Abstract
Gene expression is regulated by transcription factors that interact with cis-regulatory elements. Predicting these elements from sequence data has proven difficult. We describe here a successful computational search for elements that direct expression in a particular temporal-spatial pattern in the Drosophila embryo, based on a single well characterized enhancer model. The fly genome was searched to identify sequence elements containing the same combination of transcription factors as those found in the model. Experimental evaluation of the search results demonstrates that our method can correctly predict regulatory elements and highlights the importance of functional testing as a means of identifying false-positive results. We also show that the search results enable the identification of additional relevant sequence motifs whose functions can be empirically validated. This approach, combined with gene expression and phylogenetic sequence data, allows for genome-wide identification of related regulatory elements, an important step toward understanding the genetic regulatory networks involved in development.
[Sequence data reported in this paper have been deposited in GenBank with accession nos. AF513981 (Eve MHE) and AF513982 (Hbr DME). Supplementary material is available online at http://www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: R. Blackman]
Tightly orchestrated spatial and temporal regulation of gene transcription is critical to the proper development of all metazoans. A substantial part of this regulation results from the interaction of transcription factors (TFs) with specific cis-regulatory DNA sequences. These regulatory sequences are organized in a modular fashion, with each module (enhancer) containing one or more binding sites for a specific combination of TFs (Davidson 2001). In each cell, the available TFs derive both from that cell's developmental history and as a direct response to one or more inductive intercellular signals. These tissue-restricted and signal-activated TFs then bind to specific sites within the enhancers of particular genes, defining a combinatorial transcriptional code that facilitates the expression of those genes in a particular developmental context.
The recent availability of whole-genome DNA sequences has created the potential for identifying cis-regulatory elements via a bioinformatics approach on a genomic scale. Although computational methods have served well for purposes of finding genes and even individual exons in genome data, regulatory element prediction has proven difficult. A number of approaches have been explored (for review, see Fickett and Wasserman 2000; Ohler and Niemann 2001; Pennacchio and Rubin 2001), but many challenges remain. Methods that work well in yeast, where regulatory sequences are mainly promoter-proximal (Roth et al. 1998; Tavazoie et al. 1999; Hughes et al. 2000), are difficult to extend to higher eukaryotes where the regulatory modules are extensive and can lie many kilobases on either side of a coding region, or within an intron (Arnone and Davidson 1997). Other methods rely on models developed from the prior characterization of a large number (ten or more) of regulatory elements of similar function (Frech et al. 1997; Wasserman and Fickett 1998; Krivan and Wasserman 2001), but unfortunately, such extensive information is not available for most genes. The identification of dense clusters of known TF binding sites has also been used as the basis for computational searching (Frith et al. 2001; Berman et al. 2002; Markstein et al. 2002). However, the predictive value of these various approaches remains uncertain: Although they have been successful at recognizing the known modules used in constructing the models, little critical experimental validation of putative novel elements has been performed.
In the present study, we used the detailed functional characterization we made of a single Drosophila dorsal mesodermal enhancer, the Eve MHE (Halfon et al. 2000), to devise criteria for a model-based computational search of the Drosophila genome for similar cis-regulatory modules. This approach requires neither extensive gene expression data nor sequences from related organisms. As such, it represents a significant addition to the methods available for finding regulatory modules because it enables the identification of a specific functional class of element—a dorsal mesodermal enhancer—using a whole-genome analysis based on a single detailed model. We also demonstrate that alignment of the results from such a search can serve as a vehicle for the discovery of additional relevant sequence motifs, one example of which we have empirically validated. We suggest that this simple model-based approach, if combined with both expression and phylogenetic sequence data, will allow for a large-scale characterization of functionally-related cis-regulatory elements, a fundamental step toward understanding the genetic regulatory networks involved in development.
RESULTS
The eve MHE
We recently described a comprehensive model for the transcriptional integration of multiple intercellular signals that act together to establish the identities of a subset of muscle and cardiac progenitor cells in the dorsal mesoderm of the Drosophila embryo (Fig. 1A–C; Halfon et al. 2000). In this model, expression of the progenitor identity gene even skipped (eve) is regulated via the action of at least five TFs binding to multiple sites in a 312 bp enhancer located approximately 6 kb downstream of the eve coding region, the Muscle and Heart Enhancer (MHE). Three of the TFs, that is, dTcf, Mothers Against Dpp (Mad), and Pointed (Pnt) function downstream of and are activated by the Wingless (Wg), Decapentaplegic (Dpp), and Ras/MAP kinase signaling pathways, respectively, while the other two TFs, Twist (Twi) and Tinman (Tin), are mesodermally restricted selector proteins. The transcriptional code comprised by these five transcription factors can account for all of the signaling events and genetic data known to affect eve regulation in the mesodermal progenitors (although it is likely that additional factors remain to be identified; see below).
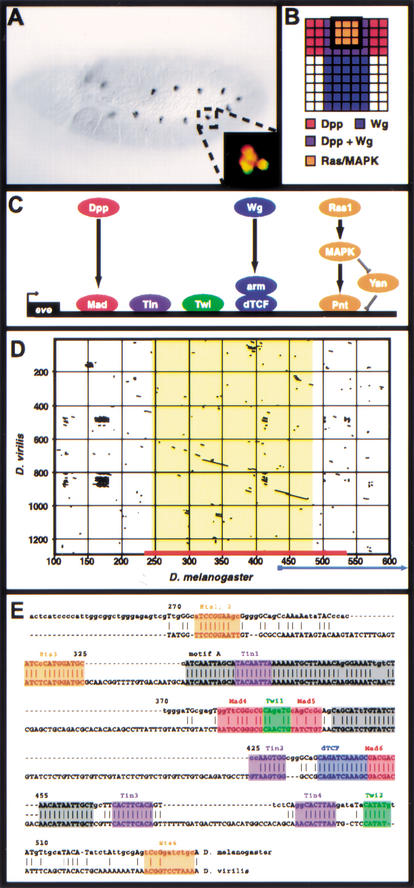
Figure 1.
