Abstract
A null mutation in the gene encoding the putative E3 ubiquitin–protein ligase Mahogunin causes spongiform neurodegeneration, a recessively transmitted prion-like disease in mice. However, no substrates of Mahogunin have been identified, and the cellular role of Mahogunin is unknown. Here, we report the identification of TSG101, a key component of the endosomal sorting complex required for transport (ESCRT)-I, as a specific Mahogunin substrate. We find that Mahogunin interacts with the ubiquitin E2 variant (UEV) domain of TSG101 via its PSAP motif and that it catalyzes monoubiquitylation of TSG101 both in vivo and in vitro. Depletion of Mahogunin by small interfering RNAs in mammalian cells disrupts endosome-to-lysosome trafficking of epidermal growth factor receptor, resulting in prolonged activation of a downstream signaling cascade. Our findings support a role for Mahogunin in a proteasome-independent ubiquitylation pathway and suggest a link between dysregulation of endosomal trafficking and spongiform neurodegeneration.
INTRODUCTION
Endocytic trafficking is crucial to the function and survival of all eukaryotic cells. A key step of endocytic trafficking of cell surface receptors occurs at the early endosome, where a sorting decision has to be made between recycling back to the plasma membrane or transport to the lysosome for degradation. Cargo proteins destined for the lysosome, such as epidermal growth factor receptor (EGFR), are incorporated into vesicles that bud into the lumen of the endosome, leading to the formation of the multivesicular body (MVB). Mature MVBs (also known as late endosomes) then fuse with lysosomes, thus allowing the degradation of cargo proteins (Mullins and Bonifacino, 2001; Katzmann et al., 2002; Gruenberg et al., 2004). Endosome-to-lysosome trafficking not only controls protein degradation but also determines the potency and duration of intracellular signaling (Waterman and Yarden, 2001; Katzmann et al., 2002).
Recent studies have provided insight into the molecular events underlying endosomal sorting and trafficking of cargo to lysosomes. It has become clear that monoubiquitylation of cargo proteins serves as a sorting signal for targeting to MVB vesicles (Katzmann et al., 2002; Gruenberg et al., 2004). Current models propose that the ubiquitylated cargo proteins are recognized by endosomal ubiquitin-binding protein hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs). Hrs then binds TSG101, a key component of the endosomal sorting complex required for transport (ESCRT)-I complex, leading to the recruitment of ESCRT-I and two other ESCRT complexes, ESCRT-II and ESCRT-III, to facilitate cargo transport into the MVB (Katzmann et al., 2001; Babst et al., 2002a,b). However, it remains unclear how the activities of these endosomal trafficking machinery components are regulated in cells.
Spongiform neurodegeneration, best known as the hallmark of prion disease, is characterized by vacuolation in neurons, neuronal cell death, and astrocytosis. Although the prion protein has been extensively studied, the pathogenic mechanisms underlying spongiform neurodegeneration remain elusive. Interestingly, a recent genetic study reveals that a null mutation in the gene encoding a novel protein called Mahogunin (Mgrn1) causes a recessively transmitted form of spongiform neurodegeneration in mice that includes many features of prion disease but without accumulation of protease-resistant prion protein (He et al., 2003). Mahogunin is a ubiquitously expressed protein that contains a C3HC4-type RING finger, a motif thought to be a key determinant of the E3 ubiquitin–protein ligase activity (Phan et al., 2002; He et al., 2003). Recombinant Mahogunin protein has been shown to exhibit E2-dependent autoubiquitylation activity in vitro, suggesting that Mahogunin functions as an E3 ubiquitin–protein ligase (He et al., 2003). To date, no Mahogunin substrates or binding partners have been identified, and the cellular role of Mahogunin remains unknown.
By sequence analysis, we find a highly conserved PSAP tetrapeptide motif in the COOH-terminal region of Mahogunin. The P(S/T)AP tetrapeptide motif (where the second position can be either a serine or threonine) was initially identified in human immunodeficiency virus type 1 (HIV-1) Gag p6 late domain (Gottlinger et al., 1991; Huang et al., 1995), and subsequently it was found in several other enveloped retroviruses (for review, see Morita and Sundquist, 2004) and in a number of proteins involved in endosomal trafficking, such as Hrs, ALG-2 interacting protein1 (AIP1), Vps37B, and Target of Myb1-like 1 (TomL1) (Lu et al., 2003; von Schwedler et al., 2003; Pornillos et al., 2003; Stuchell et al., 2004; Puertollano, 2005). The P(S/T)AP motif of these proteins binds to the NH2-terminal Ubiquitin E2 Variant (UEV) domain of TSG101, and the interaction is critically involved in retrovirus budding and MVB formation (Garrus et al., 2001; Martin-Serrano et al., 2001; Pornillos et al., 2002a,b; Katzmann et al., 2002; Gruenberg and Stenmark, 2004).
In this study, we characterized the association of Mahogunin with TSG101, and we investigated the cellular function of Mahogunin. Our results reveal for the first time that Mahogunin ubiquitylates TSG101 and plays an essential role in the regulation of endosome-to-lysosome trafficking. These findings suggest that defective endosomal trafficking may be a pathogenic mechanism underlying spongiform neurodegeneration.
MATERIALS AND METHODS
Expression Constructs
Human Mahogunin (Mgrn1) full-length cDNA (KIAA0544; GenBank accession no. BAA25470) was obtained from Kazusa DNA Institute (Chiba, Japan). Human full-length TSG101 cDNA was a generous gift from Stanley N. Cohen (Stanford University, Stanford, CA). For yeast two-hybrid analyses, the full-length Mahogunin cDNA was subcloned into pACT2 or pGBKT7, and TSG101 was subcloned into pGBT9 (Clontech, Palo Alto, CA). Mahogunin and TSG101 deletion and point mutation constructs (Figure 1, B and C) were generated by QuikChange site-directed mutagenesis kits (Stratagene, La Jolla, CA). Full-length rat Hrs was subcloned into pGBKT7. Other yeast two-hybrid constructs used in this study were generous gifts of the following investigators: pGBT9-AIP1 (H. G. Gottlinger, Harvard University, Cambridge, MA), pGAD424-TomL1-GAT-VHS (R. Puertollano, National Institutes of Health, Bethesda, MD), pGBKT7-GAG (P. Spearman, Vanderbilt University, Nashville, TN). Mahogunin and TSG101 cDNAs were subcloned into pCMV-Myc and pEGFP-C2 vector (Clontech, Mountain View, CA) to produce the Myc- and green fluorescent protein (GFP)-tagged forms. The expression vectors encoding hemagglutinin (HA)-tagged wild-type ubiquitin (Ub-wt) and mutant ubiquitin (Ub-K0; all lysine residues of ubiquitin were changed to arginines) were generously provided by Ted Dawson (Johns Hopkins University, Baltimore, MD). Ubiquitin cDNAs were subcloned into pCMV-Myc to produce Myc-tagged forms.
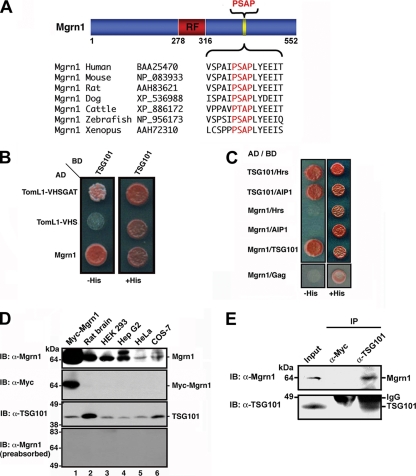
Figure 1.
Mahogunin specifically interacts with TSG101. (A) Sequence alignment of the conserved C-terminal PSAP motif in Mahogunin homologues from different species. (B) Interaction between Mahogunin and TSG101 was tested by yeast two-hybrid assays. TomL1-VHSGAT and TomL1-VHS were used as a positive and a negative control, respectively. (C) The interactions between Mahogunin and P(S/T)AP motif-containing TSG101-binding proteins were analyzed in yeast two-hybrid assays. Growth on histidine-deficient (−His) medium is indicative of an interaction. (D) Anti-Mahogunin antibody recognizes a 64-kDa protein in rat brain and several cell lines (top). Lysate from HeLa cells expressing Myc-tagged Mahogunin was included as a positive control (lane 1). The same membrane was reprobed with anti-Myc and anti-TSG101 antibodies as indicated. The specificity of the anti-Mahogunin antibody was confirmed by preabsorption with 20 μg of GST–Mahogunin fusion protein (bottom). (E) HEK293 cell lysates were subjected to immunoprecipitation with anti-TSG101 antibody or mouse IgG, followed by immunoblotting with anti-Mahogunin and anti-TSG101 antibodies. IB, immunoblot; IP, immunoprecipitation.
Antibodies
Anti-Mahogunin antibody was generated against recombinant Mahogunin protein and affinity-purified as described previously (Chin et al., 2000). Other antibodies used in this study include anti-EGFR (sc-120 and sc-03), anti-TSG101 (C-2), anti-GFP (B-2), anti-Myc (A-14), anti-Actin (C-19), and anti-Ubiquitin (P4D1 and FL76) from Santa Cruz Biotechnology (Santa Cruz, CA); anti-Ubiquitin (FK2; Affiniti Research Products, Exeter, United Kingdom), anti-lysosomal-associated membrane protein (LAMP)2 (H4B4; Developmental Studies Hybridoma Bank, Iowa City, IA); anti-early endosomal antigen (EEA)1 (BD Transduction Laboratories, Lexington, KY); anti-HA (12CA5), anti-Myc (9E10), anti-extracellular signal-regulated kinase (ERK)1/2, and anti-phospho-ERK1/2 (Cell Signaling Technology, Beverly, MA); and secondary antibodies conjugated to horseradish peroxidase, fluorescein isothiocyanate, Texas Red, or Cy5 (Jackson ImmunoResearch Laboratories, West Grove, PA).
