Abstract
Abscisic acid (ABA) is an important phytohormone regulating various plant processes, including seed germination. Although phosphorylation has been suggested to be important, the protein kinases required for ABA signaling during seed germination and seedling growth remain elusive. Here, we show that two protein kinases, SNF1-RELATED PROTEIN KINASE2.2 (SnRK2.2) and SnRK2.3, control responses to ABA in seed germination, dormancy, and seedling growth in Arabidopsis thaliana. A snrk2.2 snrk2.3 double mutant, but not snrk2.2 or snrk2.3 single mutants, showed strong ABA-insensitive phenotypes in seed germination and root growth inhibition. Changes in seed dormancy and ABA-induced Pro accumulation consistent with ABA insensitivity were also observed. The snrk2.2 snrk2.3 double mutant had a greatly reduced level of a 42-kD kinase activity capable of phosphorylating peptides from ABF (for ABA Response Element Binding Factor) transcription factors. ABA-induced expression of several genes whose promoters contain an ABA response element (ABRE) was reduced in snrk2.2 snrk2.3, suggesting that the mechanism of SnRK2.2 and SnRK2.3 action in ABA signaling involves the activation of ABRE-driven gene expression through the phosphorylation of ABFs. Together, these results demonstrate that SnRK2.2 and SnRK2.3 are redundant but key protein kinases that mediate a major part of ABA signaling in Arabidopsis.
INTRODUCTION
The phytohormone abscisic acid (ABA) is a key factor in regulating developmental and physiological processes in plants, including seed dormancy and germination and seedling growth, as well as in controlling many abiotic stress responses (Leon-Kloosterziel et al., 1996; Schroeder et al., 2001; Bray, 2002; Finkelstein and Gibson, 2002; Zhu, 2002; Assmann, 2003; Chow and McCourt, 2004;Yamaguchi-Shinozaki and Shinozaki, 2006). Forward genetic screens for ABA response mutants in seed germination have identified several mutants showing insensitivity to ABA, such as the dominant aba insensitive1 (abi1) and abi2 and recessive abi3, abi4, and abi5 mutants of Arabidopsis thaliana (Koornneef et al., 1984; Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000). Although abi1 and abi2 impair many ABA responses, including the inhibition of seed germination and seedling growth and the promotion of stomatal closure, abi3, abi4, and abi5 only show ABA insensitivity in seed germination and early seedling development (Koornneef et al., 1984; Finkelstein and Somerville, 1990; Ooms et al., 1993; Parcy et al., 1994). ABI3, ABI4, and ABI5 are transcription factors that are expressed mainly in seeds with only low levels of expression in vegetative tissues (Giraudat et al., 1992; Finkelstein et al., 1998, 2002; Finkelstein and Lynch, 2000). ABI1 and ABI2 were found to be protein phosphatases (Meyer et al., 1994; Leung et al., 1994, 1997) that negatively regulate ABA responses (Sheen, 1998), suggesting that there exists a protein kinase(s) as a positive regulator of ABA signaling. With the exception of a protein kinase that positively regulates stomatal responses to ABA (Mustilli et al., 2002; Yoshida et al., 2002), ABA-activated protein kinases as positive regulators of ABA responses have not been reported.
The importance of phosphorylation has also been indicated by analyzing the activation of ABA response element (ABRE) binding factors (ABFs; also referred to as AREBs), which are basic leucine zipper-type (bZIP) transcription factors involved in ABA signaling. ABFs, including ABF1, ABF2 (AREB1), ABF3, ABF4 (AREB2), and ABI5, bind to the ABRE, which is a conserved cis element in the promoters of many ABA-induced genes (Guiltinan et al., 1990; Yamaguchi-Shinozaki and Shinozaki, 2006), and activate transcription (Choi et al., 2000; Uno et al., 2000). These ABA-responsive genes encode, for example, putative protective proteins, enzymes required for osmolyte synthesis or transcription factors that in turn regulate still other changes in gene expression (Bray, 2002; Zhu, 2002). Consistent with the ABA-insensitive phenotype of abi5, overexpression of ABF3, ABF4, and ABI5 caused ABA hypersensitivity in germination and seedling growth (Lopez-Molina et al., 2001; Kang et al., 2002). It has also been demonstrated that the ABF proteins must themselves be activated in an ABA-dependent manner to increase the expression of target genes (Uno et al., 2000). Several lines of evidence have shown that phosphorylation is involved in the activation of ABFs. ABI5 was phosphorylated after ABA treatment (Lopez-Molina et al., 2001). Likewise, the protein kinase inhibitor staurosporine suppressed the activation of ABF2 by ABA (Uno et al., 2000). Glutathione S-transferase (GST)–fused ABF2 or ABF4 proteins could be phosphorylated in vitro by extracts from ABA-treated plants (Uno et al., 2000). Also, expression of a phosphorylation-mimicking, and presumably constitutively active, form of ABF2 caused reduced germination and seedling growth and increased the expression of ABA-responsive genes in plants under unstressed conditions (Furihata et al., 2006). These results are consistent with the existence of protein kinases that act as positive regulators of ABA signaling.
