Abstract
Asymmetric subcellular distribution of RNA is used by many organisms to establish cell polarity, differences in cell fate, or to sequester protein activity. Accurate localization of RNA requires specific sequence and/or structural elements in the localized RNA, as well as proteins that recognize these elements and link the RNA to the appropriate molecular motors. Recent advances in biochemistry, molecular biology, and cell imaging have enabled the identification of many RNA localization elements, or “zipcodes,” from a variety of systems. This review focuses on the mechanisms by which various zipcodes direct RNA transport and on the known sequence/structural requirements for their recognition by transport complexes. Computational and experimental methods for predicting and identifying zipcodes are also discussed.
Keywords: RNA zipcode, cytoplasmic RNA transport, localized RNA
INTRODUCTION
Multiple mechanisms exist for ensuring that proteins are produced in the appropriate place and time within a cell or organism. Many of these regulatory processes act at the post-transcriptional level. Localization of mRNA is commonly employed to target protein products to specific regions within a cell. Some mRNAs are localized to appropriate subcellular structures at the same time they are being translated, and it is the emerging nascent peptide that directs the localization of both the ribosome and the transcript. For example, transcripts encoding secreted proteins are localized to the endoplasmic reticulum (ER) by recruitment of the nascent signal peptide (Nicchitta 2002). In Bacillus subtilis, the transmembrane domains of the glucose transporter IICB target the ribosome–mRNA–protein complex to the cell membrane (Kawamoto et al. 2005). In other cases, such as the localization of some transcripts to mitochondria in yeast, transport is driven by sequences in the RNA itself (Corral-Debrinski et al. 2000). Asymmetric subcellular distribution of RNA is critical for proper embryo development, for establishing differences in cell fate, and for neuronal function (Chartrand et al. 2001). Unlike the examples of RNA localization to the ER, asymmetric cytoplasmic RNA localization is generally a motor-driven process and, like the transport of RNAs to mitochondria, is commonly directed by cis-acting elements. The RNA is recognized by RNA-binding protein(s) and the resulting RNA–protein complex, or mRNP, is linked either directly or indirectly to molecular motors that transport the mRNP to its final destination (Oleynikov and Singer 1998). Following transport, the mRNA is anchored at its site of translation; additional mechanisms often exist to ensure that the mRNA is not prematurely translated during transport. Examples of this process occur in organisms from yeast to mammals (Tekotte and Davis 2002).
“Zipcodes” are cis-acting motifs that direct mRNAs for transport to appropriate locations within a cell or organism (Kislauskis and Singer 1992). Zipcodes range in length from a few nucleotides to over 1 kb; however, it is possible that some of the longer zipcodes have not been reduced to their minimally sufficient lengths, making it difficult to identify key RNA determinants of transport. It is believed that zipcodes serve as binding sites for proteins that form complexes with molecular motors, thereby linking the RNA to the cellular transport machinery (Mowry 1996). In yeast, the interactions between the RNA-binding species, protein linker, and motor have been defined (Bohl et al. 2000; Long et al. 2000; Takizawa and Vale 2000). In other systems, the involvement of motors in the transport process has been established (Brendza et al. 2000; Januschke et al. 2002; Betley et al. 2004; Yoon and Mowry 2004), but the linkage of RNA-binding proteins and accessory factors to motors remains uncharacterized. Often, translation inhibitors are associated with the transport mRNP complex to ensure that the mRNA is not prematurely translated en route to its final destination. Translational control has been reviewed elsewhere (e.g., Bashirullah et al. 1998; Johnstone and Lasko 2001; Kindler et al. 2005; Sossin and Desgroseillers 2006) and will not be discussed here. Additional RNA sequences may be required to promote the assembly of a localization-competent RNP (Czaplinski and Mattaj 2006).
An essential feature of zipcodes is that they can direct localization independently of the adjacent RNA sequence: fusing a zipcode to a reporter RNA results in a subcellular distribution of the reporter similar to that observed for the native mRNA. Although zipcodes are usually located in 3′ untranslated regions (UTRs) of transported messages (e.g., Mowry and Melton 1992; Kim-Ha et al. 1993; Gavis et al. 1996a; Zhou and King 1996b; Deshler et al. 1997; Macdonald and Kerr 1997; Chan et al. 1999), in some instances they can mediate localization when placed at the 5′ end or even in the middle of a reporter RNA (Cohen et al. 2005), although not all zipcodes are position independent (Kislauskis et al. 1994). Zipcodes in bud-localized yeast RNAs, in contrast to transported RNAs in metazoans, are generally found in coding regions (Shepard et al. 2003), but can function efficiently when located in 3′ UTRs of reporter transcripts (e.g., Chartrand et al. 1999; Gonzalez et al. 1999; Hachet and Ephrussi 2004).
IDENTIFICATION OF ZIPCODES
Identifying zipcodes has been problematic for several reasons. In many cases, the essential RNA-binding protein(s) of the transport mRNP is not known a priori, since transported RNAs are often associated with large, multiprotein complexes. Therefore, it is often not possible to screen for zipcodes based on protein-binding ability (e.g., to select for RNAs that bind an immobilized protein); once mapped, however, zipcode sequences have been successfully used to identify cognate RNA-binding proteins (e.g., Gautreau et al. 1997; Hoek et al. 1998; Cote et al. 1999; Bergsten et al. 2001). Second, many zipcodes are recognized by the transport machinery on the basis of both sequence and structure. For example, the bicoid localization element contains one helical region, in which only the secondary structure but not sequence is important, as well as a region in which base identities are essential for transport (Macdonald and Kerr 1998). Furthermore, reporter sequences used to test putative zipcodes for localization activity could induce the zipcode to adopt a different structure than it does in the native RNA, thus producing a false negative result. Improper folding of zipcodes out of context may be the reason that many have not been minimized beyond several hundred nucleotides in length. In fact, differences in reporter constructs have been invoked (Huang et al. 2003) to explain why some groups observed that the full-length 3′ UTR of αCamKII localized to dendrites (Mayford et al. 1996; Blichenberg et al. 2001) and another group found that it did not (Mori et al. 2000). Thus, efforts to detect zipcodes by motif-finding algorithms may be hampered both by the inherent degeneracy of the sequence and by the availability of minimally sufficient, validated zipcodes. Third, differences in criteria for determining “localized” RNAs can affect which sequences are identified as zipcodes. For example, Ferrandon et al. (1994) reported that the dimerization domain III of the bicoid 3′ UTR is sufficient for embryonic localization based on its ability to recruit Staufen and localize to astral microtubules (a localization pattern not observed for full-length bicoid). In contrast, when Snee et al. (2005) mapped the regions required to achieve a wild-type (WT) bicoid localization pattern at the anterior of the embryo, stem–loops IV–V were required in addition to domain III. Similarly, the discrepancies in localization elements identified in αCamKII have also been attributed to localization scoring criteria (Huang et al. 2003). Two groups counted RNAs as “dendritically localized” if they were at a distance of more than one soma diameter from the cell body (Mori et al. 2000; Blichenberg et al. 2001), whereas another group scored RNAs located along the lengths of dendrites (Huang et al. 2003). Fourth, many RNAs are localized by multistep processes, with each step being governed by distinct zipcodes and potentially coupled to the preceding and/or subsequent step (see below). Therefore, multiple elements are required together to effect localization, precluding the identification of minimal subelements.
The most common method for identifying zipcodes involves monitoring the subcellular distributions of fragments derived from the localized RNA, either on their own or fused to reporter transcripts. Akin to transcriptional promoter bashing, this brute force approach identifies sequences that are sufficient for transport. Alternately, deletion of regions within the native RNA that impair localization can be used to identify essential sequences. In cases where the RNA-binding protein is known, putative zipcode sequences can be isolated rapidly and efficiently on the basis of their binding ability (Jambhekar et al. 2005); however, as noted above, the RNA-binding species in many localization pathways are not known.
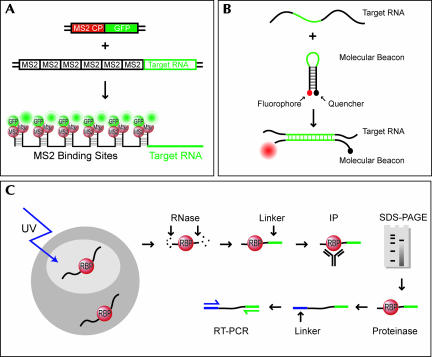
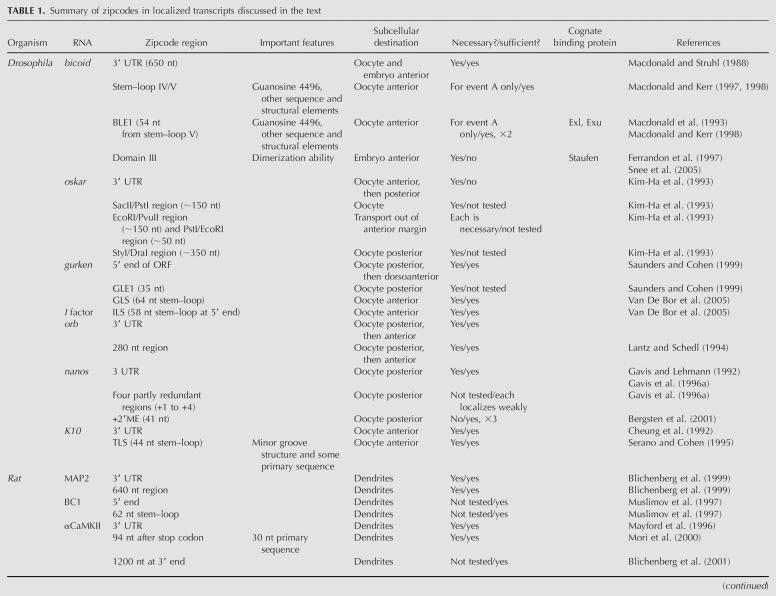
Many methods have been developed for monitoring RNA distribution in live or fixed cells. Early studies in Drosophila were conducted by detecting transcripts of interest in sections of cells or embryos (Macdonald and Struhl 1988). Traditionally, RNA distribution has been visualized in fixed cells by fluorescence in situ hybridization (FISH) to native or reporter RNAs. RNA transport and localization can be visualized in live cells by injecting fluorescently labeled RNA; while this technique works well in some oocytes and embryos, it is not feasible for smaller cells such as yeast and bacteria. Furthermore, because RNAs can be “marked” for cytoplasmic transport early on in the nucleus (Kruse et al. 2002; Kress et al. 2004), it is likely that RNAs injected in the cytoplasm may not recruit the full complement of proteins necessary for transport. And in some cases, the fluorescent tag on the RNA can compromise its structural integrity (Wilkie et al. 2001). Other techniques developed more recently allow visualization of fully processed mRNA in multiple cell types. A reporter RNA containing MS2 stem–loop aptamers can be tracked by coexpressing an MS2-coat protein–GFP fusion (Fig. 1A); this strategy is effective in both yeast and mammalian cells (Bertrand et al. 1998; Fusco et al. 2003), but the MS2 aptamers and/or bound MS2-coat protein may interfere with localization of some RNAs. Molecular beacons can also be used to visualize native RNA in live cells (Fig. 1B; Bratu et al. 2003). This technique involves injecting an oligonucleotide probe (“molecular beacon”) complementary to the native RNA. Self-complementary sequences at the 5′ and 3′ ends of the probe induce it to fold in a hairpin; a fluorophore is coupled to one end and a quencher at the other. In the folded state, the 5′ and 3′ ends are in close proximity and all fluorescence is quenched. Upon binding to the target RNA, the hairpin probe unfolds and the quencher is removed from the vicinity of the fluorophore, resulting in a fluorescent signal. A similar approach employs two molecular beacons that anneal to adjacent regions of the target RNA. One beacon is conjugated to a donor fluorophore and the other to an acceptor, resulting in a FRET signal upon specific binding of both beacons to their target (Santangelo et al. 2004). If the molecular beacons overlap with zipcodes, however, they could mask recognition by the transport machinery and preclude identification of bona fide zipcodes. In fact, masking of zipcodes by hybridization to exogenous oligonucleotides has been used to identify zipcodes in loss-of-function assays (Kislauskis et al. 1994). Alternately, binding of the transport machinery to localized transcripts may prevent hybridization of the RNA to the beacon probe, resulting in a loss of signal.