Western Blot Analysis
Rat tissues or cultured cells were homogenized in 1% SDS, and protein extracts were subjected to SDS-PAGE and then transferred onto nitrocellulose membranes. Membranes were blocked for 1 h in Tris-buffered saline containing 0.1% Tween (TBST) and 5% nonfat dried skim milk, and then they were incubated for 3 h at 4°C with the indicated antibody. After extensive washing with TBST, membranes were incubated for 1 h with the appropriate horseradish peroxidase-conjugated secondary antibody. Antibody binding was detected by using the enhanced chemiluminescence system (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Yeast Two-Hybrid Assays
Transformation of yeast CG1948 cells with indicated constructs was performed by the lithium acetate method according to the manufacturer's instructions (Clontech). The cotransformed yeast cells were isolated by growth on defined medium lacking leucine and tryptophan. Protein–protein interaction was monitored by growth on medium lacking leucine, tryptophan, and histidine.
Cell Transfection and Immunoprecipitation
Human epitheloid carcinoma (HeLa) cells, human hepatoma (HepG2) cells, human embryonic kidney (HEK)293, and African green monkey kidney cells (COS-7) were cultured in DMEM supplemented with 10% (vol/vol) fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin in 5% CO2 at 37°C. Cells were transfected using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. For immunoprecipitation, cells were lysed at 24 h posttransfection in the lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton-X 100, 1% IGEPAL, 1 mM phenylmethylsulfonyl fluoride, 1 μg/m1 leupeptin, 1 μg/m1 pepstatin, and 1 μg/m1 bestatin). Immunoprecipitations were carried out with the indicated antibodies as described previously (Chin et al., 2001).
Glutathione S-Transferase (GST) Pull-Down Assays
For GST pull down assays, the TSG101ΔC and Mgrn1ΔN3ΔC1 fragments were subcloned into pGEX-4T-2 vector (GE Healthcare) and transformed into BL21 Escherichia coli cells (Invitrogen). Twenty micrograms of purified GST protein, GST-TSG101ΔC, or GST-Mgrn1ΔN3ΔC1 was immobilized on glutathione-agarose beads (GE Healthcare) and then incubated for 3 h at 4°C with 500 μl of COS-7 cell lysates expressing the indicated Myc-tagged Mahogunin or GFP-tagged TSG101 proteins as described previously (Chin et al., 2000). Bound proteins were resolved by SDS-PAGE and detected by Western blotting using anti-Myc antibodies.
Immunofluorescence Microscopy
For immunofluorescence microscopy, untransfected or transfected HeLa cells were fixed in 4% paraformaldehyde and processed for indirect immunofluorescence microscopy as described previously (Chin et al., 2000). Fluorescence images were acquired on a Zeiss LSM510 confocal fluorescence microscope (Carl Zeiss, Jena, Germany).
In Vitro and In Vivo Ubiquitylation Assays
In vitro ubiquitylation assays were performed using a well-established reconstitution system as described previously (Shimura et al., 2000). Briefly, immunopurified GFP-TSG101 immobilized on the agarose beads was incubated at 37°C in 30-μl reaction buffer (50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 0.6 mM dithiothreitol, and 2 mM ATP) containing 10 μg of wild-type (Ub-wt) or mutant ubiquitin (Ub-K0), 200 ng of human recombinant E1, 400 ng of E2 (Ubc5a, Ubc7, or Ubc8), and 900 ng of recombinant GST-tagged wild-type or mutant Mahogunin. After incubation for 2 h, the agarose beads were washed extensively, and the reaction products were analyzed by SDS-PAGE and immunoblotting by using anti-ubiquitin and anti-GFP antibodies.
In vivo ubiquitylation assays were performed as described previously (Wheeler et al., 2001). Briefly, lysates of HeLa cells expressing the indicated Myc-tagged Mahogunin, GFP-tagged TSG101, and wild-type HA-tagged ubiquitin (Ub-wt) or mutant (Ub-K0) ubiquitin were immunoprecipitated with anti-GFP antibody. Ubiquitylated TSG101 was detected by immunoblotting with anti-HA antibodies.
For detection of multiple monoubiquitylation of TSG101, sequential immunoprecipitations were performed in three steps with anti-GFP, anti-HA, and anti-Myc antibodies from lysates of HeLa cells coexpressing GFP-tagged TSG101, HA-tagged Ub-K0, Myc-tagged Ub-K0, and FLAG-tagged Mgrn1 as described previously (Haglund et al., 2003b). Briefly, cell lysates were subjected to immunoprecipitation with anti-GFP antibodies. Immunocomplexes bound to protein A/G-Sepharose beads were denatured and eluted by incubation at 95°C in 50 mM Tris-HCl, pH 7.5, containing 2% SDS. Ten percent of the eluate was saved for immunoblot analysis. The remaining 90% was diluted 1:20 in lysis buffer and subjected to a second immunoprecipitation with anti-HA antibodies. Precipitated proteins were again denatured, diluted, and subjected to a third immunoprecipitation with anti-Myc antibodies. The precipitated proteins from the first and the third immunoprecipitation were separated by SDS-PAGE and immunoblotted using anti-GFP, anti-HA, and anti-Myc antibodies.
Small Interfering RNA (siRNA) Oligonucleotides and Transfection
For RNA interference against human Mahogunin, we used the following 2 pairs of oligonucleotides (Dharmacon, RNA Technologies, Lafayette, CO): Mgrn1 siRNA 1, sense sequence 5′-GAACUCGGCCUAUCGCUACUU-3′ and antisense sequence 5′-PGUAGCGAUAGGCCGAGUUCUU-3′, corresponding to nucleotide 57-75 of human Mahogunin; Mgrn1 siRNA 2, sense sequence 5′-AAGAUUGACUUCUCGGAAUUU-3′ and antisense sequence 5′-PAUUCCGAGAAGUCAAUCUUUU-3′, corresponding to nucleotide 529-547 of human Mahogunin. HeLa cells were transfected with the indicated siRNAs by using the TransIT-siQUEST transfection reagent (Mirus, Madison, WI) following the manufacturer's instructions. Experiments were performed 48 h after the siRNA transfection.
EGFR Endocytic Trafficking Assays
For measurement of epidermal growth factor (EGF) endocytic trafficking, human Mgrn1 siRNA-transfected HeLa cells were starved in serum-free growth medium for 6 h at 37°C. After washing with ice-cold phosphate-buffered saline (PBS), cells were incubated with the uptake medium (DMEM containing 20 mM HEPES, pH 7.5, 1% bovine serum albumin and 3 μg/ml Alex488-EGF) for 1 h at 4°C. After washing with PBS, the cells were incubated in prewarmed growth medium at 37°C. After 0-, 30-, and 180-min incubations, cells were fixed and processed for immunofluorescence microscopy.
For quantification of the amount of intracellular Alexa488-EGF, confocal images were obtained from randomly selected fields of mock or Mgrn1 siRNA-treated cells at fixed intensity settings that were below the level of saturation. Unprocessed images were used for postimaging analysis. MetaMorph imaging system software (Molecular Devices, Sunnyvale, CA) was used to integrate the pixel intensity above background for ∼20–30 cells from three separate experiments. Statistical analysis was performed by analysis of variance (ANOVA) with a Tukey's posthoc test.
EGFR Degradation Assays
Human Mgrn1 siRNA-transfected HeLa cells were starved in serum-free medium for 6 h and then incubated in medium with or without supplemented 100 ng/ml EGF (Invitrogen) for the indicated times. The cells were then lysed in 1% SDS, and the lysates were analyzed by SDS-PAGE, followed by Western blotting using anti-EGFR antibodies. Equal loading was verified using anti-Actin antibodies. The bands intensities were quantified using NIH Image version 1.63 (http://rsb.info.nih.gov/nih-image/download.html).
ERK1/2 Phosphorylation Assays
Human Mgrn1 siRNA-transfected HeLa cells were starved in serum-free medium for 6 h and then treated with 10 ng/ml EGF for 5 min at 37°C. After washing with acidic solution (150 mM NaCl, 100 mM glycine, pH 3.0) and then with PBS, cells were chased in serum-free medium for the indicated times at 37°C. Cell lysates were analyzed by SDS-PAGE, followed by immunoblotting using anti-ERK1/2 or anti-phospho-ERK1/2 antibodies.
RESULTS
Mahogunin binds TSG101, a Key Component of the ESCRT-I Complex
The finding of an evolutionarily conserved PSAP motif in Mahogunin homologues across different species (Figure 1A) prompted us to ask whether Mahogunin interacts with TSG101. We performed yeast two-hybrid interaction assays to assess the ability of full-length Mahogunin to bind TSG101. As expected from a previous report (Puertollano, 2005), TSG101 binds to the positive control TomL1-VHS GAT but not to the negative control TomL1-VHS (Figure 1B). We found that full-length Mahogunin specifically interacts with full-length TSG101, but not with other P(S/T)AP motif-containing TSG101- binding proteins, such as Hrs (Bache et al., 2003), Alix/AIP1 (von Schwedler et al., 2003), or HIV-1 Gag protein (Garrus et al., 2001) (Figure 1C).
