Abstract
The West Nile virus minus-strand 3′ terminal stem loop (SL) RNA was previously shown to bind specifically to cellular stress granule (SG) components, T cell intracellular antigen-1 (TIA-1) and the related protein TIAR. In vitro TIAR binding was 10 times more efficient than TIA-1. The 3′(−)SL functions as the promoter for genomic RNA synthesis. Colocalization of TIAR and TIA-1 with the viral replication complex components dsRNA and NS3 was observed in the perinuclear regions of West Nile virus- and dengue virus-infected cells. The kinetics of accumulation of TIAR in the perinuclear region was similar to those of genomic RNA synthesis. In contrast, relocation of TIA-1 to the perinuclear region began only after maximal levels of RNA synthesis had been achieved, except when TIAR was absent. Virus infection did not induce SGs and progressive resistance to SG induction by arsenite developed coincident with TIAR relocation. A progressive decrease in the number of processing bodies was secondarily observed in infected cells. These data suggest that the interaction of TIAR with viral components facilitates flavivirus genome RNA synthesis and inhibits SG formation, which prevents the shutoff of host translation.
Keywords: T cell intracellular antigen-related protein, T cell intracellular antigen-1, viral replication complexes
Eukaryotic cells respond to environmental stresses, such as oxidative stress, heat shock, UV irradiation, endoplasmic reticulum (ER) stress, and some viral infections, by altering the protein expression machinery (1). The expression of proteins responsible for damage repair is increased, whereas translation of constitutively expressed proteins is aborted by redirection of these mRNAs from polysomes to discrete cytoplasmic foci known as stress granules (SGs) for transient storage (2). Several cell proteins, including T cell intracellular antigen-1 (TIA-1), TIA-1-related protein (TIAR) (2), and Ras-Gap-SH3 domain-binding protein (G3BP) are involved in SG assembly (3). Phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) by protein kinase R (PKR) and other kinases prevents the assembly of the active ternary preinitiation complex eIF2-GTP-tRNAMet and inhibits translation initiation and polysome assembly (4). TIA-1 and TIAR bind to these inactive translation initiation complexes as well as to the mRNA poly(A)+ and self-aggregate, promoting the assembly of SGs (2). Processing bodies (PBs), another type of foci identified in the cytoplasm of eukaryotic cells (5), contain components of the 5′–3′ mRNA degradation pathway as well as the miRNA-dependent silencing protein GW182 (6). PBs do not require eIF2α phosphorylation for their assembly (7). Although SGs and PBs differ in their size and shape as well as in their mechanism of assembly, the two types of foci are found side by side in mammalian cells and a physical association between them was demonstrated by real-time fluorescence imaging (7). This study also showed that PBs are highly motile, whereas the positions of SGs are relatively fixed, and that SGs deliver mRNAs to associated PBs for degradation.
TIA-1 and TIAR are essential, multifunctional, nucleocytoplasmic shuttling proteins that are expressed in most types of cells and tissues (8, 9). TIA-1 and TIAR contain three N-terminal RNA recognition motifs (RRM) (10) and a glutamine-rich C-terminal domain, called prion-related domain (PRD), that shares structural and functional characteristics with the aggregation domains of mammalian and yeast prion proteins (11). A recombinant TIA-1 protein lacking the three RNA-binding domains was unable to bind poly(A)+ RNA and recruit it to SGs (2). Deletion of the TIA-1 PRD inhibited protein aggregation and SG assembly (11). PRD aggregation is regulated by the molecular chaperone heat shock protein 70 (HSP70), and overexpression of HSP70 prevented cytosolic aggregation of PRD (11). TIA-1 and TIAR also regulate translational silencing of a subset of cellular mRNAs by binding to AU-rich sequences in the 3′ noncoding regions (NCRs) of these mRNAs (12). In addition, TIA-1 and TIAR proteins interact specifically with the 3′-terminal stem loop (SL) structure of the West Nile virus minus-strand RNA [WNV3′(−)SL] (13).
WNV is classified in the family Flaviviridae, genus Flavivirus, along with a number of other human pathogens, including dengue virus (DV), Japanese encephalitis virus, yellow fever virus, and tick-borne encephalitis virus. The flavivirus genome is a single-stranded, positive sense RNA of ≈11 kb containing a single ORF and is the only viral mRNA produced during the virus replication cycle. Flavivirus replication takes place in the perinuclear region of the cytoplasm of infected cells. Three structural (capsid, membrane, and envelope) and seven nonstructural (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) viral proteins are produced by proteolytic processing of the single polyprotein by viral and cellular proteases (14, 15). The C-terminal portion of NS5 is the RNA-dependent RNA polymerase (RdRp), whereas the N-terminal part is a methyltransferase involved in RNA capping (16). The N terminus of NS3 has serine protease activity, whereas the C terminus contains helicase, NTPase, and triphosphatase activities (14, 15). The NS3, NS5, and NS1 proteins, as well as the hydrophobic NS2a, NS2b, NS4a, and NS4b proteins, were previously shown to be components of membrane bound viral RNA replication complexes located in the perinuclear region of infected cells (17–19).
In this study, both TIA-1 and TIAR were shown to colocalize with the WNV and DV NS3 protein as well as with viral dsRNA, a marker for viral replication complexes, in infected mammalian cells. The data obtained suggest that these interactions facilitate viral genomic RNA synthesis and down-regulate SG formation in infected cells.