In the regulation of stomatal aperture, SnRK2.6 (for SNF1-related protein kinase2.6/OST1) has been identified as a positive regulator of ABA signaling (Mustilli et al., 2002; Yoshida et al., 2002). SnRK2.6 is a member of the SnRK family of protein kinases. The Arabidopsis genome contains 38 SnRKs, of which 10 (SnRK2.1 to SnRK2.10) are SnRK2s (Hrabak et al., 2003). There are also 10 SnRK2 class kinases (SAPK1 to SAPK10) in rice (Oryza sativa). Although related to yeast SNF1, SnRK2s, as well as SnRK3s, appear to be plant-specific classes of kinases. Many of these kinases have been assigned different names in past reports, but the SnRK2.1 to SnRK2.10 designations are now the accepted nomenclature (Hrabak et al., 2003). Several experiments have shown that the kinase activity of SnRK2s can be activated by ABA and that SnRK2s phosphorylate ABFs in several tissues. In Vicia faba, the SnRK2-type protein kinase AAPK is activated by ABA in guard cells and regulates stomatal closure (Li et al., 2000). Of the Arabidopsis SnRK2s, SnRK2.2, SnRK2.3, SnRK2.6, SnRK2.7, and SnRK2.8 could be activated by ABA when expressed in Arabidopsis protoplasts (Boudsocq et al., 2004). Consistent with this finding, green fluorescent protein–fused SnRK2.2, SnRK2.3, SnRK2.6, SnRK2.7, and SnRK2.8 expressed in T87 cells were also activated by ABA and phosphorylated GST-fused fragments of ABF2 and ABF4 (Furihata et al., 2006; Yoshida et al., 2006). In rice, SAPK8, SAPK9, and SAPK10, which are homologous with SnRK2.2, SnRK2.3, and SnRK2.6, were activated by ABA in a protoplast system (Kobayashi et al., 2004). These SAPKs phosphorylated TRAB1, which is a rice ortholog of the Arabidopsis ABFs (Kobayashi et al., 2005). In wheat (Triticum aestivum), PKABA1, which is induced by ABA at the transcript level, phosphorylates Ta ABF (Anderberg and Walker-Simmons, 1992; Johnson et al., 2002).
The snrk2.6 mutation affected leaf water loss through the regulation of stomatal closure (Mustilli et al., 2002; Yoshida et al., 2002). However, seed dormancy and germination were not affected in snrk2.6 (Mustilli et al., 2002; Yoshida et al., 2002). It is possible that another SnRK2 protein kinase may function in seed germination and other ABA-regulated traits besides stomatal closure. Because forward genetic screens have failed to identify this kinase, there might be several redundant protein kinases that mediate ABA signaling in these processes, and a reverse genetics approach may be necessary to identify them.
SnRK2.2 and SnRK2.3 are two protein kinases most closely related to SnRK2.6 (Hrabak et al., 2003). In this study, we isolated snrk2.2 and snrk2.3 single mutants and a snrk2.2 snrk2.3 double mutant. The double mutant was insensitive to ABA in seed germination and seedling growth. These results suggest that SnRK2.2 and SnRK2.3 are the key protein kinases mediating ABA signaling during seed germination and seedling growth. We also present data indicating that the effect of these two protein kinases is mediated at least in part by phosphorylating ABFs and regulating ABA-responsive genes.
RESULTS
snrk2.2 snrk2.3 Is Insensitive to ABA in Seed Germination
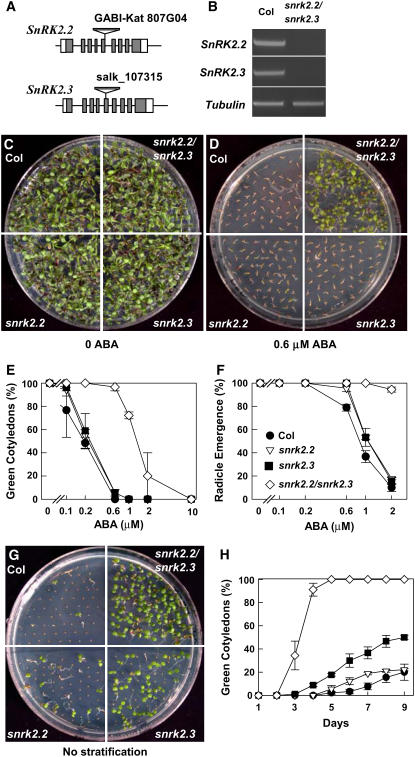
To analyze the function of SnRK2.2 and SnRK2.3, we obtained T-DNA insertion lines (Figure 1A) from the GABI and Salk T-DNA collections and isolated snrk2.2 and snrk2.3 homozygous mutants by PCR screening. The single mutants were crossed, and further PCR screening was done to obtain a snrk2.2 snrk2.3 double mutant. RT-PCR analysis confirmed that expression of both SnRK2.2 and SnRK2.3 was abolished in the double mutant (Figure 1B).
Figure 1.
Seed Germination and Dormancy Assays.
(A) Diagrams of SnRK2.2 and SnRK2.3 showing positions of the T-DNA insertions.
(B) RT-PCR analysis with SnRK2.2, SnRK2.3, and Tubulin primers using total RNA extracted from seedlings of the wild type (Columbia [Col-0]) and snrk2.2 snrk2.3 as the template.
(C) and (D) Photographs of Col-0, snrk2.2, snrk2.3, and snrk2.2 snrk2.3 seedlings on control (Murashige and Skoog [MS]) medium with 3% sucrose (C) or 0.6 μM ABA medium (D) at 9 d after the end of stratification.
(E) Quantification of the percentage of seedlings with green cotyledons after 6 d on the indicated concentrations of ABA (means ± se; n = 3). Each measurement consisted of at least 30 seeds. For symbols, see (F).
(F) Quantification of radicle emergence of each genotype at 3 d after the end of stratification (means ± se; n = 3). Medium used was MS medium without sucrose.
(G) Germination of nonstratified seeds at 7 d after sowing on MS medium with 3% sucrose.
(H) Germination time course (measured by appearance of green cotyledons) for nonstratified seeds of each genotype (means ± se; n = 3). For symbols, see (F).
When we analyzed ABA responses in snrk2.2, snrk2.3, and snrk2.2 snrk2.3, snrk2.2 snrk2.3 was found to be ABA-insensitive in seed germination and early seedling growth. This was true in assays measuring the emergence of green cotyledons (Figures 1C to 1E) as well as when germination was scored by radicle emergence (Figure 1F). In all of these assays, the snrk2.2 and snrk2.3 single mutants showed either no significant difference from the wild type or a much weaker ABA-insensitive phenotype relative to the double mutant. This finding suggests significant functional redundancy between SnRK2.2 and SnRK2.3.
