Abstract
Plant mechanical strength is an important agronomic trait. To understand the molecular mechanism that controls the plant mechanical strength of crops, we characterized the classic rice mutant brittle culm1 (bc1) and isolated BC1 using a map-based cloning approach. BC1, which encodes a COBRA-like protein, is expressed mainly in developing sclerenchyma cells and in vascular bundles of rice. In these types of cells, mutations in BC1 cause not only a reduction in cell wall thickness and cellulose content but also an increase in lignin level, suggesting that BC1, a gene that controls the mechanical strength of monocots, plays an important role in the biosynthesis of the cell walls of mechanical tissues.
INTRODUCTION
The plant cell wall, a strong fibrillar network that provides mechanical support to cells, tissues, and the entire plant body, is a highly organized composite that may contain many different polysaccharides, aromatic substances, and proteins. The structure and composition of plant cell walls are ideally suited to the functions they perform. For example, parenchyma cells, which consist of primary walls, provide the main structural support in growing regions of the plant body. Sclerenchyma cells, which have both primary walls and thick secondary walls, provide the major mechanical support in nonelongating regions of the plant body (Carpita and McCann, 2000). Cellulose usually constitutes ∼20 to 30% of the dry weight of the primary walls and ∼40 to 90% of the secondary walls, depending on the cell type (Taylor et al., 1999). In some cells, lignin may be incorporated into the cell wall, enhancing its mechanical strength. Despite extensive descriptions of the chemical and physical structures of cell walls, the mechanisms that regulate the deposition of cell wall materials and that determine cell wall strength remain to be elucidated.
To understand the mechanisms that regulate the mechanical strength of the plant body and the biosynthesis of plant cell walls, mutants defective in stem strength have been isolated and characterized. For example, the barley brittle culm (bc) mutants were first described based on the physical properties of the culms, which have an ∼80% reduction in the amount of cellulose and a twofold decrease in breaking strength compared with those of wild-type plants (Kokubo et al., 1989, 1991), indicating that cellulose content is related to the mechanical strength of the plant body. In Arabidopsis, mutants with reduced stem strength have been identified and some corresponding genes have been cloned and characterized. The irregular xylem mutants (irx1 to irx3) show a cellulose defect in secondary walls, and the stiffness of mature stems is decreased (Turner and Somerville, 1997). The IRX1 and IRX3 genes encode the catalytic subunits of the cellulose synthase isoforms CesA7 and CesA8 (Taylor et al., 1999, 2000), which are essential for the production of cellulose in the cell. The irx4 mutant is defective in a cinnamoyl-CoA reductase, resulting in a reduction in the lignin content of secondary walls and a failure to retain an upright growth habit (Jones et al., 2001). Moreover, INTERFASCICULAR FIBERLESSI, a gene that regulates interfascicular fiber differentiation and stem strength in Arabidopsis, was identified as a member of plant homeodomain/Leu zipper family (Zhong and Ye, 1999; Ratcliffe et al., 2000). FRAGILE FIBER1 (FRA1), a kinesin-like protein, is essential for the oriented deposition of cellulose microfibrils and cell wall strength (Zhong et al., 2002), and FRA2 encodes a katanin-like protein that regulates fiber cell length and wall thickness (Burk et al., 2001; Burk and Ye, 2002). These studies indicate that the genes involved in the biosynthesis and/or modifications of cell walls, especially secondary walls, are essential for plant mechanical strength and that the molecular mechanisms that determine plant mechanical strength are complex.
Recent studies suggest that covalent lipid modification is a major means of regulating the activity as well as the cellular localization of a protein. Glycosylphosphatidylinositol (GPI), for example, has been posited as a common means of anchoring membrane proteins in mammalian, yeast, and parasitic cells (Udenfriend and Kodukula, 1995). The addition of the GPI moiety is performed in the endoplasmic reticulum and implies the cleavage of a hydrophobic C-terminal peptide and the subsequent linkage of a preassembled GPI anchor via an amide bond onto the last amino acid residue after the cleavage, called the GPI attachment or ω-site. GPI-anchored proteins are required for many important physiological functions, including signal transduction (Peles et al., 1997), parasitic evasion of host immune responses (Ferguson et al., 1994), and cell-to-cell recognition and nutrient uptake (Rothberg et al., 1990). In plants, several GPI-anchored proteins have been documented (Schultz et al., 1998; Sherrier et al., 1999; Schindelman et al., 2001; Sedbrook et al., 2002). However, only COBRA, SKU5, and SOS5, which encode the GPI-anchored proteins in Arabidopsis, have been functionally studied recently (Schindelman et al., 2001; Sedbrook et al., 2002; Shi et al., 2003). The mutation in COBRA affects the orientation of cell expansion and the cellulose content of the cell walls in the root elongation zone, implying a role for COBRA in cell wall maintenance and/or biosynthesis. COBRA belongs to a multigene family consisting of 12 members in Arabidopsis, all of which are predicted to encode putative GPI-anchored proteins; they are designated COBRA-like (COBL) proteins (Roudier et al., 2002). However, the functions of these COBL proteins in plants are poorly understood.
In rice, at least six bc mutants (bc1 to bc6) have been reported, and some of them were mapped to different chromosomes using classic or molecular approaches (Kinoshita, 1995). Recently, a bc1 allelic mutant, bc1-2 (originally named fp1), was isolated from a rice indica variety based on its brittle culms, leaves, and leaf sheaths (Qian et al., 2001). Here, we report the in-depth characterization of the rice bc1 mutants and the map-based cloning of the BC1 gene as well as its spatial and temporal expression patterns. Our findings indicate that BC1 is a COBRA-like protein that functions in regulating the biosynthesis of secondary cell walls to provide the main mechanical strength for rice plants.
RESULTS
The bc1 Mutant Has a Reduction in Mechanical Properties
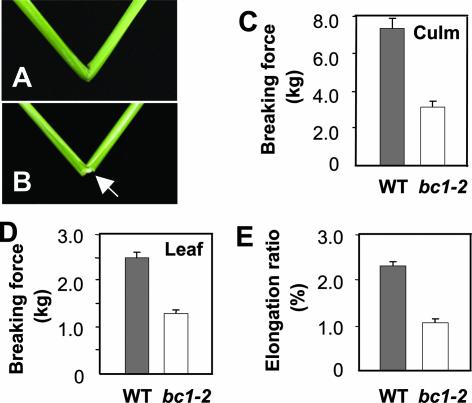
To understand the mechanism that controls plant mechanical strength in cereal crops, we performed an in-depth analysis of a classic rice bc1 mutant and its recently isolated allele, bc1-2 (Qian et al., 2001). Although morphologically, bc1-2 mutant plants are indistinguishable from wild-type plants (data not shown), they have brittle culms and leaves that can be broken easily by bending (Figures 1A and 1B). To accurately describe this phenotype, we quantitatively compared the breaking forces of bc1-2 and wild-type plants, which define the forces required to break the segments of culms or leaves, and the elongation ratios of the leaves, which reflect the elasticity of plant tissues. As shown in Figures 1C and 1D, the forces required to break the mutant culms and leaves were decreased to 43 and 52% of those required for the wild type. The elongation ratio of the bc1-2 leaves also was decreased by ∼50% compared with that of the wild-type plants (Figure 1E). The striking decreases in the breaking forces and the elongation ratio of the bc1-2 mutant indicate that the mutations in bc1 affect both mechanical strength and the elasticity that enables plant organs or cells to maintain their proper shapes and positions.