Results
TIA-1 and TIAR Colocalize with WNV and DV2 Proteins in Infected Baby Hamster Kidney (BHK) Cells.
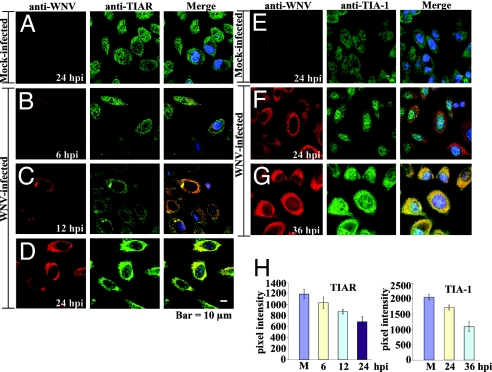
A previous study showed that in C57/BL/6 TIAR−/− MEFs, the replication of WNV was reduced by 6- to 8-fold compared with control MEFs, whereas WNV grew to the same levels in TIA-1−/− MEFs and control MEFs (13). This was consistent with the 10-fold higher binding efficiency of TIAR for the WNV3′(−)SL as compared with TIA-1 (13). To determine whether a flavivirus infection altered the distribution of these cell proteins, BHK cells were infected with WNV at MOI of 5 and at 3, 6, 12, and 24 hours after infection (hpi), the cells were fixed, permeabilized, incubated with anti-TIA-1 or anti-TIAR and then anti-WNV antibody, and visualized by confocal microscopy as described in Materials and Methods and supporting information (SI) Materials and Methods. As expected, TIAR (Fig. 1A, green) and TIA-1 (Fig. 1E, green) were evenly distributed throughout mock-infected cells (20). Under these infection conditions, WNV proteins were detected in ≈90% of the cells. WNV proteins were faintly detected by 6 hpi (Fig. 1B, red). Although at 6 hpi, TIAR was still found in both the cytoplasm and nucleus, this protein was also concentrated in a few bright foci in the cytoplasm (Fig. 1B, green). Merged images obtained at 6 hpi showed colocalization of TIAR foci, with foci of viral protein (Fig. 1B, merge). Foci of TIA-1 were not observed at this time (data not shown). By 12 hpi, the majority of TIAR was in the cytoplasm (Fig. 1C, green) and the amount of WNV protein had increased and was concentrated in the perinuclear region (Fig. 1C, red). Again, colocalization of TIAR but not TIA-1 (data not shown) with the viral proteins was observed mainly in perinuclear foci (Fig. 1C, merge). By 24 hpi, the intensity of WNV protein staining had increased significantly (Fig. 1 D and F, red), and TIAR strongly colocalized in the perinuclear region with the WNV proteins (Fig. 1D, merge). In contrast, only a few bright cytoplasmic foci of TIA-1 colocalized with the WNV proteins by 24 hpi (Fig. 1F, merge; a pattern similar to that seen at 12 hpi with TIAR). Strong colocalization of TIA-1 with the WNV proteins was observed by 36 hpi (Fig. 1G, merge). Quantification of the amount of TIAR or TIA-1 in the nucleus confirmed that the distribution of both TIA-1 and TIAR was altered with time after WNV infection but the redistribution of TIAR occurred more rapidly (Fig. 1H).
Fig. 1.
Colocalization of TIA-1 and TIAR with WNV proteins in infected BHK cells. (A–D) Laser scanning confocal microscopy of mock-infected (A) or WNV-infected (MOI of 5) (B–D) BHK cells stained with anti-WNV (red) and anti-TIAR antibody (green) at the indicated times after infection. (E–G) BHK stained with anti-WNV (red) and anti-TIA-1 antibody (green) at the indicated times after WNV (F and G) or mock (E) infection. Nuclear DNA (blue) was stained with Hoechst 33258 dye. (H) Quantification of the amount of TIAR (Left) or TIA-1 (Right) in the nucleus of mock-infected cells (M) and WNV-infected cells at the indicated times after infection. The relative pixel intensity in the nuclei of 20 cells at each time after infection was measured, and the mean values were plotted. Error bars indicate the standard deviation of the mean.
It was previously reported that the expression of TIA-1 was three times higher in TIAR−/− than in wild-type MEFs (13). To determine whether the time course of TIA-1 redistribution was altered when TIAR was not present, TIAR−/− MEFs were infected with WNV (MOI of 5), and the locations of TIA-1 and WNV proteins were analyzed at 12 and 24 hpi. TIA-1 was equally distributed in both the cytoplasm and nucleus of mock-infected TIAR−/− MEFs (SI Fig. 5A, green), but these cells were significantly larger than BHK cells. The time course of appearance, staining intensity, and distribution of the WNV proteins in these cells (SI Fig. 5 B and C, red) were similar to those observed with infected BHK cells. At 12 hpi, a few bright foci of TIA-1 that colocalized with WNV proteins were observed (SI Fig. 5B, merge), similar to what was observed by 12 hpi for TIAR in BHK cells. By 24 hpi, a significant increase in TIA-1 in the perinuclear region and strong colocalization with viral proteins was observed (SI Fig. 5C). At all times, a significant portion of TIA-1 remained in the nucleus. These results indicate that, in the absence of TIAR, colocalization of TIA-1 with the viral proteins occurred more quickly, with a time course similar to that of TIAR colocalization in cells expressing both proteins.