To verify that the Mahogunin-TSG101 interaction occurs in vivo, we generated a polyclonal anti-Mahogunin antibody against purified recombinant Mahogunin protein. Western blot analysis demonstrated that the anti-Mahogunin antibody specifically recognized the 552-amino-acid recombinant Mahogunin protein encoded by the KIAA0544 cDNA clone in transfected HeLa cells (Figure 1D, lane 1). The recombinant Mahogunin protein has an apparent molecular weight of 64 kDa, which is consistent with the predicted molecular mass. The anti-Mahogunin antibody also recognized an endogenous Mahogunin protein of 64 kDa in rat brain and several cell lines (Figure 1D, lane 2-6). In addition, the anti-Mahogunin antibody detected a second protein band with higher apparent molecular weight, which exhibits strong immunoreactivity in human hepatoma Hep G2 cells (Figure 1D, lane 4). Although its identity remains to be determined, this upper band likely represents the 576-amino acid Mahogunin isoform (GenBank accession no. NP_056061) derived from alternative splicing of human Mahogunin gene. This larger Mahogunin isoform has a 24-amino acid longer C-terminal tail compared with the 552-amino acid Mahogunin isoform (KIAA0544). Our results suggest that the 552-amino acid Mahogunin protein is the major isoform expressed in most cells, whereas the 576-amino acid Mahogunin isoform is relatively less abundant (Figure 1D). The specificity of both protein bands detected by the anti-Mahogunin antibody was confirmed by preabsorption experiments with GST-tagged recombinant Mahogunin protein (Figure 1D, bottom). Coimmunoprecipitation analysis with the anti-Mahogunin antibody revealed that endogenous Mahogunin specifically interacts with TSG101 in HEK293 cells (Figure 1E), indicating an in vivo association of Mahogunin with TSG101.
Mahogunin and TSG101 Interact in a Bimodal Manner
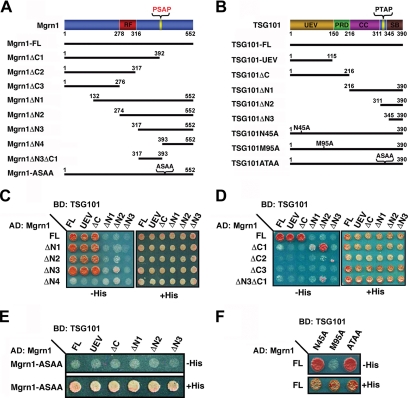
To map the binding sites mediating the interaction between Mahogunin and TSG101, we generated several deletion and site-specific mutants of Mahogunin and TSG101 (Figure 2, A and B). As shown in Figure 2C, the NH2-terminal deletions of Mahogunin (Mgrn1ΔN1-ΔN3) did not affect the ability of Mahogunin to bind full-length or the UEV domain of TSG101. In contrast, the Mahogunin deletion mutant Mgrn1ΔN4, even though it contains the COOH-terminal PSAP motif, exhibited markedly reduced binding to the full-length TSG101 and its UEV domain. These results suggest that, in addition to the COOH-terminal PSAP motif, there is another site within the amino acids 317-392 of Mahogunin that is involved in the interaction with TSG101.
Figure 2.
Identification of Mahogunin and TSG101 binding sites by deletion and site-specific mutation analyses. (A and B) Schematic representation of the human Mahogunin and TSG101 constructs used in this study. The following domains of Mahogunin and TSG101 are indicated: RF, RING finger; UEV, ubiquitin E2 variant; PRD, proline-rich domain; CC, coiled-coil; SB, steadiness box. (C and D) Yeast two-hybrid analysis of the interaction between N- or C-terminal deletion mutants of Mahogunin and various deletion mutants of TSG101. (E) Binding of the Mahogunin PSAP motif mutant, Mgrn1-ASAA, to wild-type and mutant forms of TSG101. (F) Binding of wild-type Mahogunin to site-specific mutants of TSG101. AD, activation domain; BD, binding domain.
To further understand the structural requirements for the interaction between Mahogunin and TSG101, we next assessed the interaction of the COOH-terminal deletion mutants of Mahogunin (Mgrn1ΔC1-ΔC3) with full-length and truncated forms of TSG101. As shown in Figure 2D, despite dramatically reduced interaction with full-length TSG101, Mgrn1ΔC1, which deletes the COOH-terminal PSAP motif-containing region, strongly interacted with TSG101ΔN2, but not with other TSG101 fragments. In contrast, Mgrn1ΔC2 and ΔC3 did not interact with full-length or any of the truncated forms of TSG101. We found that the residues 317-392 of Mahogunin (Mgrn1ΔN3ΔC1) were sufficient for binding TSG101ΔN2, although the binding capacity is reduced compared with Mgrn1ΔC1 (Figure 2D). Together, these data indicate that, in addition to the COOH-terminal PSAP motif and UEV domain of TSG101, the residues 317-392 of Mahogunin and the COOH-terminal residues 311-390 of TSG101 are also involved in mediating the interaction between Mahogunin and TSG101.
Previous studies have shown that the N45A and M95A point mutation in the UEV domain of TSG101 abolished the UEV-ubiquitin and UEV-P(T/S)AP interaction, respectively (Pornillos et al., 2002b; Pornillos et al., 2003). Therefore, we analyzed the effects of these mutations on the interaction between Mahogunin and TSG101 in the yeast two-hybrid interaction assays. As shown in Figure 2F, TSG101-M95A, but not TSG101-N45A mutation, selectively abolished the interaction of TSG101 with the wild-type Mahogunin. Moreover, mutation of the Mahogunin PSAP motif to ASAA abrogated the ability of Mahogunin to interact with full-length and the UEV domain of TSG101 (Figure 2E). Consistent with the results of yeast two-hybrid analysis, coimmunoprecipitation analysis showed that wild-type Mahogunin efficiently coimmunoprecipitated with GFP-tagged wild-type TSG101 (Figure 3A, lane 2), confirming that Mahogunin interacts with TSG101 in mammalian cells. The in vivo Mahogunin-TSG101 interaction was abolished by the mutation of the Mahogunin PSAP motif to ASAA (Figure 3A, lane 3) as well as by the M95A point mutation in TSG101 (Figure 3A, lane 5). These data indicate that the binding of Mahogunin to TSG101 is primarily mediated by the interaction between Mahogunin PSAP motif and TSG101 UEV domain.
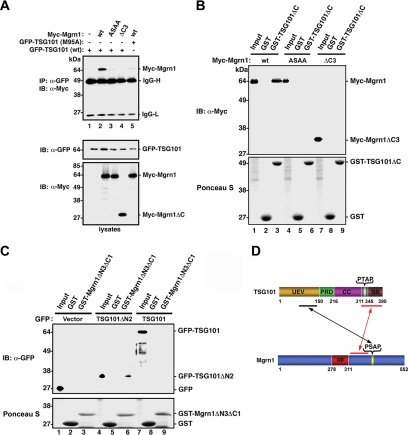
Figure 3.
In vitro and in vivo interactions between Mahogunin and TSG101. (A) Coimmunoprecipitation analysis of the interaction between Mahogunin and TSG101. HeLa cells expressing the indicated Myc-tagged Mahogunin and GFP-tagged wild-type or mutant TSG101 were immunoprecipitated with anti-GFP antibody, followed by immunoblotting with anti-Myc antibody. Cell lysates were analyzed by immunoblotting with anti-GFP (middle) and anti-Myc (bottom) antibodies. IB, immunoblot; IP, immunoprecipitation. (B) GST pull-down assays were performed by incubation of GST-fused TSG101ΔC proteins or GST with lysates from COS-7 cells expressing the indicated Myc-tagged wild-type or mutant Mahogunin. Bound proteins were detected by immunoblotting using anti-Myc antibody. (C) GST-fused Mgrn1ΔN3ΔC1 proteins or GST was incubated with lysates from COS-7 cells expressing the indicated GFP-tagged wild-type TSG101 or TSG101ΔN2. Bound proteins were detected by immunoblotting using anti-GFP antibody. (D) Model illustrating the identified binding sites of Mahogunin and TSG101.
To further characterize the Mahogunin-TSG101 interaction biochemically, we performed in vitro binding assays using GST-fusion proteins immobilized on glutathione-agarose beads. We found that GST-TSG101ΔC, which contains the UEV and PRD domain of TSG101, was able to efficiently pull down wild-type Mahogunin (Figure 3B, lane 3), but not mutant forms of Mahogunin in which the PSAP motif is mutated (Mgrn1-ASAA) or deleted (Mgrn1ΔC3) (Figure 3B, lanes 6 and 9). Moreover, in agreement with the results of yeast two-hybrid interaction analysis (Figure 2D), we found that the GST fusion protein containing residues 317-392 of Mahogunin (GST-Mgrn1ΔN3ΔC1) was able to bind the TSG101 COOH-terminal fragment containing residues 311-390 (TSG101ΔN2) (Figure 3C, lane 6), but not full-length wild-type TSG101 (Figure 3C, lane 9), suggesting that the secondary Mahogunin binding site within the residues 311-390 of TSG101 may be masked by a closed conformation of full-length TSG101. Together, our results provide evidence supporting a model that the binding between Mahogunin and TSG101 is mediated in a bimodal manner, involving a primary interaction between the Mahogunin PSAP motif and TSG101 UEV domain and a secondary interaction between the residues 317-392 of Mahogunin and the COOH-terminal residues 311-390 of TSG101 (Figure 3D).