ABA also controls seed dormancy, and we observed decreased seed dormancy in snrk2.2 snrk2.3. For the dormancy experiments, plants of each genotype tested were grown in different sections of the same pot and seeds were harvested at the same time to minimize the effect of seed maturation and storage conditions. snrk2.2 snrk2.3 seeds showed green cotyledons at 5 d after sowing, whereas almost none of the wild-type seeds had germinated at this time (Figures 1G and 1H). In this case, the single mutants, especially snrk2.3, also germinated more than the wild type but less than snrk2.2 snrk2.3.
snrk2.2 snrk2.3 Is Also Insensitive to ABA Regulation in Seedling Growth
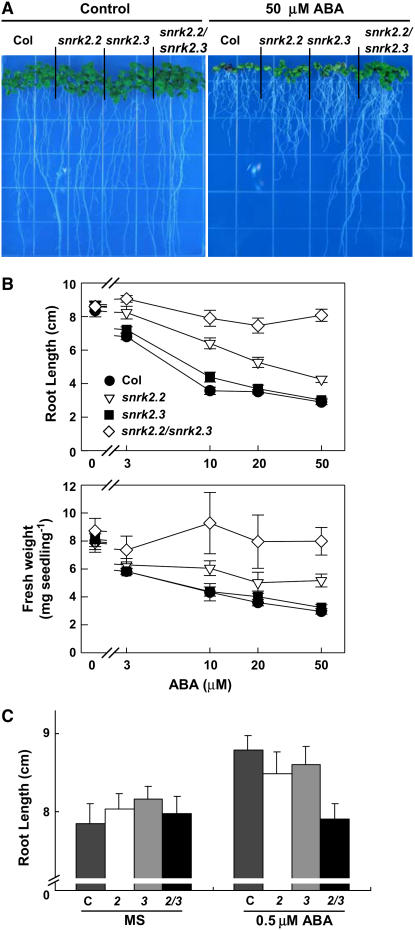
After germination is complete, ABA still can regulate seedling growth. We analyzed this aspect of ABA sensitivity in our mutants and snrk2.6 by transferring 4-d-old seedlings germinated on agar plates to the same medium with or without ABA. The length of the primary root and seedling fresh weight were then measured 14 d later. Based on both of these measures, snrk2.2 snrk2.3 grew more than the wild type on medium containing 3 to 50 μM ABA (Figures 2A and 2B). snrk2.2, but not snrk2.3, also grew more than the wild type but less than snrk2.2 snrk2.3 under these conditions. Low concentrations of ABA (<1 μM) are known to stimulate root growth (Ephritikhine et al., 1999). In our experiments, there was a slight stimulation of root elongation in the wild type in response to 0.5 μM ABA (Figure 2C). This stimulation by 0.5 μM ABA was not observed in the snrk2.2 snrk2.3 double mutant. Thus, in these traits as well, snrk2.2 snrk2.3 is insensitive to regulation by ABA, and there is redundancy between the functions of SnRK2.2 and SnRK2.3.
Figure 2.
ABA Inhibition of Seedling Growth.
(A) Photographs of seedlings at 14 d after transfer to control medium (MS medium with 3% sucrose) or medium containing 50 μM ABA. Seedlings were 4 d old at the time of transfer and had equal root lengths at that time.
(B) Quantification of root length and seedling fresh weight for seedlings treated as described for (A). For fresh weight determination, seven seedlings were weighed at one time and the result was divided by seven. Data are means ± se (n = 28 for root length and n = 4 for fresh weight).
(C) Quantification of root length for seedlings at 14 d after transfer to control medium or medium containing 0.5 μM ABA. Data are means ± se (n = 21). Col-0, snrk2.2, snrk2.3, and snrk2.2 snrk2.3 are indicated by C, 2, 3, and 2/3, respectively.
As was the case for seed germination, no difference between snrk2.6 and the wild type was observed in the sensitivity of seedling growth to ABA (see Supplemental Figure 1 online).
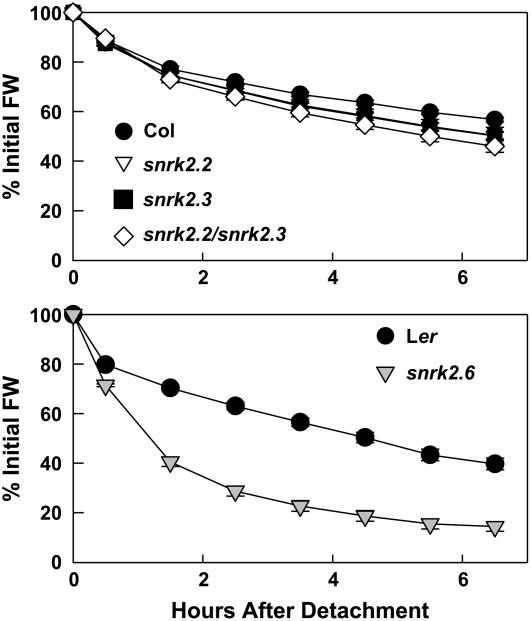
snrk2.2 snrk2.3 Is Little Affected in Leaf Water Loss, in Contrast with snrk2.6
Next, leaf water loss of the mutants was examined. Previous studies have shown that the defining phenotype of snrk2.6 is impaired stomatal closure leading to greater leaf water loss (Mustilli et al., 2002; Yoshida et al., 2002). snrk2.2 and snrk2.3 had only slight increases in leaf water loss (Figure 3). The effect on leaf water loss in snrk2.2 snrk2.3 was greater, approximately the sum of the snrk2.2 and snrk2.3 single mutant losses; however, it was still much smaller than the effect in snrk2.6 (Figure 3). Thus, these data suggested that SnRK2.2 and SnRK2.3 have only minor roles in stomatal control and further distinguished their function from that of SnRK2.6.
Figure 3.
Leaf Water Loss Assay.
Water loss was measured using detached leaves of Col-0, snrk2.2, snrk2.3, and snrk2.2 snrk2.3 (top panel) or Landsberg erecta (Ler) and snrk2.6/ost1 (bottom panel). Data are means ± se (n = 4 to 8). FW, fresh weight.
