Abstract
We have analyzed the biochemical consequences of mutations that affect viral RNA synthesis in Semliki Forest virus temperature-sensitive (ts) mutants. Of the six mutations mapping in the multifunctional replicase protein nsP2, three were located in the N-terminal helicase region and three were in the C-terminal protease domain. Wild-type and mutant nsP2s were expressed, purified, and assayed for nucleotide triphosphatase (NTPase), RNA triphosphatase (RTPase), and protease activities in vitro at 24°C and 35°C. The protease domain mutants (ts4, ts6, and ts11) had reduced protease activity at 35°C but displayed normal NTPase and RTPase. The helicase domain mutation ts1 did not have enzymatic consequences, whereas ts13a and ts9 reduced both NTPase and protease activities but in different and mutant-specific ways. The effects of these helicase domain mutants on protease function suggest interdomain interactions within nsP2. NTPase activity was not directly required for protease activity. The similarities of the NTPase and RTPase results, as well as competition experiments, suggest that these two reactions utilize the same active site. The mutations were also studied in recombinant viruses first cultivated at the permissive temperature and then shifted up to the restrictive temperature. Processing of the nonstructural polyprotein was generally retarded in cells infected with viruses carrying the ts4, ts6, ts11, and ts13a mutations, and a specific defect appeared in ts9. All mutations except ts13a were associated with a large reduction in the production of the subgenomic 26S mRNA, indicating that both protease and helicase domains influence the recognition of the subgenomic promoter during virus replication.
Semliki Forest virus (SFV), a member of the Alphavirus genus, has a positive-strand RNA genome of about 11.5 kb. The 42S RNA genome codes for a large nonstructural polyprotein (P1234) of 2,432 amino acid (aa) residues which is responsible for viral RNA replication. P1234 is autocatalytically processed to yield four nonstructural proteins, nsP1 to nsP4. The structural proteins (capsid protein and envelope proteins E1, E2, and E3) are also translated as a polyprotein from a subgenomic 26S mRNA that is identical to the 3′ third of the 42S RNA genome (reviewed in references 20 and 45).
The RNA synthesis of alphaviruses takes place in the cytoplasm in association with characteristic cytopathic vacuoles (CPVs), which are modified endo- and lysosomal structures (12). The membranes of the CPVs contain numerous small invaginations, or spherules, with a diameter of about 50 nm. Cryoimmunoelectron microscopy studies indicate that all four of the nsPs are associated with CPVs and particularly with the spherules, together with nascent RNA molecules, suggesting that the spherules represent the units of RNA replication (25). Although many of the early events of replication are poorly understood, it seems that the polyprotein stage is necessary for the membrane association, proper assembly, and endo- and lysosomal targeting of the replication complexes (39).
Several of the functions of the individual nsPs have been revealed. According to sequence homology and results with alphavirus temperature-sensitive (ts) mutants, nsP4 is the catalytic subunit of the polymerase complex (20). The specific functions of nsP3 are still unknown. The protein consists of a conserved amino-terminal domain of about 300 residues and a highly variable carboxy terminus, which is phosphorylated on several serine and threonine residues (50, 51). There is evidence that nsP3 is involved in the synthesis of minus strand RNA (7, 52). nsP1 has unique methyltransferase and guanylyltransferase activities needed for the capping of viral RNAs in the cytoplasm of infected cells (1, 20). This protein is associated with intracellular membranes primarily via a stretch of amino acids located in the middle of the protein, which forms an amphipathic alpha-helical membrane binding peptide (2, 26). As part of the P1234 polyprotein, nsP1 seems to be responsible for the membrane association of the whole replication complex (38, 39).
The largest multifunctional replicase protein, nsP2 (799 aa), has two principal domains. The N-terminal half has sequence motifs typical for RNA helicases and nucleotide triphosphatases (NTPases) (15), whereas the C-terminal half is a thiol proteinase of the papain superfamily (3). The whole nsP2 protein and the N-terminal 470 residues have NTPase activity, which is stimulated by the presence of RNA. The NTPase activity is knocked out by mutation of Lys192, located in a Walker A motif (GxxGxGKS), which also leads to loss of virus infectivity (35, 36). The N-terminal fragment also has RNA triphosphatase (RTPase) activity, which removes only the γ-phosphate from the 5′ end of the RNA (47). The RNA helicase activity has been demonstrated only for the full-length nsP2 protein (14).
Deletion analysis and site-directed mutagenesis of Sindbis virus (SIN; a related alphavirus) nsP2 have verified that the carboxy-terminal half contains the autoprotease activity of the protein (9, 18), for which a conserved cysteine (Cys 478 in SFV) and histidine (His 548 in SFV) are essential (13, 44, 49). The isolated, purified C-terminal domain, Pro39, of 341 residues is an active protease. Its substrate specificity was studied in vitro by using the cleavage sites of the SFV nonstructural polyprotein P1234. Twenty amino acids from both sides of the predicted cleavage site of each junction were expressed as a fusion protein with thioredoxin. Cleavage of the constructed nsP3-nsP4 site was efficient, whereas cleavage at site 1/2 was very inefficient (49). In vitro translation and processing of truncated nonstructural polyprotein constructs revealed that the cleavage at the 1/2 site takes place relatively slowly and in cis, but it is essential for the cleavage at site 2/3, since the release of the authentic N terminus of nsP2 is an important prerequisite for cleavage at the 2/3 site (48). The rapid cleavage at the 3/4 site, combined with the slow cleavage at the 1/2 site, results in the production of the short-lived RNA early polymerase complex (P123 plus nsP4), which is responsible for the synthesis of minus strands early in infection (23, 27, 43, 52). This complex is then converted to the stable late polymerase complex that consists of fully processed components and synthesizes positive-strand RNA.
Yet other functions have been localized to the carboxy-terminal part of nsP2. A short sequence, PRRRV651, was identified as a nuclear localization signal responsible for the transport of a large fraction of nsP2 to the nucleus. The significance of the nuclear localization is not clear, since mutations preventing the nuclear transport of nsP2 are not lethal. However, these mutants have lost their neuropathogenicity for mice (10, 35). Mutations causing a persistent-replication phenotype in mammalian cells often affect SIN nsP2 residue Pro726 (11). Many SIN RNA-negative ts mutants with defects in the regulation of the subgenomic 26S RNA synthesis have mutations in the carboxy terminus of nsP2. This has been taken as evidence that nsP2 serves as an initiation factor for the transcription of 26S RNA from the minus-strand RNA template (20).
We have recently identified the amino acid changes present in SFV ts mutants, which have defects in the early events of virus replication (30). As expected, these RNA-negative ts mutants had mutations in the nonstructural proteins, which are the virus-specific components of the RNA polymerase complex. Most of these ts mutants had a point mutation in the multifunctional nsP2 protein (six out of eight mutants). Here we have characterized the enzymatic properties of nsP2 proteins containing these single amino acid replacements in order to understand the relationships between the reactions catalyzed by nsP2 and the RNA-negative phenotype of the viruses. The respective mutant viruses have also been studied in temperature shift-up experiments in cell culture. We found that mutations in the protease domain of nsP2 always affected the processing of the replicase polyprotein precursor but did not influence the functions of the helicase domain. In contrast, each helicase domain mutation displayed individual biochemical properties and some of them also modulated protease activity.
MATERIALS AND METHODS
Viruses and cells.
The isolation and characterization of SFV ts mutants have been described previously (21, 22). The cloning and sequencing of the SFV prototype to create the infectious cDNA clone SFV4 have been previously reported (29). Individual mutations have been analyzed in the prototype (here termed the wild-type) background by using recombinant viruses designated with the prefix SFots or SFons (30). The viruses have been grown and studied in BHK21 cells (American Type Cell Culture Collection).
Protein expression and purification.