Mahogunin Colocalizes with TSG101 and Is Recruited to Endosomes via Its Interaction with TSG101
Mahogunin is a novel protein of unknown function, and the subcellular localization of Mahogunin has never been characterized. To determine whether our anti-Mahogunin antibody could be used for immunocytochemistry, we performed immunofluorescence confocal microscopic analysis of the intracellular distribution of endogenous Mahogunin in HeLa cells by using the affinity-purified anti-Mahogunin antibody. We found immunostaining of endogenous Mahogunin in the nucleus as well as in the cytoplasm with a punctate staining pattern (Figure 4, d). The immunostaining was abolished by preabsorption with excess recombinant GST-tagged Mahogunin proteins (Figure 4, a), confirming that the Mahogunin staining is specific. Moreover, double immunofluorescence labeling analysis of HeLa cells expressing Myc-tagged Mahogunin with anti-Mahogunin and anti-Myc antibodies revealed a substantial overlap in the Mahogunin and Myc staining patterns (Figure 4, j–l), further confirming that our anti-Mahogunin antibody is able to recognize Mahogunin protein by immunostaining.
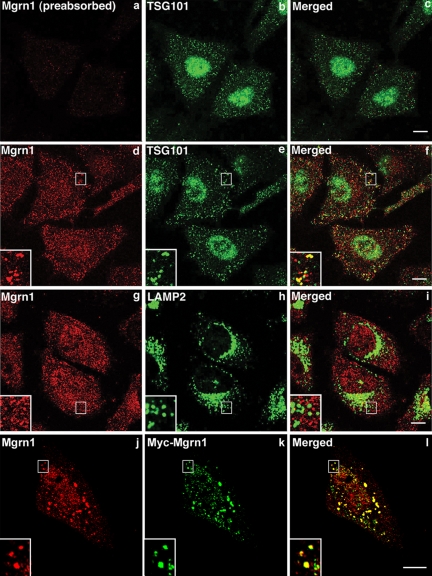
Figure 4.
Endogenous Mahogunin and TSG101 colocalize. (a–i) HeLa cells were costained with antibodies against Mahogunin and TSG101 or LAMP2 as indicated. Preabsorption with GST-tagged Mahogunin protein specifically abolished the immunostaining by anti-Mahogunin antibody (a). (j–k) HeLa cells expressing Myc-tagged Mahogunin were costained with anti-Mahogunin (j) and anti-Myc (k) antibodies. Insets show threefold magnification of the boxed areas. Bar, 10 μm.
Next, we performed double immunofluorescence labeling experiments to examine whether endogenous Mahogunin colocalizes with TSG101 in HeLa cells. We found that endogenous Mahogunin is localized to vesicular structures that are broadly distributed throughout the cytoplasm and partially colocalized with TSG101-positive endosomes (Figure 4, d–f), but not with the lysosomal marker LAMP2 (Figure 4, g–i). It was previously reported that TSG101 mainly colocalizes with LAMP1-positive late endocytic structures (Bache et al., 2003). Our finding of partial colocalization of Mahogunin with TSG101 but not with LAMP2 suggests that Mahogunin may associate with TSG101-positive endosomal intermediates in MVB formation. Interestingly, TSG101 has been shown to also localize in the nucleus and have a nonendosomal function (Xie et al., 1998; Slagsvold et al., 2006). Consistent with these studies, we found that a significant percentage of TSG101 is present in the nucleus (Figure 4, e). Similarly, we also observed nuclear staining of endogenous Mahogunin (Figure 4, d and g) as well as of Myc-tagged Mahogunin (Figure 4, k), suggesting that Mahogunin may have an additional, as yet unidentified nuclear function.
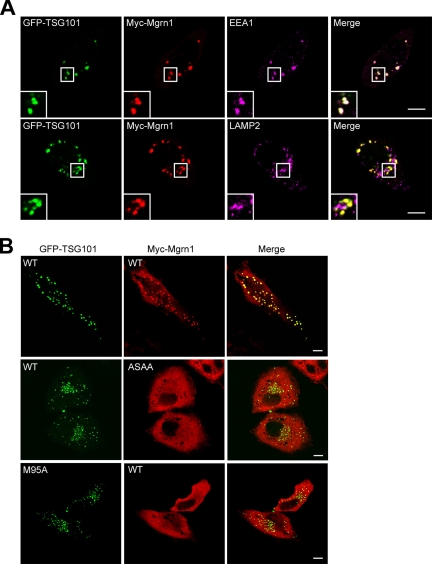
Given the observed colocalization of Mahogunin with TSG101 (Figure 4, d–f), we asked whether the endosomal localization of Mahogunin is dependent on TSG101. To test this, we examined the colocalization of transiently expressed Myc-tagged wild-type or mutant Mahogunin with GFP-tagged wild-type or mutant TSG101 in HeLa cells (Figure 5). Consistent with the previous reports (Goila-Gaur et al., 2003; Martin-Serrano et al., 2003), we found that overexpression of GFP-TSG101 induced large vesicular structures in the cytoplasm (data not shown). When coexpressed with GFP-tagged wild-type TSG101, Myc-tagged Mahogunin was recruited to the GFP-TSG101–containing vesicular structures (Figure 5A). Triple immunofluorescence labeling analysis revealed that GFP-tagged TSG101 and Myc-tagged Mahogunin colocalize on EEA1-positive early endosomes but not on LAMP2-positive late endosomes and lysosomes (Figure 5A). As shown in Figure 5B, in cells coexpressing Mahogunin and TSG101 mutants that are unable to interact, Myc-Mgrn1-ASAA and wild-type GFP-TSG101 or wild-type Myc-Mgrn1 and GFP-TSG101-M95A, there was no colocalization or recruitment of Myc-Mahogunin to GFP-TSG101–positive endosomes. These results suggest that the endosomal localization of Mahogunin is dependent on its interaction with TSG101.
Figure 5.
Colocalization of Mahogunin with TSG101 on endosomes depends on its interaction with TSG101. (A) HeLa cells coexpressing GFP-tagged TSG101 and Myc-tagged Mgrn1 were costained with anti-Myc and anti-EEA1, or anti-LAMP2 antibodies. GFP-TSG101 was directly visualized by GFP fluorescence. (B) HeLa cells coexpressing the indicated GFP-tagged wild-type or mutant TSG101 and Myc-tagged wild-type or mutant Mahogunin were immunostained with anti-Myc antibody. GFP-TSG101 was directly visualized by GFP fluorescence. Insets show twofold magnification of the boxed areas. Bar, 10 μm.
TSG101 Is a Specific Substrate of Mahogunin E3 Ubiquitin–Protein Ligase
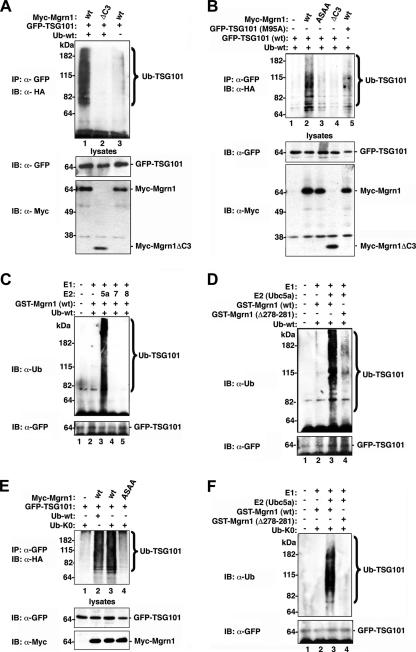
Our finding of a specific interaction between Mahogunin and TSG101 raises the possibility that TSG101 may serve as a substrate for Mahogunin E3 ubiquitin–protein ligase. To test this possibility, we first examined whether Mahogunin promotes TSG101 ubiquitylation in cells by using a well-established in vivo ubiquitylation assay (Wheeler et al., 2001). We found that expression of Myc-tagged wild-type Mahogunin (Myc-Mgrn1) resulted in robust ubiquitylation of TSG101 in HeLa cells (Figure 6A, lane 1). In contrast, expression of a Mahogunin deletion mutant (Myc-Mgrn1ΔC3) lacking the RING finger domain and COOH terminal regions had no effect on TSG101 ubiquitylation (Figure 6A, lane 2). Moreover, we found that the ability of Mahogunin to promote TSG101 ubiquitylation was dramatically reduced by the mutation of Mahogunin PSAP motif to ASAA (Figure 6B, lane 3). In addition, the Mahogunin-mediated ubiquitylation of TSG101 was also significantly decreased by the point mutation M95A of TSG101 (Figure 6B, lane 5). These results, together with the biochemical data (Figures 2 and 3), indicate that the interaction between Mahogunin and TSG101 is required for TSG101 ubiquitylation in vivo.
Figure 6.