To determine whether TIA-1/TIAR colocalization with viral proteins was also characteristic of other flavivirus infections, BHK cells were infected with DV2, (MOI of 0.1), a divergent flavivirus from a different serocomplex than WNV, and analyzed at 12, 24, 36, and 72 hpi as described in Materials and Methods. Because of the lower MOI, at 12 and 24 hpi, DV2 proteins were detected in only ≈5% of the cells (data not shown). By 36 hpi, clusters of infected cells with bright virus-specific cytoplasmic foci were observed (≈40% of the cells) (SI Fig. 6 B and D, red). At 72 hpi, in ≈60% of the cells, DV2 proteins were detected with a distribution and staining intensity (SI Fig. 6 C and E, red) similar to that observed in WNV-infected cells at 24 hpi (Fig. 1 D and F, red). Strong colocalization of both TIAR (SI Fig. 6B and C, merge) and TIA-1 (SI Fig. 6 D and E, merge) with DV2 proteins was detected at 36 and 72 hpi. This colocalization was primarily in the perinuclear region but was also observed in foci extending into the cytoplasm of the infected cells (SI Fig. 6 B–E, merge). In the uninfected cells visible in the middle panels of SI Fig. 6 B–E, TIA-1 and TIAR were equally distributed in the cytoplasm and nucleus. These results suggest that the colocalization of TIA-1/TIAR with viral proteins is a general feature of flavivirus infections.
TIA-1 and TIAR Interact with Sites of WNV and DV2 RNA Replication.
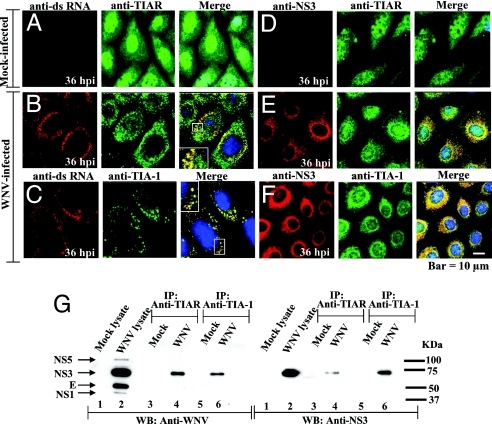
In previous in vitro RNA–protein interaction assays, the TIA-1/TIAR proteins were shown to bind specifically to the WNV3′(−)SL RNA (13). The minus-strand RNA of flaviviruses is present in infected cells only in RNA replication complexes (14, 15). An antibody to dsRNA that does not detect either cellular ribosomal RNA or tRNA was previously used to detect flavivirus replication complexes in infected cells (21). To test whether TIA-1/TIAR colocalized with viral dsRNA in infected cells, BHK cells infected with WNV (MOI of 0.1) or DV2 (MOI of 0.1) were fixed and incubated with anti-dsRNA and anti-TIA-1 or anti-TIAR antibody. A lower MOI for WNV and later time points were used to better visualize the replication complexes. Perinuclear foci stained with anti-dsRNA antibody were observed in WNV-infected cells at 36 hpi (Fig. 2 B and C, red) but not in mock-infected cells (Fig. 2A). In WNV-infected cells, strong colocalization of TIA-1/TIAR with the dsRNA was observed in bright perinuclear foci (Fig. 2 B and C, merge). Similar dsRNA perinuclear foci and similar TIA-1/TIAR colocalization with these foci were observed in DV2-infected cells at 72 hpi (SI Fig. 7 B and C), indicating that TIA-1/TIAR strongly colocalized with flavivirus RNA complexes in infected cells.
Fig. 2.
Interaction of TIA-1 and TIAR with WNV replication complex components. BHK cells were infected with WNV at a MOI of 0.1. (A–F) Laser scanning confocal microscopy of mock-infected (A and D) and WNV-infected (B, C, E, and F) cells stained with anti-dsRNA (A–C, red) or anti-NS3 (D–F, red) and then with either anti-TIAR (A, B, D, and E, green) or anti-TIA-1 (C and F, green) antibodies at 36 hpi. Enlarged views are included in the B and C merged panels. (G) Coimmunoprecipitation of WNV proteins by anti-TIA-1 and anti-TIAR antibodies. BHK cells were infected with WNV at a MOI of 5. Immunoprecipitates were visualized by Western blot using anti-WNV (Left) or anti-NS3 antibody (Right). Lane 1, mock-infected lysate; lane 2, WNV-infected lysate; lanes 3 and 5, mock-infected immunoprecipitates; lanes 4 and 6, WNV-infected immunoprecipitates. Arrows indicate the positions of the WNV proteins detected. Positions of molecular weight standards are indicated on the left.
Previous immunofluorescence studies using antibodies specific to dsRNA as well as to individual flavivirus NS proteins suggested that NS3 and NS5, as well as NS1, NS2a, and NS4a, associate with ds viral RNA in flavivirus replication complexes located in the perinuclear region of infected cells (22, 23). Broader areas of colocalization were observed with the anti-WNV (Fig. 1) than with the anti-dsRNA antibody, suggesting that TIA-1/TIAR might also interact with a viral nonstructural protein. Coimmunoprecipitation experiments were done by using lysates prepared from mock-infected or WNV-infected BHK cells at 24 hpi and anti-TIAR or anti-TIA-1 antibody, as described in Materials and Methods. Cell lysates, as well as immunoprecipitates, were then analyzed by SDS/PAGE and transferred to nitrocellulose membranes for Western blot analysis using anti-WNV (Fig. 2G Left). Four WNV proteins, presumed to be NS5, NS3, E, and NS1, were detected in lysates from WNV-infected cells (Fig. 2G Left, lane 2) but not in lysates or immunoprecipitates from mock-infected cells (Fig. 2G Left, lanes 1, 3, and 5). In immunoprecipitates from WNV-infected cells, both anti-TIAR (Fig. 2G Left, lane 4) and anti-TIA-1 (Fig. 2G Left, lane 6) antibody coimmunoprecipitated a viral protein with the expected size of NS3. To verify that this was NS3, samples precipitated with anti-TIA-1 or anti-TIAR antibodies were analyzed by Western blot analysis using an anti-WNV NS3 monoclonal antibody. NS3 was detected in lysates from WNV-infected cells after immunoprecipitation with both anti-TIAR (Fig. 2G Right, lane 4) or anti-TIA-1 (Fig. 2G Right, lane 6) antibody.
