 4 EF hands, the first of which cannot bind Ca2+. Many family members possess an N-terminal myristoyl group. b The crystal structure of human NCS-1 in the Ca2+-bound form (PDB ID code 1G8I) showing the presence of three bound Ca2+ ions (green spheres) in EF hands 2-4. c The crystal structure of rat calmodulin in Ca2+-loaded form (PDB ID code 3CLN) with four bound Ca2+ ions. d Comparison of the Ca2+ affinities of NCS proteins and that of calmodulin. The figures show schematic binding curves which are modelled on the Ca2+ -dependency of the Ca2+/myristoyl switch of hippocalcin in live cells 37 , the in vitro binding of Ca2+ to NCS-1 123 and for calmodulin is based on Ca2+ binding in vitro to calmodulin alone or with a bound peptide from its target CaMKII 107 . e A comparison of the Ca2+ -dependency of activation of retinal guanylyl cyclase in the presence of 1 mM Ca2+. The schematic binding curves are modelled on in vitro data 86 . The range of measured Ca2+ concentrations in mouse rod photoreceptors 85 is shaded in blue.
Structural insights from recoverin
Extensive structural information is available for recoverin, which was the first of the NCS proteins to be discovered 42 . Recoverin inhibits rhodopsin kinase when in its Ca2+-bound form 43 , and is therefore important in light adaptation in photoreceptors 44 . In its Ca2+-free form, the myristoyl group of recoverin is sequestered in a hydrophobic pocket within the protein45 . Sequential Ca2+ binding to EF hand 3 followed by EF hand 2 46-48 results in a significant conformational change that leads to extrusion of the myristoyl group 32 . This ‘Ca2+/myristoyl switch’ enables recoverin to associate with membranes through its myristoyl group in a Ca2+-dependent manner (see the Ca2+/myristoyl switch movie in the Online links box). As the hydrophobic residues of recoverin that are involved in the interaction with the myristoyl group are conserved in the other members of the NCS protein family, it was thought that all myristoylated NCS proteins would have a similar Ca2+/myristoyl switch32 . However, in addition to recoverin, only the class B proteins show this switch 36-38, 49 . Other members of the family instead use their myristoyl group for constitutive membrane association 38, 39, 50.
The Ca2+-induced conformational change in recoverin also exposes a hydrophobic groove, which forms the binding site for its only known target, rhodopsin kinase 51 . A similar hydrophobic surface is exposed in the structure of Ca2+-bound NCS-1 27, 28 and KChIP1 33, 52 , and so the same mechanism could be used by other NCS proteins for some of their Ca2+-dependent target interactions. This does not, however, explain all of the interactions as, for example, GCAPs do not require Ca2+ for binding to their target guanylyl cyclases and remain bound in the presence or absence of Ca2+ 50, 53 and binding sites for target proteins on NCS-1 have been mapped to domains other than the exposed hydrophobic surface 54 . In addition, early structural studies implied that KChIP1 interacted with Kv4 channels through the hydrophobic groove 33 but more recent structural analyses 52, 55 have shown that two distinct sites within the N-terminal domain of Kv4 channels can bind independently to different regions of KChIP1, and mutagenesis has demonstrated a third direct interaction with KChIPs involving the C terminus of the channel 56, 57 . All three sites of interaction affect the traffic of Kv4 channels to the plasma membrane and their regulation by KChIPs, but in distinct ways 52, 55, 57 . The differential ability of the NCS proteins to interact with specific target proteins is undoubtedly the result of differences in surface residues that are involved in the protein-protein interactions within an overall conserved structure2.
The functions of NCS proteins
The NCS proteins regulate many cellular events in neurons and in retinal photoreceptors which are listed in Table 1. The evidence for these functions originally came from a range of biochemical analyses and studies of the effects of overexpressing the proteins, but in recent years many of these proposed functions have been confirmed in genetic studies in which individual NCS proteins have been knocked out or disrupted (Table 2).
Table 2.
Summary of the phenotypes resulting from genetic alterations of NCS proteins
| NCS protein | Organism | Phenotypes |
|---|---|---|
| NCS-1 (frequenin) | S. cerevisiae | Knockout is lethal owing to requirement in Pik1 activation 21 |
| S. pombe | Growth defect at high Ca2+, decreased sporulation 147 | |
| D. discoideum | Development accelerated 76 | |
| C. elegans | Knockout impairs learning 77 | |
| D. melangaster | Overexpression increases facilitation of neurotransmission16 | |
| D. rerio | Knockdown of NCS-1a impairs development of inner ear 22 | |
| Hippocalcin | Mouse | Knockout causes impaired spatial learning and reduced neurodegeneration 138, 148 |
| Recoverin | Mouse | Knockout results in shorter responses to bright flasher in dark-adapted state and decreased sensitivity to dim light in retinal rod cells44. 58 Signal transmission downstream of phototransduction reduced 58 |
| GCAP1/2 | Mouse | Rod and cone photoreceptors with knockout of both GCAP1 and 2 show increased light sensitivity and a slower recovery following flash response 59, 60, 149, 150 |
| Expression of GCAP1 in GCAP1/2-knockout mice restores normal function 59, 60 | ||
| Expression of GCAP2 in GCAP1/2-knock-out mice is not effective in recovering normal kinetics of flash response 150 | ||
| GCAP1 | Human | Various mutations result in cone or rod/cone degeneration 151 |
| KChIP2 | Mouse | Knockout is highly susceptible to cardiac arrhythmias 93 |
| KChIP3 | Mouse | One study showed reduced responses to pain and elevated dynorphin levels in knockout 70 |
| Another study showed reduced levels of Aβ peptides, enhanced long-term potentiation and downregulation of Kv4 channels in hippocampus 70, 71 |
GCAP, guanylyl cyclase-activating protein; KChIP, Kv channel-interacting protein; NCS, neuronal Ca2+ sensor; Pik1, phosphatidylinositol kinase.
Recoverin and the GCAPs are expressed only in the retina, where both classes of proteins have specific roles in light adaptation 44, 58-60 . The only known targets for regulation by the GCAPs are retinal guanylyl cyclases23 ; recoverin interacts with rhodopsin kinase and inhibits its activity and has an additional, uncharacterized, effect on visual sensitivity unrelated to its regulation of rhodopsin kinase 58 . The other NCS proteins are expressed in neurons and have been implicated in a wider range of cellular functions 5 . The KChIPs were discovered as proteins that interacted with K+ channels of the Kv4 family 61 . KChIP3 was independently discovered as calsenilin, a protein that could interact with, and affect processing of, presenilin62 and amyloid precursor protein63 , and alter Ca2+ release from the endoplasmic reticulum64 . KChIP4 also interacts with presenilin65 . KChIP3 was also identified as the Ca2+-dependent transcriptional repressor known as downstream regulatory element antagonistic modulator (DREAM) 66 on the basis of its binding to the downstream regulator element (DRE) sequence, which is present in the promoter region of many genes. There are well characterised pathways by which an increase in intracellular Ca2+-concentration can activate transcription. In the case of DREAM, transcription is repressed when Ca2+-concentration is low and this repression is lost when DREAM binds Ca2+. More recently, KChIP3 has been found to interact not only directly with the DRE motif but also with various transcription factors67, 68 , All four KChIPs can bind to DRE sequences in DNA and inhibit transcription 69 . The analysis of two independent strains of KChIP3-knockout mouse supports the involvement of KChIP3 in all three of the proposed functions, namely regulation of Kv4 channels, repression of gene transcription and regulation of presenilin processing 70, 71 . The functions of the class B NCS proteins (visinin-like proteins (VILIPs) 1-3 and neurocalcins) are not as well characterized (but see Table 1).
A number of studies have implicated NCS proteins in disease states (in addition to the potential role of KChIPs in Alzheimer’s disease through their interaction with presenilin) based on altered expression levels of particular NCS proteins in post-mortem brain tissue from schizophrenic and epileptic individuals and Alzheimer’s disease patients 72 . Most drugs used in the treatment of schizophrenia and depression are antagonists of dopamine D2-like receptors implicating dysfunction of dopaminergic systems in these diseases but the exact underlying defects are unknown. A direct link to NCS proteins came from the discovery that NCS-1 directly interacts with D2 receptors and can inhibit receptor desensitisation which occurs due to receptor internalisation 73 . Furthermore, increased levels of NCS-1 have been observed in brain tissue from patients with schizophrenia or bipolar disorder 74 and this could conceivably result in increased neuronal excitability, increased intracellular Ca2+ concentration and subsequent cell death. In addition, a link between a single nucleotide polymorphism (SNP) in the gene for the D2 receptor has been associated with differences in nicotine dependence in human subjects and these observations have been extended to show the existence of a genetic interaction with an SNP in a non-coding region of the NCS-1 gene 75 . The significance of this SNP in NCS-1 and whether it affects NCS-1 expression remains to be determined.