Mahogunin ubiquitylates TSG101 in vivo and in vitro. (A and B) Lysates of HeLa cells expressing the indicated Myc-tagged Mahogunin, GFP-tagged TSG101, and wild-type HA-tagged ubiquitin (Ub-wt) were immunoprecipitated with anti-GFP antibody. Ubiquitylated TSG101 was detected by immunoblotting with anti-HA antibodies (top). Cell lysates were subjected to Western blot analysis to detect the expression of TSG101 (middle) and Mahogunin (bottom). (C and D) Immunopurified GFP-TSG101 was subjected to in vitro ubiquitylation in the presence of E1, E2 (Ubc5a, Ubc7, or Ubc8), wild-type ubiquitin (Ub-wt), and wild-type or mutant GST-tagged Mahogunin as indicated. Ubiquitylated TSG101 was detected by immunoblotting with anti-ubiquitin antibody. (E) HeLa cells were cotransfected with Myc-tagged wild-type or mutant Mahogunin, GFP-tagged wild- type TSG101, and HA-tagged wild-type (Ub-wt) or mutant (Ub-K0) ubiquitin. After immunoprecipitation, ubiquitylated GFP-TSG101 was detected by immunoblotting with anti-HA antibodies. (F) Immunopurified GFP-TSG101 were ubiquitylated in vitro in the presence of recombinant E1, E2 (Ubc5a), wild-type ubiquitin (Ub-wt) or K0 mutant ubiquitin (Ub-K0), and wild-type or mutant of GST-tagged Mahogunin, as indicated. IB, immunoblot; IP, immunoprecipitation.
Next, we performed in vitro ubiquitylation assays to directly assess the E3 ubiquitin–protein ligase activity of Mahogunin for ubiquitylating TSG101 by using recombinant GST-tagged wild-type and mutant Mahogunin proteins. As shown in Figure 6C, wild-type recombinant Mahogunin protein promoted robust ubiquitylation of TSG101 in the presence of E2-conjugating enzyme Ubc5a (Figure 6C, lane 3), but not in the presence of other E2, such as Ubc7 or Ubc8 (Figure 6C, lanes 4 and 5). These data indicate that Mahogunin uses Ubc5a as its cognate E2 ubiquitin-conjugating enzyme. We found that the E3 ligase activity of Mahogunin for ubiquitylating TSG101 was substantially reduced by the deletion of the residues 278-281 of Mahogunin containing the first two cysteines of RING finger (Figure 6D, lane 4), providing further evidence supporting the function of Mahogunin as an RING-type E3 ligase.
Mahogunin Mediates Monoubiquitylation of TSG101 at Multiple Sites
Proteins can be modified by a single ubiquitin moiety or by a polymeric ubiquitin chain. Monoubiquitylation and polyubiquitylation of substrates are associated with distinct cellular functions and fates of proteins (Haglund et al., 2003a; Hicke and Dunn, 2003). Thus, it is important to define the ubiquitylation mode of TSG101 to understand the cellular roles of Mahogunin-mediated ubiquitylation. To determine whether Mahogunin mediates monoubiquitylation or polyubiquitylation of TSG101, we used a polymerization-defective mutant of ubiquitin (Ub-K0), in which all lysine residues of ubiquitin were changed to arginines, and is therefore unable to form polyubiquitin chains. We found a virtually identical pattern of in vivo ubiquitylation of TSG101 mediated by Mahogunin in the presence of Ub-K0 compared with that obtained with wild-type ubiquitin (Figure 6E, compare lane 2 and lane 3), suggesting that Mahogunin promotes monoubiquitylation of TSG101 at multiple sites in cells. However, because of the high endogenous levels of ubiquitin, it is possible that the Ub-K0 mutant might be incorporated at the end of polyubiquitin chains formed by endogenous ubiquitin. To provide evidence arguing against this possibility, we performed in vitro ubiquitylation assays in a cell-free reconstitution system by using the Ub-K0 mutant as the sole source of ubiquitin (Figure 6F). We found that wild-type Mahogunin (Figure 6F, lane 3), but not by a RING finger mutant form of Mahogunin (Figure 6F, lane 4), promoted the ubiquitylation of TSG101 by Ub-K0. The resultant ubiquitylation pattern suggests that Mahogunin directly facilitates monoubiquitylation of TSG101 at multiple sites in vitro (Figure 6F).
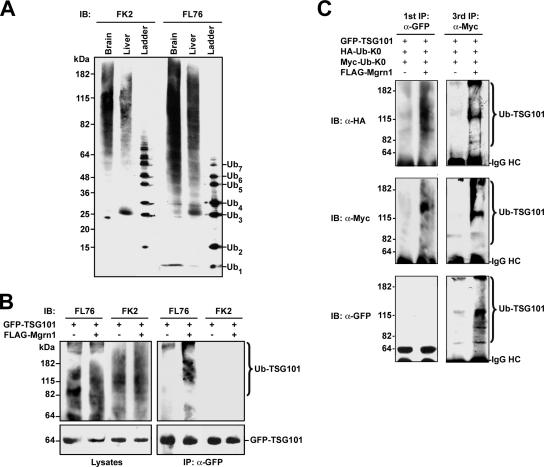
To further characterize Mahogunin-mediated ubiquitylation of TSG101 in vivo, we used anti-ubiquitin antibodies FK2 and FL76 to differentiate between monoubiquitylation and polyubiquitylation. As shown in Figure 7A, the FL76 antibody recognizes monoubiquitin as well as polyubiquitin chains, whereas the FK2 antibody only recognizes polyubiquitin chains. Using these antibodies, we found that coexpression of Mahogunin promoted modification of TSG101 by endogenous ubiquitin (Figure 7B). The ubiquitylated TSG101 was recognized by the FL76 anti-ubiquitin antibody but not by the FK2 anti-ubiquitin antibody, providing additional evidence supporting that TSG101 is monoubiquitylated at multiple sites (Figure 7B).
Figure 7.
Mahogunin mediates multiple monoubiquitylation of TSG101. (A) Homogenates from rat brain and liver were analyzed along with an ubiquitin ladder by SDS-PAGE and immunoblotting with FL76 and FK2 anti-ubiquitin antibodies as indicated. (B) HeLa cells were transfected with GFP-tagged TSG101 and FLAG-tagged Mahogunin as indicated. Cell lysates were subjected to immunoprecipitation with anti-GFP antibodies and analyzed by SDS-PAGE and immunoblotting with FL76 and FK2 anti-ubiquitin antibodies and anti-GFP antibodies. (C) HeLa cells were transfected with GFP-tagged TSG101, HA-tagged Ub-K0, Myc-tagged Ub-K0, and FLAG-tagged Mahogunin as indicated. Cell lysates were subjected to sequential immunoprecipitations with anti-GFP, anti-HA, and anti-Myc antibodies. Precipitated proteins from the first (anti-GFP) and third (anti-Myc) immunoprecipitations were analyzed by SDS-PAGE and immunoblotting with anti-GFP, anti-HA, and anti-Myc antibodies. IB, immunoblot; IP, immunoprecipitation.
To further confirm that TSG101 is modified by attachment of multiple monoubiquitin molecules instead of a polyubiquitin chain, we expressed differentially tagged, polymerization-defective mutant forms of ubiquitin, HA-tagged Ub-K0, and Myc-tagged Ub-K0, together with GFP-tagged TSG101 in HeLa cells in the absence or presence of FLAG-tagged Mahogunin. Cell lysates were subjected to sequential immunoprecipitation in three steps as described previously (Haglund et al., 2003b) by using anti-GFP, anti-HA, and anti-Myc antibodies. Before each immunoprecipitation step, samples were boiled and denatured in SDS-containing buffer to disrupt noncovalent protein–protein interactions. The sequential immunoprecipitation performed under the denaturing conditions enables the detection of multiple monoubiquitin molecules conjugated to the same TSG101 protein. Immunoblot analysis of precipitated proteins from the first and the third immunoprecipitation with anti-HA and anti-Myc antibodies revealed that TSG101 was simultaneously conjugated with both HA-tagged Ub-K0 and Myc-tagged Ub-K0 and that the incorporation of differentially tagged Ub-K0 into TSG101 was significantly increased by coexpression of Mahogunin (Figure 7C). Immunoblot analysis of the same samples with anti-GFP antibody demonstrated that a majority of GFP-TSG101 proteins precipitated by anti-GFP antibodies in the first immunoprecipitation was in the nonubiquitylated form (Figure 7C). Subsequent immunoprecipitation with anti-HA and anti-Myc antibodies enriched the ubiquitylated forms of GFP-TSG101 bearing HA-tagged Ub-K0 and Myc-tagged Ub-K0, making them detectable by immunoblotting with anti-GFP antibody (Figure 7C). The enriched ubiquitylated GFP-TSG101 species migrated as several distinct bands. The two most prominent bands detected by all three antibodies correspond to ubiquitylated forms of TSG101 with 6 and 16 monoubiquitin molecules (Figure 7C). The primary structure of TSG101 contains 27 Lys residues. Our data suggest that up to 16 of these Lys residues may be modified by Mahogunin-mediated monoubiquitylation.