FIGURE 1.
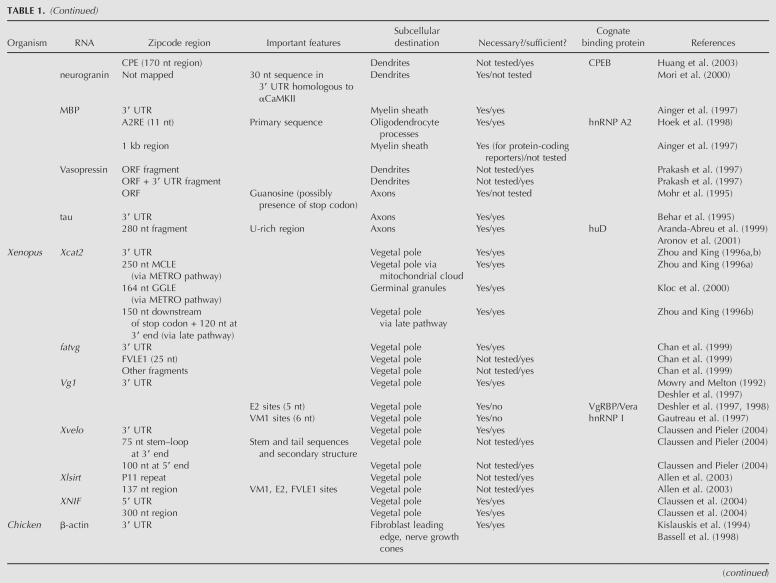
Schematics of zipcode identification strategies. (A) in vivo GFP-RNA tagging (after Bertrand et al. 1998). The RNA of interest is expressed fused to a reporter containing six tandem repeats of the MS2 stem–loop sequence. An MS2-coat protein–GFP fusion is coexpressed, resulting in a GFP-marked RNA. (B) Molecular beacons (after Bratu et al. 2003). A hairpin oligonucleotide containing sequences complementary to the target RNA is conjugated to a fluorophore at one end and a quencher at the other. In the hairpin conformation, the fluorophore is quenched. Upon injection of the molecular beacon into cells and subsequent hybridization to the target RNA, the quencher is removed from the proximity of the fluorophore and the RNA can be tracked based on a signal from the fluorophore. (C) CLIP (after Ule et al. 2005). RNA–protein complexes are cross-linked with UV, isolated, and treated with Rnase. Protein-bound RNA regions are protected from digestion. A linker is ligated to the 3′ end, and intact complexes are isolated by immunoprecipitation of the RNA-binding protein, followed by SDS-PAGE purification. The protein is then removed by digestion. A second linker is ligated to the 5′ end and the isolated RNA is amplified by reverse transcription and PCR.
The recently developed cross-linking and immunoprecipitation (CLIP) technique (Ule et al. 2003) also promises to be effective at isolating zipcode sequences (Fig. 1C). In this method, nucleic acids and proteins are first cross-linked, followed by cell lysis and RNase treatment of nucleic acids. The mRNP protein of interest is immunoprecipitated, and the bound RNA is isolated, cloned, and sequenced (Ule et al. 2005). This method has the advantage of isolating RNA–protein complexes under physiological conditions. Furthermore, because mRNP complexes are cross-linked prior to purification, the identity of the RNA-binding protein does not need to be known; in principle, any component of the mRNP can be used for immunoprecipitation. Therefore, this technique could be especially powerful for isolating zipcodes from systems in which the RNA-binding protein remains undefined; to date, however, no zipcodes have been identified by this method.
ZIPCODES ACT IN CONCERT TO DIRECT RNA LOCALIZATION
Localization of RNAs often occurs by a multistep process. A localization-competent RNP must first be assembled on the RNA, after which the complex is transported to its final destination. For a given RNA, transport can occur in multiple spatially or temporally distinct steps or can be affected by multiple redundant mechanisms. In many cases, the RNA is anchored at its final destination. Each of these steps may be directed by dedicated cis-acting signals in the transcript.
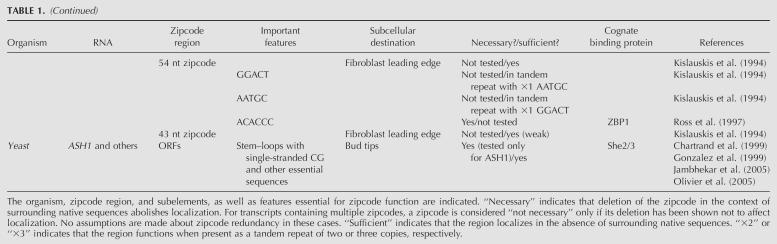
Some RNAs contain all information necessary for successful transport in a single zipcode. The non-protein-coding, dendritically localized BC1 RNA contains a 62 nucleotide (nt) zipcode at its 5′ end (Muslimov et al. 1997), which is unlikely to contain multiple, independently functioning subelements due to its short length. This zipcode may directly recruit the machinery responsible for transporting it in a single microtubule-dependent step (Cristofanilli et al. 2006), and the RNA may not have cis-acting sequences for transport by redundant mechanisms. Another dendritically localized transcript, the MAP2 mRNA, contains a 640 nt element in its 3′ UTR that is necessary and sufficient for transport (Blichenberg et al. 1999). Because this element is large and is predicted to contain multiple structural domains, it is possible that distinct subelements within the zipcode mediate individual steps of the localization process (see Table 1).
TABLE 1.
Summary of zipcodes in localized transcripts discussed in the text
Unlike BC1, many RNAs contain multiple sequence/structural motifs essential for proper localization. In oligodendrocytes, two distinct elements in the MBP RNA mediate its transport to and retention at the myelin compartment. An 11 nt element, A2RE, serves as a binding site for hnRNP A2 (Hoek et al. 1998) and is necessary and sufficient for transport (Munro et al. 1999). This element is found in other transported RNAs but not in isoforms of transcripts that are unlocalized (Ainger et al. 1997), suggesting that it might be a key determinant for targeting RNAs for transport. A second, more complex, element over 1 kb in length is essential for proper localization of a protein-coding reporter RNA following transport, and may be involved in RNA anchoring (Ainger et al. 1997).
Like MBP RNA, the Drosophila oskar (osk) and gurken (grk) mRNAs are localized in multiple, nonoverlapping steps with earlier localization events being required for later ones. osk contains three regions that direct three steps of the localization process (Kim-Ha et al. 1993): (1) movement of the RNA from nurse cells into the oocyte; (2) accumulation at the anterior margin; and (3) localization at the posterior end of the oocyte. Cis-acting determinants directing each step have been mapped in the 3′ UTR, and it was hypothesized that different combinations of cis elements could direct RNAs to different final destinations (Kim-Ha et al. 1993). The three steps of osk localization appear nonoverlapping: elimination of early events by mutation of the corresponding zipcode generally precludes later steps of the localization process (Kim-Ha et al. 1993).
gurken mRNA displays a two-step localization pattern in oocytes that is governed by two zipcodes (Saunders and Cohen 1999; Thio et al. 2000). During stages 1–6, grk localizes to the posterior of oocytes. The determinants of this localization activity are inferred to lie within the first 35 bases of the protein-coding region (termed GLE1), but this sequence has not been tested directly for zipcode activity (Saunders and Cohen 1999). During stage 8, grk relocalizes to the anterodorsal corner of the oocyte, and this localization is mediated by 64 nt downstream of GLE1. The predicted secondary structure of this element is similar to that of a zipcode derived from the I factor transcript, an LTR RNA whose localization during stages 8–9 parallels that of grk (Van De Bor et al. 2005). While Van de Bor and colleagues did not report a role for the grk 3′ UTR in directing localization (Van De Bor et al. 2005), two other studies reported that the 3′ UTR was necessary for the final stages of anterodorasal accumulation (Saunders and Cohen 1999; Thio et al. 2000). This discrepancy was attributed to differences in reporters used for assaying localization (Van De Bor et al. 2005). All three studies agreed that the 3′ UTR of grk, unlike that of several other transcripts localized in Drosophila oocytes, has no localization activity on its own (Saunders and Cohen 1999; Thio et al. 2000; Van De Bor et al. 2005).
In contrast to the above transcripts, which are transported in multiple, nonoverlapping steps, some RNAs contain multiple localization elements that mediate transport via partially redundant mechanisms. For example, two fragments within a 280 nt zipcode in the 3′ UTR of orb can independently mediate its localization to the oocyte posterior during early stages, but both are required together for accumulation at the anterior end of Drosophila oocytes during late stages (Lantz and Schedl 1994).
Localization of bicoid (bcd) mRNA to the anterior end of Drosophila oocytes also occurs in distinct steps, termed event A (which acts during early phases of oogenesis, stages 4–5) and event B (which occurs later, after stage 6). Both events are directed by 650 nt in the 3′ UTR (Macdonald and Struhl 1988). The entire UTR is nearly 900 nt in length and folds into five distinct domains (Brunel and Ehresmann 2004). Event A is mediated by stem–loops IV–V in the 3′ UTR (Macdonald and Kerr 1997), or artificially by a tandem repeat of a 53 nt sequence (called BLE1) within stem–loop V (Macdonald et al. 1993), which binds transport complex components Exl and Exu (Macdonald et al. 1995). A single-base mutation in stem–loop V (G4496U) eliminates event A, and bcd accumulation at the anterior of the oocyte occurs later via event B (Macdonald and Kerr 1997). Cis-acting determinants of event B have not been isolated, but some trans-acting factors necessary for localization during and after event B (e.g., Swallow, Exuperantia, and Staufen) have been identified (Berleth et al. 1988; Stephenson et al. 1988; St Johnston et al. 1991; Macdonald and Kerr 1997).