Specificity and range of NCS function
NCS-1: one protein, multiple functions
NCS-1 is most probably derived from the primordial NCS precursor and is expressed from yeast to man. Studies in mammalian cells have implicated NCS-1 in several functions 5 , although the only data using genetic manipulations come from lower organisms, in which very diverse phenotypes have resulted (Table 2). The S. cerevisiae orthologue frequenin (Frq1) is essential for survival because it is involved in the activation of one of the two phosphatidylinositol 4-kinases, Pik1 21 . Knockout of NCS-1 is not lethal in other organisms but leads to developmental phenotypes, including changes in the rate of development, in Dictyostelium discoideum 76 . Knockdown of ncs-1, one of the two very closely related NCS-1 genes in zebrafish, abolishes formation of the semicircular canals of the inner ear 22 . Changes in neuronal functions are shown in D. melanogaster, in which overexpression of the NCS-1 orthologue frequenin resulted in increased facilitation of neurotransmission 16 , and in C. elegans, in which knockout of NCS-1 impaired learning and memory 77 . How can a single Ca2+ sensor be involved in so many different activities? NCS-1 is expressed highly in all brain regions 20, 25 and also in many non-neuronal cell types 18, 20 . In mammalian cells, NCS-1 regulates the orthologue of the yeast Pik1, phosphatidylinositol 4-kinase IIIβ 78, 79 , which could explain some of its functional roles, but a key aspect of NCS-1 is its ability to bind to, and regulate, many target proteins that are unrelated to each other 80 (Figure 3). The interactions of NCS-1 with different targets proteins can involve distinct regions of NCS-1 54, 73 , and some interactions can be independent of Ca2+ binding 73, 80 . The link between these target interactions and the varied physiological roles of NCS-1 still remain to be fully established.
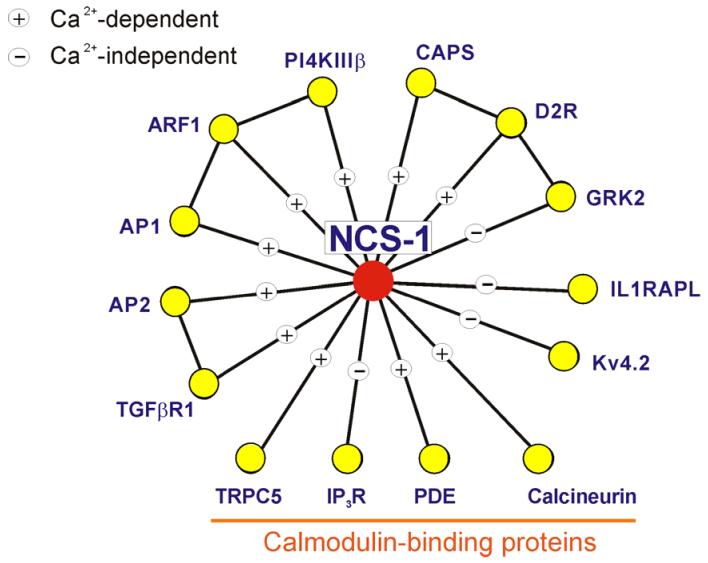
Figure 3.
A protein-protein interaction map showing the known interactions of NCS-1. The figure summarises data on the known protein interactions made by NCS-1 proteins and indicates whether these are Ca2+-dependent or -independent. AP1, clathrin adaptor protein 1 80 ; AP2, clathrin adaptor protein 2 80 ; ARF1, ADP ribosylation factor 1 79 ; CAPS, Ca2+-dependent activator protein of secretion 80 ; calcineurin 110 ; D2R, dopamine receptor type 2 73 ; GRK2, G-protein-coupled receptor kinase 2 73 ; IL1RAPL, interleukin 1 receptor associated protein-like protein 54 ; IP3R, inositol 1,4,5 trisphophate receptor 124 ; Kv4.2, Kv4.2 potassium channel subunit 125, 126 ; PDE, cyclic nucleotide phosphodiesterase 110 ; PI4K, phosphatidylinositol 4-kinase type IIIβ 78, 79 ; TGFβR1, the type I receptor for transforming growth factor β 80 ; TRPC5, transient receptor potential channel 5 127 .
The role of GCAPs in light adaptation
There are three genes that encode GCAPs in the human genome 23 . GCAPs are present only in the retina and have a specific role in light adaptation during phototransduction through their ability to regulate retinal guanylyl cyclases 1 and 2. GCAPs interact with and activate these two proteins at low concentrations of Ca2+ in the light, but at higher Ca2+ levels in the dark these GCAPs undergo an ‘activator-inhibitor’ transition 81 to become inhibitors of guanylyl cyclases 1 and 2 (Figure 2). GCAP1 and GCAP2 are present in both rod and cone photoreceptors in human retina; GCAP3 is expressed only in cones, but is not expressed in the mouse, which implies that this protein is not essential for normal vision 82 . GCAP1 and GCAP2 are present in the retina at similar concentrations and regulate both of the guanylyl cyclases83 . Expressing GCAP1 in mice that lack both GCAP1 and GCAP2 recovers almost all normal function in rods and cones 59, 60.
Different GCAPs, same targets
As the two forms of GCAP regulate the same targets in the same cell type, why is more than one GCAP required? GCAP1 and GCAP2 are only 49% identical and appear to interact with guanylyl cyclases in different ways 83, 84 . A significant aspect of GCAP function is their high Ca2+ affinity. Analysis of the Ca2+ affinities of GCAP1 and GCAP2 and the Ca2+ requirement of the GCAPs for regulation of retinal guanylyl cyclases has revealed two important issues. First, the affinities of the GCAPs for Ca2+ is set to the relevant physiological range (25-250 nM in mouse rods, for example 85) at intracellular concentrations of Mg2+ 86 The second issue is that GCAP1 and GCAP2 differ in their Ca2+-binding affinities for retinal guanylyl cyclase activation (Fig 2) by about 7-fold 83, 86 . This has led to the proposal of a ‘Ca2+ relay’ model for retinal guanylyl cyclase activation 84 in which both GCAPs would be needed to function over the physiological range of Ca2+ concentration in the retina. The fine-tuning of these different Ca2+ affinities would increase the overall sensitivity of the regulatory mechanisms, as their joint action would expand the dynamic range of responses to Ca2+ without losing the sensitivity that is contributed by the cooperative effect of Ca2+ on retinal guanylyl cylase regulation by each GCAP.
KChIPs: a large subfamily with distinct properties
Fast-inactivating A-type K currents are known to be important in controlling crucial aspects of neuronal excitability, including inhibition of the back-propagation of dendritic action potentials. These currents are also important for synaptic plasticity, and for regulation of excitability in the heart 87, 88 . Kv4 channel α-subunits are essential pore-forming determinants of A-type currents in somato-dendritic compartments such as in hippocampal pyramidal neurons 89 . Changes in the expression of Kv4 channels have been implicated in conditions such as epilepsy 90 and Kv4.2 channels in dorsal horn neurons of the spinal cord are targets for regulation of pain sensitivity 91 . The regulation of Kv4 subunits by KChIPs has therefore been studied in detail and provides clues as to why this class of NCS proteins is so diverse. The KChIP class has become the most diversified of the NCS proteins during evolution, from one KChIP in D. melanogaster, which is equally similar to all mammalian KChIPs, to mammals, which have four KChIP genes and a large number of potential splice variants 24 . Analysis in human tissues indicates that at least 16 KChIP isoforms are detectably expressed (Fig 5) although some of the KChIP2 isoforms are expressed only in the heart 92 , where KChIP2 has an established role in the regulation of Kv4 channels 93 . The conserved C-terminal EF hand-containing domain of the human isoforms are at least 70% identical to each other and the key difference between these KChIP isoforms lies in their N-terminal domains. The literature on these splice variants is confusing as the variants have been given multiple names 88, 94 and additional variants that have been studied might be expressed only in certain mammalian species92.
Three aspects of the KChIPs might be important for understanding their diversity: differential cell-type expression of the proteins; differences in their intracellular targeting and localization; and the different effects that they have on the traffic and gating properties of Kv4 channels. It is also possible that the existence of multiple genes might allow cell-type specific regulation at the level of transcription through their distinct promoters.
Different KChIPs, different intracellular localizations
When expressed in heterologous cell types the KChIPs show differences in their intrinsic targeting and localization, which might be important for their efficiency in stimulating traffic of Kv4 channels to the plasma membrane 39, 95 . KChIP1.2 is myristoylated and this allows its targeting to a population of intracellular vesicles, which increases the efficiency of trafficking of Kv4 channels to the plasma membrane 39, 40 . KChIPs 2.1, 2.2, 2.3 and 3.1 possess potential sites for palmitoylation (Fig 4) that is predicted to allow their membrane association. Palmitoylation of KChIPs 2 and 3 is required for efficient trafficking of Kv4 channels 35 . Surprisingly, however, the intrinsic targeting of KChIPs 2 and 3 differs: only palmitoylated KChIP2 associates with the plasma membrane when expressed alone 95 (Fig 4). It seems that KChIP3 only becomes membrane associated when it interacts with Kv4 channels. The other KChIPs do not possess any obvious membrane localization signals and when expressed alone are usually cytosolic 95 (Fig 4) and become membrane-associated only through their binding to Kv4 channels.
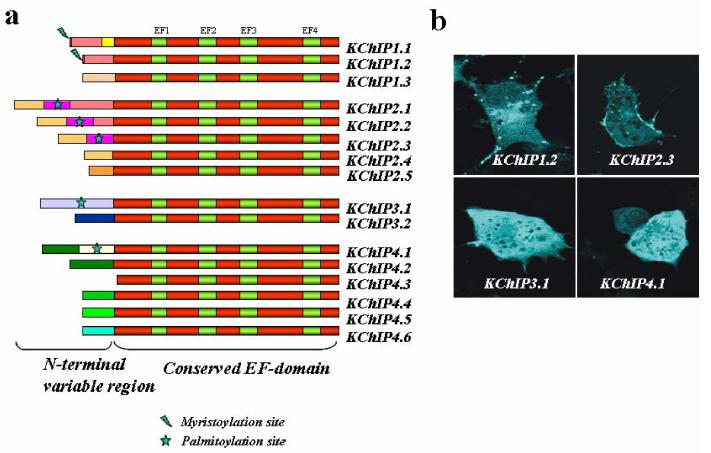
Figure 4.