Mahogunin Depletion Alters the Morphology of Endosome and Lysosome and Disrupts Eendosome-to-Lysosome Trafficking of EGFR
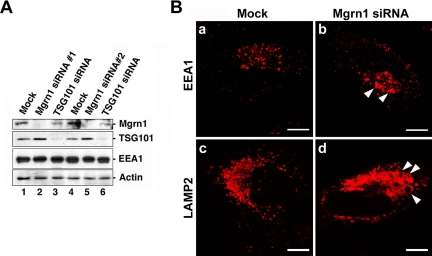
To determine the biological role of Mahogunin, we examined the cellular effects of siRNA-mediated knockdown of Mahogunin expression in HeLa cells. As shown in Figure 8A, Mgrn1 siRNA 1 and Mgrn1 siRNA 2, two distinct siRNA duplexes targeting different regions of human Mahogunin mRNA, both specifically inhibited the expression of endogenous Mahogunin, but not EEA1, TSG101, or actin (Figure 8A, lanes 2 and 5). We found that, in Mahogunin-depleted cells, EEA1-positive early endosomes were enlarged and more concentrated in the perinuclear region compared with mock-treated control cells (Figure 8B, a and b). In addition, LAMP2-positive late endosomes and lysosomes became clustered in the perinuclear region, and they often showed enlarged vacuole-like structures (Figure 8B, c and d). These changes in the endolysosomal structures induced by Mahogunin depletion are similar to the morphological changes caused by depletion of the MVB sorting machinery components, such as Hrs, Alix/AIP1, or TSG101 (Bache et al., 2003; Cabezas et al., 2005; Doyotte et al., 2005), suggesting that Mahogunin may have a role in MVB formation and/or late endosome/lysosome biogenesis.
Figure 8.
Mahogunin knockdown alters endosome and lysosome morphology. (A) HeLa cells were transfected with Mgrn1 siRNA, TSG101 siRNA, or mock treated for 48 h and then analyzed by immunoblotting with anti-Mahogunin, anti-TSG101, anti-EEA1, and anti- actin antibodies. (B) Mock (a and c) or Mgrn1 siRNA-treated (b and d) HeLa cells were immunostained using anti-EEA-1 (a and b) and anti-LAMP2 (c and d) antibodies and analyzed by immunofluorescence confocal microscopy. Bar, 10 μm.
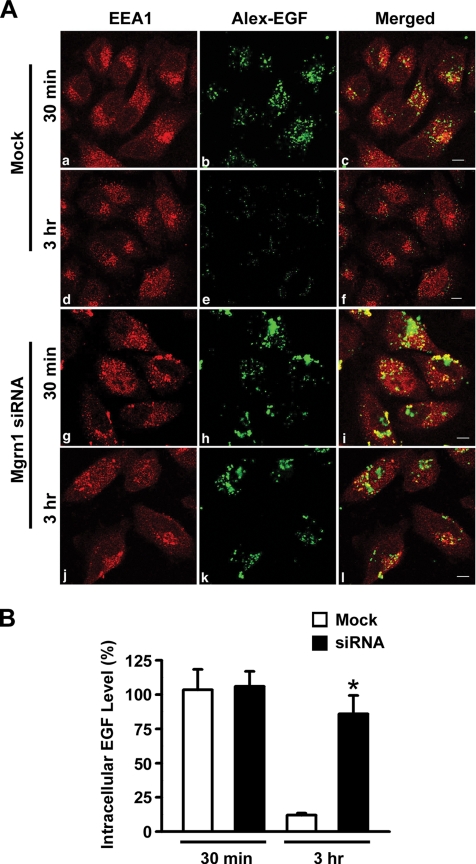
Next, we analyzed the effects of Mahogunin depletion on the endosome-to-lysosome trafficking by using a well-established EGFR trafficking assay. In this assay, mock-treated control and Mahogunin-depleted cells were first allowed to internalize Alexa488-labeled EGF (AX-EGF) and then chased for 30 min and 3 h at 37°C with unlabeled medium. The fate of the internalized EGF was monitored by confocal immunofluorescence microscopy. We found that Mahogunin-depleted cells internalized a similar amount of AX-EGF compared with mock-treated cells (Figure 9, A and B), indicating that Mahogunin depletion has little effect on ligand-induced EGFR endocytosis. After the 3-h chase, internalized AX-EGF signal almost disappeared in mock-treated cells, indicating that most of the internalized AX-EGF had been degraded (Figure 9, A and B). In comparison, Mahogunin-depleted cells retained a significant amount of the internalized AX-EGF after the same 3-h chase period (Figure 9, A and B), and the internalized EGF was accumulated in EEA1-positive enlarged endosomal structures (Figure 9A, j–l). Together, these findings indicate that the degradation of internalized EGF was impaired by Mahogunin depletion.
Figure 9.
Mahogunin knockdown disrupts endosome-to-lysosome trafficking of EGFR. (A) Mock (a–f) or Mgrn1 siRNA-transfected (g–l) HeLa cells were treated with Alexa488-EGF (green) for 1 h at 4°C. After washing, cells were incubated at 37°C for 30 min or 3 h, and then stained for EEA1 (red). Bar, 10 μm. (B) The amount of intracellular Alexa488-EGF was quantified and represented as the percentage of EGF after 30 min. The bar graph shows the results (mean ± SE) from three independent experiments. An ANOVA with a Tukey's post hoc test revealed statistically significant (p < 0.01) difference (asterisk) in the amount of Alexa488-EGF remaining in the Mock siRNA-treated cells versus that in the Mgrn1 siRNA-treated cells.
Mahogunin Is Essential for Ligand-induced EGFR Degradation and Silencing of EGFR Signaling
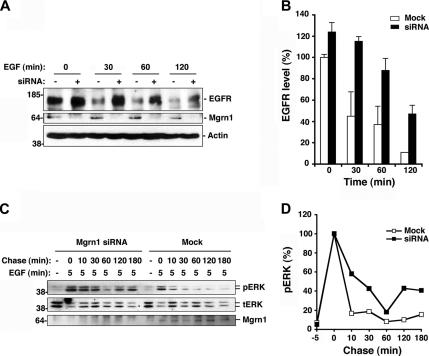
We next assessed the effects of Mahogunin depletion on the ligand-induced degradation of EGF receptors by Western blotting by using anti-EGFR antibodies. We found that depletion of Mahogunin had little effect on the steady-state levels of EGFR but that it strongly inhibited EGF-induced EGFR degradation compared with mock-treated cells (Figure 10, A and B). The extent of inhibition of EGFR degradation by Mahogunin depletion is comparable with the decrease in EGFR degradation caused by depletion of ESCRT-1 complex subunits, such as Vps37B and TSG101 (Bache et al., 2004; Doyotte et al., 2005).
Figure 10.
Depletion of Mahogunin inhibits EGFR degradation and disrupts downstream signaling of EGFR. (A) Mock or Mgrn1 siRNA-transfected HeLa cells were treated with 100 ng/ml EGF for the indicated times. Equal amounts of protein from whole cell lysates were analyzed by immunoblotting with anti-EGFR, anti-Mahogunin, and anti-actin antibodies. (B) The remaining EGFR level after EGF treatment was quantified and expressed as a percentage of the EGFR level in untreated control cells. Data are mean ± SEM (error bar) of the results from two independent experiments. (C) Mock or Mgrn1 siRNA-transfected HeLa cells were treated with 10 ng/ml EGF for 5 min followed by an acidic wash to remove cell surface EGF. Cells were then chased in serum-free medium for the indicated times and analyzed by immunoblotting using anti-phospho-ERK1/2 (top), anti-ERK 1/2 (middle), and anti-Mahogunin antibodies. (D) The level of phospho-ERK was quantified and normalized against the level of total ERK and plotted as a percentage of the relative level of phospho-ERK at chase time t = 0.
Given the critical role of the endosome-to-lysosome trafficking of EGFR in the attenuation of EGFR signaling (Waterman and Yarden, 2001), we investigated whether depletion of Mahogunin affects the mitogen-activated protein (MAP) kinase signaling pathway downstream of EGF-activated EGFR. The kinetics of EGF-dependent activation of MAP kinases (ERK1 and ERK2) was monitored over time by immunoblotting by using a phospho-specific antibody against the activated forms of ERK1/2. Consistent with previous reports (Babst et al., 2000, Bache et al., 2006), within 5 min after EGF stimulation, ERK1/2 proteins were strongly phosphorylated in both mock-treated cells (Figure 10, C and D). This onset phase of ERK1/2 phosphorylation was not significantly affected by Mahogunin depletion (Figure 10, C and D). In contrast, Mahogunin depletion dramatically altered the inactivation phase of ERK1/2 phosphorylation, leading to prolonged activation of MAP kinase signaling cascade (Figure 10, C and D). This result is similar to the sustained ERK phosphorylation observed in cells with reduced TSG101 expression (Babst et al., 2000). Together, our data demonstrate that Mahogunin is required for the degradation of EGFR and the down-regulation of the EGFR-activated MAP kinase signaling pathway.
DISCUSSION
A null mutation in the novel RING finger protein Mahogunin was recently identified as the genetic defect responsible for spongiform neurodegeneration in the mahoganoid mice. However, very little is known about the biological function of Mahogunin and how loss of Mahogunin function causes spongiform neurodegeneration. In the present study, we identified the first substrate of the Mahogunin E3 ubiquitin–protein ligase: TSG101, a key component of the endosomal sorting ESCRT machinery. Our findings reveal that Mahogunin is a novel regulator of endosome-to-lysosome trafficking of membrane cargo, such as EGFR, and they suggest that dysregulation of endosomal trafficking may be critically involved in spongiform neurodegeneration.