Figure 1.
Phenotypes and Physical Properties of Wild-Type and bc1-2 Mutant Plants.
(A) A wild-type culm.
(B) An easily broken bc1-2 culm, as indicated by the arrow.
(C) The force required to break culms.
(D) The force required to break flag leaves.
(E) The elongation ratios of leaves.
Error bars were obtained from 10 measurements for (C) and from 22 measurements for (D) and (E). WT, wild type.
bc1 Is Defective in Cell Walls of Mechanical Tissues
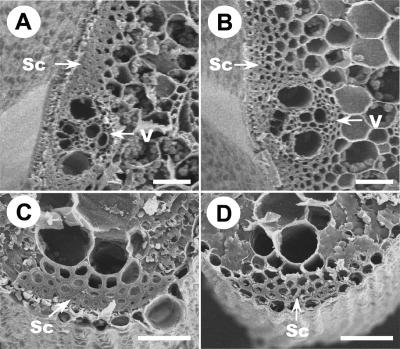
Reduction in the mechanical strength of culms and leaves may reflect alterations in cell wall structure, composition, or fiber length. Therefore, we examined cell wall morphology with scanning electron microscopy. In wild-type rice, several layers of mechanical cells, especially those around the peripheral vascular tissues and under the epidermal layer in culms and leaf veins, provide the mechanical support for the plants. Scanning electron microscopy observations revealed that the wild-type sclerenchyma cell walls were heavily thickened and that the cells were nearly completely filled up at the mature stages of culms and leaves (Figures 2A and 2C), in striking contrast to those of bc1-2 mutant plants (Figures 2B and 2D). However, no differences in cell length and width were found between the bc1-2 and wild-type plants (data not shown). These results suggest that reduction in the mechanical strength of bc1-2 plants very likely resulted from a defect in the cell wall thickening of mechanical tissues, such as sclerenchyma cells, in the mutant plants.
Figure 2.
Scanning Electron Micrographs Showing the Differences between Sclerenchyma Cells and Vascular Bundles in Wild-Type and bc1-2 Plants.
(A) Cross-section of a wild-type culm.
(B) Cross-section of a bc1-2 mutant culm.
(C) Cross-section of a wild-type leaf.
(D) Cross-section of a bc1-2 leaf.
Sc, sclerenchyma cells; V, vascular bundles. Bars = 12.5 μm.
The bc1 Plant Has an Altered Cell Wall Composition
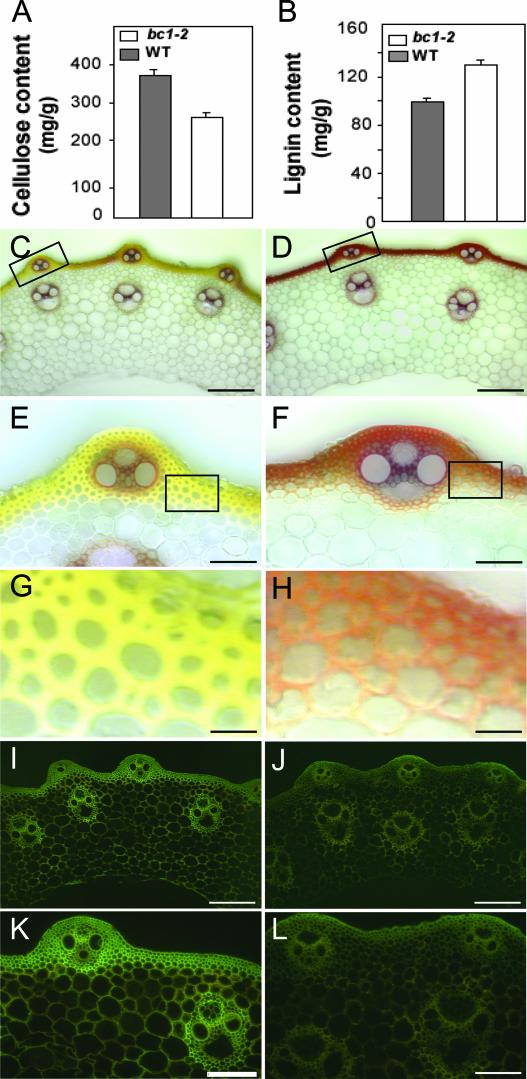
To determine whether the cellular phenotype and the reduced mechanical strength in bc1-2 mutant plants result from altered cellulose biosynthesis, we compared the crystalline cellulose contents of mutant and wild-type plants, because cellulose is the major component of plant cell walls. As shown in Figure 3A, the amount of cellulose in bc1-2 culms was reduced to ∼70% of that in the wild type, suggesting that BC1 may directly or indirectly play an important role in cellulose biosynthesis. In some cells, cell walls also may contain a proportion of lignin that contributes to mechanical strength. Therefore, the lignin content of the bc1-2 mutant plants also was assayed. As shown in Figure 3B, the Klason lignin of the bc1-2 culms increased by ∼30% compared with that of the wild-type culms, indicating that plant cells may have a sophisticated mechanism to balance the contents between cellulose and lignin.
Figure 3.
Measurement and Staining of Cellulose and Lignin in Wild-Type and bc1-2 Plants.
(A) and (B) Cellulose (A) and lignin (B) contents (milligrams per gram of total cell wall residues) of the culm segments from wild-type (WT) and bc1-2 plants. The error bars were obtained from five measurements.
(C) to (H) Wiesner's staining of the transverse culm sections of wild-type (C) and bc1-2 (D) plants, and magnified sections ([E] and [F]), showing the increased level of lignin in the walls of sclerenchyma cells and vascular bundles in the mutant culm. (G) and (H) Magnified sections of (E) and (F), showing the irregular thin and defective walls of sclerenchyma cells in the mutant culm.