The KChIP class of NCS proteins and their splice variants a Alternative splicing of the four human KChIP genes generates a series of variants with distinct N-terminal domains. Some of the isoforms possess myristoylation or palmitoylation sites (as indicated) that could confer membrane targeting and localization. The isoforms shown here have been shown to be expressed in human tissues 24 There is no generally agreed terminology for KChIP isoforms and those shown are based on the following sequences (Human KChIP1 isoforms: GenBank accession number DQ148478 (1.1), DQ148477 (1.2), DQ148476 (1.3). Human KChIP2 isoforms: NM_01491 (2.1), DQ148480 (2.2), DQ148481 (2.3), DQ148482 (2.4), DQ148483 (2.5). Human KChIP3 isoforms: DQ148485 (3.1), DQ148486 (3.2). Human KChIP4 isoforms: DQ148487 (4.1), DQ148488 (4.2), DQ148491 (4.3), DQ148489 (4.4), DQ148490 (4.5), DQ148492 (4.6)).
b When expressed in COS-7 cells as fluorescently tagged proteins, the isoforms of each of the four of the KChIP genes show different localization patterns. KChIP1.2 is present on vesicular structures, KChIP2.3 is expressed on the plasma membrane, and KCHIPs 3.1 and 4.1 are diffuse and cytosolic.
Different effects of KChIPs on Kv4 channels
The first study of the effects of KChIP on Kv4 channels compared KChIPs 1.2, 2.3, 3.161 , which were all found to have the same effects (with some quantitative differences): increasing cell surface expression of Kv4, shifting the voltage-dependency of activation, slowing of inactivation and more rapid recovery from inactivation. The N terminus of the KChIPs is not required for its interaction with Kv4 or the basic effects on channel traffic and gating properties 61 , but it has subsequently become clear that the variability in the N-terminal domains of the KChIP isoforms generates the variable effects on Kv4 channels. A comparison of the effects of KChIP1 splice variants on the rate of recovery from inactivation of Kv4 channels has shown differences in the effects of KChIP1.1 and 1.2 isoforms 96, 97 . Several studies on KChIP2 isoforms have shown differences in the efficiency of stimulation of traffic of Kv4 channels to the cell surface and in the modulation of gating properties 35, 94, 98, 99 . In fact, one KChIP2 isoform inhibits Kv4 surface expression, although this isoform is expressed only in the heart 92, 94 .One KChIP4 isoform, KChIP4.1, has similar effects to those originally reported in increasing traffic to the cell surface and slowing channel inactivation 65, 100 . By contrast, the KChIP4.4 isoform does not stimulate traffic of Kv4 channels and leads to the almost complete abolition of the fast inactivation of Kv4 channels. This effect has been attributed to the presence of a K-channel inactivation suppressor (KIS) domain in the N terminus of this isoform 65, 100.
The majority of studies on the properties of KChIPs and their influence on Kv4 channels have looked at the effect of single KChIPs. It has become apparent, however, that interaction of KChIPs with Kv4 channels results in the formation of an octomeric structure containing four KChIPs and four Kv4 channel subunits 52, 101 . As KChIPs can form homo- or hetero-oligomers 102, 103 , it raises the possible additional complexity that functional KChIP-Kv4 complexes could contain more than one KChIP isoform leading to further degrees of subtle variation in channel function.
Differential expression for different proteins
The possibility of KChIPs being components of hetero-oligomers makes consideration of the expression patterns of the KChIP proteins of considerable importance. Analysis of the expression of the four KChIP genes through the use of PCR after reverse transcription of RNA (RT-PCR) and in situ hybridization showed major regional differences in their expression in the brain 24 . The use of antisera specific for KChIP1-4, which are believed to be able to recognise all of the splice variants of each gene, indicated a marked cell-type specific expression of the KChIPs in the hippocampus, cortex striatum and cerebellum 95, 104, 105 . It would clearly be of interest to have more detailed mapping of the expression of each of the KChIP splice variants, but the information already available shows that each neuronal cell type might express only a limited repertoire of the possible set of KChIPs along with specific Kv4 channel isoforms. The detailed properties of A-type K currents are very variable between neuronal cell types or even in different regions of the same neuron 87, 88 , and it is easy to imagine that this could result in part from cell-type specific expression or localization of the KChIP isoforms to fine-tune the channels for their physiological function.
Why are many Ca2+ sensors needed?
Genetic studies have demonstrated that individual NCS proteins have essential functions that cannot be compensated for by other family members or other Ca2+ sensors in the absence of their expression. Differences in Ca2+ affinities, localization through intrinsic targeting signals, different cellular dynamics, differential cellular expression and distinct target proteins are factors in both the specialization of NCS protein function and the basis for increased diversity of the family during evolution.
Differing Ca2+ affinities
Calmodulin interacts with a large number of different target proteins to regulate a multitude of cellular processes, so the question arises as to why NCS proteins are also required to transduce Ca2+ signals in neurons. One key factor is the difference in affinity for Ca2+ shown by the NCS proteins compared to calmodulin (Fig 2). Calmodulin binds Ca2+ in vitro with a dissociation constant (KD) of ∼5-10 μM. Its affinity is increased, however, following binding of target proteins, which can reduce the off-rate for Ca2+ 106, 107 , leading to prolonged activation of target proteins and an increase in the apparent Ca2+ affinity within cells 108 . It was also found 108 .that most of the Ca2+-bound calmodulin in cells is bound to target proteins which are i excess and therefore only target proteins with high affinities for calmodulin are likely to be regulated under physiological conditions. This could explain why additional specific Ca2+ sensors are required.
The measured Ca2+-binding affinities of each of the NCS proteins varies 4 and the exact Ca2+ -binding affinities of each of the NCS proteins seems to be fine-tuned by conserved variation in the sequences of the EF-hand domains and by effects on protein structure of the N-terminal myristoyl group 48 and even the C-terminal residues of the proteins 109 so as to match their sensitivity to physiological requirements. Nevertheless, the general picture is that they are expected to be in a Ca2+ -free state under resting conditions and all have a higher affinity for Ca2+ than calmodulin does, which allows them to bind Ca2+ following small increases in concentration above resting levels. In addition, all of the NCS proteins show intrinsic cooperativity in Ca2+ binding when assayed in vitro or in assays that measure activation of their target proteins. This means that the NCS proteins usually have restricted dynamic ranges over which they can respond to changes in intracellular Ca2+ concentration to regulate their target proteins. This is exemplified by experiments on hippocalcin: analysis of its Ca2+/myristoyl switch in living cells indicated a value for half-maximal activation of 300 nM free Ca2+ and is fully activated when intracellular free Ca2+ concentration reaches 800nM Ca2+ 37 . The small concentration ranges over which the NCS proteins can respond to Ca2+ is also an explanation for the need for more than one GCAP as discussed above.
Specific subcellular and cellular localizations
As noted above, not all NCS proteins have a Ca2+/myristoyl switch and some of them, such as NCS-1 and KChIP1, are constitutively membrane associated through their myristoyl group. Despite this membrane association, NCS-1 and KChIP1 target to distinct organelles where their presence enables them to respond to very transient, local elevations in intracellular Ca2+ 39 . By contrast, NCS proteins that translocate to membranes using the Ca2+/myristoyl switch might require longer lasting and more global Ca2+ elevations for their activation and interaction with targets or could require repetitive Ca2+ signals to allow them to accumulate on membranes. This already gives some indication of how NCS proteins could mediate distinct cellular changes in response to different Ca2+ signals.
In addition to their distinct subcellular localization patterns, many NCS proteins are also expressed in a cell-type specific manner. NCS-1 seems to be expressed in most, if not all, neurons whereas other NCS proteins have distinct and more limited patterns of cellular expression in the brain 25, 104 and, as described above, the multiple KChIPs isoforms have highly cell-type specific expression patterns.
Specific target proteins
Initial work on NCS-1 showed that in vitro it could regulate the same targets as calmodulin, including calcineurin and phosphodiesterase 110 . These are not likely to be physiological targets for NCS-1, as calmodulin is present at much higher concentrations than NCS-1 in neurons and calcineurin and phosphodiesterase have a high affinity for calmodulin which is therefore much more likely to regulate these targets. It cannot be ruled out, however, that these proteins might be regulated by NCS-1 at concentrations of Ca2+ that are too low to activate calmodulin. It is now clear that NCS-1 (Figure 3) and other members of the NCS family have distinct target proteins that they regulate80 — many of these do not overlap with calmodulin and so are more likely to be the true physiological targets that contribute to the specificity of the NCS protein function.