The Eve MHE provides a model for the transcriptional integration of multiple intercellular signals. (A) Stage 11 Drosophila embryo stained with antibodies against Eve. A small cluster of cells in the dorsal mesoderm of each segment, the Eve-positive muscle and cardiac progenitors, express Eve. Anterior is to the left. Inset: Double-staining for Eve (green) and the MHE reporter construct (red) shows that the MHE is sufficient to drive expression in the Eve-positive cells (Halfon et al. 2000). (B) Signaling events required for Eve expression in the dorsal mesoderm. One hemisegment is represented, with dorsal to the top and anterior to the left. Expression of Eve is induced in cells that receive signaling from the Dpp, Wg, and Ras/MAPK pathways (Carmena et al. 1998). (C) The transcriptional code used at the MHE. The signal-responsive TFs, that is, Mad, dTcf, and Pnt bind along with the mesodermal selector proteins Twi and Tin to activate transcription. (D) Dot plot showing that the MHE sequence (x-axis, red bar) is conserved in D. virilis (y-axis). The area of extensive homology (on the diagonal) is shown in yellow; little homology exists flanking this region for several hundred base pairs. An alternate sequence recently proposed to have MHE-like activity (Knirr and Frasch 2001) is indicated by the blue arrow; note that most of this sequence lies in nonconserved regions. (E) Sequence alignment of MHE and vMHE. Known MHE binding sites are shown in color; gray boxes indicate conserved regions where functional sites have not yet been identified. Base pair numbering corresponds to that shown in D.
The MHE Sequence is Well Conserved in a Distantly Related Drosophila Species
As part of our continuing analysis of the MHE, we cloned and sequenced the corresponding region downstream of the eve gene in the distantly related (∼40 Mya; Kwiatowski et al. 1994) species Drosophila virilis. We found that the D. virilis element (vMHE) maintains extensive sequence conservation with the MHE (Fig. 1D,E), and is functional when used to drive reporter gene expression in D. melanogaster, albeit more weakly than the MHE (data not shown). Consistent with our model of MHE function, at least one representative of each relevant TF binding site can be found in the vMHE, although not every functional MHE sequence motif is conserved (e.g., Ets4, Fig. 1E; Halfon et al. 2000). Importantly, several blocks of conserved sequence can be observed in which no TF binding sites have been characterized to date (Fig. 1E, gray shading), suggesting that additional but presently unidentified factors may be involved in MHE-derived gene regulation. Of note, sequences included in a proposed variant eve mesodermal enhancer (Knirr and Frasch 2001) are not conserved in D. virilis (Fig. 1D, blue arrow).
A Computational Search for Dorsal Mesodermal Enhancers
The combinatorial Wg+Dpp+Ras/MAP kinase signaling code necessary for eve transcription appears to be necessary not only for Eve expression but also for that of other dorsal mesodermal genes (Carmena et al. 1998, 2002; Halfon et al. 2000). We thus hypothesized that the same transcriptional code—Twi, Tin, dTcf, Mad, and Pnt—might also be responsible for the regulation of other genes. Moreover, we reasoned that, were this the case, this code might act via a cis-regulatory element resembling the MHE in that it would contain clustered binding sites for all five TFs. To test this idea comprehensively, we undertook a computational search of the entire Drosophila genome to identify elements that, based on their related composition, would be predicted to function as MHE-like dorsal mesodermal enhancers (DMEs).
The genome was searched to locate regions which contain predicted binding sites for all five TFs found within the eve MHE. The search was conducted essentially as follows (see Methods): the ScanACE program (Hughes et al. 2000) was used to identify each occurrence in the genome of a predicted binding site for each of the five TFs. An algorithm was then run to detect all instances in which all five TFs are found within a 500 bp window; these regions were termed “elements.” The 500 bp window size was chosen in recognition of the ∼300 bp size of the model (the MHE) and in an effort to limit the number of positive returns from the search so as to make feasible testing of our predictions.
This initial search resulted in 647 elements (Table 1 and Supplemental Table 1 available online at http://www.genome.org). To determine how likely it was that these would occur merely by chance, given the number of each predicted binding site in the genome, Monte Carlo simulations were performed in which the predicted sites were randomly distributed, and the cooccurrence search was run (see Methods). The results from these simulations indicate that cooccurrence of the five motifs happens significantly less than expected at random (P < 0.0001; Supplemental Fig. 1 available online at http://www.genome.org). This finding suggests that such cooccurrences have functional consequences that have acted to keep the individual motifs separated in the course of evolution except when needed for the expression of specific genes.
Table 1.