Full-length nsP2 of SFV and its mutant derivatives (ts1, ts4, ts6, ts9, ts11, ts13a, and GNS), as well as the protease substrates, were expressed in Escherichia coli BL21(DE3) (Stratagene) by using pET32b and pET32c (Novagen) encoding carboxy-terminal six-His tags. The proteins were purified by Ni2+ affinity chromatography essentially as previously described (13). Bacteria were grown in 900 ml of Luria-Bertani medium containing ampicillin (100 μg/ml) at 37°C until the optical density at 600 nm reached 0.6. The culture was cooled on ice, equilibrated at 17°C, and induced with 40 μg/ml isopropyl-β-d-thiogalactopyranoside for 16 to 18 h at 17°C. The cells were then pelleted; resuspended in ice-cold buffer containing 20 mM sodium phosphate (pH 7.4), 200 mM NaCl, 0.1% Tween 20, and 1 mM phenylmethylsulfonyl fluoride; and lysed in a French press (15,400 lb/in2) at 4°C. The cell lysate was diluted to 25 ml with the same buffer and centrifuged for 30 min at 15,000 × g at 4°C. The supernatant was supplemented with imidazole (final concentration, 40 mM) and loaded into an Ni2+ chromatography column (HiTrap Chelating HP; GE Healthcare) at 4°C. After washing with 20 mM HEPES (pH 7.4)-200 mM NaCl-0.1% Tween 20-100 mM imidazole, the protein was eluted in 2.5 ml of the same buffer containing 500 mM imidazole. After the addition of EDTA (final concentration, 10 mM), the protein preparation was loaded into a desalting column (PD-10; GE Healthcare) and eluted in 3.5 ml of storage buffer containing 20 mM HEPES (pH 7.4), 200 mM NaCl, 2 mM dithiothreitol (DTT), and 20% glycerol. The protein preparation was aliquoted, frozen in liquid nitrogen, and stored at −70°C.
The mutations (30, 36) were introduced into the nsP2 expression plasmid (48) by using available restriction sites. The short thioredoxin fusion protein substrates that span the cleavage junctions have been previously described (49). The Pro-X substrate, representing a longer stretch of the 2/3 junction (341 aa of nsP2 and 170 aa of nsP3) was a kind gift of A. Lulla (University of Tartu, Tartu, Estonia). Protein concentration was determined with the Bio-Rad protein assay, and a standard curve was made by using serial dilutions of bovine serum albumin. The purity of preparations was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) with 10% gels for nsP2 and 15% gels for in vitro protease substrates.
RNA synthesis.
RNA substrates were produced by in vitro transcription of pGEM3Zf(+) (Promega) digested with EcoRI. The synthesis of 64-nucleotide RNA probes (sequence, GAA UAC UCA AGC UUG CAU GCC UGC AGG UCG ACU CUA GAG GAU CCC CGG GUA CCG AGC UCG AAU U) was carried out in the presence of 50 μl of Promega transcription buffer including 0.5 mM each NTP (Amersham Bioscience), 50 μCi of either [γ-32P]GTP or [α-32P]GTP (5,000 Ci/mmol; Amersham Bioscience), 40 U of RNasin (Promega), 40 U of SP6 RNA polymerase (Promega), and 2.5 μg of linear DNA substrate. The mixtures were incubated at 37°C for 2 h. The reaction was stopped by adding 1 U of DNase RQ (Promega) per μg of template DNA (30 min at 37°C). RNA preparations were purified by Sephadex G25 spin columns (Amersham Bioscience). The resultant RNA samples were extracted with phenol-chloroform-isoamyl alcohol (125:24:1, pH 4.7) and precipitated with ethanol in the presence of 0.3 M ammonium acetate. RNA concentration was determined by absorbance at 260 nm. The purity of the RNA was verified by PAGE in 10% gels containing 7 M urea.
Protease assay.
The in vitro protease activity of nsP2 was assayed with the Trx34 (49) or the Pro-X substrate. Purified substrates (10 to 50 μM Trx34, 8 μM Pro-X) and nsP2 preparations (1 μM) were mixed in prewarmed reaction buffer (20 mM HEPES [pH 7.2], 150 mM NaCl, 2 mM DTT) in a 50-μl final volume. The reaction mixtures were incubated at different temperatures, and aliquots were taken at the indicated times into SDS sample buffer, followed by analysis by SDS-PAGE on 15% gels. Coomassie blue-stained gels were analyzed by densitometry.
NTPase assay.
Reaction mixtures (50 μl) containing 20 mM HEPES (pH 7.4), 1 mM MgCl2, 5 mM KCl, 150 mM NaCl, 2 mM DTT, 100 to 200 μM GTP, 0.1 μCi [γ-32P]GTP (Amersham Bioscience) substrate, and 5 nM enzyme were incubated for 30 to 60 min at 24°C or 35°C. Aliquots were taken at the indicated times, and the reaction was stopped by adding EDTA to a final concentration of 250 mM. The aliquots were analyzed by thin-layer chromatography (TLC) on polyethyleneimine-cellulose plates (Merck) developed with 1 M LiCl-1 M formic acid. The reaction products were visualized by phosphorimaging. In some experiments, [γ-32P]ATP was used under the same conditions.
RTPase assay.
RTPase activity was assayed essentially as described previously (47). The reaction mixture (10 μl) contained 20 mM HEPES (pH 7.4), 1 mM MgCl2, 5 mM KCl, 150 mM NaCl, 2 mM DTT, 200 nM γ-32P-labeled RNA substrate, and 1 nM enzyme. Reactions were carried out for 30 min at 24°C or 35°C. Aliquots were taken at the indicated times, and the reactions were stopped with EDTA. Samples were analyzed by TLC, as in the NTPase assay. In competition experiments, the concentration of the RNA substrate was 1 μM, that of the enzyme was 5 nM, and 2 mM MgCl2 was used. The reaction mixture included increasing amounts of GTP, from 25 μM to 1.5 mM, or 25 μM [γ-S]GTP (Sigma).
Protein labeling and immunoprecipitation.
Confluent BHK cells in 60-mm dishes were infected at 28°C with wild-type or mutant SFV at 50 PFU/cell. At 5 h postinfection (p.i.), the medium was replaced with methionine-free medium and the incubation was continued at either 28°C or 39°C for 30 min, followed by a 10-min or 30-min pulse with 150 or 50 μCi/ml [35S]methionine, respectively. Thereafter, the cultures were chased with medium containing 2 mM unlabeled methionine (and 100 μg/ml cycloheximide when indicated). Cells were collected in 600 μl of 1% SDS, and the lysate was passed through a 25-gauge needle 20 times to shear the DNA. The samples were boiled for 2 min and stored at −70°C.
For immunoprecipitation, samples were diluted 10-fold in NET buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% NP-40) and precleared with a protein A-Sepharose CL-4B slurry (1:1 in NET buffer), followed by incubation with continuous mixing for 60 min at 4°C. After removal of the beads, the supernatant was exposed to rabbit polyclonal antisera against SFV nsP1, nsP2, nsP3, and nsP4 in combinations (39). After overnight incubation at 4°C, protein A-Sepharose slurry was added and the samples were incubated for 1 h at 4°C. The beads were washed four times with NET buffer containing 0.4 M NaCl, and the bound antigen was eluted by boiling the beads for 2 min in SDS sample buffer. The labeled proteins were analyzed by SDS-PAGE in 7.5% gels. After electrophoresis, the gels were fixed for 15 min in a solution containing 10% acetic acid and 40% methanol, dried on 3MM paper, and exposed to phosphorimaging plates. To reach quantitative immunoprecipitation conditions, the required amount of each anti-nsP antiserum, as well as the volume of protein A-Sepharose bead slurry, was determined by titration.
Preparation of the P15 fraction and nsP2 immunoprecipitation.
Confluent BHK cells in 140-mm dishes were infected (100 PFU/cell) with either wild-type SFV or Fots9 at 28°C. At 8 h p.i., cells were washed in ice-cold phosphate-buffered saline, scraped, and collected by centrifugation at 900 × g for 5 min at 4°C. The pellets were resuspended in RS buffer (10 mM Tris-HCl [pH 8.0], 10 mM NaCl), and the cells were allowed to swell for 15 min on ice and were disrupted with a tight-fitting Dounce homogenizer (20 strokes). The nuclei were removed by sedimentation at 900 × g for 5 min at 4°C, and the supernatant was centrifuged at 15,000 × g for 20 min at 4°C (34). The resulting pellet (P15) was resuspended in 190 μl of ice-cold NET buffer and processed for immunoprecipitation with 10 μl of rabbit antiserum against nsP2-50 μl of protein A-Sepharose slurry (1:1 in NET buffer). After 3 h of incubation, the beads were collected by a short centrifugation and washed three times with 500 μl of ice-cold NET buffer containing 400 mM NaCl and three times with 500 μl of ice-cold NTPase buffer. The beads were finally resuspended in 50 μl of NTPase buffer and tested for NTPase activity as described above.
Viral RNA analysis.