Future perspectives
There have been significant advances in our understanding of the functional roles of the NCS and other neuronal Ca2+ sensor proteins and the way in which their basic properties have been fine-tuned to enable them to regulate different cellular processes by coupling Ca2+ signals to the regulation of specific target proteins. The diversity of the NCS family is clearly important in allowing subtle and neuron-specific patterns of Ca2+ -dependent regulation to occur. Further mapping of the cellular expression of all the family members, including the various isoforms, will be valuable. We also need experimental approaches to examine the relationship between particular Ca2+ signals and activation of the NCS protein complement within different neurons to provide information on when and where these proteins are switched on during neuronal activity. We know in only a few cases the molecular link between regulation of an identified target protein and the physiological outcome at a cellular or organism level and this link needs to be explored in more detail through molecular manipulations of target protein interactions. Finally, we still do not know the full range of the physiological functions of the NCS proteins and, for some of them, very little functional information is available. Future work will need to examine the functions of the less well-studied NCS proteins through targeted gene disruption in animals and the use of gene knockdown approaches in isolated neurons.
Box 2 Other neuronal Ca2+ sensor proteins.
The NCS proteins are not the only EF-hand Ca2+-binding proteins with predominantly neuronal expression. In recent years another EF-hand subfamily known as the Ca2+-binding proteins (CaBPs) 112 or caldendrins 113 has been discovered, which consists of 5 genes in humans and which emerged during evolution in vertebrates. CaBP1 has three splice variants, a long form originally called caldendrin 113 and two shorter forms known as CaBP1 long and short 112 . CaBPs are expressed in neurons 114 — or in some cases in the retina (CaBP2, 4 and 5) 112 — and have been shown to have roles in the regulation of intracellular targets such as inositol trisphosphate (InsP3) receptors 115-117 as well as L- and P/Q-type voltage-gated Ca2+ channels 118, 119 and transient receptor potential (TRP) channels120 . These proteins seems to overlap in their target interactions with calmodulin to a greater extent than the NCS proteins (Fig 4) but a non-redundant role for CaBP4 in the regulation of Ca(v)1.4 Ca2+ channels in photoreceptors has been established using Cabp4-knockout mouse 121 ; mutations in the gene encoding CaBP4 result in autosomal recessive night blindness in humans 122 . For information on other members of the calmodulin superfamily of EF-hand Ca2+-binding proteins see reference 2.
Acknowledgements
I would like to thank Neil Venn for supplying some of the images used in Figure 4 and Professor Alan Morgan for comments on the manuscript. Work in the author’s lab was supported by the Wellcome Trust.
Glossary
- Ca2+ nanodomain
A local Ca2+ signal generated within around 20nm of an open Ca2+ channel
- Ca2+ microdomain
A local Ca2+ signal generated within less than 1μm of an open Ca2+ channel
- Long term potentiation (LTP)
A long-lasting increase in the efficiency of neurotransmission, induced by high frequency stimulation, usually studied in hippocampal neurons and seen as an increase in the magnitude of post-synaptic potentials.
- Long term depression (LTD)
A long-lasting decrease in the efficiency of neurotransmission, induced by low frequency stimulation, usually studied in hippocampal and cerebellar neurons and seen as an decrease in the magnitude of post-synaptic potentials.
- Myristoylation
A post-translational modification that adds a covalently-linked myristoyl group to glycine at position 2 of a protein via the action of the enzyme N-myristoyl transferase. The sequence requirement for myristoylation is well established. This lipid modification can then allow the protein to become membrane associated through lipid insertion of the myristoyl group. The functional significance of myristoylation for protein localisation can be tested by mutation of the required glycine.
- Nuclear factor of activated T cells (NFAT)
A transcription factor that in unstimulated cells resides in the cytoplasm as a highly phosphorylated protein. Elevation Ca2+ activates calcineurin through calmodulin. Calcineurin dephosphorylates NFAT which can then translocate into the nucleas and regulate gene transcription until it is rephosphorylated.
- Palmitoylation
A post-translational modification that links a palmitoyl group to an internal cysteine of a protein through a thioester linkage. This lipid modification can then allow the protein to become membrane associated through lipid insertion of the palmitoyl group. The functional significance of palmitoylation for protein localisation can be tested by mutation of the relevant cysteine residue.
- Phosphatidylinositol 4-kinase
An enzyme that converts phosphatidylinositol to phosphatidylinositol 4-phosphate which is then converted to phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2). (PI(4,5)P2) is required for many cellular events including membrane traffic events and is the target for phospholipase C which breaks down PI(4,5)P2 to release the second messengers diacylglycerol and inositol 1,4,5 trisphosphate.
- Single nucleotide polymorphism (SNP)
A nucleotide within the coding or non-coding region of a gene that can be variable between different members of the population. The presence of different nucleotides may have no effect or could change a single amino acid within the protein or alternatively lead to differences in gene expression or its regulation.
Footnotes
About the author. Robert Burgoyne is Professor of Physiology and Head of the School of Biomedical Sciences in the University of Liverpool, Liverpool, UK. He received his PhD from the University of Birmingham in 1977 working on vaccinia virus and subsequently moved his interests to the study of neurons and neuroendocrine cells. Since establishing his research in Liverpool in 1983, his laboratory has been concerned with regulation of neuronal signalling by Ca2+ including the mechanisms that underlie hormone and neurotransmitter release. He has studied the roles of proteins involved in the machinery for vesicle exocytosis and in more recent years the roles of neuronal calcium sensors in neuronal signalling pathways.
FURTHER INFORMATION
Ca2+/myristoyl switch movie: http://www.liv.ac.uk/physiology/ncs/conform.html
FURTHER INFORMATION
Burgoyne home page:
http://www.liv.ac.uk/physiology/root/department%20of%20physiology/research/burgoyne/index.htm
NCS family home page:
http://www.liv.ac.uk/physiology/ncs/index.html
AfCS Molecule pages:
NCS-1
http://www.signaling-gateway.org/molecule/query?afcsid=A000957
KChIP1
http://www.signaling-gateway.org/molecule/query?afcsid=A001308
KChIP2
http://www.signaling-gateway.org/molecule/query?afcsid=A001309
KChIP3
http://www.signaling-gateway.org/molecule/query?afcsid=A001310
References
- 1. Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. An excellent review that introduces general aspects of calcium signalling.
- 2. Ikura M, Ames JB. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: two ways to promote multifunctionality. Proc Natl Acad Sci U S A. 2006;103:1159–64. doi: 10.1073/pnas.0508640103. An overview of the diversity of EF-hand containing calcium binding proteins
- 3.Fernandez-Chacon R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 4.Burgoyne RD, Weiss JL. The neuronal calcium sensor family of Ca2+-binding proteins. Biochem. J. 2001;353:1–12. [PMC free article] [PubMed] [Google Scholar]
- 5.Burgoyne RD, O′Callaghan DW, Hasdemir B, Haynes LP, Tepikin AV. Neuronal calcium sensor proteins: multitalented regulators of neuronal function. Trends Neurosci. 2004;27:203–209. doi: 10.1016/j.tins.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Sabatini BL, Oertner TG, Svoboda K. The life cycle of Ca2+ ions in dendritic spines. Neuron. 2002;33:439–452. doi: 10.1016/s0896-6273(02)00573-1. [DOI] [PubMed] [Google Scholar]
- 7.Augustine GJ, Santamaria F, Tanaka K. Local calcium signaling in neurons. Neuron. 2003;40:331–346. doi: 10.1016/s0896-6273(03)00639-1. [DOI] [PubMed] [Google Scholar]
- 8.Berridge MJ. Neuronal calcium signalling. Neuron. 1998;21:13–26. doi: 10.1016/s0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- 9.Sabatini BL, Regehr WG. Timing of neurotransmission at fast synapses in the mammalian brain. Nature. 1996;384:170–172. doi: 10.1038/384170a0. [DOI] [PubMed] [Google Scholar]
- 10.Yang S-N, Tang Y-G, Zucker RS. Selective induction of LTP and LTD by postsynaptic [Ca2+]i elevation. J. Neurophysiol. 1999;81:781–787. doi: 10.1152/jn.1999.81.2.781. [DOI] [PubMed] [Google Scholar]
- 11.Gomez TM, Zheng JQ. The molecular basis for calcium-dependent axon pathfinding. Nat Rev Neurosci. 2006;7:115–25. doi: 10.1038/nrn1844. [DOI] [PubMed] [Google Scholar]
- 12.Bito H, Deisseroth K, Tsien RW. Ca2+-dependent regulation in neuronal gene expression. Current Opinion in Neurobiology. 1997;7:419–429. doi: 10.1016/s0959-4388(97)80072-4. [DOI] [PubMed] [Google Scholar]
- 13.Hara MR, Snyder SH. Cell Signaling and Neuronal Death. Annu Rev Pharmacol Toxicol. 2007;47:117–41. doi: 10.1146/annurev.pharmtox.47.120505.105311. [DOI] [PubMed] [Google Scholar]
- 14.Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3:175–90. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- 15.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–32. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 16.Pongs O, et al. Frequenin - A novel calcium-binding protein that modulates synaptic efficacy in the drosophila nervous system. Neuron. 1993;11:15–28. doi: 10.1016/0896-6273(93)90267-u. [DOI] [PubMed] [Google Scholar]
- 17.Nef S, Fiumelli H, de Castro E, Raes MB, Nef P. Identification of a neuronal calcium sensor (NCS-1) possibly involved in the regulation of receptor phosphorylation. J Recept Signal Transuct Res. 1995;15:365–378. doi: 10.3109/10799899509045227. [DOI] [PubMed] [Google Scholar]
- 18.McFerran BW, Graham ME, Burgoyne RD. NCS-1, the mammalian homologue of frequenin is expressed in chromaffin and PC12 cells and regulates neurosecretion from dense-core granules. J. Biol. Chem. 1998;273:22768–22772. doi: 10.1074/jbc.273.35.22768. [DOI] [PubMed] [Google Scholar]
- 19.Kapp-Barnea Y, Melnikov S, Shefler I, Jeromin A, Sagi-Eisenberg R. Neuronal calcium sensor-1 and phosphatidylinositol 4-kinase β regulate IgE receptor-triggered exocytosis in cultured mast cells. J. Immunol. 2003;171:5320–5327. doi: 10.4049/jimmunol.171.10.5320. [DOI] [PubMed] [Google Scholar]
- 20.Gierke P, et al. Expression analysis of members of the neuronal calcium sensor protein family: combining bioinformatics and Western Blot analysis. Biochem. Biophys. Res. Comm. 2004;323:38–43. doi: 10.1016/j.bbrc.2004.08.055. [DOI] [PubMed] [Google Scholar]
- 21. Hendricks KB, Wang BQ, Schnieders EA, Thorner J. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nature Cell Biology. 1999;1:234–241. doi: 10.1038/12058. This paper demonstrated the presence of an NCS-1 orthologue in yeast for the first time, showed that it was essential for survival and also identified its target protein. The interaction with phosphatidylinositol-4-OH kinase was later confirmed for the mammalian proteins.