Selected Subset of Elements
| Element | Closest gene(s) | Chromosome | bp location | Expression data1 | in mesoderm2 |
|---|---|---|---|---|---|
| DME1 | eve/TER94 | 2R | 5014107 | Y | Y3 |
| DME2 | stumps (hbr) | 3R | 10352207 | Y | Y3 |
| DME3 | dmcf2 | 2R | 4951127 | Y | Y3 |
| DME4 | mcso18E/CG12531 | X | 19459844 | Y | Y3 |
| DME5 | CG15391/dpp | 2L | 2352035 | Y | Y |
| DME6 | fru | 3R | 14282221 | Y | Y |
| DME7 | rst/CG4116 | X | 2797776 | Y | Y |
| DME8 | Btk29A | 2L | 8173348 | Y | Y |
| DME9 | CG17588/TpnC41C | 2R | 278747 | Y | N |
| DME10 | SRPK/Mtk | 2R | 10364562 | Y | N |
| DME11 | CG13740/hig | 2R | 4256438 | Y | N |
| DME12 | Acp76A/CG3797 | 3L | 18944209 | Y | N |
| DME13 | pav/CG15010 | 3L | 4220351 | Y | N |
| DME14 | Acp76A/CG3797 | 3L | 18940434 | Y | N |
| DME15 | ara/caup | 3L | 12514566 | Y | N |
| DME16 | nAcRα-96Aa/b | 3R | 20193510 | Y | N |
| DME17 | CG14506/Cnx99A | 3R | 25025121 | Y | N |
| DME18 | beat | 2L | 15897130 | Y | N |
| DME19 | CG6634/CG14020 | 2L | 5391354 | N | n/a |
| DME20 | CG5833/CG13133 | 2L | 9990955 | N | n/a |
| DME21 | CG14006/11147 | 2L | 5649514 | N | n/a |
| DME22 | CG10030 | 2L | 3629606 | N | n/a |
| DME23 | RfeSP/10871 | 2L | 1623318 | N | n/a |
| DME24 | CG12511/CG7236 | 2L | 5630916 | N | n/a |
| DME25 | CG15357/CG7312 | 2L | 1876200 | N | n/a |
| DME26 | sif | 3L | 5644285 | N | n/a |
| DME27 | CG3746/CG6664 | 3L | 16908965 | N | n/a |
| DME28 | CG10632 | 3L | 12484307 | N | n/a |
| DME29 | CG6738/CG13830 | 3L | 18842876 | N | n/a |
| DME30 | CG5214/KP78b | 3R | 7086876 | N | n/a |
| DME31 | CG9458/CG7921 | 3R | 5628192 | N | n/a |
| DME32 | CG7920/CG7921 | 3R | 25722579 | N | n/a |
| DME33 | sp2/CG14277 | 2L | 8142925 | N | n/a |
| DME34 | CG4546/CG9625 | 3R | 11607475 | N | n/a |
Based on annotations present in Flybase.
Includes all annotations for mesoderm.
Known to have expression in embryonic dorsal mesoderm.
cis-Regulatory elements directing dorsal mesodermal expression should map in proximity to mesodermally expressed genes. We used the gene-expression annotations present in Flybase (Flybase 1999; http://flybase.bio.indiana.edu) to assess how many of the predicted elements were located either within the introns of or adjacent to genes with known mesodermal expression. The Flybase annotations indicate a strong bias for mesodermally expressed genes mapping near the elements identified by our search (P = 0.0001; Supplemental Table 2 available online at http://www.genome.org). This enrichment for mesodermal genes is consistent with our hypothesis that cooccurrence of the five TFs might be predictive for dorsal mesodermal regulatory elements.
cis-Regulatory modules frequently contain multiple occurrences of a given TF binding site (Arnone and Davidson 1997). As a first step in the analysis of our search results, therefore, we filtered the elements to select only those that occur in noncoding sequence and in which each TF binding motif is present at least twice, with the exception of that for dTcf, which is found only once in the model sequence, the MHE. We refer to this set as the “selected subset.” The selected subset contains 33 putative DMEs, in addition to the Eve MHE, for a total of 34 elements (Table 1).
DME-2 Functions as a Dorsal Mesodermal Enhancer for the hbr Gene
Element DME-2 maps to an intron of the heartbroken (hbr) gene (also known as stumps and dof; Michelson et al. 1998; Vincent et al. 1998; Imam et al. 1999). As this gene is known to be expressed in the embryonic dorsal mesoderm under control of the Wg, Dpp, and Ras signaling pathways (data not shown; Halfon et al. 2000; Carmena et al. 2002), we focused first on this element. Hbr is initially expressed broadly in the embryonic mesoderm, but at early stage 11 its mesodermal expression is dorsally restricted and undergoes dynamic modulation in subsets of dorsal cells, including the Eve-expressing muscle and heart progenitors (Fig. 2A,B; Halfon et al. 2000; Carmena et al. 2002). So as not to inadvertently delete potentially important sequences, we selected approximately 1.5 kb of intronic sequence, centered on the region identified by our search, and used it to drive reporter gene expression in transgenic embryos. This sequence element, hereafter referred to as the Hbr DME, drove expression of the reporter gene in a number of embryonic dorsal mesodermal cells (Fig. 2C; data not shown). No expression was detected in nonmesodermal Hbr-positive cells (i.e., the ectodermally derived tracheal precursors; Fig. 2C), although some expression was detected in epidermal cells overlying the reporter-expressing mesodermal cells (data not shown). Not all Hbr-positive mesodermal cells expressed the reporter gene, suggesting that mesodermal regulatory elements in addition to the DME are required to govern other aspects of mesodermal Hbr expression. Double-labeling with antibodies to Eve and to the DME-driven β-galactosidase revealed that at least one of the two Eve-positive mesodermal progenitors expressed the DME reporter (Fig. 2D). Expression in the other Eve progenitor was occasionally, although not consistently, detected (data not shown).
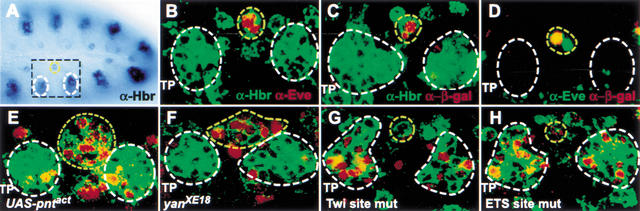
Figure 2.