Confluent BHK cell monolayers in 35-mm dishes were infected as for protein studies and labeled with 50 to 100 μCi of [3H]uridine for 60 min. Cell cultures were kept at 28°C for 6 h and then transferred to 39°C in the presence of 2 μg/ml actinomycin D and 100 μg/ml cycloheximide. Cells were labeled from 7 to 8 h p.i. at 39°C and collected in 1 ml of Trizol reagent (Invitrogen). Parallel infected cultures were incubated at 28°C for 6 h, shifted up to 39°C for 2 h, and thereafter shifted back to 28°C. After 1 h of temperature equilibration, the cells were labeled and lysed in Trizol as described above. RNAs were purified according to the manufacturer's instructions and stored at −70°C. RNA samples were denatured by glyoxal in dimethyl sulfoxide and analyzed by electrophoresis on 0.8% agarose gels in sodium phosphate, pH 7.0, with a buffer recirculation system. Gels were then soaked for 30 min in Amplify (Amersham Bioscience) and dried at 55°C, and radioactivity was detected by fluorography on preflashed films. Quantitation was performed by densitometry or by scintillation counting (Optiphase Supermix; Perkin-Elmer) of the radioactivity recovered from the bands after cutting and dissolving with Optisolv (Perkin-Elmer).
Data analysis and quantification.
Data were analyzed densitometrically with the Aida Image Analyzer program (version 3.44.035) and a Fuji BAS-1500 phosphorimager in the case of radioactive samples from in vitro experiments. Care was taken to avoid intensity saturation, and signal linearity was checked by serial dilutions. For the polyprotein-processing experiments, the intensities of all of the polyprotein precursor bands (P1234, P123, P12, and P34) and individual nsP1-to-nsP4 bands were quantitated, and their sum total at each time point was taken as 100%. The results for each protein are expressed as a percentage of this total, and they thus describe the relative rates of processing. For the in vitro assays, complete cleavage of the substrates was achieved by an excess of enzyme in three parallel independent experiments. The released products were quantified, and a value of 100% was given to the average of the intensities. All of the data shown in the figures and tables are averages of two independent experiments. The absolute values of the two data points differed, in each case, less than 10% in the in vitro experiments and less than 15% in the experiments carried out with cells.
RESULTS
To better understand the roles of the enzymatic activities of nsP2 in the replication cycle of SFV, we have here systematically analyzed the properties of nsP2 proteins which contain individual point mutations derived from SFV ts strains (Fig. 1A and B). In some experiments, an NTPase- and helicase-inactivating mutant (K192N, termed GNS) was used as an additional control. Wild-type nsP2 and its mutant derivatives were produced in E. coli as carboxy-terminally histidine-tagged proteins and purified by Ni2+ affinity chromatography as shown for the wild-type protein in Fig. 1C. All of the mutant proteins were purified by the same protocol, giving similar preparations (Fig. 1D). The derivatives ts9 and, to a lesser extent, ts13a displayed a somewhat lower level of binding to the affinity resin, resulting in a lower yield of the target proteins and a consequently increased relative amount of impurities.
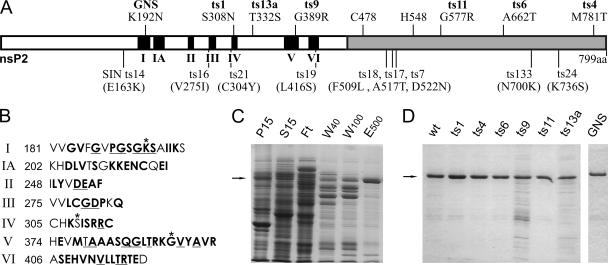
FIG. 1.
nsP2, its mutant derivatives, and their purification. (A) Schematic representation of nsP2. In the helicase domain (white), the conserved helicase motifs (6, 15, 24) are in black and numbered. The protease domain is in gray. Above, the locations and amino acid changes of the SFV ts mutations are indicated. The names of the mutants studied are in bold. The location of the NTPase-helicase active-site mutation (GNS) is also marked, and the protease active-site cysteine and histidine residues are shown. Below, the locations of SIN ts mutations (8, 17) are shown for comparison. The amino acid changes for SIN are in parentheses to indicate that the amino acid numbers cannot be directly compared to those of SFV, as in most cases there is a difference of a few amino acids between the structurally corresponding residues of SFV and SIN. (B) Amino acid sequences of the helicase motifs of SFV. The motif and the number of the first amino acid are indicated on the left. Amino acids conserved in alphaviruses are in bold, and amino acids that are generally most conserved in the wider alphavirus-like superfamily (part of the consensus sequences, as defined by Koonin and Dolja [24]) are underlined. The residues mutated in this study are marked with asterisks in motifs I (GNS), IV (ts1), and V (ts9). (C) Purification pattern of E. coli-expressed wild-type nsP2 with a six-His tag at the carboxy terminus. Purification by Ni2+ affinity chromatography was monitored by SDS-PAGE in a 10% gel. Lanes: P15 and S15, insoluble and soluble fractions, respectively, after centrifugation of bacterial extract at 15,000 × g; Ft, column flowthrough fraction; W40 and W100, column washes with buffer containing 40 or 100 mM imidazole; E500, fraction eluted with 500 mM imidazole. (D) SDS-PAGE analysis of each nsP2 mutant derivative after purification as described for panel C. wt, wild type.
Protease activity of nsP2 in vitro.
As the substrate for the purified nsP2 proteases, we used the recombinant protein Trx34, which contains approximately 20 residues on both sides of the cleavage site between the nsP3 and nsP4 domains of P1234, fused to the thioredoxin carrier (Fig. 2A). Trx34 was also histidine tagged and purified by Ni2+ affinity chromatography. After 60 min of incubation, the reaction mixtures were analyzed by SDS-PAGE. Under these conditions, the substrate and the larger cleavage product could be easily visualized after staining with Coomassie blue (Fig. 2B). Densitometric analysis was used to quantitate the amounts of substrate and product in samples withdrawn after different times of incubation.
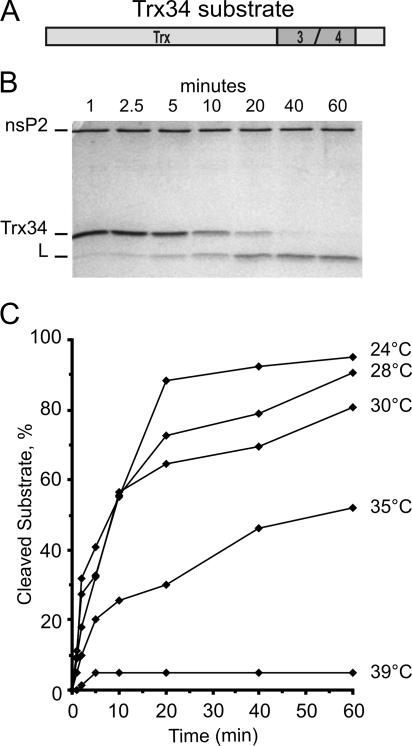
FIG. 2.
Temperature dependence of nsP2 protease activity. (A) Schematic representation of the protease assay substrate Trx34. The amino-terminal 115 aa of thioredoxin are followed by 37 aa from the cleavage region between nsP3 and nsP4 in the SFV replicase polyprotein. The carboxy terminus contains a six-His tag. (B) Electrophoresis on a 15% polyacrylamide gel after a protease assay performed at 24°C with wild-type nsP2. The time course of the reaction is presented. The positions of nsP2, the uncleaved substrate (Trx34), and the large cleavage product (L) are indicated. The smaller cleavage product has migrated out of the gel. (C) Protease time course assays as in panel B were performed at different temperatures with the wild-type nsP2 enzyme. The data plotted here and in the subsequent figures are the averages of two reproducible experiments, as indicated in Materials and Methods.
First we assayed the wild-type nsP2 protease activity at different temperatures (Fig. 2C). The purified recombinant protein had very low activity at 39°C, which is the restrictive temperature for the replication of the ts mutants, becoming inactive after 5 min. Since significant activity, resulting in 50% cleavage of the substrate in 60 min, could be detected at 35°C, we selected 24°C and 35°C as permissive and restrictive temperatures, respectively, for further experiments with the mutant proteins. Thus, the temperature difference was 11°C, the same as that used in the in vivo studies, but the range was shifted by 4°C.