- 22.Blasiole B, et al. Neuronal calcium sensor-1 gene ncs-1 is essential for semicircular canal formation in zebrafish inner ear. J. Neurobiol. 2005;64:285–297. doi: 10.1002/neu.20138. [DOI] [PubMed] [Google Scholar]
- 23.Palczewski K, Sokal I, Baehr W. Guanylate cyclase-activating proteins: structure, function and diversity. Biochem. Biophys. Res. Comm. 2004;322:1123–1130. doi: 10.1016/j.bbrc.2004.07.122. [DOI] [PubMed] [Google Scholar]
- 24. Pruunsild P, Timmusk T. Structure, alternative splicing, and expression of the human and mouse KCNIP gene family. Genomics. 2005;86:581–93. doi: 10.1016/j.ygeno.2005.07.001. A comprehensive analysis of the expression of KChIP splice variants in human and mouse brain.
- 25.Paterlini M, Revilla V, Grant AL, Wisden W. Expression of the neuronal calcium sensor protein family in the rat brain. Neuroscience. 2000;99:205–216. doi: 10.1016/s0306-4522(00)00201-3. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi M, Takamatsu K, Saitoh S, Miura M, Noguchi T. Molecular cloning of hippocalcin, a novel calcium-binding protein of the recoverin family exclusively expressed in hippocampus. Biochem. Biophys. Res. Commun. 1992;189:511–517. doi: 10.1016/0006-291x(92)91587-g. [DOI] [PubMed] [Google Scholar]
- 27.Bourne Y, Dannenberg J, Pollmann V, Marchot P, Pongs O. Immunocytochemical localisation and crystal structure of human frequenin (neuronal calcium sensor 1) J. Biol. Chem. 2001;276:11949–11955. doi: 10.1074/jbc.M009373200. [DOI] [PubMed] [Google Scholar]
- 28.Ames JB, et al. Structure and calcium-binding properties of Frq1, a novel calcium sensor in the yeast Saccharomyces cerevisiae. Biochemistry. 2000;39:12149–12161. doi: 10.1021/bi0012890. [DOI] [PubMed] [Google Scholar]
- 29.Vijay-Kumar S, Kumar VD. Crystal structure of recombinant bovine neurocalcin. Nature Structural Biology. 1999;6:80–88. doi: 10.1038/4956. [DOI] [PubMed] [Google Scholar]
- 30.Ames JB, Dizhoor AM, Ikura M, Palczewski K, Stryer L. Three-dimensional structure of guanylyl cyclase activating protein-2, a calciumsensitive modulator of photoreceptor guanylyl cyclases. J. Biol. Chem. 1999;274:19329–19337. doi: 10.1074/jbc.274.27.19329. [DOI] [PubMed] [Google Scholar]
- 31.Stephen R, Palczewski K, Sousa MC. The crystal structure of GCAP3 suggests molecular mechanism of GCAP-linked cone dystrophies. J Mol Biol. 2006;359:266–75. doi: 10.1016/j.jmb.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ames JB, et al. Molecular mechanics of calcium-myristoyl switches. Nature. 1997;389:198–202. doi: 10.1038/38310. A classic paper on the structural characterisation of myristoylated recoverin in its calcium bound-form which illuminated the basis of the calcium-myristoyl switch.
- 33.Zhou W, Qian Y, Kunjilwar K, Pfaffinger PJ, Choe S. Structural insights into the functional interaction of KChIP1 with shal-type K+ channels. Neuron. 2004;41:573–586. doi: 10.1016/s0896-6273(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 34.Scannevin RH, et al. Two N-terminal domains of Kv4 K+ channels regulate binding to and modulation by KChIP1. Neuron. 2004;41:587–598. doi: 10.1016/s0896-6273(04)00049-2. [DOI] [PubMed] [Google Scholar]
- 35.Takimoto K, Yang E-K, Conforti L. Palmitoylation of KChIP splicing variants is required for efficient cell surface expression of Kv4.3 channels. J. Biol.Chem. 2002;277:26904–26911. doi: 10.1074/jbc.M203651200. [DOI] [PubMed] [Google Scholar]
- 36.Spilker C, Dresbach T, Braunewell K-H. Reversible translocation and activity-dependent localisation of the calcium-myristoyl switch protein VILIP-1 to different membrane compartments in living hippocampal neurons. J. Neurosci. 2002;22 doi: 10.1523/JNEUROSCI.22-17-07331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O′Callaghan DW, Tepikin AV, Burgoyne RD. Dynamics and calcium-sensitivity of the Ca2+-myristoyl switch protein hippocalcin in living cells. J. Cell Biol. 2003;163:715–721. doi: 10.1083/jcb.200306042. Analysis of the calcium-myristoyl switch of hippocalcin in living cells through the use of confocal imaging and elevation of intracellular calcium by photolysis of caged calcium.
- 38.O′Callaghan DW, et al. Differential use of myristoyl groups on neuronal calcium sensor proteins as a determinant of spatio-temporal aspects of Ca2+-signal transduction. J. Biol.Chem. 2002;277:14227–14237. doi: 10.1074/jbc.M111750200. [DOI] [PubMed] [Google Scholar]
- 39.O′Callaghan DW, Hasdemir B, Leighton M, Burgoyne RD. Residues within the myristoylation motif determine intracellular targeting of the neuronal Ca2+ sensor protein KChIP1 to post-ER transport vesicles and traffic of Kv4 K+ channels. J. Cell Sci. 2003;116:4833–4845. doi: 10.1242/jcs.00803. [DOI] [PubMed] [Google Scholar]
- 40.Hasdemir B, Fitzgerald DJ, Prior IA, Tepikin AV, Burgoyne RD. Traffic of Kv4 K+ channels mediated by KChIP1 is via a novel post-ER vesicular pathway. J Cell Biol. 2005;171:459–469. doi: 10.1083/jcb.200506005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O′Callaghan DW, Haynes LP, Burgoyne RD. High-affinity interaction of the N-terminal myristoylation motif of the neuronal calcium sensor protein hippocalcin with phosphatidylinositol 4,5-bisphosphate. Biochem J. 2005;391:231–238. doi: 10.1042/BJ20051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dizhoor AM, et al. Recoverin: a calcium sensitive activator of retinal rod guanylate cyclase. Science. 1991;251:915–918. doi: 10.1126/science.1672047. [DOI] [PubMed] [Google Scholar]
- 43.Chen CK, Inglese J, Lefkowitz RJ, Hurley JB. Ca2+-dependent interaction of recoverin with rhodopsin kinase. J. Biol. Chem. 1995;270:18060–18066. doi: 10.1074/jbc.270.30.18060. [DOI] [PubMed] [Google Scholar]
- 44.Makino CL, et al. Recoverin regulates light-dependent phosphodiesterase activity in retinal rods. J. Gen. Physiol. 2004;123:729–741. doi: 10.1085/jgp.200308994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka T, Ames JB, Harvey TS, Stryer L, Ikura M. Sequestration of the membrane targeting myristoyl group of recoverin in the calcium-free state. Nature. 1995;376:444–447. doi: 10.1038/376444a0. [DOI] [PubMed] [Google Scholar]
- 46.Senin II, et al. Ca2+-myristoyl switch in the neuronal calcium sensor recoverin requires different functions of Ca2+ binding sites. J.Biol. Chem. 2002;277:50365–50372. doi: 10.1074/jbc.M204338200. [DOI] [PubMed] [Google Scholar]
- 47.Ames JB, Hamasaki N, Molchanova T. Structure and calcium-binding studies of a recoverin mutant (E85Q) in an allosteric intermediate state. Biochemistry. 2002;41:5776–87. doi: 10.1021/bi012153k. [DOI] [PubMed] [Google Scholar]
- 48.Weiergraber OH, Senin II, Philippov PP, Granzin J, Koch K-W. Impact of N-terminal myristoylation on the Ca2+-dependent conformational transition in recoverin. J.Biol. Chem. 2003;278:22972–22979. doi: 10.1074/jbc.M300447200. [DOI] [PubMed] [Google Scholar]
- 49.Spilker C, Braunewell K-H. Calcium-myristoyl switch, subcellular localisation, and calcium-dependent translocation of the neuronal calcium sensor protein VILIP-3, and comparison with VILIP-1 in hippocampal neurons. Mol. Cell. Neurosci. 2003;24:766–778. doi: 10.1016/s1044-7431(03)00242-2. [DOI] [PubMed] [Google Scholar]
- 50.Olshevskaya EV, Hughes EE, Hurley JB, Dizhoor AM. Calcium binding, but not calcium-myristoyl switch, controls the ability of guanyl cylcase-activating protein GCAP-2 to regulated photoreceptor guanyl cyclase. J. Biol. Chem. 1997;272:14327–33. doi: 10.1074/jbc.272.22.14327. [DOI] [PubMed] [Google Scholar]
- 51.Ames JB, Levay K, Wingard JN, Lusin JD, Slepak VZ. Structural Basis for Calcium-induced Inhibition of Rhodopsin Kinase by Recoverin. J Biol Chem. 2006;281:37237–45. doi: 10.1074/jbc.M606913200. [DOI] [PubMed] [Google Scholar]
- 52. Pioletti M, Findeisen F, Hura GL, Minor DL. Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer. Nat Struct Mol Biol. 2006;13:987–995. doi: 10.1038/nsmb1164. Structure for a complex between KChIP1 and the N-terminus of a Kv4.3 channel that demonstrates two sites of interaction. This paper presents a structure that differs from that published earlier and may have been artefactual. See also reference 55 for a similar structure.