Anti-WNV NS3 monoclonal antibody was next used to study the colocalization of TIA-1/TIAR with NS3 in BHK cells infected with WNV or DV2 at 36 or 72 hpi, respectively. NS3 was previously shown to be broadly distributed around the nucleus and to colocalize with dsRNA foci in flavivirus-infected cells (21). In the merged images, both TIA-1 and TIAR strongly colocalized with NS3 in WNV-infected cells (Fig. 2 E and F, merge). Bright focal areas, as well as more diffuse areas of colocalization, were observed. Similar results were observed in DV2-infected cells at 72 hpi (SI Fig. 7 E and F).
WNV and DV2 Interfere with SG Formation in Infected BHK Cells.
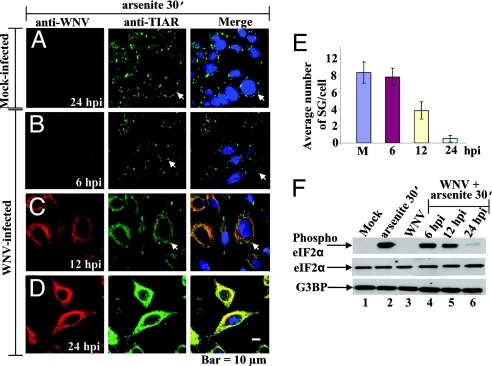
TIA-1/TIAR were previously reported to be required for the formation of SGs in cells after treatment with arsenite (2), and both proteins were shown to colocalize with viral nonstructural proteins and replication complexes in flavivirus-infected cells. To determine whether a flavivirus infection would interfere with cellular SG formation, BHK cells were mock-infected or infected with WNV and exposed to oxidative stress by treatment with 0.5 mM arsenite for 30 min. Infected cells were treated at 6, 12, or 24 hpi. The cells were then fixed and stained with anti-WNV (Fig. 3 A–D, red) and anti-TIAR antibody (Fig. 3 A–D, green). At 6 hpi, when only a small amount of colocalization of TIAR with WNV proteins was observed (Fig. 3B, merge), the number of SGs induced by the arsenite was slightly reduced (Fig. 3B, green; Fig. 3E) as compared with mock-infected cells (Fig. 3A, green; Fig. 3E). At 12 hpi, SG formation further decreased (Fig. 3C, green; Fig. 3E) as colocalization of TIAR and when WNV proteins increased (Fig. 3C, merge). Few, if any, SGs were detected at 24 hpi (Fig. 3D, green; Fig. 3E) in cells showing strong colocalization between TIAR and WNV proteins (Fig. 3D, merge). WNV infection alone did not induce SG formation at any time after infection (Fig. 1 B–D, green). The effect of DV2 infection on SG formation was also investigated. BHK cells infected with DV2 for 36 or 72 h were treated with 0.5 mM arsenite for 30 min and immunostained with anti-DV2 and anti-TIAR (SI Fig. 8). At both times, strong colocalization of TIAR and DV2 proteins was observed. Few SGs were observed in infected cells (SI Fig. 8 B and C, green; SI Fig. 8D), whereas many SGs were observed in uninfected cells (SI Fig. 8A, green).
Fig. 3.
WNV infection interferes with SG formation. (A–D) Laser scanning confocal microscopy of mock-infected (A) and WNV-infected (MOI of 5) (B–D) BHK cells treated with 0.5 mM arsenite for 30 min at the indicated times after infection. The cells were fixed, permeabilized, and stained with anti-WNV (A–D, red) and with the SG marker, anti-TIAR (A–D, green). Arrows indicate representative SG. (E) Quantification of the number of SGs in mock-infected (M) and WNV-infected cells at the indicated times after infection. Visible SGs in 70 cells were counted for each experimental condition, and the average number of SG per cell was plotted. Error bars indicate the standard deviation of the mean. (F) Western blot analysis of phospho-eIF2α in BHK cells mock-infected or infected with WNV for 24 h or in infected cells treated with 0.5 mM arsenite for 30 min at the indicated times after infection. Lane 1, mock-infected BHK cells; lane 2, cells treated with 0.5 mM arsenite for 30 min; lane 3, cells infected with WNV (MOI of 5); lanes 4–6, cells infected with WNV and then treated with arsenite for 30 min at the indicated times. Phospho-eIF2α (S51) (Top), total eIF2α (Middle), and G3BP (Lower) were detected with specific antibodies (SI Materials and Methods).