Like events A and B in bcd localization, two pathways—message transport organizer (METRO) and the late pathway—effect transport of RNAs to the vegetal pole of Xenopus oocytes. Recently, the two pathways were shown to have some transport complex components in common, indicating a partial overlap in function (Claussen et al. 2004; Choo et al. 2005); accordingly, several RNAs are substrates of both pathways (see below), suggesting functional redundancy. The METRO pathway acts during early stages of oogenesis and transports RNAs via the mitochondrial cloud. Substrates of this pathway include Xcat-2 (Forristall et al. 1995) and Xlsirt (Kloc et al. 1993) RNAs. During transport via the METRO pathway, Xcat-2 localizes to germinal granules within the mitochondrial cloud (Kloc et al. 1998), and, like the MBP 3′ UTR, it contains distinct elements governing its localization to appropriate cellular structures. A 250 nt mitochondrial cloud localization element (MCLE) at the 5′ end of the 3′ UTR directs the RNA to the mitochondrial cloud (Zhou and King 1996a), and specific localization to the germinal granules is mediated by a downstream 164 nt germinal granule localization element (GGLE) (Kloc et al. 2000). Xcat-2 can also localize in stage IV embryos via the late pathway, and the sequences mediating localization by this pathway map to the 5′ and 3′ ends of the 3′ UTR (Zhou and King 1996b). These determinants overlap partially with both the MCLE and the GGLE, yet the exact sequences required for localization via the late pathway are distinct from those required for the METRO pathway. Another vegetally localized mRNA, fatvg, also localizes by both the METRO and late pathways and contains multiple localization elements in its 3′ UTR (Chan et al. 1999, 2001). A 25 nt sequence from the 5′ end of the UTR (the fatvg localization element, FVLE1) is sufficient for localizing RNA via the late pathway (Chan et al. 1999), and it is likely that some of the other zipcodes mediate localization by the METRO pathway. Like Xenopus, ascidians also employ two different pathways of localizing RNAs in embryos, with each pathway requiring distinct zipcode elements (Sasakura and Makabe 2002).
The studies in Drosophila and Xenopus suggest that zipcodes are modular and that different combinations of elements can direct different localization programs, but this principle has not been extensively tested. Differentially localized transcripts rarely contain shared elements, indicating that zipcodes are not used in a modular manner in vivo. Few attempts have been made at engineering RNAs with novel subcellular destinations by incorporating appropriate zipcodes. In one case, however, addition of the GGLE to the Xlsirt MCLE did localize the hybrid RNA to germinal granules (Kloc et al. 2000). Identification of additional zipcodes in localized transcripts, as well as efforts to engineer localized RNAs, will provide more information about the modularity of zipcode function.
ZIPCODE ELEMENTS ACT SYNERGISTICALLY TO TARGET RNAS TO THE TRANSPORT MACHINERY
Not only can multiple elements mediate multiple steps in the localization process, elements can also act synergistically to promote individual steps. In some cases, multimers of a single motif promote localization synergistically; and in other cases, different motifs cooperate to ensure localization. Many transcripts employ a combination of both strategies in order to ensure efficient and specific transport.
The Vg1 localization element, which encompasses >300 nt in the Vg1 3′ UTR (Mowry and Melton 1992; Deshler et al. 1998) and is responsible for localizing the RNA to the vegetal pole of Xenopus embryos, contains four sequence elements, E1–E4, in two to four copies each (Deshler et al. 1997) as well as a VM1 element in three copies (Gautreau et al. 1997). Deletion of all copies of E1, E2, E3, or E4 compromises localization efficiency, with E2 deletions showing the strongest defect. Element E2 binds Vera/VgRBP, an endoplasmic reticulum RNA-binding protein that is proposed to link Vg1 RNA to the ER for transport (Deshler et al. 1997, 1998). Recently, bud localization of ASH1 mRNA in yeast was also shown to require association of the transport complex with endoplasmic reticulum, suggesting that this mechanism of transport may be conserved between species (Schmid et al. 2006). However, because the E2 elements in the fatvg 3′ UTR are dispensable for transport, it seems that this element alone is not sufficient for vegetal localization in Xenopus oocytes (Chan et al. 1999). The VM1 element is a binding site for hnRNP I, and a fragment containing two copies of the VM1 element can direct localization when present as a tandem repeat (Gautreau et al. 1997). However, a native cluster of five VM1 elements in the 3′ UTR of Xenopus borealis Vg1 cannot support localization (Lewis et al. 2004), suggesting that the sequence or structural context of the elements either affects recognition by hnRNP I or is necessary for other events following hnRNP I binding. It has been proposed that the Vg1 zipcode function requires clusters of VM1 and E2 sites together (Lewis et al. 2004).
Nanos RNA contains four elements (+1 to +4) in its 3′ UTR, each of which localizes weakly to the posterior of Drosophila oocytes; in concert, the four elements confer full localization ability (Gavis et al. 1996a). A 41 nt region within the +2′ element (termed +2′ME) shows high-sequence conservation in Drosopholia melanogaster and Drosophlia virilis and is bound by a 75 kD protein of unknown function (Bergsten et al. 2001). Three tandem copies of +2′ME localize as efficiently as the entire +2′ element, which is reminiscent of the localization of bicoid RNA mediated by two tandem copies of BLE1. Surprisingly, when localization-impaired versions of the +2′ME nanos localization element are combined with the WT +1 element, the composite RNA localizes less efficiently than the +1 element alone, indicating that mutations in the +2′ME region impair the activity of the +1 element. This result led the authors to suggest that long-range interactions between localization elements govern transport of nanos RNA (Bergsten et al. 2001), but the mechanisms by which these interactions occur and are detected by the cellular transport machinery remain unknown.
A 54 nt zipcode in the 3′ UTR of chicken β-actin mRNA also contains multiple motifs that direct localization synergistically to the leading edge of chicken fibroblasts (Kislauskis et al. 1994). Two motifs, GGACT and AATGC, are found in both the 54 nt zipcode and in a separate 43 nt region that shows weak localization ability. Although each motif alone (in the absence of most of the other zipcode sequences) localizes poorly, synthetic constructs containing tandem repeats of one motif along with one copy of the other show enhanced localization ability (Kislauskis et al. 1994). In addition, an AC-rich region between the two motifs of the 54 nt zipcode is essential for its activity (Kislauskis et al. 1997). Two ACACCC sequences in this region each bind to ZBP1, a protein that contains 4 KH domains and bears homology with hnRNP proteins (Ross et al. 1997) and is also involved in RNA transport (Farina et al. 2003). The stronger of the two β-actin binding sites is predicted to be single stranded (Ross et al. 1997). The same sequence motif occurs in the 3′ UTR of α3 integrin mRNA, and this motif is necessary for its localization to adhesion complexes at the periphery of human cells in culture (Adereth et al. 2005). Whether this motif functions as a zipcode out of context of either the β-actin or α3 integrin transcripts is not known.
Like the aforementioned examples, some transported mRNAs contain multiple redundant zipcodes. However, there are at least two examples of systems where multimerization of single elements or combinations of distinct elements are not required to direct localization. In Xenopus, the fatvg zipcode (FVLE1) displays efficient localization when present in a single copy (Chan et al. 1999). In addition, one bud-localized RNA in yeast, ASH1, contains four zipcodes (three in the coding region and one in the 3′ UTR), each of which is sufficient for localizing a reporter RNA when fused to the 3′ end (Chartrand et al. 1999; Gonzalez et al. 1999). Similarly, each of the two zipcodes from bud-localized WSC2 localizes a reporter construct with full efficiency (Jambhekar et al. 2005). In context of the native ASH1 RNA, however, all four zipcodes are necessary for full levels of localization (Gonzalez et al. 1999). Because the reporter mRNAs used in these studies were likely not translated, it is not clear whether the requirement for multiple zipcodes reflects long-range interactions with surrounding sequences or whether passage of ribosomes along the mRNA impairs zipcode activity (see below).
Although repeats of minimal motifs (e.g., VM1, BLE1, +2′ME) can often substitute for intact zipcodes in localizing RNAs, it remains unclear exactly how these repeated sequences compensate for the loss of other essential zipcode elements. For example, how does multimerization of a fragment from stem–loop V of the bicoid 3′ UTR (BLE1) compensate for the loss of stem–loop IV? One possibility is that the various RNA-binding proteins needed for localizing a particular transcript can bind weakly to each other as well as to RNA. Creating a high-density cluster of one protein via multimerization of its binding site may recruit other essential proteins to the mRNP via protein–protein, rather than protein–RNA, interactions. In support of this possibility, clusters of VM1 and E2 sites in Vg1 RNA recruit 40LoVe, a component of the localizing mRNP (Czaplinski and Mattaj 2006). Arn et al. (2003) have proposed that specific RNA recognition involves multiple low-affinity and low-specificity interactions that can occur either on a complex RNA target or on tandem repeats of a minimal recognition element.
ROLE OF RNA PROCESSING IN ZIPCODE FUNCTION
It is generally believed that mRNA processing is not required for zipcode recognition by transport complexes, although some studies have indicated that the two may not be completely independent. Experiments involving injection of zipcode RNAs into oocytes suggest that processing is not required for zipcode recognition, as these RNAs localize efficiently in the absence of 5′ capping, poly(A) tail addition, or splicing (e.g., Glotzer et al. 1997). The vegetally localized Xenopus Xlsirt RNA is noncoding (Kloc et al. 1993) and likely not subject to mRNA processing. In yeast, expressing zipcode RNAs under the Pol III promoter (which eliminates standard mRNA processing events) also does not interfere with recognition by the transport complex (Long et al. 2000; Jambhekar et al. 2005; Olivier et al. 2005) or localization (Beach and Bloom 2001). Although localization of cyclin B mRNA to pole cells and somatic nuclei during Drosophila embryogenesis (Whitfield et al. 1989; Raff et al. 1990) is influenced by splicing, there does not appear to be a mechanistic relationship between the two processes: the unlocalized spliced isoform simply lacks the 3′ UTR zipcode present in the localized transcript (Dalby and Glover 1992).