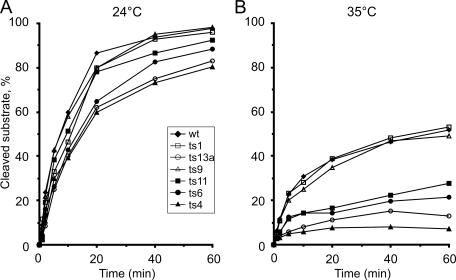
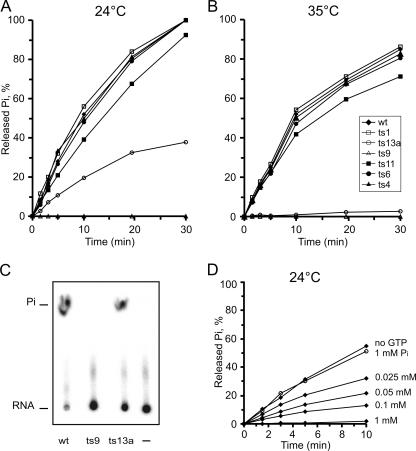
Time course experiments carried out at 24°C showed that the ts1, ts9, and ts11 mutant proteins displayed kinetics similar to those of wild-type nsP2, while ts6, ts4, and ts13a had slightly lower activities (Fig. 3A). At 35°C, the wild-type, ts1, and ts9 protease activities were almost identical, resulting in about 50% cleavage of the substrate after 60 min of incubation (Fig. 3B). In contrast, ts4, ts13a, ts6, and ts11 showed greatly reduced activity at 35°C. The ts4 mutation caused the most severe effect, since after 5 min at 35°C no further cleavage could be detected (Fig. 3B). As the mutations of ts4, ts6, and ts11 were located in the carboxy-terminal protease domain, this result could be expected. In contrast, the strong protease defect of nsP2 of ts13a, where the mutation is located in the helicase domain, was unexpected. Therefore, we also analyzed the protease activity of the NTPase-helicase active-site mutant GNS, which had wild-type protease activity at both temperatures in the assay (data not shown).
FIG. 3.
Protease activity at two temperatures. Purified Trx34 substrate and nsP2 derivatives were incubated for 60 min at 24°C (A) and at 35°C (B). Aliquots were taken at the indicated time points and analyzed by SDS-PAGE as described in the legend to Fig. 2. Wild-type (wt) nsP2 and mutant proteins with changes in the C-terminal protease domain are marked with closed symbols, whereas mutant proteins with changes in the N-terminal helicase domain are marked with open symbols.
NTPase and RTPase activities in vitro.
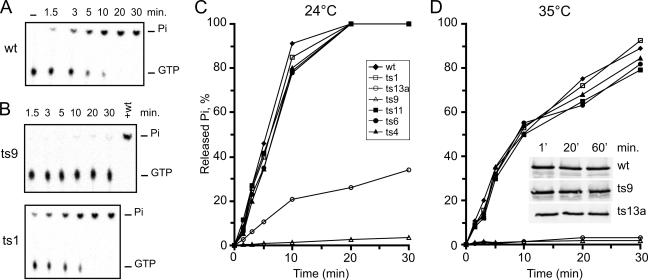
The release of Pi was analyzed in both NTPase and RTPase reactions. An example of a time course for the NTPase reaction is shown in Fig. 4A. Wild-type nsP2 was incubated with labeled [γ-32P]GTP for 30 min at 24°C. The first lane on the left shows the substrate incubated in the reaction buffer for 30 min in the absence of enzyme. The conditions were optimized for wild-type nsP2 at 24°C and 35°C. Time course studies were carried out at both temperatures with the wild-type and all of the mutant nsP2 proteins (Fig. 4C and D). At 24°C, ts1, ts4, ts6, and ts11 showed activity identical to that of the wild type while ts13a had greatly reduced activity. Interestingly, ts9 showed only a trace of activity at this permissive temperature (Fig. 4B and C). When the assay was carried out at 35°C, the overall activity of the wild type was reduced compared to that obtained by incubation at 24°C, as with the protease assay. At 35°C, again the wild type, ts1, ts4, ts6, and ts11 were virtually identical while ts9 and ts13a had essentially no activity (Fig. 4D). Thus, out of the three proteins with mutations in the helicase domain, only ts1 (S308N) had no defect in GTPase activity (Fig. 4B). The same results were obtained when ATP was used as the substrate (data not shown).
FIG. 4.
GTPase activity at two temperatures. [γ-32P]GTP substrate and nsP2 derivatives were incubated for 30 min at 24°C or 35°C. Aliquots from each reaction mixture were taken at the indicated time points, and the released Pi was separated from the uncleaved substrate by TLC. The reaction products were visualized by phosphorimaging and quantified by densitometry. (A) Typical time course assay performed at 24°C with wild-type (wt) nsP2. The first lane on the left represents a negative control without added enzyme. (B) The same reaction performed at 24°C with ts9 and ts1. Complete cleavage is reached by the addition of an equivalent amount of wild-type nsP2 (lane marked +wt in the ts9 panel). (C and D) Results of Pi release time course experiments are plotted for all of the nsP2 derivatives at the two temperatures, as indicated. The proteins indicated in the inset in panel D were incubated under the assay conditions at 35°C for the times indicated and were analyzed by Western blotting with anti-nsP2 antibodies.
In the RTPase assay, we used a 64-nucleotide-long RNA with 5′ γ-32P-labeled G as described in Materials and Methods. At 24°C, nsP2 proteins with mutations of ts1, ts4, ts6, and ts11 had activity profiles almost identical to that of wild-type nsP2. Again, ts9 showed no detectable activity at 24°C while the ts13a mutant protein had significantly lower activity (Fig. 5A and C). At 35°C, the maximum release of Pi by wild-type nsP2 was about 80% of that detected during incubation at 24°C. ts1, ts4, ts6, and ts11 had close to the same activity as the wild type, whereas ts13a showed very low activity and ts9 was completely inactive (Fig. 5B). The low GTPase and RTPase activities of nsP2 proteins carrying the ts9 and ts13a mutations cannot be due to nonspecific inactivation of these purified proteins, since the same preparation of ts9 showed wild-type-like activity when used for the in vitro protease assays; ts13a was also more active as a protease than NTPase and RTPase (Fig. 3). The mutant proteins were stable under the NTPase assay conditions, even in extended incubations (Fig. 4D, inset), showing that inactivity is not caused by proteolytic degradation.
FIG. 5.
RTPase activity at two temperatures. (A and B) γ-32P-labeled RNA substrate and nsP2 derivatives were incubated for 30 min at 24°C or 35°C. Aliquots from each reaction mixture were taken at the indicated time points, and the released Pi was separated from the uncleaved substrate by TLC. The time course experiments are plotted for all of the nsP2 derivatives. (C) Representative reactions at the 20-min time point as catalyzed by wild-type (wt) nsP2, ts9, and ts13a at 24°C. A negative control without added enzyme is shown on the right. (D) Competition assay. The indicated amount of GTP (closed symbols) or Pi (open symbols) was present in the RTPase reaction mixture, which was performed at 24°C with wild-type nsP2.
NTPase and RTPase share the same active site in nsP2.
Interestingly, the activity profiles of GTPase and RTPase were very similar for the wild type and all of the nsP2 mutants. The same active site is used for both RTPase and NTPase reactions in some viral replicase proteins (4, 19). To investigate this possibility, we carried out the triphosphatase reaction in the presence of unlabeled GTP (Fig. 5D). To overcome any chelating effect of the GTP, the concentration of Mg2+ was raised from 1 to 2 mM. Increasing the concentration of GTP resulted in a progressive inhibition of the triphosphatase reaction, such that at 1 mM GTP the triphosphatase reaction was virtually undetectable. This effect was not due to the product inhibition of phosphate released from the GTP, as 1 mM phosphate had no significant inhibitory effect (Fig. 5D). The presence of 25 μM [γ-S]GTP, a nonhydrolyzable analog of GTP in the reaction mixture, completely inhibited the triphosphatase reaction (data not shown). Thus, the results of these inhibition experiments further support the hypothesis that the same reaction center is also involved in catalyzing the GTPase and triphosphatase reactions in the alphavirus nsP2 protein. The GNS mutation also inactivates both NTPase and RTPase (36, 47).
NTPase activity of nsP2 derived from infected cells.