(I) to (L) Calcofluor staining of the transverse culm sections of wild-type (I) and bc1-2 (J) plants, and magnified sections (K) and (L), showing the decreased level of cellulose in the cell walls of sclerenchyma cells and vascular bundles in the mutant culm.
Bars = 160 μm in (C), (D), (I), and (J), 40 μm in (E) and (F), 10 μm in (G) and (H), and 80 μm in (K) and (L).
To determine whether the alterations of cellulose and lignin are localized in particular cells, transverse sections of the culms of wild-type and mutant plants were histochemically stained with Wiesner and calcofluor solutions. Wiesner stain is known to react with cinnamaldehyde residues in lignin, and the color intensity approximately reflects the total lignin content. The color differences in mechanical tissues, especially in the sclerenchyma cells below the epidermis, between wild-type (Figures 3C and 3E) and mutant (Figures 3D and 3F) plants were clear, indicating an apparent increase in lignin quantity in mutant plants. On the other hand, calcofluor stains cellulose, callose, and other β-glucans. As shown in Figures 3I to 3L, much stronger fluorescent signals were observed in the sclerenchyma cells and vascular bundles in the wild type (Figures 3I and 3K) than in the bc1-2 mutant (Figures 3J and 3L), demonstrating a significantly high level of ordered cellulose in the mechanical tissues and cells in wild-type plants. In addition, compared with the smooth and thickened walls of sclerenchyma cells of the wild type (Figure 3G), those of mutant plants were irregularly thin and uneven (Figure 3H). This finding is consistent with the scanning electron microscopy observations (Figure 2) and indicates that the bc1 mutant is deficient mainly in the secondary cell walls.
Therefore, we further compared the cell wall monosaccharide composition between bc1-2 mutant and wild-type culms by gas chromatography. As shown in Table 1, glucose content was reduced significantly in the bc1-2 mutant, consistent with the results obtained from cellulose assays. By contrast, the contents of xylose and arabinose were increased in the bc1-2 mutant, whereas other analyzed sugars were not altered significantly. Because glucose and xylose are the two major sugars that constitute cellulose and hemicellulose, respectively, in the secondary cell walls, it is very likely that the bc1 mutation mainly affects the biosynthesis of secondary cell walls, which explains the histochemical staining observation (Figure 3) and the difference in mechanical strength between wild-type and bc1-2 plants (Figure 1).
Table 1.
Comparison of the Contents of Cell Wall Sugars between bc1-2 and Wild-Type Culms
| Sugar | bc1-2 Mutanta | Wild Typea | Difference | P Value b |
|---|---|---|---|---|
| Glucose | 433 ± 11.1 | 612 ± 15.9 | −179 | <0.001 |
| Xylose | 149 ± 5.64 | 107 ± 3.94 | 42 | <0.001 |
| Arabinose | 21.3 ± 0.94 | 18.0 ± 0.83 | 3.3 | 0.022 |
| Galactose | 11.9 ± 0.60 | 11.5 ± 0.69 | 0.4 | 0.693 |
| Mannose | 1.74 ± 0.07 | 1.78 ± 0.08 | −0.04 | 0.696 |
| Rhamnose | 3.66 ± 0.21 | 3.58 ± 0.21 | 0.08 | 0.786 |
The sugar contents of cell walls of bc1-2 mutant and wild-type culms are given as means ± se of eight independent assays. Each wall component was calculated as milligrams per gram of total cell wall residues.
Determined using Student's two-sample t test.
Positional Cloning of BC1
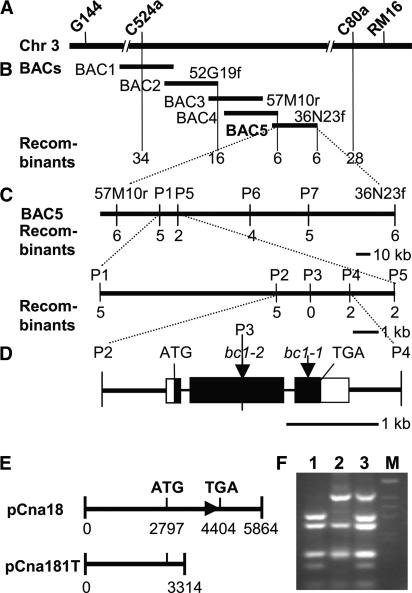
Our previous studies placed the BC1 gene in an interval between the RM16 and C524a markers in the centromeric region of chromosome 3 (Qian et al., 2001). To fine-map the BC1 locus, we generated a large F2 mapping population derived from a cross between bc1-2 and C-Bao, a polymorphic rice japonica variety. Of ∼30,000 F2 plants, 7068 segregants showing the bc1-2 mutant phenotype were used for fine-mapping and the BC1 gene was located between the two cleaved amplified polymorphic sequence (CAPS) markers C524a and C80a (Figure 4A). Within this region, the molecular markers and contigs of BAC or YAC clones had not been reported; therefore, we screened a rice BAC library using C524a and C80a as probes, developed new molecular markers from the ends of the BAC contig identified with the C524a probe (Table 2), and fine-mapped the BC1 gene onto a single BAC, OSJNBa0036N23 (Figure 4B). To further narrow the BC1 locus, we sequenced OSJNBa0036N23 using a shotgun strategy, developed seven additional markers, P1 to P7 (Table 2), and located BC1 in an interval of a 3.3-kb DNA fragment between the P2 and P4 markers (Figure 4C). Within this region, only a single opening reading frame (ORF) was predicted, which appears to encode a protein that shows a strong similarity to Arabidopsis COBRA, a protein required for oriented cell expansion and the proper deposition of cellulose in the root elongation zone (Schindelman et al., 2001).
Figure 4.
Cloning and Confirmation of the BC1 Gene.
(A) The BC1 locus was mapped in the chromosome 3 (Chr 3) centromeric region between markers C524a and RM16.
(B) A BAC contig covering the BC1 locus. The numerals indicate the number of recombinants identified from 7068 bc1-2 F2 plants. BAC1, OSJNBa0006H20; BAC2, OSJNBa0052G19; BAC3, OSJNBa0007B10; BAC4, OSJNBa0057M10; BAC5, OSJNBa0036N23.
(C) Fine mapping of the BC1 locus with the markers (P1 to P7) developed based on the OSJNBa0036N23 sequence. The BC1 locus was narrowed to a 3.3-kb genomic DNA region between CAPS markers P2 and P4 and cosegregated with marker P3.