A detailed analysis of osk localization, however, revealed a mechanistic role for splicing in directing transport. osk transgenes, expressed in a background in which no endogenous osk is produced, required splicing at the first of three exons for localization to the posterior during late stages of oogenesis (Hachet and Ephrussi 2004). Localization was independent of intron sequence, as replacement of the first intron sequence with that of the third intron supported localization. Although exon–junction complexes are presumed to be assembled at each of the three introns in osk, assembly is required only at the first junction for localization. Interestingly, only splicing at the first intron supports colocalization of osk RNA with Y14 (Hachet and Ephrussi 2004), a component of the exon–junction complex and an essential factor in osk transport (Hachet and Ephrussi 2001). Surprisingly, the requirement for splicing seems to function in trans: a LacZ reporter fused to the 3′ UTR of osk (which contains zipcode regions) (Kim-Ha et al. 1993) localizes efficiently only when endogenous osk is expressed (Hachet and Ephrussi 2004). In support of the role of splicing in mediating localization, components of exon–junction complexes (e.g., Y14 and mago) (Hachet and Ephrussi 2001; Le Hir et al. 2001; Mohr et al. 2001), as well as factors involved in nonsense-mediated decay (e.g., eIF4AIII) (Palacios et al. 2004), are required for osk localization. It is not clear, however, whether these proteins play dual roles in splicing or transport, or whether splicing is simply a prerequisite for nuclear export and therefore cytoplasmic localization. In a separate study, insertion of a zipcode (the TLS region from the anteriorly localized Drosophila K10 transcript) near a 5′ splice site supported localization but impaired splicing (Cohen et al. 2005). Whether this result reflects simple steric interference between the localization and splicing machineries or a more complex mechanistic relationship remains unknown. It is likely that other localized transcripts also require mRNA processing factors for transport.
In addition to the connection between splicing and localization, there is evidence that mRNAs are “marked” for transport prior to nuclear export. hnRNP I, a component of the Vg1 and VegT transport mRNPs, associates with both mRNAs in the nucleus and remains bound to them in the cytoplasm. Prrp and Xstau, other essential components of the transport complex, are recruited to the RNAs in the cytoplasm, presumably via their interactions with hnRNP I (Kress et al. 2004). Thus, binding of shuttling factors to mRNA in the nucleus can guide the assembly of a transport complex in the cytoplasm. In yeast, She2 may perform a function similar to hnRNP I. She2 shuttles between the nucleus and cytoplasm, and its nuclear export requires binding to RNA (Kruse et al. 2002), suggesting that transported RNAs emerge from the nucleus with She2 already bound. She2 could then recruit She3 and Myo4 to effect transport (Kruse et al. 2002). In addition to the core–She complex components, Loc1, a nuclear protein that binds double-stranded RNA nonspecifically, is also essential for bud localization of ASH1 (Long et al. 2001). Loc1 was recently shown to be important for rRNA processing as well as nuclear export of 60S ribosomal subunits, and it was hypothesized that aberrant nuclear-cytoplasmic shuttling or defects in translation may impair ASH1 localization (Urbinati et al. 2006).
Cytoplasmic mRNA processing may also affect zipcode activity. In yeast, anchoring of ASH1 to the bud tip requires translation (Gonzalez et al. 1999). Surprisingly, the translation and localization processes seem to act antagonistically. All four ASH1 zipcodes are necessary for full levels of localization of the native RNA (Gonzalez et al. 1999) or translatable forms of the RNA with zipcodes ectopically located in the UTR (Chartrand et al. 2002). However, only one zipcode is sufficient for localizing a nontranslatable reporter (Beach and Bloom 2001; Jambhekar et al. 2005), suggesting that translation impairs zipcode activity. In support of this finding, deletion of associated translation inhibitors Khd1 or Puf6 impairs localization (Irie et al. 2002; Gu et al. 2004). Because yeast zipcodes generally lie in coding regions, it is possible that ribosomes compete with the She complex for zipcode occupancy; the presence of four zipcodes in ASH1 may ensure that at least one remains bound to the She complex at any time. The secondary structure of the zipcodes delays passage of ribosomes along the RNA (Chartrand et al. 2002), thus ensuring that the RNA is not translated prior to its localization in the bud. A similar translation-based mechanism may inhibit dendritic localization of RNAs in neural cells: transport to dendrites appears to be the “default” state, and association with ribosomes restricts RNAs to the cell body (Lu et al. 1998). Analogously, a 1 kb region in the MBP 3′ UTR is required (in addition to the A2RE-containing zipcode) to localize a protein-coding reporter RNA to the myelin compartment of oligodendrocytes. However, the A2RE element is sufficient for localizing a non-protein-coding construct (Ainger et al. 1997). In these cases, it is not clear how zipcodes (which are generally located in the 3′ UTR) can overcome translation-dependent inhibition of transport.
Translation also influences the localization of vasopressin RNA, which is localized to both dendrites and axons of neurons. Two fragments, one entirely in the coding region and the other overlapping the ORF and 3′ UTR, act synergystically to localize the RNA to dendrites (Prakash et al. 1997). Alleles of the gene containing a single base (guanosine) deletion in the downstream zipcode impair axonal but not dendritic localization (Mohr et al. 1995). Zipcodes mediating axonal localization of this RNA have not been identified. It is possible that the downstream dendritic targeting signal also mediates axonal localization, and that the guanosine deletion impairs recognition specifically by the axonal transport complex. The authors, however, proposed that the loss of a stop codon caused by the frameshift mutation impaired release of the transcript from ribosomes, thus preventing recognition by the transport machinery (Mohr et al. 1995). Translational regulation is critical for localization in several other systems, although the mechanisms likely involve trans-acting factors rather than direct interaction of ribosomes and RNA zipcodes (see, e.g., Wilhelm et al. 2003; Yano et al. 2004).
Factors promoting RNA stability may also play a role in localization. In the case of tau mRNA, a U-rich region in the 3′ UTR that binds HuD and increases RNA stability (Aranda-Abreu et al. 1999) is also required for axonal localization (Aronov et al. 2001). However, it is not clear whether HuD is an essential component of the transport complex, or whether, in the absence of HuD, the RNA is simply degraded before it can be transported. At this time it is not known whether transported RNAs are generally more or less stable than their nontransported counterparts, or even whether they are degraded by similar mechanisms.
Regulation of zipcode activity
With a few notable exceptions (see below), zipcodes generally function constitutively, providing the cytoskeletal structure is intact and appropriate trans-acting factors are present. In yeast, disruption of actin filaments causes symmetric distribution of bud-localized RNAs (Jansen et al. 1996; Long et al. 1997; Takizawa et al. 1997, 2000), and dissolution of actin cables late in mitosis causes bud–tip localized RNAs to migrate to the bud neck (Beach et al. 1999). β-actin RNA localization to fibroblasts also requires actin microfilaments (Sundell and Singer 1991), while RNA transport in Drosophila (Pokrywka and Stephenson 1995) and Xenopus (Yisraeli et al. 1990) oocytes and β-actin RNA localization in nerve growth cones (Bassell et al. 1998) require microtubules. Actin filaments have also been implicated in anchoring localized RNAs in yeast, Xenopus, and fibroblasts (Yisraeli et al. 1990; Sundell and Singer 1991; Beach and Bloom 2001; Liu et al. 2002). Therefore, cytoskeletal rearrangements during cellular development can alter the distribution of RNAs, although cytoskeletal regulation may not be sufficient for proper RNA targeting in all cases (Theurkauf and Hazelrigg 1998).
Surprisingly, some zipcodes can function in cellular contexts different from those in which they are normally active, indicating that there is minimal negative regulation of zipcode function. This phenomenon has been demonstrated most extensively in Drosophila. RNAs that are asymmetrically localized in oocytes accumulate at the apical region when injected into embryos, in a pattern similar to pair-rule transcripts (Bullock and Ish-Horowicz 2001). These transcripts localize identically in embryos even though they exhibit different localization programs in oocytes. Furthermore, cis-acting mutations that impair localization in oocytes also impair apical transport in embryos (Bullock and Ish-Horowicz 2001; Snee et al. 2005), suggesting that the localization machinery in both settings recognizes similar zipcodes. However, proteins known to be part of the ovarian bicoid transport complex (e.g., Swallow, Modulo, poly[A]-binding protein, Smooth, Nod) (Arn et al. 2003) were not detected in a fractionated bicoid-binding embryonic extract (Snee et al. 2005). It has been proposed that zipcode recognition by the transport complex results in each case from multiple low-affinity and low-specificity interactions (Arn et al. 2003; Snee et al. 2005). However, there do not generally appear to be combinatorial control mechanisms for repressing zipcode activity as there are for repressing DNA promoter elements (Istrail and Davidson 2005).
One exception is the localization of β-actin mRNA, which is subject to positive and negative control mechanisms. In fibroblasts, this mRNA is localized to the leading edge by zipcode(s) located in the 3′ UTR (see above). Localization was stimulated by PDGF treatment or by serum addition following starvation. Both chemical inhibition and activation of PKA or PKC, as well as decreasing cellular ATP levels, reduced localization (Latham et al. 1994). The investigators proposed that the cellular phosphorylation status regulates β-actin mRNA localization. Similarly, β-actin mRNA localization to nerve growth cones is regulated by extracellular signals. Addition of neurotrophin-3 (NT-3) stimulated localization, as did activation of adenylate cyclase (Zhang et al. 2001). In both systems, it is not clear how signaling pathways regulate β-actin mRNA transport. The simplest explanation is that trans-acting factors required for transport are regulated by phosphorylation. Given the rapid (within 2 min) localization of β-actin mRNA in response to the appropriate stimuli (Latham et al. 1994) and the fact that β-actin mRNA is transported to leading edges in the presence of translation inhibitors (Sundell and Singer 1990), it is unlikely that de novo synthesis of mRNP components is required for transport. It is possible, however, that repressors of transport are bound to negatively acting RNA elements in situations where the RNA is not localized, and that extracellular stimuli promote zipcode function by removing these repressors. Identifying the entire complement of β-actin mRNA-bound proteins under localizing and nonlocalizing conditions in the two cell types will help to elucidate how localization of this transcript is regulated.
The localization of αCaMKII also appears to be under positive and negative regulation, and cis-acting elements responsible for this regulation have been defined. The zipcode for this transcript was mapped to the 3′ UTR (Mayford et al. 1996), and subsequent fine-mapping studies yielded three positive cis-acting regions. Blichenberg et al. (2001) identified an ∼1200 nt fragment from the latter half of the 3′ UTR as being sufficient for localizing an EGFP reporter. Another zipcode was mapped to the distal 170 nt of the UTR, which contains cytoplasmic polyadenylation elements (CPEs) (Huang et al. 2003). Zipcode activity of this region required WT cytoplasmic polyadenylation element-binding protein (CPEB). Because many transcripts (both localized and nonlocalized) contain CPEs, the investigators proposed that the structural context of the CPE may determine whether it functions as a zipcode. Furthermore, because the CPE mediated localization when placed in a polylinker sequence, the investigators concluded that the CPE may be a default transport signal that is masked by additional negative-regulatory cis-acting signals in nonlocalized transcripts (Huang et al. 2003). A third zipcode was reported within the first 94 nt of the 3′ UTR. A 30 nt sequence within this region showed high homology with the 3′ UTR of neurogranin (another dendritically localized RNA), and this motif was necessary for dendritic localization of 3′ UTR sequences derived from both αCaMKII and neurogranin (Mori et al. 2000).