- 53.Palczewski K, Polans A, Baehr W, Ames JB. Ca2+-binding proteins in the retina: structure, function and the etiology of human visual diseases. BioEssays. 2000;22:337–350. doi: 10.1002/(SICI)1521-1878(200004)22:4<337::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 54.Bahi N, et al. IL1 receptor accessory protein like, a protein involved in X-linked mental retardation, interacts with Neuronal Calcium Sensor-1 and regulates exocytosis. Hum Mol Genet. 2003;12:1415–1425. doi: 10.1093/hmg/ddg147. [DOI] [PubMed] [Google Scholar]
- 55. Wang H, et al. Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits. Nat Neurosci. 2007;10:32–9. doi: 10.1038/nn1822. Structure for a complex between KChIP1 and the N-terminus of a Kv4.3 channel that demonstrates two sites of interaction. This paper presents a structure that differs from that published earlier and may have been artefactual. See also reference 52 for a similar structure.
- 56.Callsen B, et al. Contribution of N- and C-terminal Kv channel domians to KChIP interaction. J. Physiol. 2005;568:397–412. doi: 10.1113/jphysiol.2005.094359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han W, Nattel S, Noguchi T, Shrier A. C-terminal domain of Kv4.2 and associated KChIP2 interactions regulate functional expression and gating of Kv4.2. J Biol Chem. 2006;281:27134–44. doi: 10.1074/jbc.M604843200. [DOI] [PubMed] [Google Scholar]
- 58.Sampath A, et al. Recoverin improves rod-mediated vision by enhancing signal transmission in the mouse retina. Neuron. 2005;46:413–420. doi: 10.1016/j.neuron.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 59.Howes KA, et al. GCAP1 rescues rod photoreceptor response in GCAP1/GCAP2 knockout mice. EMBO Journal. 2002;21:1545–1554. doi: 10.1093/emboj/21.7.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pennesi ME, Howes KA, Baehr W, Wu SM. Guanylate cyclase-activating protein (GCAP) 1 rescues cone recovery kinetics in GCAP1/GCAP2 knockout mice. Proc. Natl. Acad. Sci. USA. 2003;100:6783–6788. doi: 10.1073/pnas.1130102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. An WF, et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. The first demonstration of a interaction between KChIPs 1-3 and Kv4 potassium channels and analysis of the functional consequences for surface expression and channel gating properties.
- 62.Buxbaum JD, et al. Calsenilin: A calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nature Medicine. 1998;4:1177–1181. doi: 10.1038/2673. [DOI] [PubMed] [Google Scholar]
- 63.Jo DG, Jang J, Kim BJ, Lundkvist J, Jung YK. Overexpression of calsenilin enhances γ-secretase activity. Neurosci Lett. 2004;378:59–64. doi: 10.1016/j.neulet.2004.12.078. [DOI] [PubMed] [Google Scholar]
- 64.Leissring MA, et al. Calsenilin reverses presenilin-mediated enhancement of calcium signalling. Proc. Natl. Acad. Sci. USA. 2000;97:8590–8593. doi: 10.1073/pnas.97.15.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morohashi Y, et al. Molecular cloning and characterisation of CALP/KChIP4, a novel EF-hand protein interacting with presenilin 2 and voltage-gated potassium channel subunit kv4. J. Biol. Chem. 2002;277:14965–14975. doi: 10.1074/jbc.M200897200. [DOI] [PubMed] [Google Scholar]
- 66. Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. This paper describes the discovery of DREAM (KChIP3) as a repressor of transcription of specific genes containing the DRE element including prodynophin and c-fos. It shows that the repression is lost when DREAM binds calcium as the calcium-bound form can not bind to the DRE element.
- 67.Rivas M, Mellstrom B, Naranjo JR, Santisteban P. Transcriptional repressor DREAM interacts with thyroid transcription factor-1 and regulates thyroglobulin gene expression. J.Biol. Chem. 2004;279:33114–33122. doi: 10.1074/jbc.M403526200. [DOI] [PubMed] [Google Scholar]
- 68.Zaidi NF, et al. Calsenilin interacts with transcriptional co-repressor C-terminal binding protein(s) J Neurochem. 2006;98:1290–301. doi: 10.1111/j.1471-4159.2006.03972.x. [DOI] [PubMed] [Google Scholar]
- 69.Link WA, et al. Day-night changes in downstream regulatory element antagonist aodulator/potassium channel interacting protein activity contribute to circadian gene expression in pineal gland. J Neurosci. 2004;24:5346–5355. doi: 10.1523/JNEUROSCI.1460-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cheng H-YM, et al. DREAM is a critical transcriptional repressor for pain modulation. Cell. 2002;108:31–43. doi: 10.1016/s0092-8674(01)00629-8. A knock-out mouse study that established physiological functions of KChIP3/DREAM/calsenilin.
- 71. Lilliehook C, et al. Altered Aβ formation and long-term potentiation in a calsenilin knock-out. J. Neurosci. 2003;23:9097–9106. doi: 10.1523/JNEUROSCI.23-27-09097.2003. A knock-out mouse study that established physiological functions of KChIP3/DREAM/calsenilin.
- 72.Braunewell K-H. The darker side of Ca2+ signaling by neuronal Ca2+-sensor proteins: from Alzheimer′s disease to cancer. Trends Pharmacol. Sci. 2005;26:345–351. doi: 10.1016/j.tips.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 73.Kabbani N, Negyessy L, Lin R, Goldman-Rakic P, Levenson R. Interaction with the neuronal calcium sensor NCS-1 mediates desensitization of the D2 dopamine receptor. J. Neurosci. 2002;22:8476–8486. doi: 10.1523/JNEUROSCI.22-19-08476.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koh PO, et al. Up-regulation of neuronal calcium sensor-1 (NCS-1) in the prefrontal cortex of schizophrenic and bipolar patients. Proc. Natl. Acad. Sci. U.S.A. 2003;100:313–317. doi: 10.1073/pnas.232693499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dahl JP, et al. Interaction between variation in the D2 dopamine receptor (DRD2) and the neuronal calcium sensor-1 (FREQ) genes in predicting response to nicotine replacement therapy for tobacco dependence. Pharmacogenomics J. 2006;6:194–9. doi: 10.1038/sj.tpj.6500358. [DOI] [PubMed] [Google Scholar]
- 76.Coukell B, Cameron A, Perusini S, Shim K. Disruption of the NCS-1/frequenin-related ncsA gene in Dictyostelium discoideum accelerates development. Dev Growth Differ. 2004;46:449–458. doi: 10.1111/j.1440-169x.2004.00761.x. [DOI] [PubMed] [Google Scholar]
- 77.Gomez M, et al. Ca2+ signalling via the neuronal calcium sensor-1 regulates associative learning and memory in C.elegans. Neuron. 2001;30:241–248. doi: 10.1016/s0896-6273(01)00276-8. [DOI] [PubMed] [Google Scholar]
- 78.Zhao X, et al. Interaction of neuronal calcium sensor-1 (NCS-1) with phosphatidylinositol 4-kinase beta stimulates lipid kinase activity and affects membrane trafficking in COS-7 cells. J. Biol. Chem. 2001;276:40183–40189. doi: 10.1074/jbc.M104048200. [DOI] [PubMed] [Google Scholar]
- 79.Haynes LP, Thomas GMH, Burgoyne RD. Interaction of neuronal calcium sensor-1 and ARF1 allows bidirectional control of PI(4) kinase and TGN-plasma membrane traffic. J.Biol. Chem. 2005;280:6047–6054. doi: 10.1074/jbc.M413090200. [DOI] [PubMed] [Google Scholar]
- 80. Haynes LP, et al. Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin. Proteomics. 2006;6:1822–1832. doi: 10.1002/pmic.200500489. A demonstration of the range, diversity and specificity of interacting proteins for members of the NCS and CaBP protein families.