Because phosphorylation of eIF2α at S51 also regulates SG formation, the effect of WNV and DV2 infections on eIF2α phosphorylation was assayed. BHK cells were infected with either virus, and, at various times after infection, cells were treated for 30 min with 0.5 mM arsenite. Control infected cells were not treated with arsenite. Proteins in cell lysates were analyzed by Western blot using a polyclonal antibody specific for eIF2α phosphorylated at S51 (Fig. 3F). Under these experimental conditions, phosphorylation of eIF2α was not detected in mock-infected cells or 24 h after WNV (Fig. 3F, lane 3) or 36 h after DV2 (SI Fig. 8E, lane 1) infection. High levels of eIF2α phosphorylation were observed in extracts of mock-infected cells treated with arsenite (Fig. 3F, lane 2). Reduced levels of phospho-eIF2α were detected in WNV-infected extracts by 12 hpi and were further decreased by 24 hpi (Fig. 3F, lanes 4–6). In the DV2 extracts, decreased phosphorylation of eIF2α was observed at later times after infection (SI Fig. 8E, lanes 2–5). The large number of uninfected cells present in the DV2-infected cultures are the source of the higher levels of phospho-eIF2α observed until 6 days after infection (dpi). The levels of total eIF2α and of another protein involved in SG formation, G3BP, remained constant under all conditions tested and after infection with both viruses (Fig. 3F and SI Fig. 8E). Although neither WNV nor DV2 infection induced SG formation in BHK cells, cells infected with both flaviviruses became progressively more resistant to SG induction by arsenite with time after infection.
WNV and DV2 Infections Interfere with PB Assembly.
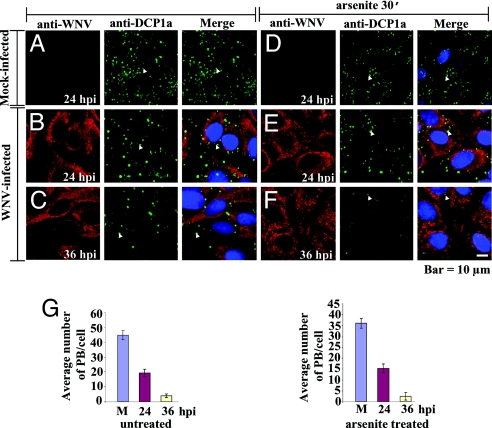
SGs and PBs share some components and are dynamically linked by their role in regulating cytoplasmic mRNA storage and/or degradation (7, 24). To test whether WNV and DV2 infections also interfere with PB assembly, BHK cells mock-infected or infected for various times with either WNV or DV2 were treated or not treated for 30 min with 0.5 mM arsenite. The cells were then fixed and incubated with antibody to the PB marker DCP1a and anti-WNV (Fig. 4) or anti-DV2 (SI Fig. 9). PBs were detected in untreated cells (Fig. 4 A–C, green; SI Fig. 9 A–C, green) as well as in arsenite-treated cells (Fig. 4 D–F, green; SI Fig. 9 D–F, green). In uninfected, WNV-infected, and DV2-infected cells, PBs were evenly distributed throughout the cytoplasm (Fig. 4 and SI Fig. 9). In both uninfected and infected cells treated with arsenite, the number of PBs was decreased (Fig. 4G; SI Fig. 9G), and the remaining PBs were mainly located around the nucleus (Fig. 4 D–F, green; SI Fig. 9 D–F, green). In WNV-infected cells, a noticeable reduction in PB formation was observed by 24 hpi in untreated (Fig. 4B, green; Fig. 4G Left) and arsenite-treated (Fig. 4E, green; Fig. 4G Right) cells as compared with mock-infected cells (Fig. 4 A and D, green; Fig. 4G). A further reduction in PB formation was observed in both arsenite-untreated (Fig. 4C, green; Fig. 4G Left) and treated (Fig. 4F, green; Fig. 4G Right) WNV-infected cells at 36 hpi. In DV2-infected cells, a reduction in PB formation was observed at later times after infection (SI Fig. 9 B, C, E, and F, green; SI Fig. 9G), consistent with the slower replication kinetics of DV2. These results indicate that both WNV and DV2 infections interfere with PB assembly in BHK cells.
Fig. 4.
WNV infection interferes with PB assembly. (A–F) Laser scanning confocal microscopy of mock-infected (A and D) and WNV-infected (MOI of 5) (B, C, E, and F) cells that were treated (Right) or not treated (Left) with arsenite for 30 min at the indicated times. The cells were stained with anti-WNV (A–F, red) and anti-DCP1a (a PB marker) (A–F, green) antibody. Arrowheads indicate representative PBs. (G) Quantification of the number of PBs in mock-infected (M) and WNV-infected cells treated (Right) or not treated (Left) with arsenite. PBs in 50 cells were counted for each experimental condition at the indicated times after infection, and the average number of PBs per cell was plotted. Error bars indicate the standard deviation of the mean.
Discussion
In normal cells that are not undergoing stress, TIA-1 and TIAR are distributed fairly evenly between the cytoplasm and nucleus. In response to stress, TIA-1 and TIAR aggregate with cell mRNAs in SGs (2). In this study, a unique distribution pattern for these proteins was observed in cells infected with two different flaviviruses, WNV and DV2. First TIAR and then TIA-1 relocated to the perinuclear region in flavivirus-infected cells. In WNV-infected cells, perinuclear foci of TIAR were detected by 6 hpi, and a significant amount of TIAR was observed in this region by 12 hpi. With TIA-1, perinuclear foci were not detected until 24 hpi, and significant concentrations of TIA-1 in this region were not seen until 36 hpi. Interestingly, the initiation of TIA-1 relocation to the perinuclear region in WNV-infected cells did not begin until the majority of TIAR had already relocated to this region.