Two potential repressors of the 5′ 94 nt zipcode have been reported. One repressor was proposed to lie between nucleotides 94 and 725 (as the first 725 nt did not show zipcode activity), and an enhancer was postulated to lie between nucelotides 725 and 831 (as nucleotides 1–831 did localize to dendrites) (Blichenberg et al. 2001). A simpler explanation for these contradictory findings may be that truncating the 3′ UTR at 725 nt causes the 94 nt zipcode to adopt a conformation that is refractory to recognition by the transport complex. Another repressor was proposed to lie between nucleotides 831 and 1497 of the 3′ UTR, based on the finding that the first 831 nt localized to dendrites, while the first 1497 did not. Depolarizing neurons by addition of KCl relieved inhibition and restored dendritic localization of nucleotides 1–1497 of the 3′ UTR (Mori et al. 2000). This finding was unexpected because localization of full-length αCaMKII (unlike that of BDNF and TrkB, whose dendritic localization is sensitive to depolarization) (Tongiorgi et al. 1997) is constitutive (Burgin et al. 1990). In the native RNA, the zipcodes identified by Blichenberg et al. (2001.) and Huang et al. (2003) may bypass the requirement for depolarization. A Y-element in the protein-coding region of the transcript, which is essential for transport and binds TB-RBP (a protein implicated in both localization and translation inhibition) (Severt et al. 1999), may also relieve repression. These positive elements were not present in the KCl-sensitive 1–1497 construct. It is not clear whether depolarization causes a conformational change in the RNA itself, a change in the expression/activity of any trans-acting factors, or both. Analogous to the putative transport repressors in αCaMKII, the VM1 sites in the Vg1 zipcode have been proposed to repress transport, since injection of VM1-containing repeat RNA (which titrates out hnRNP I) enhanced Vg1 localization (Czaplinski and Mattaj 2006). Conclusive demonstration of cis-acting inhibitors of transport awaits isolation of sequences that can prevent localization out of context of their native RNAs.
Sequence and structural basis of RNA recognition
Despite the large number of zipcodes isolated in a variety of systems, no clear patterns in zipcode sequence or structure have emerged. Some zipcodes appear to be recognized by multiple transport complexes. For example, a 300 nt zipcode from the vegetally localized Xenopus XNIF mRNA (Claussen et al. 2004), as well as a 137 nt zipcode from Xlsirt (Allen et al. 2003), are recognized by proteins that participate in both the METRO and late transport pathways. The K10 zipcode, as well as three other transcripts that are transported in Drosophila oocytes, localize apically when injected in blastoderm embryos (Bullock and Ish-Horowicz 2001). Conversely, some transport complexes recognize multiple zipcodes. Xcat-2 and Xlsirt, for example, are both localized by the METRO pathway (see above), but the 137 nt zipcode in Xlsirt is not found in Xcat-2 (Allen et al. 2003). Few zipcodes have been minimized sufficiently to allow detailed analysis of the exact requirements for activity. The dendritic targeting element in MAP2 mRNA, which is localized to dendrites of neurons, comprises a 640 nt region in the 3′ UTR, which is predicted to fold into multiple stem–loops (Blichenberg et al. 1999). However, it is not known which sequence/structural subelements are necessary for localization. Furthermore, the RNA-binding proteins that recognize zipcodes remain unknown in many cases, making analysis of the physical RNA–protein interactions impossible.
An attractive model for zipcode recognition is that trans-acting factors recognize a specific secondary structure in the RNA, often a hairpin stem–loop structure, along with a small number of specific nucleotides (Chartrand et al. 1999). This model has largely proven to be correct in the case of the TLS element (Cohen et al. 2005), a zipcode present in both the orb and K10 transcripts of Drosophila (Serano and Cohen 1995). The native zipcode is predicted to fold in a stem–loop consisting of a 17 base pair (bp) stem interrupted by two single-base bulges and an 8 base loop (Cohen et al. 2005). Fine-mapping studies showed that increasing loop size or decreasing the length of the stem interfered with transport/localization. Interestingly, some compensatory mutations in the stem supported localization while others did not. Mutations that altered the stereochemistry of the minor groove (e.g., U·A → C·G or G·C) impaired localization, but those that preserved minor groove stereochemistry (e.g., U·A → A·U) had no effect. The only exception was at the fifth position of the stem, where the U·A → A·U mutation caused subtle defects in localization. It appears, therefore, that the TLS element is recognized on the basis of hydrogen-bond patterns in the minor groove of the helix, with a small contribution from the major groove of base-pair 5 (Cohen et al. 2005).
Like the TLS element, the dendritic targeting signal at the 5′ end of the BC1 RNA (Muslimov et al. 1997) also folds into a single stem–loop (Rozhdestvensky et al. 2001). This finding was surprising because, based on sequence similarity to tRNA, BC1 was expected to adopt a cloverleaf structure. Although tRNAs are also localized to dendrites, some of them remain to function in the cell body. It was hypothesized that the difference in structure allows BC1 to be transported more efficiently into dendrites than are tRNAs (Rozhdestvensky et al. 2001). The specific sequence and structural properties of BC1 that allow its transport remain undefined. Therefore, it is not clear whether any hairpin bearing structural similarity to BC1 is competent for localization.
A 75 nt zipcode from the 3′ UTR of Xvelo1, a transcript vegetally localized by the late pathway in Xenopus oocytes, is also predicted to fold into a hairpin structure (Claussen and Pieler 2004). Detailed analysis of this zipcode, however, revealed an RNA-recognition strategy more complicated than simple hairpin binding. Deletion of a largely single-stranded 5′ tail abolished localization, indicating that the hairpin alone is insufficient for localization. Second, compensatory mutations in the stem region (which maintain the predicted secondary structure of the RNA) did not support WT levels of localization. Because the compensatory mutations maintained the architecture of the minor groove of the helix, it appears that the Xvelo1 zipcode is not recognized by a TLS-like mechanism. It is possible that nucleotide identities in the stem are important for recognition of Xvelo1 by the transport machinery, or that these bases contribute to forming a specific three-dimensional structure necessary for recognition.
Two transcripts that are localized to the dorsoanterior corner of Drosophila oocytes, grk and I factor, share similar hairpin zipcodes that function during stage 8 of oogenesis (see above) (Van De Bor et al. 2005). While both zipcodes contain three stem regions separated by two internal bulged loops, the precise stem lengths, bulge sizes and locations, and terminal loop lengths differ, as do the nucleotide identities. Because detailed mutagenesis studies have not been performed on these zipcodes, the molecular mechanism by which any shared trans-acting factors could recognize such a diversity of substrates, yet maintain specificity for their targets, remains unclear.
Zipcodes mediating bud localization of mRNAs in yeast have, perhaps, been most well characterized (Chartrand et al. 1999; Gonzalez et al. 1999; Jambhekar et al. 2005; Olivier et al. 2005). These sequences, like the stem–loop IV–V zipcode in the Drosophila bicoid RNA (Macdonald and Kerr 1998), reveal a primary sequence as well as structural requirements. The isolation of multiple short zipcodes recognized by a single transport complex (She2/3) (Jambhekar et al. 2005) has provided insight into the rules as well as the exceptions governing She2/3 recognition of target RNAs. A core single-stranded CG dinucleotide (Jambhekar et al. 2005), often present as a CGA triplet (Olivier et al. 2005), appears essential for transport, while a stretch of downstream adenosines contributes to, but is not essential for, She2/3 recognition (Jambhekar et al. 2005). Although the primary sequences of the stem regions of different zipcodes vary, these sequences contribute to recognition in some (but not all) cases (Jambhekar et al. 2005). In some cases, compensatory mutations in stem regions maintain activity (Chartrand et al. 1999; Gonzalez et al. 1999; Jambhekar et al. 2005; Olivier et al. 2005), whereas in other cases they do not (Jambhekar et al. 2005). A nonessential bulged cytosine also enhances zipcode recognition when present (Jambhekar et al. 2005; Olivier et al. 2005), and this enhancement depends on the distance between the cytosine and CG dinucleotide (Olivier et al. 2005). The variable contributions of different sequence and secondary structural features suggest that the She complex recognizes hairpin zipcodes on the basis of tertiary structures as well as primary sequence and secondary structure. Although the hairpin mechanism initially proposed for recognition of She2-dependent zipcodes was attractive, subsequent analyses reveal a more complex recognition strategy. Because many nonlocalized transcripts are also predicted to fold into multiple hairpin structures (A. Jambhekar and J.L. DeRisi, unpubl.), the secondary-structure-recognition model is not sufficient to explain the specific recognition of zipcodes by She2/3. Furthermore, it has not been possible to engineer novel zipcodes by incorporating the essential primary sequence and secondary structural motifs elucidated by comparative and mutational analyses (A. Jambhekar and J.L. DeRisi, unpubl.), indicating that these requirements alone do not define a She2-dependent zipcode. Although the crystal structure of She2 revealed residues important for RNA binding and dimerization (Niessing et al. 2004), it is not clear how the She complex might recognize its target zipcodes with high affinity and specificity. A three-dimensional structure of the RNA–protein complex will be necessary to elucidate the roles of various RNA sequence and structural features in binding.
Although analysis of most zipcodes has focused on sequence and secondary structural elements, the higher-order structure of the bicoid 3′ UTR is essential for its embryonic localization, suggesting that complex structures also may be important for other zipcodes. An intermolecular dimerization event generates a quaternary structure that directs recognition and localization to astral microtubules during early stages of embryogenesis by Staufen (Ferrandon et al. 1997). Dimerization of the UTR initiates via base pairing between single-stranded loops in domain III, and is then stabilized by surrounding sequences (Wagner et al. 2001). Staufen contains five double-stranded RNA-binding domains (dsRBDs), of which three bind RNA and two do not (Micklem et al. 2000). The NMR structure of dsRBD3 complexed with RNA reveals binding of the domain to an RNA monomer (Ramos et al. 2000); it is not clear whether the bcd 3′ UTR dimer is recognized by one or more intact molecules of Staufen. RNA dimerization may serve to nucleate higher-order RNA–protein “packages” that function as the substrates for efficient mRNP transport. The characterization of a mechanism for RNA dimerization provides a good starting point for examining the formation and structures of transported RNA packages.
Predicting zipcodes
Because of the complexity of and variations in RNA–protein recognition strategies employed by different transport systems, faithful in silico predictions of zipcode regions have proven to be quite difficult. Furthermore, the lack of sufficient numbers of zipcodes recognized by a given transport system, combined with the difficulties of predicting higher-order RNA structures, have impeded progress in this area.