Mahogunin Is a Novel Endosomal E3 Ligase That Monoubiquitylates TSG101
Although Mahogunin was initially reported to contain only the RING finger domain (Phan et al., 2002; He et al., 2003), we found a TSG101 UEV domain-binding PSAP tetrapeptide motif that is evolutionarily conserved in Mahogunin (Figure 1A). We have confirmed that the PSAP motif in Mahogunin is essential for its interaction with the TSG101 UEV domain. Furthermore, our deletion analysis reveals that the Mahogunin–TSG101 association involves an additional interaction between the RING finger downstream region (residues 317-392) of Mahogunin and the COOH-terminal region (residues 311-390) of TSG101 (Figures 2 and 3). This bivalent binding mode (Figure 3D) is similar to the bimodal interaction observed between TSG101 and other P(S/T)AP motif-containing TSG101-binding proteins, such as Hrs, Alix/AIP1, and Vps37B (Lu et al., 2003; Pornillos et al., 2003; Strack et al., 2003; Bache et al., 2003; Stuchell et al., 2004). These data, together with our findings that Mahogunin does not interact with the TSG101-binding proteins Hrs, Alix/AIP1, and Gag (Figure 1C), suggest that Mahogunin may compete with these other P(S/T)AP motif-containing cellular or viral proteins for binding TSG101. Our immunofluorescence confocal microscopic analyses reveal that endogenous Mahogunin is localized to TSG101-positive endosomes but not to LAMP2-positive late endosomes and lysosomes (Figure 4). Furthermore, the recruitment of Mahogunin to TSG101-positive endosomes depends on the ability of Mahogunin to bind TSG101 (Figure 5). These results suggest that the specific interaction of Mahogunin with TSG101 occurs on TSG101-positive endosomes.
Our in vivo and in vitro ubiquitylation studies demonstrate that the binding of TSG101 to Mahogunin targets the substrate TSG101 for ubiquitylation by Mahogunin E3 ligase in cooperation with its cognate E2 enzyme Ubc5a (Figure 6). Furthermore, our results indicate that Mahogunin promotes multimonoubiquitylation instead of polyubiquitylation of TSG101 (Figures 6 and 7). It has become clear that, unlike polyubiquitylation, which targets proteins for degradation by the proteasome, monoubiquitylation acts in a manner analogous to phosphorylation for modulating protein activity, location, and interactions (Haglund et al., 2003a; Hicke and Dunn, 2003). Thus, it is conceivable that the Mahogunin-mediated monoubiquitylation of TSG101 modulates the ability of TSG101 to interact with cargo proteins and/or other components of the ESCRT sorting machinery and thereby regulates endosomal sorting and trafficking of proteins to the lysosome for degradation.
Mahogunin Functions in the Regulation of Endosome-to-Lysosome Trafficking
Consistent with the results of the biochemical characterization and subcellular localization studies of Mahogunin, our functional studies provide direct evidence that Mahogunin plays an essential role in regulation of endosome-to-lysosome trafficking. We found that siRNA-mediated depletion of Mahogunin in HeLa cells causes enlargement and clustering of EEA1-positive endosomes and LAMP2-positive late endosomes/lysosomes (Figure 8B) and inhibits the endosomal trafficking of internalized EGF–EGFR complexes to lysosomes for degradation (Figures 9 and 10, A and B). These results are strikingly similar to the phenotypes that resulted from depletion of TSG101 or other cellular proteins involved in endosomal sorting and trafficking, such as Hrs, Golgi-localized γ-ear–containing, Arf-binding 3, ubiquitin-specific processing protease Y, or Vps24 (Bache et al., 2003, 2006; Puertollano and Bonifacino, 2004; Doyotte et al., 2005; Row et al., 2006), suggesting that Mahogunin is a novel regulator of endosomal sorting and MVB formation.
Endocytic trafficking of cell surface receptors has emerged as a major mechanism for controlling the diversity, potency, and duration of intracellular signaling (Di Fiore and De Camilli, 2001). Binding of ligands (e.g., EGF) to their receptors (e.g., EGFR) not only activates the signaling pathways at the plasma membrane but also triggers the rapid endocytosis of ligand–receptor complexes. After internalization, the ligand–receptor complexes are transported to early endosomes, where they continue to bind and phosphorylate downstream effector proteins, leading to activation of signaling pathways that may be distinct from those originated at the cell surface (Ceresa and Schmid, 2000). Sorting of activated receptors into MVB internal vesicles prevents signaling of the receptors to their downstream effector proteins, thereby attenuating signal transduction (Yarden, 2001; Katzmann et al., 2002). Consistent with a role of Mahogunin in regulation of endosomal trafficking of EGFR to the MVB pathway, we found that siRNA-mediated depletion of Mahogunin not only inhibits EGFR degradation but also leads to prolonged activation of EGFR downstream MAP kinase signaling (Figure 10). Our findings indicate that Mahogunin is critically involved in the control of endosome-to-lysosome trafficking and intracellular signaling.
Recently, it was reported that TSG101 can be monoubiquitylated by Tal, an 80-kDa RING-type E3 ubiquitin–protein ligase that contains a tandem P(S/T)AP tetrapeptide motif at its C terminus (Amit et al., 2004). Interestingly, siRNA-mediated depletion of Tal causes accelerated EGFR degradation (Amit et al., 2004), an effect that is opposite to the inhibition of EGFR degradation observed when Mahogunin is depleted (Figures 9 and 10). There are at least two possibilities that could explain the different effects of Mahogunin and Tal E3 ligases on EGFR trafficking. First, Mahogunin and Tal are localized to different subcellular compartments and thereby may play distinct roles in regulation of endocytic trafficking. Tal, also known as RING Finger-Leucine-Rich Repeat Containing Protein, was originally identified as a cytosolic protein that regulates cell adhesion in PC12 cells (Li et al., 2003). The subcellular localization of endogenous Tal protein remains virtually unknown. Subcellular fraction analysis reveals the presence of exogenously expressed Tal in the nuclear and cytosol fractions but not in the membrane fraction (Li et al., 2003). Immunostaining studies show that a majority of exogenously expressed Tal is diffusely distributed in the cytoplasm and a significant fraction of Tal is localized to a “submembranal ring” in close apposition with the plasma membrane (Amit et al., 2004). Tal-mediated ubiquitylation of TSG101 has been proposed to change the conformation of TSG101 from a membrane-bound active form to an inactive soluble form (Amit et al., 2004). In contrast, our data indicate that endogenous as well as exogenous Mahogunin colocalizes with TSG101 on early endosomes (Figures 4 and 5) and suggest that Mahogunin-mediated ubiquitylation of TSG101 plays an active role in facilitating endosome-to-lysosome trafficking of EGFR (Figures 8–10). Second, TSG101 contains 27 lysine residues that can potentially be ubiquitylated. Mahogunin and Tal may mediate ubiquitylation of TSG101 at distinct lysine residues, leading to different functional consequences. Interestingly, a recent study shows that coexpression of Tal promotes the degradation of wild-type TSG101 but not mutant TSG101 in which the lysines in the VPS28-binding region have been mutated (McDonald and Martin-Serrano, 2006). These data suggest that Tal mediates polyubiquitylation of the lysine residues in the VPS28-binding region of TSG101, leading to subsequent degradation of TSG101. The Tal-induced degradation of TSG101 could explain the observed effect of Tal depletion on EGFR degradation. In contrast, coexpression of Mahogunin has no effect on the degradation of TSG101 (Figures 3A and 6E; additional data not shown).
Dysregulation of Endosome-to-Lysosome Trafficking May be a Pathogenic Mechanism Underlying Spongiform Neurodegeneration
Although the mahoganoid mice share many neuropathological features with prion disease, there is no accumulation of protease-resistant prion protein in these mice (He et al., 2003). The lack of protein aggregates in the mahoganoid mice indicates that spongiform neurodegeneration can occur without protein aggregation. The identification of a null mutation in the E3 ubiquitin–protein ligase Mahogunin as the genetic defect for the mahoganoid mice (He et al., 2003) points to a direct link between aberrant ubiquitylation and spongiform neurodegeneration. The findings reported in this study support a role for Mahogunin in a proteasome-independent ubiquitylation pathway and suggest a novel mechanism by which defective ubiquitylation could lead to spongiform neurodegeneration.
Our study reveals that Mahogunin is a ubiquitously expressed E3 ligase that monoubiquitylates TSG101 and regulates endosome-to-lysosome trafficking in mammalian cells. Thus, a null mutation in Mahogunin would cause the loss of Mahogunin function and consequent dysregulation of endosome-to-lysosome trafficking in many cell types. The spongiform neurodegeneration phenotype of the Mahogunin null mouse suggests that, compared with other cell types, neurons are particularly vulnerable to defects in the endosomal–lysosomal system. Consistent with this notion, aberrant endosomal trafficking has recently been implicated in a number of neurodegenerative diseases. For example, endosomal abnormalities are among the earliest pathological features of Alzheimer's disease and Down syndrome (Nixon, 2005). Mutations in endosomal fusion regulators Rab5 guanine nucleotide exchange factor alsin (Yang et al., 2001) and Vps54 (Schmitt-John et al., 2005) cause motor neuron diseases in mammals. The recent finding that mutations in the ESCRT-III complex subunit CHMP2B cause frontotemporal dementia (Skibinski et al., 2005) provides a direct link between the dysfunction of the endosomal sorting machinery and neurodegeneration. Furthermore, overexpression or knockout of the ESCRT sorting machinery components has been shown to affect neuronal cell viability. For example, overexpression of Alix/AIP1, an ESCRT-II complex subunit that interacts with TSG101, induced cell death in both postmitotic cerebellar neurons and chicken neuroepithelial cells (Trioulier et al., 2004; Mahul-Mellier et al., 2006). In mice, targeted disruption of the gene encoding the Hrs-binding protein signal transducing adaptor molecule (STAM) 1 or associated molecule with the Src homology 3 domain of STAM (AMSH) results in extensive neuronal cell death in the hippocampus and cerebral cortex (Ishii et al., 2001; Yamada et al., 2001). These different lines of evidence indicate that dysregulation of endosome-to-lysosome trafficking may be a common pathogenic mechanism in a number of neurodegenerative diseases, including spongiform neurodegeneration. Further elucidation of the pathogenic pathway by which the endosomal–lysosomal system dysfunction leads to neurodegeneration should facilitate the development of novel rational therapies for treating prion disease and other neurodegenerative disorders.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants NS-047575, NS-047199, NS-050650, and AG-021489.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0787) on January 17, 2007.