The Hbr DME. In all panels, white circles mark the developing tracheal cells, and a yellow circle denotes the mesodermal progenitors. (A) Stage 11 embryo stained with antibodies against Hbr. Expression can be seen in the ectodermally derived tracheal pits and the mesodermal progenitors. The portion of each hemisegment shown in the remaining panels is indicated by a black box. (B) Double-labeling for Hbr (green) and Eve (red) shows that the two proteins are found in the same cells in the dorsal mesoderm. Note that Hbr is membrane-associated, whereas Eve is nuclear. Additional mesodermal cells expressing Hbr but not Eve can be seen on both sides of the Eve progenitors. (C) The Hbr DME reporter (nuclear β-galactosidase, red) is expressed in a subset of the Hbr-positive mesodermal progenitors (green). The cells of the developing trachea do not express the reporter. (D) Double-labeling for Eve (green) and the DME reporter (red) show that the DME is expressed in at least one of the Eve progenitors. (E,F) Ectopic activation of the Ras/MAPK pathway through either mesodermal expression of activated Pnt (E) or loss-of-function of the repressor Yan (F) is accompanied by an expanded number of both Hbr- (green) and DME- (red) expressing cells. (G) Mutation of the Twi binding sites in the DME results in a loss of mesodermal reporter gene activity and the acquisition of expression in the developing trachea. (H) Mutation of the Ets binding sites in the DME causes the acquisition of expression in the developing trachea but has only a minimal effect on mesodermal expression. TP, tracheal pits. All panels show anterior to the left and dorsal up.
Ectopic mesodermal activation of the Ras/MAP kinase pathway causes an increased number of cells to express Hbr (Carmena et al. 2002). We therefore examined the activity of the Hbr DME in embryos that expressed a constitutively activated form of the Ras nuclear effector Pnt in the mesoderm. As predicted, activated Pnt induced Hbr expression in an expanded number of cells, and all of these cells expressed the DME reporter gene (Fig. 2E). A similar result was observed when the DME reporter construct was crossed into embryos mutant for the Ras-inactivated repressor, Yan (Fig. 2F). The DME is thus sufficient to drive reporter gene expression in a manner that recapitulates endogenous Hbr expression not only in wild-type conditions but also when the Hbr expression pattern has been altered by experimental manipulation of the pathways known to regulate this gene.
The Hbr DME was identified by virtue of its having predicted binding sites for a number of TFs necessary for inducing dorsal mesodermal expression, including both signal-activated factors such as Pnt and tissue-specific factors such as Twi. To determine whether these predicted sequences are functionally required for DME activity, the sites were mutated in the context of the entire DME. Mutation of the Twi binding sites led to a severe decrease in DME activity in the mesoderm, with the concomitant appearance of reporter gene expression in the Hbr-expressing cells of the developing trachea (Fig. 2G). Mutation of the Ets domain (i.e., Pnt and Yan) binding sites, which are predicted to mediate responses to Ras signaling, also caused an apparent derepression of reporter gene expression in tracheal tissue (Fig. 2H). DME-dependent mesodermal expression was not substantially affected by elimination of the Ets sites, although an occasional reduction/loss of reporter gene activity was observed (Fig. 2H). Both the Twi and Ets binding sites thus play a functional role in the DME, although unlike in the Eve MHE where the Ets binding sites are essential for enhancer activation, those in the DME appear to be required mainly for repression in ectodermally derived tissues. These data validate both our model of transcriptional regulation of dorsal mesodermal gene expression and our computational prediction of binding sites.
To further confirm that the Hbr DME is a genuine cis-regulatory element, the corresponding region from D. virilis was cloned and sequenced. A high degree of sequence conservation was observed spanning an approximately 800 bp portion of the 1.5 kb DME sequence, with complete conservation of many of the binding site motifs used in our search (Fig. 3 and Supplemental Fig. 2 available online at http://www.genome.org). The DME thus meets all of the requirements of a dorsal mesodermal enhancer element that functions in a manner similar to the Eve MHE: It drives reporter gene expression in the embryonic dorsal mesoderm in a pattern that overlaps that of the MHE; the reporter gene expression responds predictably to ectopic activation of signaling pathways; binding sites for TFs predicted to play a role in mediating DME-driven expression are functionally required; and the DME sequence has been conserved through evolution.
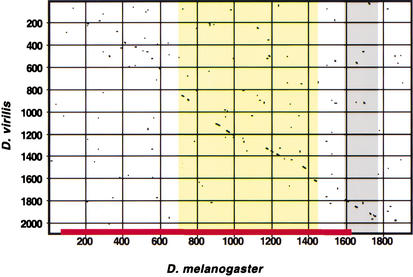
Figure 3.
Conservation of the Hbr DME. Extensive regions of conservation can be detected between D. melanogaster (x-axis) and D. virilis (y-axis) in the region of the DME (red bar). Conservation within the DME is indicated by yellow shading; gray shading indicates a conserved region not required for DME activity that may represent part of a tracheal-specific enhancer (M. Halfon, unpubl.). Little conservation is evident in the adjacent sequences (see Supplemental Fig. 3 for details. Online at http://www.genome.org).