(D) BC1 gene structure, showing the mutated sites of the two bc1 alleles. The start codon (ATG) and the stop codon (TGA) are indicated. Closed boxes indicate the coding sequence, open boxes indicate the 5′ and 3′ untranslated regions, and lines between boxes indicate introns. Mutation sites in bc1-1 and bc1-2 also are shown.
(E) Complementation constructs. The construct pCna18 contains the entire BC1 gene, including a 2795-bp upstream sequence and a 1459-bp downstream sequence. The plasmid pCna181T contains a partial BC1 gene that encodes the first 173 amino acid residues.
(F) Identification of transgenic plants. The deletion of 4 bp in bc1-2 destroys a BstNI site that is used for the CAPS marker P3. Lane 1, wild type; lane 2, bc1-2; lane 3, the pCna18-transformed rice line 1; lane M, 1-kb marker.
Table 2.
List of the PCR-Based Molecular Markers Developed in This Study
| Marker | Primer Pairsa | Fragment Sizeb | Restriction Enzyme |
|---|---|---|---|
| 36N13f | F, 5′-AATCTTCTCTTACTCCACTCGC-3′; R, 5′-ATGGGAACTACTGACTAAACCG-3′ | C = 489, b = 0 | |
| 57M10r | F, 5′-AGGAACAGATGGAATCTCAGG-3′; R, 5′-TGTCTCCTGTCCCAAACTAGC-3′ | C = 340, b = 0 | |
| 52G19f | F, 5′-GCACGCATTTAAGAAGCAGG-3′; R, 5′-TTATATGACCGTATGGCAGG-3′ | 502 | MnlI |
| P1 | F, 5′-TTGCGTATGTCTGTCAACTGC-3′; R, 5′-TTCCAACATCTGAACCCTCG-3′ | C = 1580, b = 0 | |
| P2 | F, 5′-GTTGCTCATCGTCACCATCG-3′; R, 5′-AGGTATAGAGCGAGCGGTAGC-3′ | 1037 | NsiI |
| P3 | F, 5′-GGTAGTTGAAGTTTGTGATGGC-3′; R, 5′-ATGCTCTCTCCTCGCTCTGC-3′ | 1110 | BstNI |
| P4 | F, 5′-TACTACAACGACCTGCTTATGG-3′ R, 5′-TGGAGAGCAGTGTAGGTAGGG-3′ | 828 | FokI |
| P5 | F, 5′-AGCAGCAGTAGTCGCAGTCG-3′; R, 5′-AACATAGCCATCTGGGGTCC-3′ | 1762 | MnlI |
| P6 | F, 5′-TAGAAACCAGGGTCAAGTAGGC-3′; R, 5′-GCTGAGTTTGAGGTCGGTGG-3′ | 1470 | MseI |
| P7 | F, 5′-AGTGCCTCATCTATCGCTTCG-3′; R, 5′-GTTCCACAAACCGATCTAGCG-3′ | 2046 | ScaI |
F, forward primer; R, reverse primer.
Numbers indicate the size (in bp) of amplified fragments. b, bc1-2 mutant; C, C-Bao.
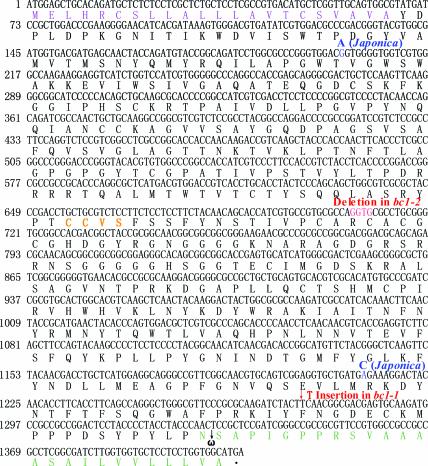
To define the molecular lesions of bc1 mutants, the genomic DNA sequences corresponding to the putative BC1 gene from the bc1-1 and bc1-2 alleles were amplified by PCR and sequenced. Comparison of the sequences of the wild-type and mutant allelic genes revealed that the bc1-1 allele carries a single nucleotide insertion in codon 425 (TTC → TTTC) in the third exon, which produces a larger protein that disrupts the GPI-attached site and the hydrophobic C terminus, and that the bc1-2 allele contains a 4-bp deletion in codons 236 and 237 (AGGTGC → AC) in the second exon, which results in a frameshift that produces premature translational products (Figures 4D and 5), suggesting that the ORF represents the BC1 gene.
Figure 5.
BC1 cDNA and Predicted Amino Acid Sequences.
Numbers at left refer to the positions of nucleotides. Red letters indicate the 4-bp deletion in bc1-2 and the 1-bp insertion in bc1-1; blue letters indicate different nucleotides between indica and japonica subspecies; purple letters indicate the N-terminal signal; orange letters indicate the conserved CCVS motif; and green letters indicate the C terminus, including the predicted ω-site and the hydrophobic tail.
The identity of BC1 was confirmed by genetic complementation analysis. The plasmid pCna18, containing the entire ORF, and pCna181T, containing a partial coding region of the ORF (Figure 4E), were introduced into the bc1-2 mutant, and 80 and 62 independent transgenic lines were obtained from the two constructs, respectively. Nearly all 80 lines of pCna18 showed a complementation of the bc1-2 phenotype, whereas all 62 lines of pCna181T failed to rescue the bc1-2 mutant. Additionally, the deletion in the bc1-2 allele disrupts the BstNI site in the genomic DNA, which can be used as a CAPS marker to determine the bc1-2 mutant background in the complementation test (Figure 4F). Therefore, we conclude that we have cloned the BC1 gene in rice.
BC1 Encodes a COBRA-Like Protein
Sequence analysis of the products from reverse transcription (RT)–PCR and rapid amplification of cDNA ends–PCR indicated that the BC1 cDNA is 1818 bp long, with an ORF of 1407 bp, a 90-bp 5′ untranslated region, and a 321-bp 3′ untranslated region (Figures 4D and 5). Sequence comparison between genomic DNA and cDNA showed that BC1 is composed of three exons that encode a 468–amino acid protein. Interestingly, we found no difference in the BC1 amino acid sequence between indica and japonica subspecies, although there are two synonymous base pair changes in the BC1 coding region between the two subspecies (Figure 5).
Basic Local Alignment Search Tool (BLAST) analysis revealed that the rice BC1 protein shares the highest identity (>72% over the entire length) with COBL4, a predicted member of the COBRA family proteins in Arabidopsis (Roudier et al., 2002). COBRA recently was identified as a putative GPI-anchored protein required for the orientation of cell expansion and the cellulose deposition of cell walls in the Arabidopsis root elongation zone (Schindelman et al., 2001). Predicted by its sequence characteristics, BC1 appears to be a member of the COBRA family, because it contains all of the conserved features of the COBRA family, including a CCVS motif, an N-terminal signal peptide sequence for secretion, a highly hydrophobic C terminus, and specific features around the ω-site required for processing (Figure 5) (Udenfriend and Kodukula, 1995).