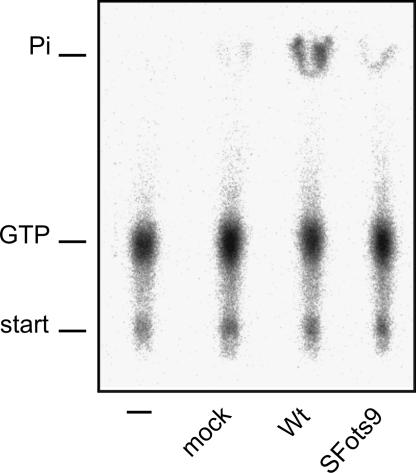
To study further the unexpected phenotype of ts9, which was almost completely inactive as an NTPase and RTPase when assayed in vitro, we wanted to purify nsP2 from infected cells. This was achieved by immunoprecipitation with specific polyclonal antibodies under nondenaturing conditions, as detailed in Materials and Methods. The postnuclear supernatant from cells infected with wild-type SFV and SFots9 at 28°C was first fractionated to obtain crude cytoplasmic membranes, sedimenting at 15,000 × g. This P15 fraction contains essentially all SFV-specific replication complexes as measured by polymerase activity (34). nsP2 was immunoprecipitated from the P15 fraction and used in the NTPase assay while still bound to Sepharose beads. Very little GTPase activity was detected from noninfected cells, in contrast to the material immunoprecipitated from cells infected with the wild-type virus (Fig. 6). To be sure that the immune serum does not inhibit the GTPase activity, we subjected the purified recombinant nsP2 protein to a similar immunoprecipitation. Compared with the unprecipitated starting material, virtually all of the GTPase activity was recovered in the precipitate. In samples derived from cells infected with SFots9 and kept at the permissive temperature, at which the virus replicates as well as the wild type, the activity was slightly above the background, representing ∼5% of the wild-type nsP2 activity (Fig. 6). SDS-PAGE analysis of the immunoprecipitates showed that roughly equal amounts of the wild-type and ts9 proteins were present in the reaction mixtures (not shown). Thus, the ts9 mutant nsP2 protein from infected cells also appears to be highly defective already at the permissive temperature.
FIG. 6.
GTPase activity of nsP2 derived from infected cells. GTPase activity was analyzed as for Fig. 4 by incubating reaction mixtures at 24°C for 60 min in the presence of 200 μM GTP. The nsP2 proteins assayed were obtained by immunoprecipitation under nondenaturing conditions (see Materials and Methods) from cells infected with wild-type (Wt) SFV or SFots9 at 28°C, as indicated at the bottom. Noninfected cells (mock sample) were analyzed in the same fashion. The first lane on the left contained no added protein in the reaction mixture.
Effects of nsP2 mutations on the processing of nonstructural polyprotein in vivo.
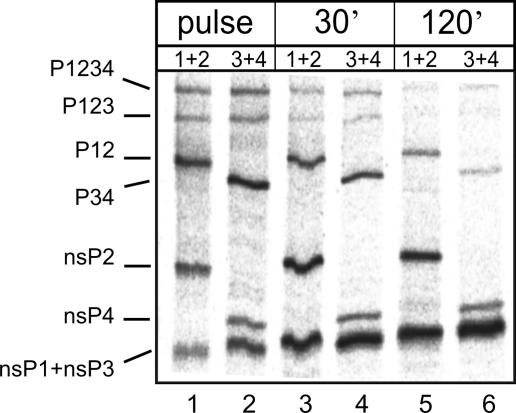
To study the processing of the nonstructural polyprotein, infection of BHK cells was continued at the permissive temperature of 28°C for 5 h, whereafter the cultures were shifted to the restrictive temperature of 39°C. The cells were labeled, after 30 min of equilibration at 39°C, with [35S]methionine for 30 min, followed by chases of 30 and 120 min in the presence of excess unlabeled methionine. Cultures infected and labeled in the same way but maintained at 28°C served as controls. After cell collection, the denatured lysates were subjected to immunoprecipitation by monospecific antisera against the individual nsPs. To resolve nsP1and nsP3, which migrate together on SDS-PAGE, we used combinations of nsP1 plus nsP2 and nsP3 plus nsP4 antisera for the immunoprecipitation. After SDS-PAGE, the gels were exposed to phosphorimaging plates. Quantitative immunoprecipitation allowed the estimation of the molar ratios of all of the nsPs and their precursors. In cells infected with the wild-type virus, the largest precursors, P1234 and P123, were detectable with both antibody combinations after the 30-min pulse. The two smaller precursors, P12 and P34, and the four mature nsPs were identifiable by the two antibody combinations (Fig. 7). After 30- and 120-min chase periods, the precursors had almost disappeared and most of the radioactivity was in the mature proteins (Fig. 7, lanes 3 to 6). Notably, there was less nsP4 than the other three nsPs.
FIG. 7.
Cleavage of nonstructural polyprotein in infected cells. BHK cells were infected with wild-type SFV. After 5 h at 28°C, the culture was shifted up to 39°C. After 30 min of equilibration, the cultures were pulse-labeled with [35S]methionine for 30 min, followed by chase periods of 30 and 120 min. The cells were collected at these time points as indicated at the top, and lysates were used for immunoprecipitation with combinations of monospecific antisera against each of the nsPs. The combinations of antibodies against nsP1 plus nsP2 and nsP3 plus nsP4 (as indicated at the top) were used to distinguish between nsP1 and nsP3, which migrate in the same position on gels. Following SDS-PAGE, the gels were dried and the radioactive bands were visualized by phosphorimaging. The positions of the polyprotein precursors and the mature nsPs are marked on the left.
The situation was different with the nonstructural proteins in the mutant-virus-infected cells, where the processing of the precursor proteins was much slower at 39°C. To analyze the processing more closely, we quantified all of the bands from phosphorimaging data and calculated their relative intensities as percentages of the total intensity of all of the proteins (P1234, P123, P12, P34, and nsP1 to nsP4). The distributions of the precursor proteins are shown in Table 1. For the wild type, the results at both 39°C and 28°C are presented. After the 30-min pulse at 39°C, close to 60% of the radioactivity was in the precursor proteins, mostly in P12 and P34, while after a 2-h chase only traces of the precursors remained.
TABLE 1.
Distribution of wild-type and mutant polyproteins during the long pulse-chase experiment
| Protein(s) and treatment | % of total nonstructural proteins
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild type at:
|
SFons1 at 39°C | SFots4 at 39°C | SFots6 at 39°C | SFots9 at 39°C | SFots11 at 39°C | SFons13a at 39°C | ||
| 28°C | 39°C | |||||||
| Precursors | ||||||||
| P1234 | ||||||||
| Pulse | 3.4 | 6.8 | 7.6 | 28.2 | 18 | 9.5 | 17.4 | 16.1 |
| 30-min chase | 0.7 | 2.8 | 2.2 | 4.7 | 7 | 0.8 | 6.1 | 7.6 |
| 120-min chase | 0.9 | 1.4 | 0.4 | 1.7 | 0.7 | 0.3 | 0.5 | 2 |
| P123 | ||||||||
| Pulse | 7.6 | 6.4 | 6.1 | 18.9 | 18.4 | 28.3 | 17.4 | 18.6 |
| 30-min chase | 3 | 1.9 | 1.7 | 15.1 | 6.2 | 4.8 | 6.1 | 7.6 |
| 120-min chase | 1.1 | 1.3 | 0.4 | 3.3 | 0.9 | 0.8 | 1.4 | 2.4 |
| P12 | ||||||||
| Pulse | 29.7 | 22.4 | 20.2 | 15.5 | 19 | 17.2 | 18.2 | 16.1 |
| 30-min chase | 12 | 8.9 | 7.9 | 20.4 | 16.9 | 5.2 | 14.6 | 16 |
| 120-min chase | 1.1 | 5 | 3.5 | 8.9 | 7.3 | 1 | 4.8 | 7.5 |
| P34 | ||||||||
| Pulse | 16.9 | 23.5 | 22.1 | 28.6 | 18.4 | 10.5 | 17.4 | 15 |
| 30-min chase | 3.4 | 10.4 | 9.8 | 26.4 | 9 | 1.6 | 6.7 | 6.4 |
| 120-min chase | 1.5 | 1 | 0.9 | 29.2 | 0.9 | 0.6 | 1.2 | 1.9 |
| Total precursors | ||||||||
| Pulse | 57.6 | 59.1 | 56 | 91.2 | 73.8 | 65.5 | 70.4 | 65.8 |
| 30-min chase | 19.1 | 24 | 21.6 | 66.6 | 39.1 | 12.4 | 27.4 | 37.6 |
| 120-min chase | 4.6 | 8.7 | 5.2 | 43.1 | 9.8 | 2.7 | 7.9 | 13.8 |
SFots4 exhibited the most severe general protease defect. After the pulse, about 90% of the total radioactivity was in the precursors and close to 50% was in the two largest proteins, P1234 and P123, which were slowly processed during the chase. The amount of radioactivity, close to 30%, in the P34 band was almost the same throughout the chase (Table 1). A similar general but somewhat milder protease defect was found in SFots6-, SFots11-, and SFons13a-infected cells at 39°C, in accordance with the results of the in vitro protease assays (Fig. 3). The processing of the precursors in SFons1-infected cells was similar to that of the wild type. With all of mutants, the processing of the nsPs at the permissive temperature was like that in wild-type-infected cells (data not shown).