Additional Elements Map Close to Known and Novel Mesodermally Expressed Genes
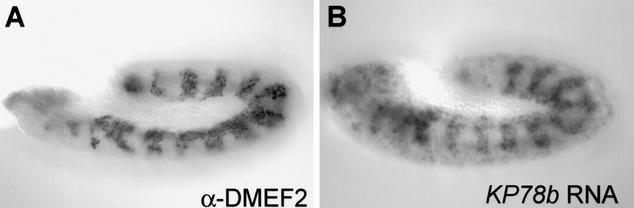
Several additional elements in the selected subset map in proximity to genes with mesodermal expression. These include the known mesodermal genes dmef2 (DME-3), meso18e (DME-4), and rst (DME-7), as well as at least one gene, KP78b (DME-30), for which mesodermal expression has not been reported (Fig. 4; data not shown; Lilly et al. 1994; Taylor 2000; Strunkelnberg et al. 2001). We tested the ability of these elements, as well as elements DME-25 and DME-31, which map near to uncharacterized genes, to function as dorsal mesodermal enhancers in transgenic embryos. Surprisingly, none of these elements appeared able to drive reporter gene expression (data not shown). We cannot rule out the possibility that some or all of these elements are true DMEs and that our assay was insufficient to detect their activities—for instance, we may have failed to incorporate all of the necessary sequences into the reporter constructs, or the elements may be highly sensitive to specific promoter-enhancer interactions and thus fail in our construct. However, the simplest explanation is that despite their lying close to genes with the expected mesodermal expression pattern, these are, as has been seen in other computational regulatory element prediction approaches, false-positive results.
Figure 4.
Additional elements identified in the search map in proximity to mesodermally expressed genes. Pictured is antibody staining showing mesodermal expression of DMEF2 (A) and whole mount in situ hybridization of KP78b RNA (B), a gene not previously known to be expressed in the mesoderm.
Identification of Additional Sequence Motifs
We next sought to determine whether we could identify additional sequence motifs that might represent binding sites for previously unrecognized TFs necessary for the generation of dorsal mesodermal gene expression. Even given the inclusion of false-positive elements, the presence of true DMEs among the search results should be sufficient to enable the identification of additional motifs common to a large number of the putative elements. We used the AlignACE program (Roth et al. 1998; Hughes et al. 2000) to look for such additional motifs within the selected subset of elements (see Methods).
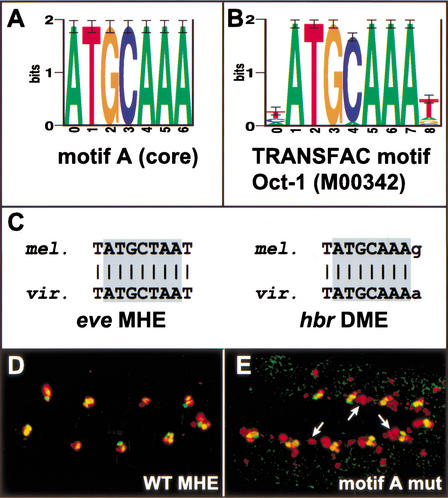
AlignACE successfully identified four of the five motifs used to conduct the search; only the Mad motif, which is a highly degenerate sequence of low information content (see Supplemental Table 3, available online at http://www.genome.org) was not found. In addition, a large number of new motifs were identified (Supplemental Fig. 3, available online at http://www.genome.org; data not shown). In keeping with the expectation that important motifs are evolutionarily conserved, we screened the set of new motifs for their presence in phylogenetically conserved regions of either the Eve MHE or Hbr DME (see Methods). Twenty-five motifs, which cluster into 14 related groups and include the known dTcf, ETS, Tin, and Twi motifs, passed this filter. A search of the TRANSFAC database (Wingender et al. 2001; http://www.gene-regulation.com) revealed that one of these motifs, motif A, matched the binding site for the POU/homeodomain TF Oct-1 (Fig. 5A–C).
Figure 5.
Identification of a new functional motif using AlignACE. (A) Sequence logo (Schneider and Stephens 1990) showing the motif A core as identified by AlignACE. (B) Sequence logo for the POU/homeodomain TF Oct-1 from TRANSFAC (Wingender et al. 2001; accession #M00342). (C) Alignments of motif A from the Eve MHE and Hbr DME with the corresponding sequences from D. virilis. (D) Wild-type and (E) motif A site mutated Eve MHE reporter gene expression (red) along with endogenous Eve expression (green). The motif A mutation leads to an expansion of reporter gene expression along the anteroposterior axis (arrows).
To test whether this motif represents a functional TF binding site, it was mutated in the context of an otherwise wild-type MHE. Analysis of transgenic embryos bearing this mutated MHE revealed an increased number of cells expressing the reporter gene, suggesting that this site is required for the binding of a transcriptional repressor (Fig. 5E). Although the actual repressor which binds to this site remains to be identified, preliminary results from a yeast one-hybrid screen suggest that the sequence is able to bind homeobox-containing TFs, consistent with the TRANSFAC data (S. Gisselbrecht and A. Michelson, unpubl.). In addition, available genetic data are compatible with the possibility that the Ladybird homeodomain proteins could act through this site (Jagla et al. 1997). These data show that analysis of the initial search output, even when confounded by the presence of false-positive results, can lead to the identification of previously unidentified binding sites important for dorsal mesodermal transcriptional regulation.
DISCUSSION
We have undertaken a computational search for cis-regulatory elements that drive transcription in the Drosophila embryonic dorsal mesoderm. The search was based on a specific model derived from the Eve MHE, a transcriptional enhancer that requires the binding of at least five distinct transcription factors to activate gene expression in a tightly defined cluster of muscle and cardiac progenitor cells. We succeeded in identifying at least one additional dorsal mesodermal enhancer, the Hbr DME, demonstrating that a search such as ours, constructed around a single detailed model of a regulatory element, provides a valid and useful means of discovery for additional elements that regulate gene expression in similar temporal and spatial patterns. Moreover, through alignment of the search results, we were able to identify previously unrecognized sequence motifs, one of which was shown to be important for the generation of the expression pattern of interest.