BC1 Is a Spatially and Temporally Expressed Gene
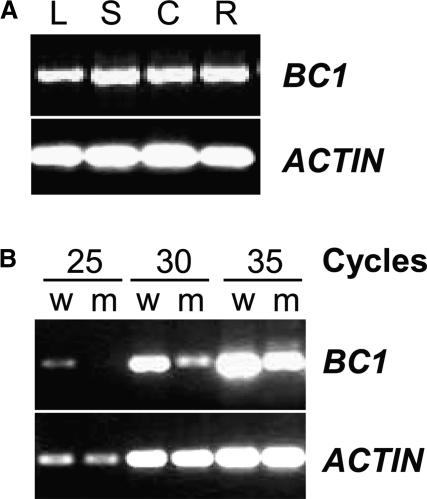
Although the steady state levels of BC1 mRNA were undetectable by RNA gel blot analysis when whole plants were sampled, its expression could be examined by RT-PCR. As shown in Figure 6A, BC1 was expressed universally throughout the wild-type plant organs, including leaves, stems, and roots, consistent with the brittleness phenotype of bc1 mutant plants. It should be noted that RT-PCR analysis demonstrated a dramatic reduction in the abundance of transcripts in the homozygous bc1-2 seedlings (Figure 6B), indicating that the mutation in bc1-2 plants may affect not only the BC1 protein function but also the steady state level of the transcript.
Figure 6.
BC1 Expression Analysis.
(A) RT-PCR analysis of BC1. Total RNA was isolated from leaves (L), leaf sheaths (S), culms (C), and roots (R) of wild-type plants. Amplification of actin cDNA was used to ensure that approximately equal amounts of cDNA were loaded.
(B) Comparison of BC1 transcripts between bc1 mutant (m) and wild-type (w) plants with RT-PCR.
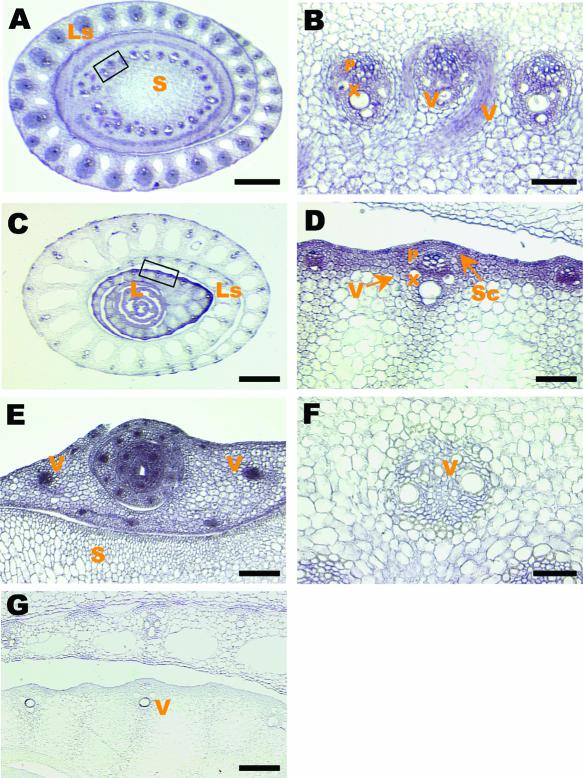
The precise expression patterns of BC1 were explored further by RNA in situ hybridization. As shown in Figures 7A to 7D, BC1 was expressed mainly in the cells of mechanical tissues, including the vascular bundles, and in the subepidermal regions where sclerenchyma cells differentiate, explaining the phenotypes observed with scanning electron microscopy (Figure 2) and histochemical staining (Figures 3C to 3L). The much stronger expression of BC1 in tiller buds (Figure 7E) than in the developing culms (Figure 7F) and in young leaf sheaths (Figure 7D) than in old ones (Figure 7C) indicate that BC1 expression also was regulated developmentally, suggesting that the biosynthesis of cell walls of mechanical tissues is temporally dynamic.
Figure 7.
Expression Patterns of the BC1 Gene Revealed by in Situ Hybridization in Transverse Sections of Wild-Type Rice Plants.
(A) Young stem and leaf sheath.
(B) A magnified section from (A), showing the strong expression of BC1 in the developing vascular bundles.
(C) Leaves and leaf sheath, showing the intense expression of BC1 in young leaves and sheaths.
(D) A magnified section from (C), showing strong signals in the developing vascular and other mechanical tissues.
(E) Tiller bud, showing strong signals in vascular bundles.
(F) Mature vascular bundles, showing the sharply decreased expression level of BC1 compared with that shown in (E).
(G) Background control, in situ hybridization of a young leaf sheath with a sense probe.
L, leaf; Ls, leaf sheath; P, phloem; S, stem; Sc, sclerenchyma cells; V, vascular bundles; X, xylem. Bars = 70 μm in (E) and (G), 450 μm in (A) and (C), and 35 μm in (B), (D), and (F).
BC1 Belongs to a Multimember Gene Family in Rice
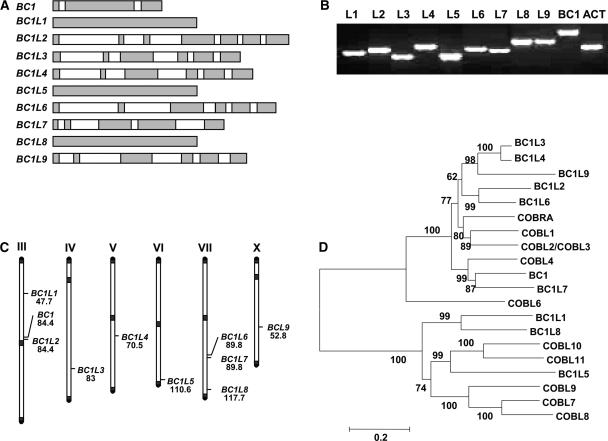
A genome-wide search of DNA and protein databases identified nine BC1-like (BC1L) genes and one pseudogene, BC1L-p1, in the rice genome (Table 3); their overall structures are illustrated in Figure 8A. All of the BC1 family proteins except BC1L2 and BC1L9 contain an N-terminal signal peptide sequence for secretion, a highly hydrophobic C terminus, the conserved CCVS domain, and a potential ω-cleavage site (Table 3). However, BC1L2 and BC1L9 lack the predicted ω-cleavage sites in the deduced protein sequences (Table 3).