- 81.Dizhoor AM, Hurley JB. Inactivation of EF-hands makes GCAP-2 (p24) a constitutive activator of photoreceptor guanulyl cyclase by preventing a Ca2+-induced “activator-to-inhibitor” transition. J. Biol. Chem. 1996;271:19346–19350. doi: 10.1074/jbc.271.32.19346. [DOI] [PubMed] [Google Scholar]
- 82.Imanishi Y, et al. Characterisation of retinal guanylate cyclase-activating protein 3 (GCAP3) from zebrafish to man. Eur. J. Neurosci. 2002;15:63–78. doi: 10.1046/j.0953-816x.2001.01835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hwang J-Y, et al. Regulatory modes of rod outer segment membrane guanylate cyclase differ in catalytic efficiency and Ca2+-sensitivity. Eur. J. Biochem. 2003;270:3814–3821. doi: 10.1046/j.1432-1033.2003.03770.x. [DOI] [PubMed] [Google Scholar]
- 84.Koch K-W. GCAPs, the classical neuronal calcium sensors in the retina. A Ca2+-relay model of guanylate cyclase activation. Calcium Binding Proteins. 2006;1:3–6. [Google Scholar]
- 85.Woodruff ML, et al. Measurement of cytoplasmic calcium concentration in the rods of wild-type and transducin knock-out mice. J Physiol. 2002;542:843–54. doi: 10.1113/jphysiol.2001.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Peshenko IV, Dizhoor AM. Guanylyl cyclase-activating proteins (GCAPs) are Ca2+/Mg2+ sensors. J. Biol.Chem. 2004;279:16903–16906. doi: 10.1074/jbc.C400065200. A study that reveals how the calcium sensitivity of GCAPs matches physiological concentrations only when assayed in the presence of a physiological concentration of magnesium.
- 87.Birnbaum SG, et al. Structure and function of Kv4-family transient potassium channels. Physiol. Rev. 2004;84:803–833. doi: 10.1152/physrev.00039.2003. [DOI] [PubMed] [Google Scholar]
- 88.Jerng HH, Pfaffinger PJ, Covarrubias M. Molecular physiology and modulation of somatodendritic A-type potassium channels. Mol. Cell. Neurosci. 2004;27:343–369. doi: 10.1016/j.mcn.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 89.Lauver A, et al. Manipulating Kv4.2 identifies a specific component of hippocampal pyramidal neuron A-current that depends upon Kv4.2 expression. J Neurochem. 2006;99:1207–1223. doi: 10.1111/j.1471-4159.2006.04185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bernard C, et al. Acquired dendritic channelopathy in temporal lobe epilepsy. Science. 2004;305:532–5. doi: 10.1126/science.1097065. [DOI] [PubMed] [Google Scholar]
- 91.Hu H-J, et al. The Kv4.2 Potassium channel subunit is required for pain plasticity. Neuron. 2006;50:89–100. doi: 10.1016/j.neuron.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 92.Patel SP, Campbell DL, Strauss HC. Elucidating kChIP effects on Kv4.3 inactivation and recovery kinetics with a minimal KChIP2 isoform. J. Physiol. 2002;545:5–11. doi: 10.1113/jphysiol.2002.031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuo H-C, et al. A defect in the Kv channel-interacting protein 2 (KChIp2) gene leads to a complete loss of Ito and confers susceptibility to ventricular tachycardia. Cell. 2001;107:801–813. doi: 10.1016/s0092-8674(01)00588-8. [DOI] [PubMed] [Google Scholar]
- 94.Decher N, Barth AS, Gonzalez T, Steinmeyer K, Sanguinetti MC. Novel KChIP2 isoforms increase functional diversity of transient outward potassium currents. J Physiol. 2004;557:761–72. doi: 10.1113/jphysiol.2004.066720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shibata R, et al. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J. Biol. Chem. 2003;278:36445–36454. doi: 10.1074/jbc.M306142200. [DOI] [PubMed] [Google Scholar]
- 96.Boland LM, et al. Functional properties of a brain-specific NH2-terminally spliced modulator of Kv4 channels. Am J Cell Physiol. 2003;285:C161–C170. doi: 10.1152/ajpcell.00416.2002. [DOI] [PubMed] [Google Scholar]
- 97.Van Hoorick D, Raes A, Keysers W, Mayeur E, Snyders DJ. Differential modulation of kv4 kinetics by KCHIP1 splice variants. Mol Cell Neurosci. 2003;24:357–366. doi: 10.1016/s1044-7431(03)00174-x. [DOI] [PubMed] [Google Scholar]
- 98.Patel SP, Parai R, Parai R, Campbell DL. Regulation of Kv4.3 voltage-dependent gating kinetics by KChIP2 isoform. J. Physiol. 2004;557:19–41. doi: 10.1113/jphysiol.2003.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deschenes I, et al. Regulation of Kv4.3 current by KChIP2 splice variants: a component of native cardiac I(to) Circulation. 2002;106:423–9. doi: 10.1161/01.cir.0000025417.65658.b6. [DOI] [PubMed] [Google Scholar]
- 100.Holmqvist MH, et al. Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc. Natl. Acad. Sci. USA. 2002;99:1035–1040. doi: 10.1073/pnas.022509299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim LA, et al. Three-dimensional structure of Ito: Kv4.2-KChIP2 Ion channels by electron microscopy at 21 A resolution. Neuron. 2004;41:513–519. doi: 10.1016/s0896-6273(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 102.Osawa M, et al. Calcium-regulated DNA binding and oligomerization of the neuronal calcium sensing protein, calsenilin/DREAM/KChIP3. J. Biol. Chem. 2001;276:41005–41013. doi: 10.1074/jbc.M105842200. [DOI] [PubMed] [Google Scholar]
- 103.Savignac M, et al. Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. EMBO J. 2005;24:3555–3564. doi: 10.1038/sj.emboj.7600810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rhodes KJ, et al. KChIPs and Kv4αsubunits as integral components of A-type potassium channels in mammalian brain. J. Neurosci. 2004;24:7903–7915. doi: 10.1523/JNEUROSCI.0776-04.2004. A study describing the cell type-specific expression of KChIPs1-4.
- 105.Strassle BW, Menegola M, Rhodes KJ, Trimmer JS. Light and electron microscopic analysis of KChIP and Kv4 localisation in rat cerebellar granule cells. J. Comp. Neurol. 2005;484:144–155. doi: 10.1002/cne.20443. [DOI] [PubMed] [Google Scholar]
- 106.Johnson JD, Snyder C, Walsh M, Flynn M. Effects of myosin light chain kinase and peptides in Ca2+ exchange with the N- and C-terminal Ca2+binding sites of calmodulin. J. Biol.Chem. 1996;271:761–767. doi: 10.1074/jbc.271.2.761. [DOI] [PubMed] [Google Scholar]
- 107.Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB. Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proc Natl Acad Sci U S A. 2006;103:13968–73. doi: 10.1073/pnas.0606433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Persechini A, Cronk B. The relationship between the free concentrations of Ca2+ and Ca2+-calmodulin in intact cells. J.Biol.Chem. 1999;274:6827–6830. doi: 10.1074/jbc.274.11.6827. [DOI] [PubMed] [Google Scholar]
- 109.Weiergraber OH, et al. Tuning of a neuronal calcium sensor. J Biol Chem. 2006;281:37594–375602. doi: 10.1074/jbc.M603700200. [DOI] [PubMed] [Google Scholar]
- 110.Schaad NC, et al. Direct modulation of calmodulin targets by the neuronal calcium sensor NCS-1. Proc. Natl. Acad. Sci. USA. 1996;93:9253–9258. doi: 10.1073/pnas.93.17.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Falke JJ, Drake SK, Hazard AL, Peersen OB. Molecular tuning of ion binding to calcium signaling proteins. Q. Rev. Biophys. 1994;27:219–90. doi: 10.1017/s0033583500003012. [DOI] [PubMed] [Google Scholar]
- 112.Haeseleer F, et al. Five members of a novel Ca2+ binding protein (CABP) subfamily with similarity to calmodulin. J. Biol. Chem. 2000;275:1247–1260. doi: 10.1074/jbc.275.2.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seidenbecher CI, et al. Caldendrin, a novel neuronal calcium-binding protein confined to the somato-dendritic compartment. J. Biol. Chem. 1998;273:21324–21331. doi: 10.1074/jbc.273.33.21324. [DOI] [PubMed] [Google Scholar]
- 114.Laube G, et al. The neuron-specific Ca2+-binding protein caldendrin: gene structure, splice isoforms, and expression in the rat central nervous system. Mol. Cell. Neurosci. 2002;19:459–475. doi: 10.1006/mcne.2001.1078. [DOI] [PubMed] [Google Scholar]
- 115.Haynes LP, Tepikin AV, Burgoyne RD. Calcium Binding Protein 1 is an inhibitor of agonist-evoked, inositol 1,4,5-trisphophate-mediated calcium signalling. J. Biol.Chem. 2004;279:547–555. doi: 10.1074/jbc.M309617200. [DOI] [PubMed] [Google Scholar]