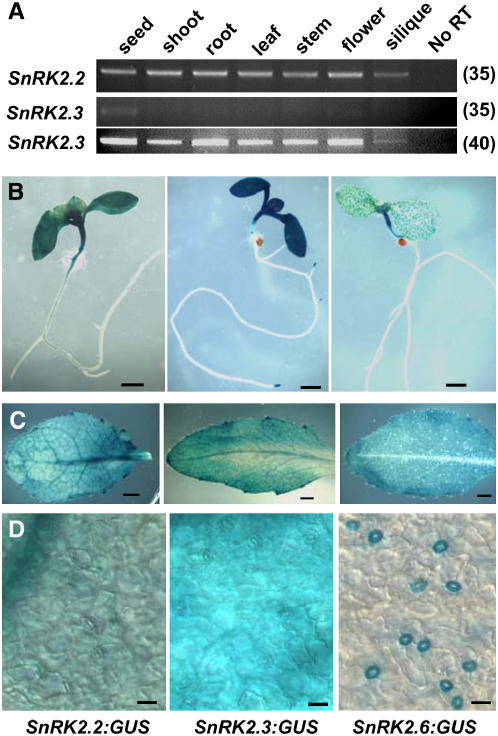
SnRK2.2 and SnRK2.3 Are Expressed in Various Tissues
SnRK2.6, which plays a role in stomatal regulation, is expressed specifically in guard cells (Mustilli et al., 2002; Yoshida et al., 2002). Because snrk2.2 snrk2.3 showed impairment in various aspects of ABA responses, the expression patterns of SnRK2.2 and SnRK2.3 were analyzed.
We examined the expression of SnRK2.2 and SnRK2.3 using RT-PCR and promoter:β-glucouronidase (GUS) fusions. RT-PCR analysis showed the expression of SnRK2.2 in all tissues examined (Figure 4A). SnRK2.3 was expressed at a lower level but still could be detected in all tissues by increasing the number of PCR cycles (Figure 4A). For a more detailed analysis of the expression patterns, promoter fragments covering 2 kb upstream of the translational start site were cloned for SnRK2.2, SnRK2.3, and SnRK2.6, fused to GUS, and transformed into Col-0 wild-type plants. GUS staining of the transgenic plants revealed SnRK2.2 and SnRK2.3 expression throughout cotyledon and leaf tissues, with similar or lower levels of expression in stems, roots, flowers, and siliques (Figures 4B and 4C; see Supplemental Figure 2 online). SnRK2.3 also showed particularly strong expression in root tips (Figure 4B; see Supplemental Figure 2 online). Examination of leaf tissues at higher magnification confirmed that the SnRK2.2 and SnRK2.3 promoter:GUS lines had staining in all cell types in the epidermis (Figure 4D). Consistent with previous reports (Mustilli et al., 2002), staining was detected only in guard cells of SnRK2.6 promoter:GUS lines. This was true in leaf tissue (Figures 4B to 4D) as well as in stems and sepals (see Supplemental Figure 2 online). Thus, the widespread expression in various tissues is consistent with a role of SnRK2.2 and SnRK2.3 in regulating various aspects of ABA response, whereas SnRK2.6 functions specifically in guard cell regulation.
Figure 4.
Expression of SnRK2.2 and SnRK2.3.
(A) RT-PCR analysis of tissue distribution of SnRK2.2 and SnRK2.3 expression. Numbers at right indicate the number of PCR cycles performed.
(B) to (D) GUS staining of plants with promoter:GUS expression driven by 2-kb promoter fragments of SnRK2.2, SnRK2.3, and SnRK2.6 (arranged from left to right in each row of photographs). Bars in (B), (C), and (D) = 1 mm, 1 mm, and 20 μm, respectively.
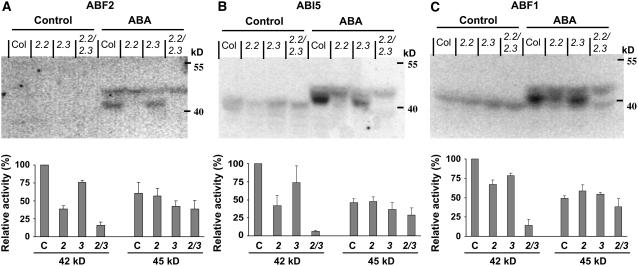
snrk2.2 snrk2.3 Is Impaired in ABA-Inducible Phosphorylation of ABFs in Vitro
The activation status of SnRK2s in vivo can be detected by in-gel kinase assay using Arabidopsis crude extracts (Yoshida et al., 2002; Boudsocq et al., 2004). In-gel kinase assays using histone as a substrate showed that an ∼44-kD ABA-induced kinase activity, but not a 42-kD activity, was missing in snrk2.6 (Yoshida et al., 2006). The phosphorylatable fragment (Gly-73 to Gln-119) of ABF2 can also be used as a substrate of SnRK2.2 and SnRK2.3 (Furihata et al., 2006). We used the snrk2 mutants to determine whether the SnRK2.2 and SnRK2.3 phosphorylation activity could be activated by ABA in vivo and whether ABFs were possible substrates. Protein extracts were prepared from Col-0, snrk2.2, snrk2.3, and snrk2.2 snrk2.3 seedlings grown under control conditions or after exposure to 100 μM ABA for 30 min. Recombinant GST-fused fragments of ABF2 (Gly-73 to Gln-119), ABI5 (Arg-132 to Gln-190), and ABF1 (Gly-83 to Glu-131) were used for in-gel kinase activity assays. Similar kinase activity was detected for each of these substrates (Figures 5A to 5C). Two bands of ABA-induced phosphorylation activity were detected (Figures 5A to 5C). The stronger band of kinase activity was at 42 kD, and this activity was reduced in all three of the mutants. snrk2.2 had a greater reduction in the 42-kD activity than did snrk2.3, and the reduction was additive in the double mutant. These results suggest that the 42-kD activity is derived from SnRK2.2 and SnRK2.3. In contrast with the 42-kD kinase activity, the 45-kD activity was weaker and was relatively unaffected in snrk2.2 or snrk2.3. It was, however, somewhat weaker in snrk2.2 snrk2.3. SnRK2.2 and SnRK2.3 may indirectly affect the 45-kD activity, which likely corresponds to SnRK2.6 (Yoshida et al., 2006).
Figure 5.
In-Gel Kinase Assay.