Table 3.
Rice BC1 Family Genes
| Locus | Accession No. | Gene Position | EST Accession No. | Amino Acid a | ω-Site |
|---|---|---|---|---|---|
| BC1 | AAAA01003837 | 16545 to 18148 | D47139 | 468 | N (443) |
| BC1L1 | AAAA01008372 | 1321 to 3336 | AU030042 | 671 | N (644) |
| BC1L2 | AAAA01003837 | 12631 to 15956 | 477 | None | |
| BC1L3 | AAAA01000310 | 31175 to 33981 | AU225729; C74834 | 456 | N (429) |
| BC1L4 | AAAA01006032 | 1439 to 4300 | C91883; AU096812 | 457 | N (430) |
| BC1L5 | AAAA01005547 | 9426 to 11477 | 683 | A (658) | |
| BC1L6 | AAAA01000918 | 6284 to 9445 | C97636 | 446 | N (419) |
| BC1L7 | AAAA01000918 | 10062 to 12498 | AU173066 | 477 | A (459) |
| BC1L8 | AAAA01019714 | 756 to 2774 | AU1738855; AU1738866 | 672 | S (644) |
| BC1L9 | AAAA01005473 | 10047 to 12814 | 683 | None | |
| BC1L-p1 | AL606587 | 73953 to 75957 |
Amino acid residue number in each predicted BC1L protein.
Figure 8.
The BC1 Gene Family in Rice.
(A) Intron/exon structure of rice BC1 family genes. Exons are indicated with gray boxes, and introns are indicated with white boxes.
(B) Expression of rice BC1 family genes in aerial parts of rice seedlings revealed by RT-PCR. Amplification of actin (ACT) transcript was used as an internal control. L1 to L9, BC1L1 to BC1L9.
(C) Chromosomal positions of BC1 family genes. The chromosomes are labeled with roman numerals. The number below the gene symbol represents the genetic position of each putative gene (centimorgans).
(D) Neighbor-joining tree of rice BC1L proteins and Arabidopsis AtCOBRA family members. The numbers at each node represent the bootstrap support (percentage). The scale bar is an indicator of genetic distance based on branch length.
Although ESTs of BC1L2, BC1L5, and BC1L9 are not found in any known EST database, all of the BC1 family genes identified in the rice genome are expressed, as revealed by RT-PCR analysis with total RNA isolated from aerial organs of rice seedlings (Figure 8B). This finding indicated that all BC1 family genes are transcriptionally active and may have different functions.
Phylogenetic Analysis of BC1 Family Genes
To determine the evolutionary relationships of rice BC1 family and Arabidopsis COBRA family members, an unrooted tree was built using the neighbor-joining method (Figure 8D). Phylogenetic analysis indicated that the rice BC1 family and the Arabidopsis COBRA family belong to a single gene superfamily, the COBRA family. As determined by phylogenetic analysis, the COBRA family is divided into two subfamilies, which is in agreement with the exon/intron organization of each subfamily member. Members in one subfamily have multiple introns/exons, whereas those in the other subfamily have a single exon (Figures 8A and 8D). BC1 and COBRA belong to two divergent clades, implying that they have related but distinct functions. This distinction was reflected by the different phenotypes of bc1 and cobra mutants. BC1 and BC1L7 in rice and COBL4 in Arabidopsis form a monophyletic clade with 99% bootstrap support. BC1 and BC1L7 are located on chromosomes 3 and 7 in rice, respectively (Figure 8C). They resulted from a gene duplication event that took place after the divergence of monocot and dicot phyla, suggesting that they might have partially redundant functions.
DISCUSSION
We have described the molecular genetic characterization of the classic rice mutant bc1, its positional cloning, and its temporally and spatially regulated expression in the cells of mechanical tissues. BC1 encodes a protein that is homologous with Arabidopsis COBRA family members and represents a 10-member family in rice. Deficiency in BC1 causes the altered biosynthesis of cellulose, hemicellulose, and lignin, leading to a reduction in the secondary cell wall thickness and the mechanical strength of rice plants.
Mutation in BC1 Alters the Formation of Secondary Cell Walls
Our results clearly demonstrate that mutations in BC1 affect the biosynthesis of secondary cell walls and result in alterations in the contents of cellulose and lignin. The prominent differences between the wild type and the bc1-2 mutant are localized mainly in the mechanical tissues, such as sclerenchyma cells and vascular bundles (Figures 2 and 3). It has known that deficiency in cellulose biosynthesis in the primary cell walls often leads to a globally altered morphology, sterility, or even lethality (Arioli et al., 1998). By contrast, plants with mutations that affect the formation of secondary cell walls usually grow relatively normally (Taylor et al., 1999, 2000). The similarities of the plant morphology and the sclerenchyma and parenchyma cell sizes between wild-type and bc1-2 plants suggest that BC1 very likely plays a role in regulating the biosynthesis of secondary cell walls. This notion was substantiated further by comparing the cell wall sugar contents of mutant and wild-type culms. Compared with wild-type plants, the bc1-2 mutant showed a dramatic alteration in glucose and xylose, the two major sugars that constitute cellulose and hemicellulose in the secondary cell walls, respectively.
The mechanism that regulates the biosynthesis of cell walls is complicated and requires the coordination of a number of metabolic pathways. Recent studies with Arabidopsis mutants defective in cellulose production have demonstrated that cellulose biosynthesis is affected if a mutation occurs either in enzymes involved in cellulose biosynthesis or in other proteins, such as 1,4-β-endoglucanase, mannose-1-phosphate guanylyltransferase, a katanin-like protein, or membrane proteins (Arioli et al., 1998; Nicol et al., 1998; Taylor et al., 1999, 2000; Fagard et al., 2000; Burk et al., 2001; Lukowitz et al., 2001; Schindelman et al., 2001; Pagant et al., 2002). In all cases, these defects cause abnormal plant growth and development, suggesting the functional importance of highly dynamic cell wall biosynthesis. The finding that mutations in BC1 cause abnormalities of the cellulose and lignin contents and the defective formation of secondary cell walls of mechanical tissues suggests that BC1 may act as an auxiliary protein to regulate secondary cell wall biosynthesis in rice. The in situ hybridization study showed that the BC1 gene is expressed mainly at the young, elongating regions of organs (Figure 7). However, at this developmental stage, sclerenchyma cells below the epidermis and bundle sheath fiber cells are not yet undergoing secondary wall thickening.