By 6 hpi, newly translated WNV proteins were detectable in the cytoplasm by immunoflourescence, and, by 12 hpi, the amount of viral protein had increased and was concentrated in the perinuclear region. Colocalization of bright foci of TIAR and of viral proteins was detected by 6 hpi and the amount of colocalization progressively increased thereafter. These bright perinuclear foci also contained dsRNA, identifying them as viral-replication complexes. Previous in vitro RNA–protein-binding assays showed that TIAR bound 10 times more efficiently than TIA-1 to the WNV 3′(−)SL RNA (13). The different kinetics of TIA-1 and TIAR redistribution in flavivirus-infected cells could be due to the more efficient interaction between cytoplasmic TIAR and the 3′ ends of viral minus strand RNAs. The 3′(−)SL is thought to contain promoter elements for genomic RNA initiation, and it was previously proposed that TIAR functions as a transcription factor for genomic RNA initiation (13). This hypothesis was recently supported by data showing that mutagenesis of the mapped TIA-1/TIAR-binding sites on the WNV 3′(−)SL RNA in a WNV infectious clone negatively affected genomic RNA synthesis (M.M.E. and M.A.B., unpublished data). Previous analyses of the kinetics of flavivirus RNA synthesis showed an initial low peak of genomic RNA synthesis between 6 and 10–12 hpi, followed by a switch to exponentially increasing genomic RNA synthesis (14, 25). The level of genomic RNA synthesis in infected cells correlated temporally with the extent of TIAR relocation to the perinuclear region.
It was shown that the yield of WNV produced by TIAR−/− MEFs, which express three times more TIA-1 than wild-type MEFs, was reduced by only 6- to 8-fold as compared with TIA-1−/− or control MEFs (13). These data suggested that TIA-1 could also function as a viral transcription factor when TIAR was not present. Consistent with this hypothesis, colocalization of TIA-1 and WNV proteins in infected TIAR−/− MEFs occurred in a time course similar to that observed with TIAR in cells expressing both proteins. These data also suggest that the ability of TIA-1 to relocate to the perinuclear region is not being actively prevented in infected cells. However, the possibility that TIA-1 is interacting with a cell protein partner and so is not available for interaction with viral components until later times after infection cannot be ruled out.
The data suggest the possibility that TIA-1/TIAR may also interact with a viral nonstructural protein as well as with the 3′(−)SL RNA. WNV NS3 was coimmunoprecipatated by both anti-TIA-1 and anti-TIAR antibodies. The four hydrophobic nonstructural proteins (NS2a, NS2b, NS4a, and NS4b) have been reported to be tightly complexed with NS3 in dense membrane fractions (26), whereas the RdRp NS5 was more loosely associated (27). Although the results indicate that viral replication complexes were coimmunoprecipitated by anti-TIA-1 and anti-TIAR antibodies, they do not provide evidence of a direct interaction between TIA-1/TIAR and a particular viral NS protein. Flaviviruses initiate RNA synthesis de novo. The interaction of TIA-1 or TIAR with a protein in a flavivirus RdRp complex might enhance the preferential recognition of minus strand templates by these complexes. TIA-1/TIAR do not bind to the 3′-terminal SL RNA of the genomic (+) RNA template (M.M.E. and M.A.B., unpublished data). As the number of TIA-1/TIAR-containing complexes progressively accumulates in infected cells, the synthesis of genomic RNA would be expected to be significantly and preferentially amplified over minus-strand RNA. The binding of TIA-1/TIAR to the 3′(−)SL RNA might stabilize it, allowing other necessary factors to assemble to facilitate genomic RNA synthesis. It is currently not known whether the self-aggregating domains of TIA-1/TIAR play a role in these protein–protein and RNA–protein interactions.
Flaviviruses prevent rather than facilitate the shutoff of host cell translation. Flaviviruses replicate at significantly slower rates than most other positive-strand RNA viruses in mammalian and avian cells, and rapid shutoff of cell protein synthesis with the resulting negative effect on cell viability would have a deleterious effect on flavivirus yields. That flavivirus infections do not induce SG formation and that infected cells become increasingly resistant to the induction of SG by arsenite treatment with time after infection has not been previously reported. These data suggest that flavivirus infections actively inhibit SG formation. The time course of the resistance to SG formation coincided with that of the relocation of TIAR to the perinuclear region rather than with the relocation of TIA-1 or with decreased phosphorylation of eIF2α. These observations suggest that the sequestration of TIAR through its interaction with the viral 3′(−)SL RNA and possibly also a viral nonstructural protein is the initial reason for the observed decrease in SG formation in flavivirus-infected cells. A previous study showed that although eIF2α was phosphorylated in both TIA−/− and TIAR−/− MEFs, SG formation was impaired in TIA−/− MEFs but not in TIAR−/− MEFs, suggesting that TIAR alone could not form SGs (11). The observation that SG formation was inhibited to some extent when TIAR, but not TIA-1, was sequestered in the perinuclear region suggests the possibility that additional components necessary for SG formation may also be sequestered in the perinuclear region of flavivirus-infected cells. If self-aggregation of TIAR in the perinuclear region occurs, this might attract additional SG components. In the previous study that identified TIAR as a WNV 3′(−)SL RNA-binding protein, three additional cell proteins (p50, p60, and p108) were shown to bind to this viral RNA (13). Some of these viral RNA-binding proteins could also be components of SGs. The assembly of pseudoSG complexes around the viral minus-strand RNA replication complexes might be part of the remodeling of the perinuclear membranes by virus infection to create an environment for efficient genomic RNA synthesis and encapsidation.