In-gel kinase assay with proteins extracted from the wild type (Col-0), snrk2.2, snrk2.3, and snrk2.2 snrk2.3 seedlings under control conditions or 30 min after 100 μM ABA treatment. The ABF2 fragment (amino acids Gly-73 to Gln-119 [A]), ABI5 fragment (Arg-132 to Gln-190 [B]), and ABF1 fragment (Gly-83 to Glu-131 [C]) were used as substrates. The graphs at bottom indicate relative radioactivity (mean ± se; n = 4 for ABF2, n = 3 for ABI5 and ABF1) of the 42- and 45-kD bands after ABA treatment normalized relative to the 42-kD activity of the wild type. In the graphs, Col-0, snrk2.2, snrk2.3, and snrk2.2 snrk2.3 are indicated by C, 2, 3, and 2/3, respectively.
In all lanes, regardless of ABA treatment, strong bands of ∼69 kD were detected and weak bands of ∼39 kD were sometimes detected besides the 42- and 45-kD ABA-inducible activities (data not shown). Together with previous studies (Boudsocq et al., 2004; Furihata et al., 2006; Yoshida et al., 2006), these data demonstrate that SnRK2.2 and SnRK2.3 have ABA-inducible kinase activity and together are likely to play a major role in phosphorylating and activating ABF2, ABI5, ABF1, and possibly other ABFs.
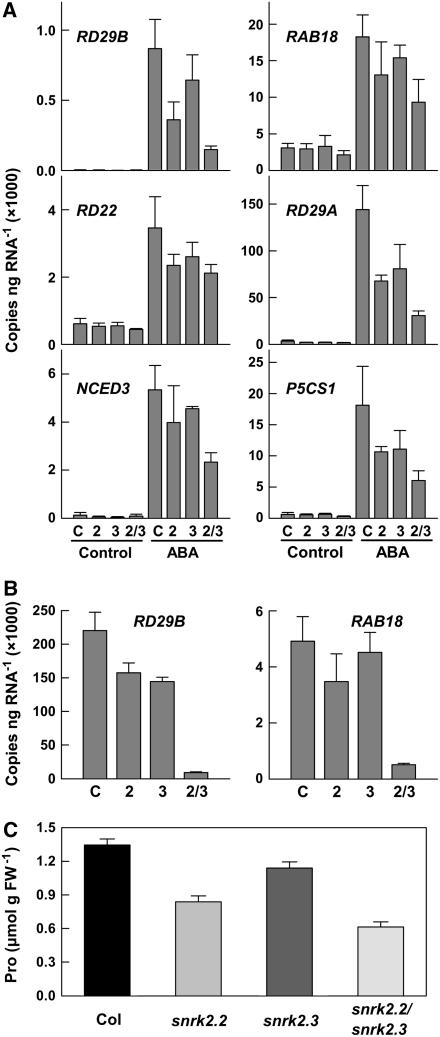
SnRK2.2 and SnRK2.3 Are Required for ABA-Induced Changes in Gene Expression and Pro Accumulation
ABA induces the expression of many genes that are important for adaptation to stress. Some well-documented examples include RESPONSIVE TO DESSICATION29B (RD29B), RD22, RD29A, RESPONSIVE TO ABA18 (RAB18), NCED3 (for 9-cis-Epoxycarotenoid Dioxygenase3), and P5CS1 (for Δ1-Pyrroline-5-Carboxylate Synthetase1) (Yamaguchi-Shinozaki and Shinozaki, 2006). Based on their activity in phosphorylating the ABF2 fragment and the other phenotypes observed above, we hypothesized that SnRK2.2 and SnRK2.3 play a role in ABA-induced gene expression changes. To examine the expression of these genes, quantitative RT-PCR was performed. In the wild type, treatment with 100 μM ABA for 3 h induced all of these genes (Figure 6A). Induction of these genes was consistently less in all three of the mutants, although the extent to which induction by ABA was blocked varied among the genes examined. As in the other phenotypes examined above, it was also consistently observed that the snrk2.2 snrk2.3 double mutant had the greatest reduction in ABA-induced gene expression.
Figure 6.
Expression of ABA-Regulated Genes and ABA-Induced Pro Accumulation.
(A) Expression of the ABA-upregulated genes RD29B, RAB18, RD22, RD29A, NCED3, and P5CS1 assayed by quantitative RT-PCR in Col-0 (C), snrk2.2 (2), snrk2.3(3), and snrk2.2 snrk2.3 (2/3) seedlings under control conditions or after 3 h of exposure to 100 μM ABA. Data are means ± se (n = 3). In some cases, such as with RD29B, expression in control seedlings was too low to be visible on the graphs.
(B) Expression of RD29B and RAB18 in seeds imbibed for 24 h (means ± se; n = 3).
(C) Pro contents of seedlings after transfer to plates containing 10 μM ABA for 96 h. Data are means ± se (n = 6 to 8). FW, fresh weight.
Some of these same ABA-regulated genes are also expressed in seeds. We found that expression of RD29B and RAB18 was greatly reduced in imbibed seeds of snrk2.2 snrk2.3 (Figure 6B). This finding confirmed that SnRK2.2 and SnRK2.3 are also responsible for regulating the expression of genes with an ABRE in their promoter during seed dormancy and germination. Other genes examined in imbibed seeds were expressed at very low levels (NCED3, RD22, and RD29A) and did not differ between any of the genotypes tested (data not shown).
Consistent with their role in ABA induction of the Pro biosynthesis gene P5CS1, we found that SnRK2.2 and SnRK2.3 also affected ABA-induced Pro accumulation. In seedlings exposed to 10 μM ABA for 96 h, Pro accumulation in snrk2.2, snrk2.3, and snrk2.2 snrk2.3 was less than that in wild-type Col-0. snrk2.2 had a greater effect than snrk2.3, and the effect was additive in the double mutant (Figure 6C). Pro content in control seedlings was low (0.15 μmol/g fresh weight) and did not differ between the wild type and the mutants. By contrast, Pro accumulation was not reduced in snrk2.6 seedlings exposed to 10 μM ABA (1.74 ± 0.17 μmol/g fresh weight for Ler versus 2.18 ± 0.15 μmol/g fresh weight for snrk2.6).