The MHE Model is Generalizable
The fact that we were able to identify a functionally related enhancer based on the MHE model shows that similar regulatory strategies can be used by coexpressed genes. As such, it may be possible to use variations on our model to discover enhancers for mesodermal genes subject to Wg and Ras but not Dpp control, or for nonmesodermal genes that also respond to combined Wg, Dpp, and Ras signaling. The MHE model is predicated on the presence of binding sites for the signal-responsive TFs dTcf, Mad, and Pnt, with tissue specificity provided by the mesodermal selector proteins, Twi and Tin (Halfon et al. 2000). Searching for the presence of the signal-activated factors in combination with selector proteins for other tissues may therefore be sufficient to detect signal-responsive genes in these other cell types. Furthermore, as additional DMEs are confirmed from analysis of our search results, shared structural features of the enhancers, such as a specific number, order, orientation, or spacing of binding sites may become apparent.
Interestingly, enhancers associated with a number of genes known to be coexpressed with eve and hbr in the dorsal mesoderm, such as heartless (Shishido et al. 1993) and Krüppel (Hoch et al. 1990) were not identified in the search, suggesting that at least some of these regulatory elements may depend on different mechanisms than those used by eve and hbr for their dorsal mesodermal expression. Thus, different regulatory mechanisms may lead to similar outcomes. A more complete understanding of these alternate strategies must await the isolation of regulatory elements for some of these additional genes.
Evaluating and Reducing the False-Positive Rate
Like those obtained with other approaches to the computational prediction of regulatory elements (e.g., Wasserman and Fickett 1998; Gailus-Durner et al. 2001; Krivan and Wasserman 2001; Berman et al. 2002; Markstein et al. 2002), our results include a number of false positives. These results are instructive, however; despite our analysis of the search results leading to the identification of a gene (KP78b) previously not known to have dorsal mesodermal expression, the correct cis-regulatory sequences for this gene were not correctly predicted. Indeed, four of the six demonstrated false positives mapped near genes with expression in the dorsal mesoderm. The presence of a computationally identified element near a gene with the expected pattern of expression thus does not guarantee that the element represents a true cis-regulatory module. Our data underscore the importance of empirical testing of computational predictions, which has not been performed comprehensively in other studies, and suggest that the efficacy of cis-regulatory prediction algorithms in general may be lower than initially estimated.
An important future refinement of our method will be to incorporate ways to better and more easily eliminate false-positive results. All of the elements we tested weredrawn from the selected subset of elements containing at least two of each TF binding site motif (except for the dTcf motif). Other filtering criteria may lead to a lower false-positive rate. A number of recent studies used a more general requirement that a minimum number of binding sites be present, without mandating a specific number for any particular site (Frith et al. 2001; Berman et al. 2002). However, it is not clear that adapting such an approach would significantly impact our success rate, as the site density of our apparent false positives is not significantly different from that of the true positives. Moreover, the fact that both of the elements we analyzed, the MHE and the hbr DME, contain only a single binding site for dTcf suggests that a simple minimum site number requirement such as that employed by Markstein et al. (2002) may not be widely applicable. We note as well that methods that are highly dependent on site number are highly sensitive to the information content of the given site motifs. Short or somewhat degenerate motifs will occur more frequently in the genome, meaning that elements composed of such binding sites will tend to be predicted at higher rates than those containing only longer or more invariant motifs. Use of a combinatorial strategy such as that described here that described here helps to reduce the leverage that low-information-content motifs have on the search results.
A considerable advantage will be gained by the ability to include additional types of information in the search algorithm. Both the Eve MHE and Hbr DME show significant degrees of sequence conservation in a distantly related species, and identification of conserved noncoding sequences should help in recognizing true-positive results (see also Loots et al. 2000; Wasserman et al. 2000). Extensive interspecific sequence comparisons are already possible for mammalian and nematode genomes, and a second Drosophila species will be sequenced in the near future. A second way of decreasing the number of false-positive results in future searches will be to incorporate genomic expression data from microarray, SAGE, high-throughput in situ hybridization, or similar expression profiling methods. A number of studies have already demonstrated that expression data, even when gleaned from separate experiments, provide a useful way of identifying potentially coregulated genes (Jensen and Knudsen 2000; Bussemaker et al. 2001; Pilpel et al. 2001).
Toward the Comprehensive Identification of cis-Regulatory Sequences
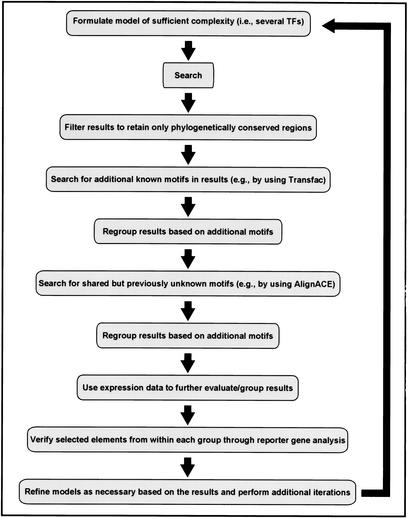
We were able to rapidly characterize the Hbr DME by starting with a single model and evaluating the search results with a combination of gene expression data, conserved sequence data, and alignment-mediated motif extraction. We propose that use of these several components together in an iterative searching strategy (Fig. 6), taking advantage of the rapidly growing availability of sequence, binding, and expression data in public databases, will provide an optimal approach to genomic-scale regulatory module discovery.