- 116.Yang J, et al. Identification of a family of calcium sensors as protein ligands of inositol trisphosphate receptor Ca2+ release channels. Proc. Natl. Acad. Sci. USA. 2002;99:7711–7716. doi: 10.1073/pnas.102006299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kasri NN, et al. Regulation of InsP3 receptor activity by neuronal Ca2+-binding proteins. EMBO J. 2004;23:312–21. doi: 10.1038/sj.emboj.7600037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou H, et al. Ca2+-binding protein-1 facilitates and forms a postsynaptic complex with Cav1.2 (L-type) Ca2+ channels. J. Neurosci. 2004;24:4698–4708. doi: 10.1523/JNEUROSCI.5523-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee A, et al. Differential modulation of Cav2.1 channels by calmodulin and Ca2+-binding protein 1. Nature Neuroscience. 2002;5:210–217. doi: 10.1038/nn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kinoshita-Kawada M, et al. Inhibition of TRPC5 channels by Ca2+ binding protein 1 in Xenopus oocytes. Pflugers Arch. 2005;450:345–354. doi: 10.1007/s00424-005-1419-1. [DOI] [PubMed] [Google Scholar]
- 121.Haeseleer F, et al. Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator in photoreceptor synaptic function. Nature Neurosci. 2004;7:1079–1087. doi: 10.1038/nn1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zeitz C, et al. Mutations in CABP4, the gene encoding the Ca2+-binding protein 4, cause autosomal recessive night blindness. Am J Hum Genet. 2006;79:657–67. doi: 10.1086/508067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cox JA, et al. Cation binding and conformational changes in VILIP and NCS-1, two neuron-specific calcium-binding proteins. J. Biol. Chem. 1994;269:32807–32814. [PubMed] [Google Scholar]
- 124.Schlecker C, et al. Neuronal calcium sensor-1 enhancement of InsP(3) receptor activity is inhibited by therapeutic levels of lithium. J Clin Invest. 2006;116:1668–1674. doi: 10.1172/JCI22466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guo W, Malin SA, Johns DC, Jeromin A, Nerbonne JM. Modulation of Kv4-encoded K+ currents in the mammalian myocardium by neuronal calcium sensor-1. J. Biol.Chem. 2002;277:26436–26443. doi: 10.1074/jbc.M201431200. [DOI] [PubMed] [Google Scholar]
- 126.Nakamura TY, et al. A role for frequenin, a Ca2+ binding protein, as a regulator of Kv4 K+ currents. Proc. Natl. Acad. Sci. 2001;98:12808–12813. doi: 10.1073/pnas.221168498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hui H, et al. Calcium-sensing mechanism in TRPC5 channels contributing to retardation of neurite outgrowth. J Physiol. 2006;572:165–72. doi: 10.1113/jphysiol.2005.102889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sippy T, Cruz-Martin A, Jeromin A, Schweizer FE. Acute changes in short-term plasticity at synapses with elevated levels of neuronal calcium sensor-1. Nature Neurosci. 2003;6:1031–1038. doi: 10.1038/nn1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weiss JL, Archer DA, Burgoyne RD. NCS-1/frequenin functions in an autocrine pathway regulating Ca2+ channels in bovine adrenal chromaffin cells. J. Biol. Chem. 2000;275:40082–40087. doi: 10.1074/jbc.M008603200. [DOI] [PubMed] [Google Scholar]
- 130.Tsujimoto T, Jeromin A, Saitoh N, Roder JC, Takahashi T. Neuronal calcium sensor 1 and activity-dependent facilitation of P/Q-type calcium channel currents at presynaptic nerve terminals. Science. 2002;295:2276–2279. doi: 10.1126/science.1068278. [DOI] [PubMed] [Google Scholar]
- 131.Koizumi S, et al. Mechanisms underlying the neuronal calcium sensor-1 evoked enhancement of exocytosis in PC12 cells. J. Biol.Chem. 2002;277:30315–30324. doi: 10.1074/jbc.M201132200. [DOI] [PubMed] [Google Scholar]
- 132.Wang C-Y, et al. Ca2+ binding protein frequenin mediates GDNF-induced potentiation of Ca2+ channels and transmitter release. Neuron. 2001;32:99–112. doi: 10.1016/s0896-6273(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 133.Nakamura TY, et al. Novel role of neuronal Ca2+ sensor-1 as a survival factor up-regulated in injured neurons. J Cell Biol. 2006;172:1081–91. doi: 10.1083/jcb.200508156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mercer WA, et al. NAIP interacts with hippocalcin and protects neurons against calcium-induced cell death through caspase-3-dependent and - independent pathways. EMBO J. 2000;19:3597–3607. doi: 10.1093/emboj/19.14.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Palmer CL, et al. Hippocalcin functions as a calcium sensor in hippocampal LTD. Neuron. 2005;47:487–94. doi: 10.1016/j.neuron.2005.06.014. A study implicating hippocalcin in long term depression. The authors identified hippocalcin interaction with the β2 clathrin adaptor protein and suggested that hippocalcin could be involved in the endocytosis of GluR2 AMPA receptors.
- 136.Nagata K, et al. The Map kinase kinase kinase MLK2 co-localizes with activated JNK along microtubultes and associates with kinesin superfamily motor KIF3. EMBO J. 1998;17:149–158. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oh DY, Yon C, Oh KJ, Lee KS, Han J-S. Hippocalcin increases phospholipase D2 expression through extracellular signal-regulated kinase activation and lysophosphatidic acid potentiates the hippocalcin-induced phospholipase D2 expression. J Cell Biochem. 2006;97:1052–1065. doi: 10.1002/jcb.20665. [DOI] [PubMed] [Google Scholar]
- 138.Kobayashi M, et al. Hippocalcin-deficient mice display a defect in cAMP response element-binding protein activation associated with impaired spatial and associative memory. Neuroscience. 2005;133:471–484. doi: 10.1016/j.neuroscience.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 139.Krishnan A, Venkataraman V, Fik-Rymarkiewicz E, Duda T, Sharma RK. Structural, biochemical and functional characterisation of the calcium sensor neurocalcin δ in the inner retinal neurons and its linkage with the rod outer segment membrane guanylate cyclase transduction system. Biochemistry. 2004;43:2708–2723. doi: 10.1021/bi035631v. [DOI] [PubMed] [Google Scholar]
- 140.Brackmann M, Schuchmann S, Anand R, Braunewell KH. Neuronal Ca2+ sensor protein VILIP-1 affects cGMP signalling of guanylyl cyclase B by regulating clathrin-dependent receptor recycling in hippocampal neurons. J Cell Sci. 2005;118:2495–505. doi: 10.1242/jcs.02376. [DOI] [PubMed] [Google Scholar]
- 141.Lin L, et al. The calcium sensor protein visinin-like protein-1 modulates the surface expression and agonist sensitivity of the α4β2 nicotinic acetylcholine receptor. J.Biol. Chem. 2002;277:41872–41878. doi: 10.1074/jbc.M206857200. [DOI] [PubMed] [Google Scholar]
- 142.Dai FF, et al. The neuronal Ca2+ sensor protein visinin-like protein-1 is expressed in pancreatic islets and regulates insulin secretion. J Biol Chem. 2006;281:21942–53. doi: 10.1074/jbc.M512924200. [DOI] [PubMed] [Google Scholar]
- 143.Lautermilch NJ, Few AP, Scheuer T, Catterall WA. Modulation of Cav2.1 channels by the neuronal calcium-binding protein visinin-like protein-2. J. Neurosci. 2005;25:7062–7070. doi: 10.1523/JNEUROSCI.0447-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li H, Guo W, Mellor RL, Nerbonne JM. KChIP2 modulates the cell surface expression of Kv1.5-encoded K+ channels. J. Mol. Cell. Cardiol. 2005;39:121–132. doi: 10.1016/j.yjmcc.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 145.Jo D-G, et al. Pro-apoptotic function of calsenilin/DREAM/KChIP3. FASEB J. 2001;15:589–591. doi: 10.1096/fj.00-0541fje. [DOI] [PubMed] [Google Scholar]
- 146.Lilliehook C, et al. Calsenilin enhances apoptosis by altering endoplasmic reticulum calcium signalling. Mol Cell Neurosci. 2002;19:552–559. doi: 10.1006/mcne.2001.1096. [DOI] [PubMed] [Google Scholar]
- 147.Hamasaki-Katagiri N, Molchanova T, Takeda K, Ames JB. Fission yeast homologue of neuronal calcium sensor-1 (Ncs1p) regulates sporulation and confers calcium tolerance. J. Biol.Chem. 2004;279:12744–12754. doi: 10.1074/jbc.M311895200. [DOI] [PubMed] [Google Scholar]
- 148.Korhonen L, et al. Hippocalcin protects against caspase-12-induced and age-dependent neuronal degeneration. Mol Cell Neurosci. 2004;28:85–95. doi: 10.1016/j.mcn.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 149.Burns ME, Mendez A, Chen J, Baylor DA. Dynamics of cyclic AMP synthesis in retinal rods. Neuron. 2002;36:81–91. doi: 10.1016/s0896-6273(02)00911-x. [DOI] [PubMed] [Google Scholar]
- 150.Mendez A, et al. Role of guanylate cyclase-activating proteins (GCAPs) in setting the flash sensitivity of rod photoreceptors. Proc Natl Acad Sci U S A. 2001;98:9948–53. doi: 10.1073/pnas.171308998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Payne AM, et al. A mutation in guanylate cyclase activator 1A (GUCA1A) in an autosomal dominant cone dystrophy pedigree mapping to a new locus on chromosome 6p21.1. Hum Mol Genet. 1998;7:273–7. doi: 10.1093/hmg/7.2.273. [DOI] [PubMed] [Google Scholar]