DISCUSSION
ABA regulates many important plant processes, including seed germination, dormancy, seedling growth, and stomatal aperture. Although many genes have been reported to be involved in ABA signaling, some key components are still missing, and ABA signaling appears to involve a highly branched network. SnRK2.6 has been identified as a positive regulator of ABA signaling but functions only in guard cell ABA responses (Mustilli et al., 2002; Yoshida et al., 2002), whereas ABI1 and ABI2 function as negative regulators (Sheen, 1998) in seed germination, seedling growth, and stomatal closure (Leung et al., 1997). This raises the question of which protein kinases might positively regulate ABA signaling in seed germination and seedling growth.
We have used a reverse genetics approach to study the functions of SnRK2.2 and SnRK2.3. snrk2.2 snrk2.3 plants are insensitive to ABA in seed germination, indicating that SnRK2.2 and SnRK2.3 are the protein kinases that positively regulate ABA signaling in seed germination. SnRK2.2 and SnRK2.3 also have highly redundant functions in seed dormancy, inhibition of seedling growth by ABA, ABA-induced Pro accumulation, and ABA-induced gene expression. This redundancy likely explains why SnRK2.2 and SnRK2.3 were not identified in the extensive forward genetic screening for ABA-insensitive mutants that has been conducted by a number of laboratories.
snrk2.2 snrk2.3 is only slightly defective in leaf water loss compared with snrk2.6. The very distinctive phenotype of snrk2.2 snrk2.3 compared with snrk2.6 raises the question of the basis for this specificity. Such differences could be caused by a difference in gene expression or their substrate specificity or activation. As described above, experiments to date have not yet found a difference in substrate specificity among the SnRK2s: SnRK2.2, SnRK2.3, SnRK2.6, and SnRK2.8 were all able to phosphorylate ABF2 (Furihata et al., 2006). Likewise, ABA activation of all of these kinases was observed (Boudsocq et al., 2004; Furihata et al., 2006). This leaves differences in expression as the most likely basis for the specific roles of SnRK2.2 and SnRK2.3 versus SnRK2.6 in different ABA responses. Consistent with this hypothesis, our results show widespread expression of SnRK2.2 and SnRK2.3, particularly in leaf and stem tissues, whereas SnRK2.6 is largely confined to the guard cells. SnRK2.2 and SnRK2.3 do have a small effect on leaf water loss, and this is consistent with their low level of expression in guard cells.
Our experiments showed a >80% reduction in snrk2.2 snrk2.3 of a 42-kD ABA-activated kinase activity capable of phosphorylating an ABF2, ABI5, or ABF1 fragment in vitro. This finding is consistent with several previous studies that have shown activation of SnRK2 activity, including SnRK2.2, SnRK2.3, and SnRK2.6, by ABA and/or osmotic stress (Boudsocq et al., 2004; Kobayashi et al., 2004; Furihata et al., 2006; Yoshida et al., 2006). However, the activation mechanism of SnRK2s is still unclear. The C-terminal domain of rice SAPK8 can confer ABA responsiveness in protoplasts. Likewise, the C-terminal region of SnRK2.6 is important for ABA response (Yoshida et al., 2006) and has been shown to bind to ABI1 in a yeast two-hybrid assay. This interaction appears to be important in vivo, as SnRK2.6 is not fully activated by ABA in abi1-1 (Yoshida et al., 2006). The molecular mechanism by which SnRK2s are activated is not known but must involve a posttranslational modification resistant to SDS treatment, because the activation status of endogenous SnRK2s is maintained after SDS-PAGE. Many kinases are themselves activated by autophosphorylation or phosphorylation by upstream kinase(s), and the hyperosmotic stress activation of rice SAPK1 and SAPK2 is mediated by phosphorylation (Kobayashi et al., 2004). The identification of SnRK2.2 and SnRK2.3 as ABA-activated kinases important in ABA signaling sets the stage for further experiments to identify upstream kinases or other regulatory mechanisms.
Our data suggest that the ABF bZIP family transcription factors are downstream targets of SnRK2.2 and SnRK2.3 in ABA signaling. The substantial loss of ABF phosphorylation activity in snrk2.2 snrk2.3 also suggests that SnRK2.2 and SnRK2.3 may be the predominant kinases that phosphorylate the transcription factors in response to ABA in seedlings. Because ABI5 and ABF1 could also be substrates of SnRK2.2 and SnRK2.3 (Figures 5B and 5C), other ABFs may also be phosphorylated by SnRK2.2 and SnRK2.3. It is possible that other SnRK2.2 or SnRK2.3 substrates outside of the ABF family may also be involved in ABA signaling.
Most of the ABA-induced genes for which we observed reduced ABA responsiveness in snrk2.2 snrk2.3 contain ABREs in their promoters. RD29A, RD29B, and P5CS1 have all been reported to contain ABREs in their promoters (Yamaguchi-Shinozaki and Shinozaki, 1994; Yoshiba et al., 1999). Our own examination of promoter sequences also revealed an ABRE ∼75 bp upstream of the transcriptional start site of NCED3 and ∼300 bp upstream of the start site in the RAB18 promoter. Thus, SnRK2.2 and SnRK2.3 are likely to affect the expression of these genes through phosphorylation of one or more ABFs, thus influencing ABF binding to the ABRE. However, RD22 has no distinct ABRE in its promoter region and instead has been reported to be regulated by MYC and MYB transcription factors (Abe et al., 1997). RD22 expression is also reduced in snrk2.2 snrk2.3. Thus, there is a question regarding whether this phenotype can be accounted for by an indirect mechanism (i.e., that SnRK2.2 and SnRK2.3 regulate the expression of a gene that in turn regulates RD22 expression) or whether there is a more direct, as yet unknown, mechanism by which SnRK2s can regulate gene expression independently of the ABRE.
METHODS
T-DNA Insertion Lines
The seeds of Arabidopsis thaliana snrk2.2 and snrk2.3 T-DNA insertion lines (GABI-Kat 807G04 and Salk_107315) were obtained from the Max Planck Institute for Plant Breeding Research (Rosso et al., 2003) and the ABRC (Alonso et al., 2003), respectively. Homozygous plants were obtained by PCR screening using primers designed by the I-Sect website (http://signal.salk.edu/cgi-bin/tdnaexpress; primer sequences are given Supplemental Table 1 online). The snrk2.6 mutant used here is the original ost1 mutant (Mustilli et al., 2002) and is in the Ler background. For clarity in the comparison with snrk2.2 and snrk2.3, we refer to ost1 as snrk2.6 throughout this report.