Figure 6.
An iterative search strategy for discovery of cis-regulatory elements based on an experimentally validated model. (See text for details).
METHODS
Fly Stocks and Reporter Gene Construction
Fly stocks and methods for reporter gene construction and mutagenesis are as described by Halfon et al. (2000). A minimum of three independent transgenic lines were analyzed for each construct. The following sequences were cloned by PCR and used for the reporter constructs: hbr/DME-2, GenBank accession no. AE003705, 113573-115127; DME-3, AE003831, 74207-74696; DME-4, AE003513, 21993-22696; DME-7, AE003426, 103046-103969; DME-25, AE003584, 51023-51921; DME-30, AE003690, 206275-207950; DME-31, AE003684, 115123-116875. For the motif A mutation, the sequence TATGCTAAT was changed to TATTATCAC.
D. virilis Cloning
Sequences from D. virilis were obtained by screening a genomic library (Blackman and Meselson 1986) with probes from the D. melanogaster Eve or Hbr coding regions (details available on request). Isolated clones were then digested and screened by Southern analysis for cross-hybridization with the MHE or DME sequences, respectively; cross-hybridizing bands were subcloned and sequenced. These sequences have been submitted to GenBank with accession nos. AF513981 (Eve MHE) and AF513982 (Hbr DME).
Sequence Alignments
Dot-plot alignments were created using MacVector (Accelrys Bioinformatics) with hash value = 1 and window = 10. Sequence alignments were performed using VISTA (Mayor et al. 2000) followed by manual adjustment for optimal alignment.
Computational Search
Binding sites for each of the five TFs were culled from the literature and used to construct position weight matrices (PWMs; Supplemental Table 3, available online at http://www.genome.org). Only sites with both in vitro binding and in vivo function were included. The program ScanACE (Hughes et al. 2000) was used to scan release 2 of the Drosophila melanogaster genome (Adams et al. 2000; http://www.fruitfly.org/sequence/download.html). Those sequences with matches that scored at a level equal to or greater than the lowest scoring functional sites in the Eve MHE were selected. We then ran a program (cooccur_scan.pl; available at http://arep.med.harvard.edu/Halfon_Grad_etal/supplemental.html) to determine regions of cooccurrence of predicted binding sites for all five TFs within at least 500 bp. Two special cases were addressed: (1) Both Twi and MAD sites can form palindromes; when this was observed, only one of the palindromes was included in further analysis. (2) Mad sites may match the initial bases of Ets sites; in these cases, the Ets sites were included to the exclusion of the overlapping Mad sites, as the Mad PWM has lower information content than the Ets PWM. The algorithm extends the windows for as long as the condition that binding sites for all five TFs appear within 500 bp holds. Using Drosophila genome annotations (http:// www.fruitfly.org/sequence/download.html), the program also assigns locations of cooccurrence regions with respect to known and predicted gene coding sequence. The results of this analysis were termed “elements.” A subset of the elements was then sorted to identify those in noncoding sequence with at least two predicted sites for each of the TFs except dTcf (termed the “selected subset”).
Monte Carlo Methods
To estimate the number of cooccurrences expected by chance given the number of motifs identified by ScanACE, a variant of the cooccur_scan.pl program was used. This program maintained the number of motifs found in the genome scan, randomized the locations of each motif, and then searched for cooccurrences as in the coccur_scan.pl program. As the D. melanogaster genome sequence is available in several FASTA format files, each representing a chromosomal arm or chromosome, the scans were performed per individual FASTA file and then summed to represent the whole genome. Gaps in the sequences denoted by strings of Ns greater than 50 letters were taken into account in location randomizations to ensure that the overall space for motif locations was nearly identical.
Motif Discovery and Evaluation
Sequence corresponding to the 34 elements in the selected subset was extracted from Release 2 of the Drosophila genome and searched for overrepresented subsequences using AlignACE (Roth et al. 1998; Hughes et al. 2000). AlignACE identified a total of 755 motifs from several runs using combinations of parameters intended to sample extensively the sequence space (all variations of –gcback 0.42, 0.45, 0.48 and –numcols 7, 10, 13). The ACE package clustering program revealed that the motifs cluster into 375 motif groups at the 0.8 correlation coefficient level.
The top 100 sites in the 34 elements for each of the 755 motifs were determined by running ScanACE with default parameters and –s 100 on the set of predicted element sequences. Using a Perl script, these sites were evaluated for phylogenetic conservation as determined by D. melanogaster versus D. virilis sequence alignments of the Eve MHE and Hbr DME. Twenty-five motifs mapped to perfectly conserved sites. Clustering using the ACE package clustering software at the 0.8 correlation coefficient level organized the 25 motifs into 14 motif groups. All of the 755 initial motifs were compared against TRANSFAC (Wingender et al. 2001) using CompareACE (Hughes et al. 2000). The ACE programs are available for download at http://arep.med.harvard.edu.
WEB SITE REFERENCES
http://www.fruitfly.org/sequence/download.html; Berkeley Drosophila Genome Project sequence downloads.
http://flybase.bio.indiana.edu; Central database on genetics of Drosophila.
http://arep.med.harvard.edu; Harvard-Lipper Center for Compuational Genetics. The ACE family of programs and cooccurrence program are available at this site.
http://www.gene-regulation.com; TRANSFAC database of transcription factor binding sites.