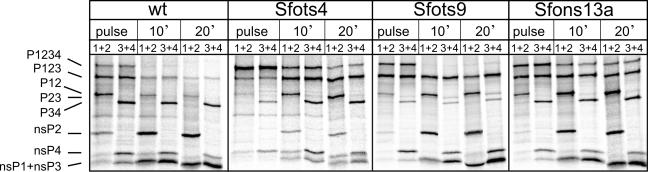
An interesting transient accumulation of only P123 was reproducibly observed in SFots9-infected cells at 39°C (Table 1). To study this specific defect more closely, we performed experiments on a shorter time scale with selected mutants. The generally defective SFots4 mutant and the other helicase domain mutant, SFons13a, were included in the experiment together with the wild-type control. The cells were labeled at the restrictive temperature for 10 min and chased in the presence of cycloheximide for 10 and 20 min before analysis. After the pulse, most of the radioactivity was in the precursor proteins (Fig. 8). After a 10-min chase, the largest precursor, P1234, had disappeared in the wild type and SFots9 but was still present in SFots4 and SFons13a. In SFots9, considerable amounts of P123 and P12 were present throughout the chase whereas the amount of P34 was clearly less compared with that in the wild-type-infected cells (Fig. 8). Thus, it appeared that the release of nsP4 from P1234 and P34 was efficient in SFots9-infected cells but the slower processing of P123 and P12 suggested a specific alteration of the protease activity of SFots9 nsP2. The precursor P23, which was extremely weak in the other mutants or the wild type, also uniquely appeared in SFots9 (Fig. 8).
FIG. 8.
Cleavage of nonstructural polyprotein during a short time course. In an experiment similar to that in Fig. 7, cells were pulsed with [35S]methionine for 10 min, followed by 10- and 20-min chase periods (as indicated at the top). The migration positions of the polyprotein precursors and the mature nsPs are marked on the left. The virus used is indicated at the top, and antibody combinations are marked as 1+2 (nsP1 plus nsP2) or 3+4 (nsP3 plus nsP4). The P23 precursor is the faint band (migrating just below P12) which appears in all lanes for SFots9, as P23 is recognized by both antibody combinations. wt, wild type.
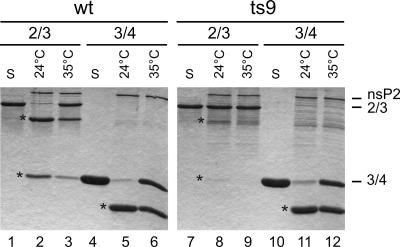
To further investigate the ts9 protease, we wanted to study it in vitro with substrates other than Trx34. As the Trx23 substrate is poorly cleaved by purified nsP2 (48), we used a longer recombinant fragment, Pro-X, representing the 2/3 site and consisting of the C-terminal 341 aa of nsP2 and 170 aa from the N terminus of nsP3. The recombinant purified nsP2 proteins of the wild type and ts9 were analyzed for the ability to cleave the purified substrates Trx34 and Pro-X (Fig. 9). As shown already in Fig. 3, ts9 nsP2 behaved like the wild-type protein in the cleavage of the Trx34 substrate at 35°C, cleaving about 50% of the substrate (Fig. 9, lanes 6 and 12). However, very little or no cleavage of the 2/3 site of Pro-X by ts9 was seen at both 24°C and 35°C (lanes 8 and 9) whereas wild-type nsP2 cleaved the same site at both temperatures (Fig. 9, lanes 2 and 3). We also wanted to study the processing of 1/2 site substrate Trx12, but it was cleaved very poorly in trans (49) already by the wild type at 24°C, and so it was not possible to evaluate the differences between the wild type and ts9. Thus, the purified ts9 nsP2 protein has a specific protease defect in vitro affecting the 2/3 site (and possibly also the 1/2 site). This defect also appeared in vivo, since P123 and also some P23 could be identified in the short pulse-chase experiment (Fig. 8).
FIG. 9.
Comparison of 2/3 and 3/4 site cleavages. The wild-type (wt) and ts9 mutant recombinant purified nsP2 proteins were incubated with substrates representing the 2/3 site (Pro-X) and the 3/4 site (Trx34; Fig. 2) at 24°C or 35°C for 1 h, as indicated at the top. The cleavage products were analyzed by SDS-PAGE on 15% gels and stained with Coomassie blue. Lanes S contain only the substrate incubated in the absence of the proteases. The positions of the protease nsP2 and the substrates are indicated on the right, and the cleavage products are marked with asterisks placed on the left side of the corresponding bands. For the 2/3 substrate, both cleavage products are visible, but for the 3/4 substrate, the smaller product has migrated out of the gel.
Effects of nsP2 mutations on subgenomic RNA synthesis.
Subgenomic RNA synthesis was first studied in a type of temperature shift-up experiment similar to that used for the protease. After a shift to 39°C at 6 h p.i. (controls were maintained at 28°C), protein synthesis was shut off by addition of cycloheximide and DNA-dependent RNA synthesis was shut off by addition of actinomycin D. The cells were labeled with [3H]uridine from 7 to 8 h p.i., with the results shown in Table 2. The labeled RNAs were extracted and separated by electrophoresis and quantified by densitometry and/or scintillation counting of excised bands. As reported previously, the ratio of genomic 42S RNA to subgenomic 26S RNA was temperature dependent. At 28°C, the synthesis of 26S RNA is predominant, whereas at higher temperatures more 42S RNA is synthesized at the expense of 26S RNA (30, 40, 46). The changes in the molar ratios of 42S RNA to 26S RNA of the different nsP2 mutants reveal several mutants with reduced 26S RNA synthesis at the restrictive temperature (SFons1, SFots4, SFots6, SFots9, and SFots11; Table 2). SFons13a had a mild effect on 26S RNA synthesis at both temperatures. The SFons1 defect could be regarded as constitutive, already manifesting itself at 28°C (Table 2). SFots4 and SFots11 had a typical wild-type pattern at 28°C with a significant reduction of 26S RNA synthesis at 39°C. SFots6 and Fots9 already had reduced subgenomic RNA synthesis at 28°C, which was accentuated significantly after a shift to 39°C. In some cases, the cells were kept for 2 h at 39°C (6 to 8 h p.i.) and then transferred back to 28°C and exposed to [3H]uridine from 9 to 10 h p.i. to study the reversibility of the subgenomic RNA synthesis defect. There was a substantial reversion of 26S RNA synthesis in the wild type and SFons13a, but SFots4 and SFots9 also displayed reversion (Table 2).
TABLE 2.
Temperature dependence of 42S/26S RNA molar ratio
| Strain | 42S/26S RNA molar ratio
|
||
|---|---|---|---|
| 28°C | 39°C | Back to 28°C | |
| Wild type | 0.22 | 0.48 | 0.28 |
| Sfons1 | 0.59 | 1.45 | NDa |
| Sfots4 | 0.25 | 1.80 | 0.37 |
| Sfots6 | 0.43 | 1.49 | ND |
| Sfots9 | 0.40 | 1.55 | 0.70 |
| Sfots11 | 0.27 | 1.10 | ND |
| Sfons13a | 0.32 | 0.79 | 0.25 |
ND, not determined.