Production of Transgenic Plants and GUS Staining
Fragments covering 2 kb upstream of the translational start sites of SnRK2.2, SnRK2.3, and SnRK2.6 were amplified by PCR using genomic DNA as templates. For SnRK2.2 and SnRK2.3, this region contains endogenous HindIII or BamHI sites. Therefore, two different fragments covering the 2-kb region were amplified, with each fragment having different portions of HindIII and BamHI sites on each end such that mixing, denaturing, and reannealing the fragments generated heterodimers with HindIII/BamHI cohesive ends. These fragments were cloned into HindIII/BamHI-digested pBI101. For the 2-kb fragment upstream of SnRK2.6, the PCR product was cloned into pGEM-T easy vector, digested with PstI, and blunted. After digestion with BamHI, the fragment was subcloned into pBI101 vector at blunted HindIII and BamHI sites. Primers used are given in Supplemental Table 1 online. The resulting plasmids were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, Col-0 plants were transformed by floral dip infiltration, and kanamycin-resistant transgenic seedlings of T2 plants and other tissues of T1 plants were used for GUS staining. For GUS staining, tissues were incubated in 5 mM K4Fe(CN)6, K3Fe(CN)6, 0.3% Triton X-100, 0.1 M phosphate buffer (a mixture of KH2PO4 and K2HPO4, pH 7.0), and 3 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (cyclohexylammonium salt; Gold Bio Tech) at 37°C for 16 h and incubated in 70% ethanol at 37°C for 16 h.
Kinase Assays
The ABF2, ABI5, and ABF1 fragments used for in vitro kinase assays were obtained by reverse transcription and amplification of the portion of the ABF2, ABI5, or ABF1 cDNA corresponding to amino acids Gly-73 to Gln-119 of ABF2, Arg-132 to Gln-190 of ABI5, and Gly-83 to Glu-131 of ABF1. The template for cloning was total RNA extracted from 12-d-old seedlings or genomic DNA of Col-0. The PCR product was digested with BamHI and XhoI (ABF2 and ABF1) or EcoRI (ABI5) and subcloned into pGEX4T1 vector (Amersham) for the production of GST fusion protein using Escherichia coli Rosetta cells (Novagen). Fusion proteins were purified using glutathione–agarose beads (Sigma-Aldrich) and dialyzed with 10 mM Tris-HCl, pH 8.8, for 16 h.
In-gel kinase assays were performed according to the protocol of Furihata et al. (2006) with some modifications. Plant materials used consisted of 10-d-old seedlings grown on control medium (MS agar with 1% sucrose) or seedlings sprayed with ABA and harvested 3 h later. Proteins were extracted in 5 mM EDTA, 5 mM EGTA, 2 mM DTT, 25 mM NaF, 1 mM Na3VO4, 50 mM β-glycerophosphate, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail (Sigma-Aldrich), and 50 mM HEPES-KOH, pH 7.5. Proteins (40 μg/lane) were separated on a SDS-PAGE gel containing 0.5 mg/mL GST-tagged substrate peptide. The gel was washed for 3 × 30 min with 0.5 mM DTT, 5 mM NaF, 0.1 mM Na3VO4, 0.5 mg/mL BSA, 0.1% Triton X-100, and 25 mM Tris-HCl, pH 7.5, and proteins were renatured with 1 mM DTT, 5 mM NaF, 0.1 mM Na3VO4, and 25 mM Tris-HCl, pH 7.5, for 2 × 30 min and 16 h at 4°C. After 30 min of incubation in reaction solution (2 mM EGTA, 12 mM MgCl2, 1 mM DTT, 0.1 mM Na3VO4, and 25 mM Tris-HCl, pH 7.5) at room temperature, the gel was incubated in 12 mL of reaction solution supplemented with 50 μCi of [γ-32P]ATP and 250 nM cold ATP for 90 min at room temperature. The gel was washed with 5% TCA and 1% sodium pyrophosphate more than five times for 30 min each, incubated with 10% glycerol, and dried. Radioactivity was quantified using a Typhoon 9410 imager (Molecular Dynamics).
Physiological Assays
Soil-grown plants and seedlings on agar plates were routinely grown under continuous light (∼75 μmol·m−2·s−1) at 23°C. For germination assays, seeds were plated on MS medium containing sucrose and ABA as indicated in the text and figure legends. Seeds were then stratified at 4°C for 4 d, and radicle emergence or the presence of green cotyledons was scored after the indicated time intervals.
For Pro analysis, seeds were plated onto half-strength MS medium with 6 mM MES, pH 5.7, without the addition of sucrose or other sugars, according to the protocols of Verslues et al. (2006), and stratified for 4 d at 4°C. ABA treatment for the analysis of Pro accumulation was performed by transferring 6-d-old seedlings to half-strength MS plates without sugar but with 10 μM ABA added to the medium. Pro was assayed using a ninhydrin-based colorimetric assay (Bates et al., 1973).
For leaf water loss measurements, fully expanded leaves were removed from 4- to 5-week-old plants and incubated under the same conditions used for seedling growth, and each sample (consisting of three to four individual leaves) was weighed at the indicated times.
RT-PCR and Quantitative PCR Analysis of Gene Expression
Total RNA was purified from seedlings, leaves, stems, and inflorescences using Trizol (Invitrogen) according to the manufacturer's instructions or from imbibed seeds and green siliques according to Penfield et al. (2005). In either case, the extracted total RNA was dissolved in 100 μL of RNase-free water and purified using the RNAeasy kit (Qiagen) according to the manufacturer's instructions, including DNase treatment. Reverse transcription reactions were performed using 0.05 μg of total RNA and SuperScriptII reverse transcriptase (Invitrogen). PCR was then performed for 20 cycles and one-twentieth of the initial PCR sample was used to set up a second PCR sample, which was amplified for an additional 15 or 20 cycles. The total number of PCR cycles used for both reactions is indicated in the figures.