Acknowledgments
We thank D. Gilbert, C. Mungall, and the staff at both Flybase and the BDGP for invaluable assistance with genome annotation, R. Blackman for the D. virilis library, L. Raj and J. Oborski for assistance with library screening and fly genetics, L. Ma for encouraging this approach, Y. Pilpel for helpful advice, and J. Aach, J. Schendure, and M. Bulyk for comments on the manuscript. This work was supported by NSF, DOE, and NHBLI-PGA grants (G.M.C.), the American Cancer Society (M.S.H.), and the Howard Hughes Medical Institute, of which A.M.M. is an Associate Investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL michelson@rascal.med.harvard.edu; FAX (617) 738-5575.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.228902.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Arnone MI, Davidson EH. The hardwiring of development: Organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman RK, Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986;188:499–515. doi: 10.1016/s0022-2836(86)80001-8. [DOI] [PubMed] [Google Scholar]
- Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- Carmena A, Gisselbrecht S, Harrison J, Jiménez F, Michelson AM. Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes & Dev. 1998;12:3910–3922. doi: 10.1101/gad.12.24.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A, Buff E, Halfon MS, Gisselbrecht S, Jimenez F, Baylies MK, Michelson AM. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol. 2002;244:226–242. doi: 10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Genomic regulatory systems. San Diego: Academic Press; 2001. [Google Scholar]
- Fickett JW, Wasserman WW. Discovery and modeling of transcriptional regulatory regions. Curr Opin Biotechnol. 2000;11:19–24. doi: 10.1016/s0958-1669(99)00049-x. [DOI] [PubMed] [Google Scholar]
- Flybase. The FlyBase database of the Drosophila Genome Projects and community literature. The FlyBase Consortium. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech K, Danescu-Mayer J, Werner T. A novel method to develop highly specific models for regulatory units detects a new LTR in GenBank which contains a functional promoter. J Mol Biol. 1997;270:674–687. doi: 10.1006/jmbi.1997.1140. [DOI] [PubMed] [Google Scholar]
- Frith MC, Hansen U, Weng Z. Detection of cis -element clusters in higher eukaryotic DNA. Bioinformatics. 2001;17:878–889. doi: 10.1093/bioinformatics/17.10.878. [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V, Scherf M, Werner T. Experimental data of a single promoter can be used for in silico detection of genes with related regulation in the absence of sequence similarity. Mamm Genome. 2001;12:67–72. doi: 10.1007/s003350010219. [DOI] [PubMed] [Google Scholar]
- Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jiménez F, Baylies MK, Michelson AM. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- Hoch M, Schroder C, Seifert E, Jackle H. cis-acting control elements for Kruppel expression in the Drosophila embryo. EMBO J. 1990;9:2587–2595. doi: 10.1002/j.1460-2075.1990.tb07440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Imam F, Sutherland D, Huang W, Krasnow MA. stumps, a Drosophila gene required for fibroblast growth factor (FGF)-directed migrations of tracheal and mesodermal cells. Genetics. 1999;152:307–318. doi: 10.1093/genetics/152.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagla K, Frasch M, Jagla T, Dretzen G, Bellard F, Bellard M. Ladybird, a new component of the cardiogenic pathway in Drosophila required for diversification of heart precursors. Development. 1997;124:3471–3479. doi: 10.1242/dev.124.18.3471. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Knudsen S. Automatic discovery of regulatory patterns in promoter regions based on whole cell expression data and functional annotation. Bioinformatics. 2000;16:326–333. doi: 10.1093/bioinformatics/16.4.326. [DOI] [PubMed] [Google Scholar]
- Knirr S, Frasch M. Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev Biol. 2001;238:13–26. doi: 10.1006/dbio.2001.0397. [DOI] [PubMed] [Google Scholar]
- Krivan W, Wasserman WW. A predictive model for regulatory sequences directing liver-specific transcription. Genome Res. 2001;11:1559–1566. doi: 10.1101/gr.180601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatowski J, Skarecky D, Bailey K, Ayala FJ. Phylogeny of Drosophila and related genera inferred from the nucleotide sequence of the Cu,Zn Sod gene. J Mol Evol. 1994;38:443–454. doi: 10.1007/BF00178844. [DOI] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: A MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine M. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Michelson AM, Gisselbrecht S, Buff E, Skeath JB. Heartbroken is a specific downstream mediator of FGF receptor signalling in Drosophila. Development. 1998;125:4379–4389. doi: 10.1242/dev.125.22.4379. [DOI] [PubMed] [Google Scholar]
- Ohler U, Niemann H. Identification and analysis of eukaryotic promoters: Recent computational approaches. Trends Genet. 2001;17:56–60. doi: 10.1016/s0168-9525(00)02174-0. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido E, Higashijima S, Emori Y, Saigo K. Two FGF-receptor homologues of Drosophila—One is expressed in mesodermal primordium in early embryos. Development. 1993;117:751–761. doi: 10.1242/dev.117.2.751. [DOI] [PubMed] [Google Scholar]
- Strunkelnberg M, Bonengel B, Moda LM, Hertenstein A, de Couet HG, Ramos RG, Fischbach KF. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development. 2001;128:4229–4239. doi: 10.1242/dev.128.21.4229. [DOI] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Taylor MV. A novel Drosophila, mef2-regulated muscle gene isolated in a subtractive hybridization-based molecular screen using small amounts of zygotic mutant RNA. Dev Biol. 2000;220:37–52. doi: 10.1006/dbio.2000.9608. [DOI] [PubMed] [Google Scholar]
- Vincent S, Wilson R, Coelho C, Affolter M, Leptin M. The Drosophila protein Dof is specifically required for FGF signaling. Mol Cell. 1998;2:515–525. doi: 10.1016/s1097-2765(00)80151-3. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Fickett JW. Identification of regulatory regions which confer muscle-specific gene expression. J Mol Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nat Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhauser R, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]