Because the expression of BC1 is decreased dramatically in nonelongating organs, in which sclerenchyma cells undergo secondary wall formation (Figure 7), it appears that BC1 is not involved directly in secondary wall formation. Arabidopsis COBRA family members are likely to be important players at the plasma membrane–cell wall interface (Roudier et al., 2002). It is very possible that BC1, as a putative GPI-anchored protein, is located at the cell wall–plasma membrane interface, where cellulose deposition takes place. This hypothesis is strengthened by our findings that BC1 is expressed mainly and functions in the cells of developing mechanical tissues. Clearly, additional studies, particularly of the biochemical nature of BC1 and its targets, will be essential to fully understand its function during cell wall biosynthesis.
BC1 Is a New Member of the COBRA Family
BC1 encodes a putative GPI-anchored COBRA-like protein. Based on hydrophobicity analysis using a method described by Kyte and Doolittle (1982), BC1 has a hydrophobic N-terminal signal peptide and a highly hydrophobic peptide at its C terminus and is predicted to be GPI anchored based on the big-PI Predictor (Eisenhaber et al., 2000) and PSORT (Nakai and Horton, 1999) programs. Normally, at the ω-site, only amino acid residues such as Ser, Asn, Ala, Gly, Asp, or Cys may be present, whereas at the ω+2-site, only Ala, Gly, Thr, or Ser are commonly found, and the ω+2-residue usually is followed by a spacer of five to seven amino acid residues rich in charged and/or Pro residues preceding a stretch of 10 to ∼30 hydrophobic residues (Udenfriend and Kodukula, 1995). The predicted BC1 protein meets all of these requirements. The bc1-1 allele was frameshifted by a single nucleotide insertion in codon 425, which results in a translational product without the GPI attachment site and the hydrophobic tail. The mutation in bc1-2 contains a 4-bp deletion in codons 236 and 237, which results in a frameshift mutation and produces a truncated protein product that lacks the C terminus. Both bc1-1 and bc1-2 are likely to be loss-of-function mutations, because the GPI attachment site and the hydrophobic C-terminal peptide required for post-translational processing are destroyed in their aberrant protein products; therefore, BC1 processing is likely to be disrupted.
Genomic analysis identified nine additional BC1L genes in the rice genome. Phylogenetic analysis indicates that BC1L, COBL, and homologs from other plants form a gene superfamily, the COBRA gene family. The COBRA gene family is an ancient family that arose before the divergence of monocot and dicot phyla. Determining the functions of all members of this family is a major challenge. Based on phylogenetic analysis and intron/exon structure, the COBRA family can be divided into two subfamilies. BC1 and COBRA belong to the same subfamily. However, BC1 and COBRA are located on two different clades (Figure 8), suggesting that BC1 and COBRA might have different functions in the cell.
The function of the GPI-anchored proteins is not understood fully. Recent studies on COBRA (Schindelman et al., 2001), SKU5 (Sedbrook et al., 2002), and SOS5 (Shi et al., 2003) have shown the importance of GPI-anchored proteins in cell expansion, proper cell wall structure, and cell surface adhesion. The fact that the rice BC1 protein shares the highest identity with Arabidopsis COBL4 suggests that members of the COBRA family may have different functions in plants and that BC1 represents a subgroup that is essential for secondary cell wall biosynthesis and plant mechanical strength.
Brittleness is one of the most important agronomic traits that affect not only grain production but also the usefulness of cereal straws as animal forage. As an important player regulating rice brittleness, BC1 (and its orthologs in other cereals) could make a significant contribution to the future improvement of these crops.
METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa) brittle culm mutant bc1-2 was isolated from the indica cultivar Shuang Ke Zao irradiated with γ-rays (Qian et al., 2001), and the bc1-1 mutant was renamed for the previously reported bc1 mutant derived from a japonica cultivar (Kinoshita, 1995). bc1-2 and C-Bao, a polymorphic japonica variety, were crossed to generate a large F2 mapping population. Rice plants were cultivated in the experimental field at the China National Rice Research Institute in the natural growing season.
Measurements of Physical Properties
The breaking force and elongation ratio of rice culms or leaves were measured with a universal force/length testing device (model DC-KZ300; Kaiming, Sichuan, China). To avoid inaccuracies from sampling, the first internodes of culms and flag leaves were used for an immediate measurement. The elongation ratio (%) was defined by the formula 100 × (L1 − L2)/L2, where L1 represents the length of the leaf segments at breaking and L2 stands for the original length of the leaf segments.
Scanning Electron Microscopy
Samples were prepared as described previously (Mou et al., 2000) with some modifications. Briefly, rice tissues were excised with a razor and immediately placed in 70% ethanol, 5% acetic acid, and 3.7% formaldehyde for 18 h. Samples were critical point dried, sputter-coated with gold in an E-100 ion sputter (Mito City, Japan), and observed with a scanning electron microscope (S570; Hitachi, Tokyo, Japan).
Carbohydrate and Lignin Measurement
Carbohydrate was assayed according to the methods described previously (Updegraff, 1969; Hoebler et al., 1989). Briefly, the first internodes of culms were ground into fine powder in liquid nitrogen. The powder was washed in phosphate buffer (50 mM, pH 7.2) three times, extracted twice with 70% ethanol at 70°C for 1 h, and dried under vacuum. The dried cell wall materials were assayed for cellulose content with the anthrone reagent with Whatman 3MM paper as the standard. For the measurement of cell wall sugars, the dried cell wall materials were hydrolyzed by incubation in 72% (w/w) H2SO4 at room temperature for 1 h and then in 2 M H2SO4 at 121°C for 1 h. The sugar alditol acetates were analyzed by gas chromatography. To measure lignin content, the first internodes of culms were ground into fine powder and extracted four times with methanol. After vacuum drying, lignin content was quantified according to the method described by Kirk and Obst (1988).
Histochemical Staining
For histochemical localization of lignin, a Wiesner reaction was performed according to a standard protocol (Strivastava, 1966). Fresh hand-cut sections (∼20 μm thick) from rice culms were incubated for 2 min in phloroglucin solution (2% in ethanol:water [95:5, v/v]; Sigma), mounted in 50% HCl, and photographed using a 3CCD (charge-coupled device) color video camera (DXC-390P; Sony, Tokyo, Japan). For cellulose staining, paraffin-embedded sections (10 μm thick) were stained with a 0.005% aqueous solution of calcofluor (fluorescent brightener 28; Sigma) for 2 min and visualized with a fluorescent microscope (Leica, Wetzlar, Germany).
DNA Isolation and DNA Gel Blot Analysis
The rice genomic DNA preparation and DNA gel blot analysis were performed as described (Mou et al., 2000; Qian et al., 2001). Briefly, rice genomic DNA (20 μg) was digested completely with restriction enzymes and then separated by 0.8% agarose gel electrophoresis. The DNA then was transferred onto a Hybond N+ membrane (Amersham) and hybridized with a 32P-labeled probe under high-stringency hybridization conditions.