DISCUSSION
We have previously characterized the properties of SFV replicase proteins with wild-type nsPs. Here we have taken advantage of ts mutants with defects in viral RNA synthesis giving them an RNA-negative phenotype. Sequencing of several previously isolated SFV RNA-negative mutants revealed the causative amino acid replacements in the replicase proteins (30). Here we analyzed the known activities of six mutants whose lesions are localized in the nsP2 protein. Three of them (ts1, ts13a, and ts9) were localized in the helicase region, and the other three (ts11, ts6, and ts4) were localized within the carboxy-terminal protease domain (Fig. 1A). To study the effects of nsP2 mutations, we produced the mutated proteins in E. coli and used the purified proteins for protease, NTPase, and RTPase assays. Polyprotein processing and RNA synthesis were studied in infected cells with recombinant viruses containing the mutated nsP2 protein in the wild-type virus background.
Protease defects.
The in vitro protease assay using purified nsP2 proteins and purified thioredoxin fusion protein Trx34 containing the polyprotein cleavage site between nsP3 and nsP4 showed that all of the protease domain mutants caused a ts protease defect. Similar results have been obtained for the isolated protease domain, Pro39, of these mutants (13). An unexpected result was obtained with ts13a nsP2, where the mutation in the helicase domain caused a severe protease defect at 35°C (Fig. 3). To study the processing of the nonstructural polyprotein P1234 in infected cells, the recombinant viruses were allowed to start replication at 28°C and the cultures were shifted to 39°C and studied at this restrictive temperature. The same four virus mutants (SFots4, SFots6, SFots11, and SFons13a) were also generally defective in nonstructural polyprotein processing during infection (Table 1). Both in vivo and in vitro, the ts4 mutation caused the strongest phenotype and all of the sites seemed to be affected by this mutation. The recently solved structure of the Venezuelan equine encephalitis virus protease shows that the alphavirus nonstructural protease consists of two subdomains, an N-terminal core protease domain that contains the catalytic residues and a C-terminal domain resembling methyltransferases that also participates in substrate binding (37). Of the mutations studied here, the ts11 lesion is localized in the core protease domain and ts4 and ts6 are in the methyltransferase-like domain, further supporting the view that the C-terminal domain is also essential for protease activity.
An intriguing observation was made in the labeling of cells infected with recombinant virus containing the ts9 mutation. SFots9 showed a unique pattern of polyprotein processing with accumulation of P123 but not P1234, which was confirmed by a shorter labeling period. In vitro, there was no defect in the cleavage of the Trx34 substrate, but ts9 nsP2 was inactive in cleaving a model 2/3 site substrate (Fig. 9). Thus, we concluded that this mutation in the helicase region affects the protease activity in a site-specific manner. The accumulation of some P23 only in ts9-infected cells supports the in vitro protease results. It has been established that the 2/3 site processing has special requirements compared to the other sites; the correct amino terminus of nsP2 is need for 2/3 site cleavage (48) but has no influence on 3/4 site processing. A specific effect of the ts9 mutation on 2/3 site processing may therefore take place as a result of disturbing the amino terminus of nsP2. However, the ts13a protease defect that does affect the 3/4 site suggests yet another type of contact between the helicase and protease domains. Even though there are contacts between the domains, which are influenced in specific ways by different mutations, the NTPase-helicase activity is not directly involved in protease activity, as the helicase active-site mutant GNS had a wild-type protease phenotype in vitro. SIN ts mutants with lesions in the helicase domain do not have protease defects (8). It is possible that a specific subdomain of the helicase is in contact with the protease domain; SFV ts13a and ts9 are located relatively close to each other in the linear sequence, and all of the SIN ts mutations are located either before or after this area (Fig. 1A).
Regulation of the subgenomic promoter.
All three of the protease domain mutant viruses produced less 26S RNA at 39°C than the wild-type virus (Table 2). The combination of polyprotein processing and 26S RNA defects in the alphavirus protease domain mutants has been noted previously (8, 22, 46). It can be hypothesized that the protease and 26S RNA activation are destroyed in the mutants simultaneously by a conformational alteration that affects both the protease (active site or substrate recognition) and 26S RNA synthesis (binding to the RNA or to another protein involved in subgenomic promoter activation). These two aspects can be affected to different extents, depending on the specific mutation. For 26S RNA synthesis, this alteration is at least partially reversible, as complexes inactivated at the restrictive temperature can reactivate 26S RNA synthesis when returned to the permissive temperature (Table 2) (40, 46).
SFons1, SFots9, and, to a lesser extent, SFons13a also had defects in 26S RNA synthesis, and thus the helicase domain is deeply involved in this process. This is in contrast to the previous SIN studies, which did not associate 26S RNA regulation with any of the helicase domain mutants (8, 17, 46). ts9 and ts13a affected the protease in different ways, but ts1 had a wild-type-like protease activity, although it was still strongly defective in subgenomic RNA synthesis. This suggests that there are several mechanisms by which the different mutants cause their 26S RNA phenotypes. ts1 may form a unique class in this respect. The ts1 mutation is located in helicase motif IV, which is involved in RNA binding (6), and therefore ts1 could influence 26S RNA synthesis through an effect on RNA binding or on helicase activity.
It is not known whether nsP2 carries out its functions as a monomer or as an oligomer. It has recently been realized that several helicases can be active as monomeric proteins (28, 31). Yet most helicases form dimers or hexamers, and even helicases that do not form structural dimers can be stimulated functionally by other helicase molecules (33). In the alphavirus-like plant viruses, there is both genetic and biochemical evidence for protein-protein interactions of the nsP2-like helicase domains (32), and in one case a hexameric helicase-like assembly has been visualized by electron microscopy (16). One interpretation of replication complex structure suggests that it contains multiple helicase-like proteins as structural components (41). Since almost all SFV and SIN ts mutants with changes located in the protease domain have defects in 26S RNA synthesis, we propose that the protease domain is the direct mediator of nsP2 participation in 26S promoter activation. If the basic form of nsP2 in the replication complex is an oligomer due to helicase domain interactions, some helicase domain mutations may influence 26S RNA synthesis by affecting the oligomerization of nsP2.
Roles of NTPase, RTPase, and RNA helicase.
We propose here that the NTPase active site is also the RTPase active site for alphaviruses, since NTPs can compete with the RNA substrate and ts mutations similarly affect both activities. The confluence of these two activities has been recently suggested for the flavivirus helicase-NTPase-RTPase NS3 and the coronavirus helicase nsp13, also based on competition and mutagenesis experiments (4, 5, 19). Therefore, all of the major groups of animal positive-sense RNA viruses that make capped RNAs (alphaviruses, flaviviruses, and coronaviruses) seem to utilize the NTPase active site that fuels the helicase also to catalyze the RTPase reaction. The proteins encoded by these viruses share the conserved Walker NTPase motifs and other helicase motifs, but they are not closely related (15, 24). They belong to different helicase superfamilies (nsP2 and nsp13 are in superfamily I, and NS3 is in superfamily II) and have different polarities of helicase action (nsp13 unwinds RNA in the 5′-to-3′ direction, and NS3 does so in the 3′-to-5′ direction) (42). This double action of the helicase domain represents an efficient use of virus coding capacity but poses interesting questions regarding the organization of the replication apparatus. Since the helicase may be required continuously in assisting the polymerase but the triphosphatase needs to act only once at the 5′ end of each positive-sense RNA, there may be different molecules of these proteins dedicated to each of the two tasks.
Based on the GNS mutation in helicase motif I (Walker A motif), the NTPase-RTPase activities appear to be essential for alphavirus replication (35). The lack of these activities could therefore account for the ts replication of ts9 and ts13a. However, recombinant nsP2 containing the ts9 mutation had very little NTPase and RTPase activity, even at the low temperature. This does not seem to be an artifact associated with the production of ts9 nsP2 in bacterial cells, as nsP2 purified from ts9-infected cells by immunoprecipitation also demonstrated low activity. These cells had been kept at the permissive temperature, at which ts9 replicates as well as the wild-type virus. While it can be speculated that low levels of NTPase (helicase) and RTPase activities could be sufficient for full replication, this is not the only possible explanation. Alternatively, we propose that the ts property of ts9 could manifest itself only in the early stages of replication, which involve the formation of replication complexes and minus strand synthesis. Temperature shift-up experiments analyzing virus yield support the concept of an early defect for ts9. If the cells are maintained for 6 h at the permissive temperature before a shift up, this is sufficient to give essentially normal virus yields 12 h later (data not shown). At the early stage of replication, nsP2 is present in the polyprotein P123. It is conceivable that the ts9 polyprotein would be fully active in all of its functions (including NTPase-helicase) at the permissive temperature but inactive in some of the essential nsP2 domain-mediated functions at the restrictive temperature. In contrast, after ts9 nsP2 is cleaved from the polyprotein, it would always have low NTPase activity, even at the permissive temperature (but it would still be capable of carrying out its functions in positive-strand synthesis). In this hypothesis, the ts9 polyprotein would be the actual ts component. Further studies of the ts9 mutant are likely to be informative with regard to the roles of the NTPase-helicase and RTPase in negative- versus positive-strand RNA synthesis. The ts9 mutation is located in helicase motif V, which is involved in mediating a complex network of interactions among the NTP, the RNA substrate, and different helicase subdomains (6). Testing of the polyprotein hypothesis necessitates comparison of the wild-type and ts9 polyproteins, which we have not attempted, as expression of the 1,818-aa P123 polyprotein in sufficient quantities is rather challenging (39). The coordination of different enzymatic activities within nsP2 is an important area for future study. In the replication complex, it is likely that the other nsPs and possibly host proteins further regulate, stabilize, and modify the behavior of nsP2. Such influences have been recently described, for instance, for the flavivirus helicase-RTPase NS3 (53).