Predicting zipcodes that can be defined only by primary sequence has been the most successful. For example, a dendritic targeting sequence in neurogranin RNA was identified based on sequence similarity to that of α-CaMKII mRNA (Mori et al. 2000). In Xenopus, clusters of CAC motifs have been identified as zipcode regions (Betley et al. 2002). The CAC motif was first identified in the context of an E2 element, which constitutes a Vera binding site (Deshler et al. 1998), and later in the VegT RNA, which also localizes in a Vera-dependent manner (Kwon et al. 2002). A search for repeated motifs in the 3′ UTRs of nine other vegetally localized RNAs generated short motifs containing CAC triplets, with statistically significant enrichments of the motifs in zipcode regions when compared with the surrounding sequences (Betley et al. 2002). CAC-containing regions from Xpat and Xcat-2 3′ UTRs, as well as from vegetally localized RNAs in ascidian embryos, were necessary and sufficient for localizing reporter constructs in Xenopus eggs. Surprisingly, the CAC zipcodes were found in transcripts localized by either the METRO or late pathways, suggesting that the motif was not pathway specific but may be recognized by a protein that functions in both pathways. Although Vera binds to zipcodes from transcripts localized by both the early and late pathways (Choo et al. 2005), the binding is dependent on CAC motifs in only some cases (Betley et al. 2002). The exact mechanism of CAC zipcode recognition in the early and late pathways remains unclear.
Identifying zipcodes that are recognized on the basis of structure as well as sequence has proven more difficult. One powerful method of identifying functional RNA elements is to search for evolutionarily conserved RNA secondary structures in related species, since essential secondary structures are predicted to be conserved while primary sequences will vary. Thus, the presence of compensatory mutations in related RNAs will signify structures under selective pressure. Algorithms such as FOLDALIGN (Gorodkin et al. 1997) and X2s (Juan and Wilson 1999), as well as manual inspection of sequences, have been used to identify functional elements in HuR target mRNAs and noncoding telomerase RNAs (Romero and Blackburn 1991; Dandjinou et al. 2004; Lopez de Silanes et al. 2004). Such analyses have been less successful in identifying zipcode regions. Many algorithms, such as CONSTRUCT (Luck et al. 1999), require good sequence alignments for accurate structure prediction, making it difficult to identify short zipcode elements buried in long UTRs or coding regions. Manual comparison of the predicted secondary structures of bicoid 3′ UTRs from six different species of Drosophila yielded globally similar structures (Macdonald 1990). The details of stem and loop lengths and sequences, however, varied between species, making it difficult to pinpoint putative transport complex binding sites (Macdonald 1990). At the same time, the low sequence divergence between the species precluded using a phylogenetic approach due to insufficient numbers of compensatory mutations (Ferrandon et al. 1997). Comparison of the nanos 3′ UTRs from D. melanogaster and D. virilis revealed several conserved sequence elements both within and outside the four zipcode regions, as well as a translational control element (TCE) that showed both sequence and structural conservation (Gavis et al. 1996a,b; Crucs et al. 2000). Mapping of zipcodes in both bicoid and nanos was initially achieved by testing various 3′ UTR fragments for localization, presumably because the sequence and structural similarities between the available sequences of homologous genes were not sufficient to identify candidate zipcode regions (Macdonald 1990; Gavis et al. 1996a).
In addition to comparing RNA sequences and structures between related species, analysis of multiple targets of a transport complex from a single species also has the potential to elucidate essential zipcode features. In Xenopus, both the VegT and Vg1 RNAs are vegetally localized by the late pathway. The 3′ UTRs of both transcripts reveal clusters of E2 and VM1 sites (Bubunenko et al. 2002) that bind Vera and hnRNP I, respectively. These clusters are similar in length and are located at similar positions in the 3′ UTRs, although the exact numbers and locations of each type of site vary between the two UTRs. It was proposed that clusters of E2 and VM1 sites define a vegetal localization element (Bubunenko et al. 2002). A similar analysis was attempted in yeast, using four She complex target zipcodes from the ASH1 gene (Olivier et al. 2005). After deriving rules for She-complex recognition, a search for novel zipcodes in 23 other known bud-localized RNAs yielded four additional putative zipcodes, of which two localized in vivo. Given that bona fide zipcodes fitting the search criteria, such as the one validated from WSC2 (Jambhekar et al. 2005), were missed, it is clear that the available tools for RNA structure prediction are not sufficiently accurate to reliably identify zipcodes within large data sets of sequences. This finding is not surprising, given that analysis of additional She-complex-dependent zipcodes revealed that variations in sequence/structure can be tolerated in some contexts (Jambhekar et al. 2005), which indicates that the initial rules derived for She-complex recognition (Olivier et al. 2005) do not accurately describe the range of zipcodes that can be recognized by this system.
Despite the recent advancements in techniques for visualizing RNA distribution, zipcode identification and characterization remain a tedious task. Elucidating essential zipcode features remains problematic because it is often impossible to test sequence and structural requirements independently. Although large zipcode-containing regions function out of context, the detailed requirements for activity often appear context dependent; thus, it is likely that the majority of zipcodes are recognized on the basis of tertiary structural features. Because motif-finding algorithms have not proven to be efficient at identifying zipcodes, the only genome-wide method for identifying RNA targets is RNA-IP, which requires prior knowledge of the RNA-binding protein (Takizawa et al. 2000). To date, there is no available three-dimensional structure of a transport complex component bound to its native RNA target. Better knowledge of the precise sequences and higher-order structures necessary for zipcode activity in various systems, as well as the development of algorithms that predict such structures, will aid in identifying novel localized transcripts and in elucidating the spatial and temporal regulation of RNAs from the time of their synthesis in the nucleus to their degradation in the cytoplasm.
ACKNOWLEDGMENTS
We thank Tracy Kress for critical reading of the manuscript.
Footnotes
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.262607.
REFERENCES
- Adereth, Y., Dammai, V., Kose, N., Li, R., Hsu, T. RNA-dependent integrin α3 protein localization regulated by the muscle blind-like protein MLP1. Nat. Cell Biol. 2005;7:1240–1247. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainger, K., Avossa, D., Diana, A.S., Barry, C., Barbarese, E., Carson, J.H. Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, L., Kloc, M., Etkin, L.D. Identification and characterization of the Xlsirt cis-acting RNA localization element. Differentiation. 2003;71:311–321. doi: 10.1046/j.1432-0436.2003.7106003.x. [DOI] [PubMed] [Google Scholar]
- Aranda-Abreu, G.E., Behar, L., Chung, S., Furneaux, H., Ginzburg, I. Embryonic lethal abnormal vision-like RNA-binding proteins regulate neurite outgrowth and τ expression in PC12 cells. J. Neurosci. 1999;19:6907–6917. doi: 10.1523/JNEUROSCI.19-16-06907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arn, E.A., Cha, B.J., Theurkauf, W.E., Macdonald, P.M. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA-binding proteins. Dev. Cell. 2003;4:41–51. doi: 10.1016/s1534-5807(02)00397-0. [DOI] [PubMed] [Google Scholar]
- Aronov, S., Aranda, G., Behar, L., Ginzburg, I. Axonal τ mRNA localization coincides with τ protein in living neuronal cells and depends on axonal targeting signal. J. Neurosci. 2001;21:6577–6587. doi: 10.1523/JNEUROSCI.21-17-06577.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah, A., Cooperstock, R.L., Lipshitz, H.D. RNA localization in development. Annu. Rev. Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- Bassell, G.J., Zhang, H., Byrd, A.L., Femino, A.M., Singer, R.H., Taneja, K.L., Lifshitz, L.M., Herman, I.M., Kosik, K.S. Sorting of β-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach, D.L., Bloom, K. ASH1 mRNA localization in three acts. Mol. Biol. Cell. 2001;12:2567–2577. doi: 10.1091/mbc.12.9.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach, D.L., Salmon, E.D., Bloom, K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- Behar, L., Marx, R., Sadot, E., Barg, J., Ginzburg, I. cis-acting signals and trans-acting proteins are involved in τ mRNA targeting into neurites of differentiating neuronal cells. Int. J. Dev. Neurosci. 1995;13:113–127. doi: 10.1016/0736-5748(95)00001-w. [DOI] [PubMed] [Google Scholar]
- Bergsten, S.E., Huang, T., Chatterjee, S., Gavis, E.R. Recognition and long-range interactions of a minimal nanos RNA localization signal element. Development. 2001;128:427–435. doi: 10.1242/dev.128.3.427. [DOI] [PubMed] [Google Scholar]
- Berleth, T., Burri, M., Thoma, G., Bopp, D., Richstein, S., Frigerio, G., Noll, M., Nusslein-Volhard, C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S.M., Singer, R.H., Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Betley, J.N., Frith, M.C., Graber, J.H., Choo, S., Deshler, J.O. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 2002;12:1756–1761. doi: 10.1016/s0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- Betley, J.N., Heinrich, B., Vernos, I., Sardet, C., Prodon, F., Deshler, J.O. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 2004;14:219–224. doi: 10.1016/j.cub.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Blichenberg, A., Schwanke, B., Rehbein, M., Garner, C.C., Richter, D., Kindler, S. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J. Neurosci. 1999;19:8818–8829. doi: 10.1523/JNEUROSCI.19-20-08818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blichenberg, A., Rehbein, M., Muller, R., Garner, C.C., Richter, D., Kindler, S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the α subunit of Ca2+/calmodulin-dependent protein kinase II. Eur. J. Neurosci. 2001;13:1881–1888. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- Bohl, F., Kruse, C., Frank, A., Ferring, D., Jansen, R.P. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratu, D.P., Cha, B.J., Mhlanga, M.M., Kramer, F.R., Tyagi, S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl. Acad. Sci. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza, R.P., Serbus, L.R., Duffy, J.B., Saxton, W.M. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 2000;289:2120–2122. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel, C., Ehresmann, C. Secondary structure of the 3′ UTR of bicoid mRNA. Biochimie. 2004;86:91–104. doi: 10.1016/j.biochi.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Bubunenko, M., Kress, T.L., Vempati, U.D., Mowry, K.L., King, M.L. A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev. Biol. 2002;248:82–92. doi: 10.1006/dbio.2002.0719. [DOI] [PubMed] [Google Scholar]