Real-time quantitative PCR analysis was performed with a 7700 sequence detection system (Applied Biosystems) using primers and TaqMan probes designed using Primer Express software (Applied Biosystems). BHQ/FAM-labeled TaqMan probes were obtained from Biosearch Technologies. Total RNA was extracted from control or 100 μM ABA–treated seedlings using the RNAeasy kit (Qiagen) including DNase treatment. Reverse transcription and PCR (10 μL) were performed with 0.1 μg of total RNA using the One-Step RT-PCR kit (Qiagen). To quantify the copy number of each RNA, the Ct (threshold cycle) value was compared with a standard curve generated using PCR products for each gene that had been purified and quantified by UV light absorbance (Zhu et al., 2005). Three biological and three technical replicates were performed for each experiment. Primers used for both RT-PCR and quantitative RT-PCR are given in Supplementary Table 1 online.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: SnRK2.2, At3g50500; SnRK2.3, At5g66880; SnRK2.6, At4g33950.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ABA Sensitivity of Germination and Seedling Growth of snrk2.6.
Supplemental Figure 2. GUS Staining of Flowers, Stems, and Siliques of SnRK2.2, SnRK2.3, and SnRK2.6 Promoter:GUS Transgenic Lines.
Supplemental Table 1. Oligonucleotides Used in This Study.
Supplementary Material
Acknowledgments
We thank Rebecca Stevenson for technical assistance. We also thank the ABRC and the Max Plank Institute for Plant Breeding Research for providing the T-DNA insertion mutants. This work was supported by National Institutes of Health Grant R01 GM-059138 and National Science Foundation Grant IBN-0420152 to J.-K.Z. P.E.V. was supported by National Institutes of Health Postdoctoral Fellowship 5F32 GM-074445.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian-Kang Zhu (jian-kang.zhu@ucr.edu).
Online version contains Web-only data.
References
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Anderberg, R.J., and Walker-Simmons, M.K. (1992). Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc. Natl. Acad. Sci. USA 89 10183–10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann, S.M. (2003). OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 8 151–153. [DOI] [PubMed] [Google Scholar]
- Bates, L.S., Waldren, R.P., and Teare, I.D. (1973). Rapid determination of free proline in water-stress studies. Plant Soil 39 205–207. [Google Scholar]
- Boudsocq, M., Barbier-Brygoo, H., and Lauriere, C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279 41758–41766. [DOI] [PubMed] [Google Scholar]
- Bray, E.A. (2002). Abscisic acid regulation of gene expression during water-deficit stress in the era of the Arabidopsis genome. Plant Cell Environ. 25 153–161. [DOI] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275 1723–1730. [DOI] [PubMed] [Google Scholar]
- Chow, B., and McCourt, P. (2004). Hormone signaling from a developmental context. J. Exp. Bot. 55 247–251. [DOI] [PubMed] [Google Scholar]
- Ephritikhine, G., Fellner, M., Vannini, C., Lapous, D., and Barbier-Brygoo, H. (1999). The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 18 303–314. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(suppl.): S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Gibson, S.I. (2002). ABA and sugar interactions regulating development. Cross-talk or voices in a crowd? Curr. Opin. Plant Biol. 5 26–32. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Somerville, C.R. (1990). Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 94 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan, M.J., Marcotte, W.R., Jr., and Quatrano, R.S. (1990). A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250 267–271. [DOI] [PubMed] [Google Scholar]
- Hrabak, E.M., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.R., Wagner, R.L., Verhey, S.D., and Walker-Simmons, M.K. (2002). The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 130 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.Y., Choi, H.I., Im, M.Y., and Kim, S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Murata, M., Minami, H., Yamamoto, S., Kagaya, Y., Hobo, T., Yamamoto, A., and Hattori, T. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44 939–949. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Yamamoto, S., Minami, H., Kagaya, Y., and Hattori, T. (2004). Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C.M. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61 377–383. [Google Scholar]
- Leon-Kloosterziel, K.M., Gil, M.A., Ruijs, G.J., Jacobsen, S.E., Olszewski, N.E., Schwartz, S.H., Zeevaart, J.A., and Koornneef, M. (1996). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10 655–661. [DOI] [PubMed] [Google Scholar]
- Leung, J., Bouvier-Durand, M., Morris, P.C., Guerrier, D., Chefdor, F., and Giraudat, J. (1994). Arabidopsis ABA response gene ABI1. Features of a calcium-modulated protein phosphatase. Science 264 1448–1452. [DOI] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Wang, X.Q., Watson, M.B., and Assmann, S.M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287 300–303. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K., Leube, M.P., and Grill, E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264 1452–1455. [DOI] [PubMed] [Google Scholar]
- Mustilli, A.C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms, J., Leon-Kloosterziel, K.M., Bartels, D., Koornneef, M., and Karssen, C.M. (1993). Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana. A comparative study using abscisic acid-insensitive abi3 mutants. Plant Physiol. 102 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development. Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S., Josse, E.M., Kannangara, R., Gilday, A.D., Halliday, K.J., and Graham, I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15 1998–2006. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Schroeder, J.I., Kwak, J.M., and Allen, G.J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410 327–330. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1998). Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc. Natl. Acad. Sci. USA 95 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues, P.E., Agarwal, M., Katiyar-Agarwal, S., Zhu, J., and Zhu, J.K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45 523–539. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. [DOI] [PubMed] [Google Scholar]
- Yoshiba, Y., Nanjo, T., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Stress-responsive and developmental regulation of Δ1-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 261 766–772. [DOI] [PubMed] [Google Scholar]
- Yoshida, R., Hobo, T., Ichimura, K., Mizoguchi, T., Takahashi, F., Aronso, J., Ecker, J.R., and Shinozaki, K. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43 1473–1483. [DOI] [PubMed] [Google Scholar]
- Yoshida, R., Umezawa, T., Mizoguchi, T., Takahashi, S., Takahashi, F., and Shinozaki, K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281 5310–5318. [DOI] [PubMed] [Google Scholar]
- Zhu, J.H., Verslues, P.E., Zheng, X.W., Lee, B., Zhan, X.Q., Manabe, Y., Sokolchik, I., Zhu, Y.M., Dong, C.H., Zhu, J.K., Hasegawa, P.M., and Bressan, R.A. (2005). HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl. Acad. Sci. USA 102 9966–9971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu, J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.