Acknowledgments
We thank Andrey Golubtsov for advice and Aleksei Lulla for the gift of reagents and for sharing unpublished data.
This work was supported by the European Union 5th framework program project SFVECTORS, as well as by Academy of Finland grant 211121 and by the Sigrid Juselius Foundation.
Footnotes
Published ahead of print on 3 January 2007.
REFERENCES
- 1.Ahola, T., and L. Kääriäinen. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 92:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola, T., A. Lampio, P. Auvinen, and L. Kääriäinen. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett, A. J., and N. D. Rawlings. 2001. Evolutionary lines of cysteine peptidases. Biol. Chem. 382:727-733. [DOI] [PubMed] [Google Scholar]
- 4.Bartelma, G., and R. Padmanabhan. 2002. Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology 299:122-132. [DOI] [PubMed] [Google Scholar]
- 5.Benarroch, D., B. Selisko, G. A. Locatelli, G. Maga, J. L. Romette, and B. Canard. 2004. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology 328:208-218. [DOI] [PubMed] [Google Scholar]
- 6.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 7.Dé, I., C. Fata-Hartley, S. G. Sawicki, and D. L. Sawicki. 2003. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J. Virol. 77:13106-13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dé, I., S. G. Sawicki, and D. L. Sawicki. 1996. Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J. Virol. 70:2706-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding, M. X., and M. J. Schlesinger. 1989. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology 171:280-284. [DOI] [PubMed] [Google Scholar]
- 10.Fazakerley, J. K., A. Boyd, M. L. Mikkola, and L. Kääriäinen. 2002. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J. Virol. 76:392-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolov, I., E. Agapov, T. A. Hoffman, B. M. Prágai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golubtsov, A., L. Kääriäinen, and J. Caldentey. 2006. Characterization of the cysteine protease domain of Semliki Forest virus replicase protein nsP2 by in vitro mutagenesis. FEBS Lett. 580:1502-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez de Cedrón, M., N. Ehsani, M. L. Mikkola, J. A. García, and L. Kääriäinen. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 448:19-22. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 16.Goregaoker, S. P., and J. N. Culver. 2003. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J. Virol. 77:3549-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn, Y. S., E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 63:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kääriäinen, L., and T. Ahola. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 71:187-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keränen, S., and L. Kääriäinen. 1974. Isolation and basic characterization of temperature-sensitive mutants from Semliki Forest virus. Acta Pathol. Microbiol. Scand. Sect. B 82:810-820. [DOI] [PubMed] [Google Scholar]
- 22.Keränen, S., and L. Kääriäinen. 1979. Functional defects of RNA-negative temperature-sensitive mutants of Sindbis and Semliki Forest viruses. J. Virol. 32:19-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, K. H., T. Rumenapf, E. G. Strauss, and J. H. Strauss. 2004. Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts. Virology 323:153-163. [DOI] [PubMed] [Google Scholar]
- 24.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 25.Kujala, P., A. Ikäheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kääriäinen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lampio, A., I. Kilpeläinen, S. Pesonen, K. Karhi, P. Auvinen, P. Somerharju, and L. Kääriäinen. 2000. Membrane binding mechanism of an RNA virus-capping enzyme. J. Biol. Chem. 275:37853-37859. [DOI] [PubMed] [Google Scholar]
- 27.Lemm, J. A., T. Rümenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin, M. K., Y.-H. Wang, and S. S. Patel. 2004. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 279:26005-26012. [DOI] [PubMed] [Google Scholar]
- 29.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lulla, V., A. Merits, P. Sarin, L. Kääriäinen, S. Keränen, and T. Ahola. 2006. Identification of mutations causing temperature-sensitive defects in Semliki Forest virus RNA synthesis. J. Virol. 80:3108-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanduri, B., A. K. Byrd, R. L. Eoff, A. J. Tackett, and K. D. Raney. 2002. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc. Natl. Acad. Sci. USA 99:14722-14727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Reilly, E. K., Z. Wang, R. French, and C. C. Kao. 1998. Interactions between the structural domains of the RNA replication proteins of plant-infecting RNA viruses. J. Virol. 72:7160-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel, S. S., and I. Donmez. 2006. Mechanisms of helicases. J. Biol. Chem. 281:18265-18268. [DOI] [PubMed] [Google Scholar]
- 34.Ranki, M., and L. Kääriäinen. 1979. Solubilized RNA replication complex from Semliki Forest virus-infected cells. Virology 98:298-307. [DOI] [PubMed] [Google Scholar]
- 35.Rikkonen, M. 1996. Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2. Virology 218:352-361. [DOI] [PubMed] [Google Scholar]
- 36.Rikkonen, M., J. Peränen, and L. Kääriäinen. 1994. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 68:5804-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russo, A. T., M. A. White, and S. J. Watowich. 2006. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure 14:1449-1458. [DOI] [PubMed] [Google Scholar]
- 38.Salonen, A., T. Ahola, and L. Kääriäinen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salonen, A., L. Vasiljeva, A. Merits, J. Magden, E. Jokitalo, and L. Kääriäinen. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J. Virol. 77:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawicki, D. L., L. Kääriäinen, C. Lambek, and P. J. Gomatos. 1978. Mechanism for control of synthesis of Semliki Forest virus 26S and 42S RNA. J. Virol. 25:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 42.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauss, E. G., R. J. de Groot, R. Levinson, and J. H. Strauss. 1992. Identification of the active site residues in the nsP2 proteinase of Sindbis virus. Virology 191:932-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. (Erratum, 58:806.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suopanki, J., D. L. Sawicki, S. G. Sawicki, and L. Kääriäinen. 1998. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J. Gen. Virol. 79:309-319. [DOI] [PubMed] [Google Scholar]
- 47.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kääriäinen. 2000. Identification of a novel function of the alphavirus capping apparatus: RNA 5′-triphosphatase activity of nsP2. J. Biol. Chem. 275:17281-17287. [DOI] [PubMed] [Google Scholar]
- 48.Vasiljeva, L., A. Merits, A. Golubtsov, V. Sizemskaja, L. Kääriäinen, and T. Ahola. 2003. Regulation of the sequential processing of Semliki Forest virus nonstructural polyprotein. J. Biol. Chem. 278:41636-41645. [DOI] [PubMed] [Google Scholar]
- 49.Vasiljeva, L., L. Valmu, L. Kääriäinen, and A. Merits. 2001. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J. Biol. Chem. 276:30786-30793. [DOI] [PubMed] [Google Scholar]
- 50.Vihinen, H., T. Ahola, M. Tuittila, A. Merits, and L. Kääriäinen. 2001. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J. Biol. Chem. 276:5745-5752. [DOI] [PubMed] [Google Scholar]
- 51.Vihinen, H., and J. Saarinen. 2000. Phosphorylation site analysis of Semliki Forest virus nonstructural protein 3. J. Biol. Chem. 275:27775-27783. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Y.-F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 68:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yon, C., T. Teramoto, N. Mueller, J. Phelan, V. K. Ganesh, K. H. M. Murthy, and R. Padmanabhan. 2005. Modulation of the nucleoside triphosphatase-RNA helicase and 5′-RTPase activities of Dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J. Biol. Chem. 280:27412-27419. [DOI] [PubMed] [Google Scholar]