- Bullock, S.L., Ish-Horowicz, D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414:611–616. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- Burgin, K.E., Waxham, M.N., Rickling, S., Westgate, S.A., Mobley, W.C., Kelly, P.T. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J. Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, A.P., Kloc, M., Etkin, L.D. fatvg encodes a new localized RNA that uses a 25-nucleotide element (FVLE1) to localize to the vegetal cortex of Xenopus oocytes. Development. 1999;126:4943–4953. doi: 10.1242/dev.126.22.4943. [DOI] [PubMed] [Google Scholar]
- Chan, A.P., Kloc, M., Bilinski, S., Etkin, L.D. The vegetally localized mRNA fatvg is associated with the germ plasm in the early embryo and is later expressed in the fat body. Mech. Dev. 2001;100:137–140. doi: 10.1016/s0925-4773(00)00517-7. [DOI] [PubMed] [Google Scholar]
- Chartrand, P., Meng, X.H., Singer, R.H., Long, R.M. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- Chartrand, P., Singer, R.H., Long, R.M. RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol. 2001;17:297–310. doi: 10.1146/annurev.cellbio.17.1.297. [DOI] [PubMed] [Google Scholar]
- Chartrand, P., Meng, X.H., Huttelmaier, S., Donato, D., Singer, R.H. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- Cheung, H.K., Serano, T.L., Cohen, R.S. Evidence for a highly selective RNA transport system and its role in establishing the dorsoventral axis of the Drosophila egg. Development. 1992;114:653–661. doi: 10.1242/dev.114.3.653. [DOI] [PubMed] [Google Scholar]
- Choo, S., Heinrich, B., Betley, J.N., Chen, Z., Deshler, J.O. Evidence for common machinery utilized by the early and late RNA localization pathways in Xenopus oocytes. Dev. Biol. 2005;278:103–117. doi: 10.1016/j.ydbio.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Claussen, M., Pieler, T. Xvelo1 uses a novel 75-nucleotide signal sequence that drives vegetal localization along the late pathway in Xenopus oocytes. Dev. Biol. 2004;266:270–284. doi: 10.1016/j.ydbio.2003.09.043. [DOI] [PubMed] [Google Scholar]
- Claussen, M., Horvay, K., Pieler, T. Evidence for overlapping, but not identical, protein machineries operating in vegetal RNA localization along early and late pathways in Xenopus oocytes. Development. 2004;131:4263–4273. doi: 10.1242/dev.01283. [DOI] [PubMed] [Google Scholar]
- Cohen, R.S., Zhang, S., Dollar, G.L. The positional, structural, and sequence requirements of the Drosophila TLS RNA localization element. RNA. 2005;11:1017–1029. doi: 10.1261/rna.7218905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski, M., Blugeon, C., Jacq, C. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 2000;20:7881–7892. doi: 10.1128/mcb.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote, C.A., Gautreau, D., Denegre, J.M., Kress, T.L., Terry, N.A., Mowry, K.L. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- Cristofanilli, M., Iacoangeli, A., Muslimov, I.A., Tiedge, H. Neuronal BC1 RNA: Microtubule-dependent dendritic delivery. J. Mol. Biol. 2006;356:1118–1123. doi: 10.1016/j.jmb.2005.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crucs, S., Chatterjee, S., Gavis, E.R. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol. Cell. 2000;5:457–467. doi: 10.1016/s1097-2765(00)80440-2. [DOI] [PubMed] [Google Scholar]
- Czaplinski, K., Mattaj, I.W. 40LoVe interacts with Vg1RBP/Vera and hnRNP I in binding the Vg1-localization element. RNA. 2006;12:213–222. doi: 10.1261/rna.2820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby, B., Glover, D.M. 3′ nontranslated sequences in Drosophila cyclin B transcripts direct posterior pole accumulation late in oogenesis and peri-nuclear association in syncytial embryos. Development. 1992;115:989–997. doi: 10.1242/dev.115.4.989. [DOI] [PubMed] [Google Scholar]
- Dandjinou, A.T., Levesque, N., Larose, S., Lucier, J.F., Abou Elela, S., Wellinger, R.J. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., Highett, M.I., Schnapp, B.J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., Highett, M.I., Abramson, T., Schnapp, B.J. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- Farina, K.L., Huttelmaier, S., Musunuru, K., Darnell, R., Singer, R.H. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon, D., Elphick, L., Nusslein-Volhard, C., St Johnston, D. Staufen protein associates with the 3′ UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Ferrandon, D., Koch, I., Westhof, E., Nusslein-Volhard, C. RNA–RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forristall, C., Pondel, M., Chen, L., King, M.L. Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development. 1995;121:201–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- Fusco, D., Accornero, N., Lavoie, B., Shenoy, S.M., Blanchard, J.M., Singer, R.H., Bertrand, E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau, D., Cote, C.A., Mowry, K.L. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development. 1997;124:5013–5020. doi: 10.1242/dev.124.24.5013. [DOI] [PubMed] [Google Scholar]
- Gavis, E.R., Lehmann, R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71:301–313. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- Gavis, E.R., Curtis, D., Lehmann, R. Identification of cis-acting sequences that control nanos RNA localization. Dev. Biol. 1996a;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- Gavis, E.R., Lunsford, L., Bergsten, S.E., Lehmann, R. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development. 1996b;122:2791–2800. doi: 10.1242/dev.122.9.2791. [DOI] [PubMed] [Google Scholar]
- Glotzer, J.B., Saffrich, R., Glotzer, M., Ephrussi, A. Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Curr. Biol. 1997;7:326–337. doi: 10.1016/s0960-9822(06)00156-4. [DOI] [PubMed] [Google Scholar]
- Gonzalez, I., Buonomo, S.B., Nasmyth, K., von Ahsen, U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 1999;9:337–340. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- Gorodkin, J., Heyer, L.J., Stormo, G.D. Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res. 1997;25:3724–3732. doi: 10.1093/nar/25.18.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, W., Deng, Y., Zenklusen, D., Singer, R.H. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes & Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet, O., Ephrussi, A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- Hachet, O., Ephrussi, A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Hoek, K.S., Kidd, G.J., Carson, J.H., Smith, R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- Huang, Y.S., Carson, J.H., Barbarese, E., Richter, J.D. Facilitation of dendritic mRNA transport by CPEB. Genes & Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie, K., Tadauchi, T., Takizawa, P.A., Vale, R.D., Matsumoto, K., Herskowitz, I. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 2002;21:1158–1167. doi: 10.1093/emboj/21.5.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrail, S., Davidson, E.H. Logic functions of the genomic cis-regulatory code. Proc. Natl. Acad. Sci. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar, A., McDermott, K., Sorber, K., Shepard, K.A., Vale, R.D., Takizawa, P.A., DeRisi, J.L. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2005;102:18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R.P., Dowzer, C., Michaelis, C., Galova, M., Nasmyth, K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Januschke, J., Gervais, L., Dass, S., Kaltschmidt, J.A., Lopez-Schier, H., St Johnston, D., Brand, A.H., Roth, S., Guichet, A. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 2002;12:1971–1981. doi: 10.1016/s0960-9822(02)01302-7. [DOI] [PubMed] [Google Scholar]
- Johnstone, O., Lasko, P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Juan, V., Wilson, C. RNA secondary structure prediction based on free energy and phylogenetic analysis. J. Mol. Biol. 1999;289:935–947. doi: 10.1006/jmbi.1999.2801. [DOI] [PubMed] [Google Scholar]
- Kawamoto, H., Morita, T., Shimizu, A., Inada, T., Aiba, H. Implication of membrane localization of target mRNA in the action of a small RNA: Mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes & Dev. 2005;19:328–338. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha, J., Webster, P.J., Smith, J.L., Macdonald, P.M. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- Kindler, S., Wang, H., Richter, D., Tiedge, H. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis, E.H., Singer, R.H. Determinants of mRNA localization. Curr. Opin. Cell Biol. 1992;4:975–978. doi: 10.1016/0955-0674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Kislauskis, E.H., Zhu, X., Singer, R.H. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis, E.H., Zhu, X., Singer, R.H. β-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc, M., Spohr, G., Etkin, L.D. Translocation of repetitive RNA sequences with the germ plasm in Xenopus oocytes. Science. 1993;262:1712–1714. doi: 10.1126/science.7505061. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Larabell, C., Chan, A.P., Etkin, L.D. Contribution of METRO pathway localized molecules to the organization of the germ cell lineage. Mech. Dev. 1998;75:81–93. doi: 10.1016/s0925-4773(98)00086-0. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Bilinski, S., Pui-Yee Chan, A., Etkin, L.D. The targeting of Xcat2 mRNA to the germinal granules depends on a cis-acting germinal granule localization element within the 3′ UTR. Dev. Biol. 2000;217:221–229. doi: 10.1006/dbio.1999.9554. [DOI] [PubMed] [Google Scholar]
- Kress, T.L., Yoon, Y.J., Mowry, K.L. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J. Cell Biol. 2004;165:203–211. doi: 10.1083/jcb.200309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse, C., Jaedicke, A., Beaudouin, J., Bohl, F., Ferring, D., Guttler, T., Ellenberg, J., Jansen, R.P. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J. Cell Biol. 2002;159:971–982. doi: 10.1083/jcb.200207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S., Abramson, T., Munro, T.P., John, C.M., Kohrmann, M., Schnapp, B.J. UUCAC- and vera-dependent localization of VegT RNA in Xenopus oocytes. Curr. Biol. 2002;12:558–564. doi: 10.1016/s0960-9822(02)00740-6. [DOI] [PubMed] [Google Scholar]
- Lantz, V., Schedl, P. Multiple cis-acting targeting sequences are required for orb mRNA localization during Drosophila oogenesis. Mol. Cell. Biol. 1994;14:2235–2242. doi: 10.1128/mcb.14.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham V.M., Jr, Kislauskis, E.H., Singer, R.H., Ross, A.F. β-actin mRNA localization is regulated by signal transduction mechanisms. J. Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Braun, I.C., Forler, D., Izaurralde, E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, R.A., Kress, T.L., Cote, C.A., Gautreau, D., Rokop, M.E., Mowry, K.L. Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech. Dev. 2004;121:101–109. doi: 10.1016/j.mod.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Liu, G., Grant, W.M., Persky, D., Latham V.M., Jr, Singer, R.H., Condeelis, J. Interactions of elongation factor 1α with F-actin and β-actin mRNA: Implications for anchoring mRNA in cell protrusions. Mol. Biol. Cell. 2002;13:579–592. doi: 10.1091/mbc.01-03-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.M., Singer, R.H., Meng, X., Gonzalez, I., Nasmyth, K., Jansen, R.P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Long, R.M., Gu, W., Lorimer, E., Singer, R.H., Chartrand, P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.M., Gu, W., Meng, X., Gonsalvez, G., Singer, R.H., Chartrand, P. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 2001;153:307–318. doi: 10.1083/jcb.153.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes, I., Zhan, M., Lal, A., Yang, X., Gorospe, M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z., McLaren, R.S., Winters, C.A., Ralston, E. Ribosome association contributes to restricting mRNAs to the cell body of hippocampal neurons. Mol. Cell. Neurosci. 1998;12:363–375. doi: 10.1006/mcne.1998.0723. [DOI] [PubMed] [Google Scholar]