Complete resistance to induction of SG and a significant decrease in eIF2α phosphorylation at S51 were not observed in arsenite-treated, WNV infected cells until 24 hpi, a time by which maximum levels of virus replication had been achieved. Unlike flaviviruses, both Semliki Forest virus (SFV), a positive-strand RNA alpha togavirus (28) and some strains of reovirus, a double-stranded RNA virus (29), rapidly shut off host cell protein synthesis. For both viruses, the phosphorylation of eIF2α (S51) as well as the aggregation of TIA-1/TIAR into SG were shown to be important for shut off of host cell protein translation (28, 29). In cells subjected to environmental stresses, the activation of PKR and/or other cellular kinases leads to the phosphorylation of eIF2α (20). Activation of PKR, followed by phosphorylation of eIF2α by PKR and the formation of SGs was postulated to result in the shut off of host cell translation in cells infected with SFV (28, 30) or Sindbis virus (30). The expression of P58IPK, a PKR/PERK inhibitor, was shown to be decreased in L929 cells infected with host shutoff-inducing strains of reovirus, and high levels of phospho-eIF2α were detected (29). In contrast, in cells infected with two flaviviruses, DV2 or Japanese encephalitis virus (MOI of 3), the expression of P58IPK was significantly up-regulated at 24 hpi (31). The timing of this up-regulation was exactly coincident with the decrease in eIF2α phosphorylation observed in arsenite-treated WNV-infected (MOI of 5) cells in the present study. Yu et al. (31) also showed that the up-regulation of P58IPK was the result of activation of the cellular unfolded protein response by the accumulation of several different flavivirus proteins in infected cells. The initial shut off of SG formation in flavivirus-infected cells appears to be due to the sequestration of TIAR, and possibly also of other SG components in the perinuclear region of infected cells through interaction with viral components. A direct benefit of this interaction for the virus is that it likely facilitates significant amplification of genomic RNA synthesis. An indirect benefit is that this interaction inhibits the shutoff of host protein synthesis. However, flaviviruses appear to use multiple mechanisms to inhibit SG formation, as evidenced by the up-regulation of P58IPK at later times after infection mediated by the accumulation of viral proteins.
PBs are involved in mRNA degradation and in translational repression (5). In the present study, a similar decrease in PB assembly was observed in both untreated and arsenite-treated flavivirus-infected cells. This decrease was temporally correlated with the initiation of TIA-1 relocation and the decrease in eIF2α phsophorylation in arsenite-treated cells. The mechanism by which flaviviruses interfere with PB assembly in infected cells is not yet known. The interference with PB assembly may be related to activation of the unfolded protein response and would provide an additional mechanism to prevent translational repression and protect viral RNA as well as cell mRNA from degradation.
Materials and Methods
Cells.
BHK-21, strain WI2 cells (32) were maintained as described (33). C57/BL/6 and C57/BL/6 TIAR−/− MEF cell lines were provided by Paul Anderson (Brigham and Woman's Hospital, Boston, MA) and grown in MEM supplemented with 10% FCS, 1× MEM nonessential amino acids (Invitrogen, Carlsbad, CA), 1 mM sodium pyruvate (Invitrogen), 2 mM glutamine (Invitrogen), and 10 μg/ml gentamycin.
Viruses.
A stock of WNV strain Eg101 was prepared in BHK cells (2 × 108 PFU/ml) as described (33). DV2, strain Bangkok D80-100, was provided by Walter Brandt (Walter Reed Army Institute of Research, Washington, DC). The titer of a 10% wt/vol suckling mouse brain homogenate was 2 × 106 PFU/ml. Cells were infected with WNV at a MOI of 5 or 0.1 and with DV2 at a MOI of 0.1. Virus was adsorbed to cells for 1 h at room temperature, the inoculum was removed, and the cells were washed three times with serum-free medium. MEM containing 5% FCS was added, and the infected cells were incubated at 37°C in a CO2 incubator. To induce stress, cells were treated at various times after infection with 0.5 mM sodium m-arsenite (Sigma–Aldrich St. Louis, MO) in MEM for 30 min. Virus growth experiments were done as described (13).
Western Blotting.
Western blot analysis was carried out as described (33). Membranes were blocked in 1× Tris-buffered saline (TBS) containing 5% nonfat dry milk (NFDM). Primary antibodies were diluted in 1× TBS containing 5% BSA (1:1,000 for anti-phospho-eIF2α, 1:500 for anti-eIF2α, 1:3,000 for anti-G3BP, and 1:500 for anti-WNV and anti-WNV NS3 antibodies), and the secondary antibody was diluted 1:2,000 in 1× TBS containing 5% NFDM.
Coimmunoprecipitation Assays.