REFERENCES
- Amit I., et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18:1737–1752. doi: 10.1101/gad.294904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Katzmann D. T., Estepa-Sabal E. J., Meerloo T., Emr S. D. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002a;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002b;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Babst M., Odorizzi G., Estepa E. J., Emr S. D. Mammalian tumor susceptibility gene 101(TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- Bache K. G., Brech A., Mehlum A., Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K. G., Slagsvold T., Cabezas A., Rosendal K. R., Raiborg C., Stenmark H. The growth-regulatory protein HCRP1/hVps37A is a subunit of mammalian ESCRT-1 and mediates receptor down-regulation. Mol. Biol. Cell. 2004;15:4337–4346. doi: 10.1091/mbc.E04-03-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K. G., Stuffers S., Malerod L., Slagsvold T., Raiborg C., Lechardeur D., Walchli S., Lukacs G. L., Brech A., Stenmark H. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell. 2006;17:2513–2523. doi: 10.1091/mbc.E05-10-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas A., Bache K. G., Brech A., Stenmark H. Alix regulates cortical actin and spatial distribution of endosomes. J. Cell Sci. 2005;118:2625–2635. doi: 10.1242/jcs.02382. [DOI] [PubMed] [Google Scholar]
- Ceresa B. P., Schmid S. L. Regulation of signal transduction by endocytosis. Curr. Opin. Cell Biol. 2000;12:204–210. doi: 10.1016/s0955-0674(99)00077-0. [DOI] [PubMed] [Google Scholar]
- Chin L. S., Nugent R. D., Raynor M. C., Vavalle J. P., Li L. SNIP, a novel SNAP-25-interacting protein implicated in regulated exocytosis. J. Biol. Chem. 2000;275:1191–1200. doi: 10.1074/jbc.275.2.1191. [DOI] [PubMed] [Google Scholar]
- Chin L. S., Raynor M. C., Wei X., Chen H. Q., Li L. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J. Biol. Chem. 2001;276:7069–7078. doi: 10.1074/jbc.M004129200. [DOI] [PubMed] [Google Scholar]
- Di Fiore P. P., De Camilli P. Endocytosis and signaling. an inseparable partnership. Cell. 2001;106:1–4. doi: 10.1016/s0092-8674(01)00428-7. [DOI] [PubMed] [Google Scholar]
- Doyotte A., Russell R. G., Hopkins C. R., Woodman P. G. Depletion of TSG101 forms a mammalian Class E compartment: a multicisternal early endosome with multiple sorting defects. J. Cell Sci. 2005;118:3003–3017. doi: 10.1242/jcs.02421. [DOI] [PubMed] [Google Scholar]
- Garrus J. E., et al. Tsg101 and vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Goila-Gaur R., Demirov D. G., Orenstein J. M., Ono A., Freed E. O. Defect in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol. 2003;77:6507–6519. doi: 10.1128/JVI.77.11.6507-6519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger H. G., Dorfman T., Sodoski J. G., Haseltine W. A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J., Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Haglund K., Di Fiore P. P., Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 2003a;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003b;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- He L., Lu X. Y., Jolly A. F., Eldbridge A. G., Watson A. G., Jackson P. K., Barsh G. S., Gunn T. M. Spongiform degeneration in mahoganoid mutant mice. Science. 2003;299:710–712. doi: 10.1126/science.1079694. [DOI] [PubMed] [Google Scholar]
- Hicke L., Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin binding proteins. Ann. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Huang M., Orenstein J. M., Martin M. A., Freed E. O. p6 Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N., Owada Y., Yamada M., Miura S., Murata K., Asao H., Kondo H., Sugamura K. Loss of neurons in the hippocampus and cerebral cortex of AMSH-deficient mice. Mol. Cell Biol. 2001;21:8626–8637. doi: 10.1128/MCB.21.24.8626-8637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Babst M., Emr S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–156. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Odorizzi G., Emr S. D. Receptor down regulation and multivesicular body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Li B., Su Y., Ryder J., Yan L., Na S., Ni B. RIFLE: a novel ring zinc finger-leucine-rich repeat containing protein, regulates select cell adhesion molecules in PC12 cells. J. Cell Biochem. 2003;90:1224–1241. doi: 10.1002/jcb.10674. [DOI] [PubMed] [Google Scholar]
- Lu Q., Hope L. W., Brasch M., Reinhard C., Cohen S. N. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahul-Mellier A. L., Hemming F. J., Blot B., Fraboulet S., Sadoul R. Alix, making a link between apoptosis-linked gene-2, the endosomal sorting complexes required for transport, and neuronal death in vivo. J. Neurosci. 2006;26:542–549. doi: 10.1523/JNEUROSCI.3069-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P. D. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 2001;7:1278–1280. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P. D. Role of ESCRT-1 in retroviral budding. J. Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B., Martin-Serrano J. VPS28 acts to stabilize TSG101 by preventing its degradation by TSG101-associated ligase. Proceedings of the 13th Conference on Retroviruses and Opportunistic Infections; 2006 February 5–8; Denver, CO. 2006. Abstr. 243. [Google Scholar]
- Morita E., Sundquist W. I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Mullins C., Bonifacino J. S. The molecular machinery for lysosome biogenesis. Bioessays. 2001;23:333–343. doi: 10.1002/bies.1048. [DOI] [PubMed] [Google Scholar]
- Nixon R. A. Endosome function and dysfunction in Alzheimer's disease and other neurodegenerative diseases. Neurobiol. Aging. 2005;26:373–382. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Phan L. K., Lin F., LeDuc C. A., Chung W. K., Leibel R. L. The mouse mahoganoid coat color mutation disrupts a novel C3HC4 RING domain protein. J. Clin. Investig. 2002;110:1449–1459. doi: 10.1172/JCI16131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O., Alam S. L., Davis D. R., Sundquist W. I. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 2002a;11:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- Pornillos O., Alam S. L., Rich R. L., Myszka D. R., Sundquist W. I. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002b;15:2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O., Higginson D. S., Stray K. M., Fisher R. D., Garrus J. E., Payne M., He G. P., Wang H. E., Morham S. G., Sundquist W. I. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 2003;162:425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Bonifacino J. S. Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- Puertollano R. Interaction of Tom1L1 with the multivesicular body sorting machinery. J. Biol. Chem. 2005;280:9258–9264. doi: 10.1074/jbc.M412481200. [DOI] [PubMed] [Google Scholar]
- Row P. E., Prior I. A., McCullough J., Clague M. J., Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and essential for receptor down regulation. J. Biol. Chem. 2006;281:12618–11262. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- Schmitt-John T., et al. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat. Genet. 2005;11:1213–1215. doi: 10.1038/ng1661. [DOI] [PubMed] [Google Scholar]
- Shimura H., et al. Familial Parkinson's disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- Skibinski G., et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- Slagsvold T., Pattni K., Malerod L., Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Strack B., Calistri A., Craig S., Popova E., Gottlinger H. G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Stuchell M. D., Garrus J. E., Muller B., Stray K. M., Ghaffarian S., McKinnon R., Krausslich H. G., Morham S. G., Sundquist W. I. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 2004;279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- Trioulier Y., Torch S., Blot B., Cristina N., Chatellard-Causse C., Verna J. M., Sadoul R. Alix, a protein regulating endosomal trafficking, is involved in neuronal death. J. Biol. Chem. 2004;279:2046–2052. doi: 10.1074/jbc.M309243200. [DOI] [PubMed] [Google Scholar]
- von Schwedler U. K., et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Waterman H., Yarden Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 2001;16:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- Wheeler T. C., Chin L. S., Li Y., Roudabush F. L., Li L. Regulation of synaptophysin degradation by mammalian homologues of seven in absentia. J. Biol. Chem. 2001;277:10273–10282. doi: 10.1074/jbc.M107857200. [DOI] [PubMed] [Google Scholar]
- Xie W., Li L., Cohen S. N. Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc. Natl. Acad. Sci. USA. 1998;95:1595–1600. doi: 10.1073/pnas.95.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., et al. Loss of hippocampal CA3 pyramidal neurons in mice lacking STAM1. Mol. Cell Biol. 2001;21:3807–3819. doi: 10.1128/MCB.21.11.3807-3819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat. Genet. 2001;29:160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- Yarden Y. The EGFR family and its ligands in human cancer. signaling mechanisms and therapeutic opportunities. Eur. J. Cancer. 2001;37(Suppl. 4):S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]