- Luck, R., Graf, S., Steger, G. ConStruct: A tool for thermodynamic controlled prediction of conserved secondary structure. Nucleic Acids Res. 1999;27:4208–4217. doi: 10.1093/nar/27.21.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald, P.M. bicoid mRNA localization signal: Phylogenetic conservation of function and RNA secondary structure. Development. 1990;110:161–171. doi: 10.1242/dev.110.1.161. [DOI] [PubMed] [Google Scholar]
- Macdonald, P.M., Struhl, G. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- Macdonald, P.M., Kerr, K. Redundant RNA recognition events in bicoid mRNA localization. RNA. 1997;3:1413–1420. [PMC free article] [PubMed] [Google Scholar]
- Macdonald, P.M., Kerr, K. Mutational analysis of an RNA recognition element that mediates localization of bicoid mRNA. Mol. Cell. Biol. 1998;18:3788–3795. doi: 10.1128/mcb.18.7.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald, P.M., Kerr, K., Smith, J.L., Leask, A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development. 1993;118:1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- Macdonald, P.M., Leask, A., Kerr, K. exl protein specifically binds BLE1, a bicoid mRNA localization element, and is required for one phase of its activity. Proc. Natl. Acad. Sci. 1995;92:10787–10791. doi: 10.1073/pnas.92.23.10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford, M., Baranes, D., Podsypanina, K., Kandel, E.R. The 3′-untranslated region of CaMKII α is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc. Natl. Acad. Sci. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem, D.R., Adams, J., Grunert, S., St Johnston, D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 2000;19:1366–1377. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, E., Morris, J.F., Richter, D. Differential subcellular mRNA targeting: Deletion of a single nucleotide prevents the transport to axons but not to dendrites of rat hypothalamic magnocellular neurons. Proc. Natl. Acad. Sci. 1995;92:4377–4381. doi: 10.1073/pnas.92.10.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, S.E., Dillon, S.T., Boswell, R.E. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes & Dev. 2001;15:2886–2899. doi: 10.1101/gad.927001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, Y., Imaizumi, K., Katayama, T., Yoneda, T., Tohyama, M. Two cis-acting elements in the 3′ untranslated region of α-CaMKII regulate its dendritic targeting. Nat. Neurosci. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- Mowry, K.L. Complex formation between stage-specific oocyte factors and a Xenopus mRNA localization element. Proc. Natl. Acad. Sci. 1996;93:14608–14613. doi: 10.1073/pnas.93.25.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry, K.L., Melton, D.A. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Munro, T.P., Magee, R.J., Kidd, G.J., Carson, J.H., Barbarese, E., Smith, L.M., Smith, R. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 1999;274:34389–34395. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- Muslimov, I.A., Santi, E., Homel, P., Perini, S., Higgins, D., Tiedge, H. RNA transport in dendrites: A cis-acting targeting element is contained within neuronal BC1 RNA. J. Neurosci. 1997;17:4722–4733. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta, C.V. A platform for compartmentalized protein synthesis: Protein translation and translocation in the ER. Curr. Opin. Cell Biol. 2002;14:412–416. doi: 10.1016/s0955-0674(02)00353-8. [DOI] [PubMed] [Google Scholar]
- Niessing, D., Huttelmaier, S., Zenklusen, D., Singer, R.H., Burley, S.K. She2p is a novel RNA-binding protein with a basic helical hairpin motif. Cell. 2004;119:491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Oleynikov, Y., Singer, R.H. RNA localization: Different zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- Olivier, C., Poirier, G., Gendron, P., Boisgontier, A., Major, F., Chartrand, P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol. Cell. Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, I.M., Gatfield, D., St Johnston, D., Izaurralde, E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- Pokrywka, N.J., Stephenson, E.C. Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev. Biol. 1995;167:363–370. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- Prakash, N., Fehr, S., Mohr, E., Richter, D. Dendritic localization of rat vasopressin mRNA: Ultrastructural analysis and mapping of targeting elements. Eur. J. Neurosci. 1997;9:523–532. doi: 10.1111/j.1460-9568.1997.tb01629.x. [DOI] [PubMed] [Google Scholar]
- Raff, J.W., Whitfield, W.G., Glover, D.M. Two distinct mechanisms localize cyclin B transcripts in syncytial Drosophila embryos. Development. 1990;110:1249–1261. doi: 10.1242/dev.110.4.1249. [DOI] [PubMed] [Google Scholar]
- Ramos, A., Grunert, S., Adams, J., Micklem, D.R., Proctor, M.R., Freund, S., Bycroft, M., St Johnston, D., Varani, G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, D.P., Blackburn, E.H. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Ross, A.F., Oleynikov, Y., Kislauskis, E.H., Taneja, K.L., Singer, R.H. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky, T.S., Kopylov, A.M., Brosius, J., Huttenhofer, A. Neuronal BC1 RNA structure: Evolutionary conversion of a tRNAAla domain into an extended stem–loop structure. RNA. 2001;7:722–730. doi: 10.1017/s1355838201002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo, P.J., Nix, B., Tsourkas, A., Bao, G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura, Y., Makabe, K.W. Identification of cis elements which direct the localization of maternal mRNAs to the posterior pole of ascidian embryos. Dev. Biol. 2002;250:128–144. doi: 10.1006/dbio.2002.0783. [DOI] [PubMed] [Google Scholar]
- Saunders, C., Cohen, R.S. The role of oocyte transcription, the 5′ UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol. Cell. 1999;3:43–54. doi: 10.1016/s1097-2765(00)80173-2. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Jaedicke, A., Du, T.G., Jansen, R.P. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol. 2006;16:1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Serano, T.L., Cohen, R.S. A small predicted stem–loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 1995;121:3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- Severt, W.L., Biber, T.U., Wu, X., Hecht, N.B., DeLorenzo, R.J., Jakoi, E.R. The suppression of testis–brain RNA binding protein and kinesin heavy chain disrupts mRNA sorting in dendrites. J. Cell Sci. 1999;112:3691–3702. doi: 10.1242/jcs.112.21.3691. [DOI] [PubMed] [Google Scholar]
- Shepard, K.A., Gerber, A.P., Jambhekar, A., Takizawa, P.A., Brown, P.O., Herschlag, D., DeRisi, J.L., Vale, R.D. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snee, M.J., Arn, E.A., Bullock, S.L., Macdonald, P.M. Recognition of the bcd mRNA localization signal in Drosophila embryos and ovaries. Mol. Cell. Biol. 2005;25:1501–1510. doi: 10.1128/MCB.25.4.1501-1510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin, W.S., Desgroseillers, L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7:1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- St Johnston, D., Beuchle, D., Nusslein-Volhard, C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Stephenson, E.C., Chao, Y.C., Fackenthal, J.D. Molecular analysis of the swallow gene of Drosophila melanogaster. Genes & Dev. 1988;2:1655–1665. doi: 10.1101/gad.2.12a.1655. [DOI] [PubMed] [Google Scholar]
- Sundell, C.L., Singer, R.H. Actin mRNA localizes in the absence of protein synthesis. J. Cell Biol. 1990;111:2397–2403. doi: 10.1083/jcb.111.6.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell, C.L., Singer, R.H. Requirement of microfilaments in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., Vale, R.D. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, P.A., Sil, A., Swedlow, J.R., Herskowitz, I., Vale, R.D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., DeRisi, J.L., Wilhelm, J.E., Vale, R.D. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Tekotte, H., Davis, I. Intracellular mRNA localization: Motors move messages. Trends Genet. 2002;18:636–642. doi: 10.1016/s0168-9525(02)02819-6. [DOI] [PubMed] [Google Scholar]
- Theurkauf, W.E., Hazelrigg, T.I. In vivo analyses of cytoplasmic transport and cytoskeletal organization during Drosophila oogenesis: Characterization of a multistep anterior localization pathway. Development. 1998;125:3655–3666. doi: 10.1242/dev.125.18.3655. [DOI] [PubMed] [Google Scholar]
- Thio, G.L., Ray, R.P., Barcelo, G., Schupbach, T. Localization of gurken RNA in Drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev. Biol. 2000;221:435–446. doi: 10.1006/dbio.2000.9690. [DOI] [PubMed] [Google Scholar]
- Tongiorgi, E., Righi, M., Cattaneo, A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J. Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule, J., Jensen, K.B., Ruggiu, M., Mele, A., Ule, A., Darnell, R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Ule, J., Jensen, K., Mele, A., Darnell, R.B. CLIP: A method for identifying protein–RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Urbinati, C.R., Gonsalvez, G.B., Aris, J.P., Long, R.M. Loc1p is required for efficient assembly and nuclear export of the 60S ribosomal subunit. Mol. Genet. Genomics. 2006;276:369–377. doi: 10.1007/s00438-006-0151-7. [DOI] [PubMed] [Google Scholar]
- Van De Bor, V., Hartswood, E., Jones, C., Finnegan, D., Davis, I. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev. Cell. 2005;9:51–62. doi: 10.1016/j.devcel.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Wagner, C., Palacios, I., Jaeger, L., St Johnston, D., Ehresmann, B., Ehresmann, C., Brunel, C. Dimerization of the 3′ UTR of bicoid mRNA involves a two-step mechanism. J. Mol. Biol. 2001;313:511–524. doi: 10.1006/jmbi.2001.5057. [DOI] [PubMed] [Google Scholar]
- Whitfield, W.G., Gonzalez, C., Sanchez-Herrero, E., Glover, D.M. Transcripts of one of two Drosophila cyclin genes become localized in pole cells during embryogenesis. Nature. 1989;338:337–340. doi: 10.1038/338337a0. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J.E., Hilton, M., Amos, Q., Henzel, W.J. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J. Cell Biol. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie, G.S., Zimyanin, V., Kirby, R., Korey, C., Francis-Lang, H., Van Vactor, D., Davis, I. Small bristles, the Drosophila ortholog of NXF-1, are essential for mRNA export throughout development. RNA. 2001;7:1781–1792. [PMC free article] [PubMed] [Google Scholar]
- Yano, T., Lopez de Quinto, S., Matsui, Y., Shevchenko, A., Shevchenko, A., Ephrussi, A. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev. Cell. 2004;6:637–648. doi: 10.1016/s1534-5807(04)00132-7. [DOI] [PubMed] [Google Scholar]
- Yisraeli, J.K., Sokol, S., Melton, D.A. A two-step model for the localization of maternal mRNA in Xenopus oocytes: Involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development. 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- Yoon, Y.J., Mowry, K.L. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. 2004;131:3035–3045. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- Zhang, H.L., Eom, T., Oleynikov, Y., Shenoy, S.M., Liebelt, D.A., Dictenberg, J.B., Singer, R.H., Bassell, G.J. Neurotrophin-induced transport of a β-actin mRNP complex increases β-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., King, M.L. Localization of Xcat-2 RNA, a putative germ plasm component, to the mitochondrial cloud in Xenopus stage I oocytes. Development. 1996a;122:2947–2953. doi: 10.1242/dev.122.9.2947. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., King, M.L. RNA transport to the vegetal cortex of Xenopus oocytes. Dev. Biol. 1996b;179:173–183. doi: 10.1006/dbio.1996.0249. [DOI] [PubMed] [Google Scholar]