BHK cells (2 × 106) were mock-infected or infected with WNV at a MOI of 5. At 24 hpi, cell lysates were prepared by using lysis buffer containing 50 mM sodium phosphate (pH 7.2), 150 mM NaCl, 1% Nonidet P-40, and Complete, Mini, EDTA-free protease inhibitor mixture (Roche, Indianapolis, IN). Cell lysates were incubated on ice for 30 min, sonicated, and then centrifuged at 2,000 × g for 5 min at 4°C. The supernatant (250 μl per ≈100 μg of total protein) was precleared with protein A/G-magnetic beads (New England Biolabs, Beverly, MA) for 1 h at 4°C with rotation. The clarified supernatants were incubated with 1 μg of anti-TIA-1 or anti-TIAR antibody at 4°C for 1 h with rotation, and then beads were added and incubated for 1 h. Beads were collected magnetically, washed seven times with 1 ml of lysis buffer, and proteins were eluted by boiling for 5 min. Proteins were separated by 12% SDS/PAGE, transferred to nitrocellulose membranes, and analyzed by Western blotting using either mouse anti-WNV or anti-NS3 antibody.
Detection of Intracellular and Cellular Viral Proteins by Immunofluorescence.
BHK, C57BL/6, and TIAR−/− cells (2 × 104) were seeded onto 3-mm coverslips (Fisher Scientific, Pittsburgh, PA) and grown to ≈50% confluency. At various times after infection, cells were fixed in 4% paraformaldehyde (Sigma–Aldrich) in PBS for 10 min at room temperature, permeabilized with 100% chilled methanol for 10 min at −20°C, washed three times in PBS, and incubated with blocking buffer [PBS containing 5% heat inactivated horse serum (Invitrogen)] overnight at 4°C. Primary antibodies were diluted in blocking buffer (1:1,000 for anti-TIAR, anti-TIA-1, and anti-Dcap1; 1:200 for anti-dsRNA; and 1:100 for anti-WNV, anti-DV2, and anti-WNV NS3 antibody) and incubated with cells at 37°C for 1 h. Cells were then washed three times for 10 min with PBS and incubated for 1 h at 37°C with Texas red- and FITC-labeled secondary antibodies diluted 1:300 in blocking buffer containing 0.5 μg/ml Hoechst 33258 dye (Molecular Probes, Bedford, MA) to stain the nuclear DNA. Coverslips were mounted in Prolong Gold antifade reagent (Invitrogen), and the cells were scanned with a confocal microscope LSM 510 (Zeiss, Oberkochen, Germany) using a 100× oil-immersion objective. The images compared for each experimental series were collected by using the same instrument settings, and the images were analyzed by using Zeiss software version 3.5.
Supplementary Material
Acknowledgments
We thank Birigit Neuhaus for assistance with confocal microscopy and W. G. Davis and Svetlana V. Scherbik for data discussions and technical advice. This work was supported by Public Health Service Research Grant AI048088 (to M.A.B.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviations
- BHK
baby hamster kidney
- WNV
West Nile virus
- TIA-1
T cell intracellular antigen-1
- TIAR
T cell intracellular antigen-related protein
- SG
stress granule
- PB
processing body.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703348104/DC1.
References
- 1.Kedersha N, Anderson P. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 2.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Anderson P, Kedersha N. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eulalio A, Behm-Ansmant I, Izaurralde E. Nat Rev. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 6.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. Molecular Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck AR, Medley QG, O'Brien S, Anderson P, Streuli M. Nucleic Acids Res. 1996;24:3829–3835. doi: 10.1093/nar/24.19.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin K, Li W, Nagayama T, He X, Sinor AD, Chang J, Mao X, Graham SH, Simon RP, Greenberg DA. J Neurosci Res. 2000;59:767–774. doi: 10.1002/(SICI)1097-4547(20000315)59:6<767::AID-JNR9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 10.Anderson P. Curr Top Microbiol Immunol. 1995;198:131–143. doi: 10.1007/978-3-642-79414-8_8. [DOI] [PubMed] [Google Scholar]
- 11.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson P, Kedersha N. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. J Virol. 2002;76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinton MA. Annu Rev Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- 15.Lindenbach BD, Rice CM. Flaviviridae: The Viruses and Their Replication. Philadelphia: Lippincott–Raven; 2007. [Google Scholar]
- 16.Koonin EV. J Gen Virol. 1993;74(Pt 4):733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- 17.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 18.Uchil PD, Satchidanandam V. J Biol Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 19.Vasudevan SG, Johansson M, Brooks AJ, Llewellyn LE, Jans DA. Farmaco. 2001;56:33–36. doi: 10.1016/s0014-827x(01)01014-x. [DOI] [PubMed] [Google Scholar]
- 20.Anderson P, Kedersha N. J Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 21.Miller S, Sparacio S, Bartenschlager R. J Biol Chem. 2006;281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- 22.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westaway EG, Mackenzie JM, Khromykh AA. Adv Virus Res. 2003;59:99–140. doi: 10.1016/s0065-3527(03)59004-2. [DOI] [PubMed] [Google Scholar]
- 24.Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. J Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 25.Trent DW, Swensen CC, Qureshi AA. J Virol. 1969;3:385–394. doi: 10.1128/jvi.3.4.385-394.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu PW, Westaway EG. Arch Virol. 1992;125:177–191. doi: 10.1007/BF01309636. [DOI] [PubMed] [Google Scholar]
- 27.Grun JB, Brinton MA. J Virol. 1987;61:3641–3644. doi: 10.1128/jvi.61.11.3641-3644.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. Mol Biol Cell. 2005;16:3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. J Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu CY, Hsu YW, Liao CL, Lin YL. J Virol. 2006;80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaheri A, Sedwick WD, Plotkin SA, Maes R. Virology. 1965;27:239–241. doi: 10.1016/0042-6822(65)90170-4. [DOI] [PubMed] [Google Scholar]
- 33.Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. J Virol. 2006;80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.