Genetic Analysis and Marker Development
The genetic linkage between the BC1 locus and molecular markers was determined using Mapmaker (Lander et al., 1987). To fine-map the BC1 locus, new molecular markers, especially the cleaved amplified polymorphic sequence markers, were developed (Table 2).
Construction of the BC1 BAC Contig
To construct a BAC contig covering the BC1 locus, we screened a BAC library with two flanking markers, C80a and C524a, identified the overlapped BAC clones based on their fingerprints (http://www.genome.clemson.edu), and determined the linkage with the new markers developed from the BAC ends. By repeating this process, the BC1 locus eventually was located in a single BAC, OSJNBa0036N23.
DNA Sequencing
The BAC clone OSJNBa0036N23 was sequenced using a shotgun approach. To sequence the bc1-1 and bc1-2 alleles, the entire genomic regions were amplified from the mutants (bc1-1 and bc1-2) and their corresponding wild-type plants by PCR with LA-Taq (TaKaRa, Dalian, China). The PCR program included 3 min at 94°C, followed by 40 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 2 min, and a final extension at 72°C for 10 min. PCR products were sequenced directly, and the mutations in bc1-1 and bc1-2 were identified and verified further by sequencing two additional independent PCR products. The mutation in bc1-2 also can be detected by restriction analysis with BstNI.
Complementation Test
A 5.86-kb genomic DNA fragment containing the entire BC1 coding region, the 2796-bp upstream sequence, and the 1459-bp downstream sequence was inserted into the binary vector pCAMBIA1300 to generate the transformation plasmid pCna18 for the complementation test. A control plasmid, pCna181T, containing the 3′ truncated BC1 gene that encodes the first 173 amino acid residues, also was constructed (Figure 3E) according to the strategy described previously (Li et al., 2003). The two binary plasmids were introduced into Agrobacterium tumefaciens LBA4404 by electroporation, and the rice bc1-2 mutant was transformed according to a published method (Hiei et al., 1994).
Reverse Transcription– or Rapid Amplification of cDNA Ends–PCR Analysis
Total RNA was extracted from leaves, leaf sheaths, culms, roots, and entire plants according to the method described by Wadsworth et al. (1988). For reverse transcription (RT)–PCR, first-strand cDNA was transcribed reversibly from total RNA with oligo(dT) as the primer and used as the template to amplify the transcripts with a profile (35 cycles) of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min. Primers for BC1 RT-PCR were BC1F and BC1R (5′-GGTAGTTGAAGTTTGTGATGGC-3′ and 5′-GCTCTCTCCTCGCTCGGCTCC-3′), and those for actin were ActF and ActR (5′-TCCATCTTGGCATCTCTCAG-3′ and 5′-TCCATCTTGGCATCTCTCAG-3′). The 5′ and 3′ ends of the BC1 cDNA were amplified from wild-type Shuang Ke Zao total RNA using the rapid amplification of cDNA ends (RACE) kit (TaKaRa) according to the kit manual. Primers for 5′ RACE were RACE1F (5′-CCCACGATGGACCAGATGACC-3′), the first nested primers N1F and N1R (5′-GTCACGGCGAGGAGCCGAGC-3′ and 5′-ACATCACGTTAAAGTGGGACG-3′), and the second nested primers N2F and N2R (5′-GAGCCGAGCGAGGAGAGAGC-3′ and 5′-CCAGATGTACCGGCAGATCC-3′). Primers for 3′ RACE were BC4R (5′-GGAGAGCAGTGTAGGTAGGG-3′) and the nested primer BC11R (5′-CAGTCCCCAGTTAGGCCAGC-3′). The RACE-PCR products were cloned into the pGEM-T vector (Promega, Madison, WI) and sequenced.
RNA in Situ Hybridization
RNA in situ hybridization was performed as described (Kao et al., 2002). The 3′ end of BC1 was subcloned into pGEM-T vector (Promega) and used as a template to generate RNA probes. Transverse sections (10 μm thick) were probed with digoxigenin-labeled antisense probes (DIG Northern Starter Kit; Roche, Indianapolis, IN). The slides were observed with a microscope (Leica) and photographed using a 3CCD color video camera (DXC-390P; SONY).
Sequence and Phylogenetic Analyses
BC1L genes were identified by searching the whole genome draft sequence database of rice subspecies indica (Yu et al., 2002) using the DNA or protein sequences of the rice BC1 gene and Arabidopsis COBRA family genes as queries. The japonica ortholog of each BCL1 gene was determined by searching the nonredundant and high-throughput genomic sequence databases using Basic Local Alignment Search Tool (BLASTN). The retrieved japonica BAC or PAC sequences were used to posit each BC1L gene to a particular chromosomal location based on the rice physical map (Chen et al., 2002).
Gene prediction was performed using Fgenesh (Salamov and Solovyev, 2000), and intron/exon structures were verified by alignment of the EST sequences of rice and other grass species with the genomic DNA. The signal peptide was predicted with SignalP version 2.0 (Nielsen and Krogh, 1998), and the hydrophobic profile was determined with the program provided by the Weizmann Institute of Sciences (http//bioinformatics. weizmann.ac.il). Glycosylphosphatidylinositol modification was predicted using big-PI (http://mendel.imp.univie.ac.at/gpi/index_content.html) and PSORT (http://psort.nibb.ac.jp).
Phylogenetic analysis included all rice BC1L proteins and Arabidopsis COBRA family members except AtCOBL5, because of the incompleteness of its sequence. Multiple sequence alignments were conducted using CLUSTAL X version 8.0 (Thompson et al., 1997) with the PAM matrix (Dayhoff, 1979). A neighbor-joining tree (Saitou and Nei, 1987) was built using MEGA version 2.1 (Kumar et al., 2001) adopting Poisson correction distance, and the tree was presented using TreeView (Page, 1996). Support for the tree obtained was assessed using the bootstrap method (Felsenstein, 1985). The number of bootstrap replicates was 1000. Similar topology was obtained by using the protpas program in the Phylip package (Felsenstein, 1993) to estimate maximum parsimony and the proml program in the Phylip package to estimate maximum likelihood. Alignments with the full-length sequences or only the conserved regions of each family member produced trees of similar topology.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact J. Li, jyli@genetics.ac.cn.
Accession Numbers
Accession numbers for the BC1 sequences reported in this article are AY328909 and AY328910.
Acknowledgments
We thank Jianru Zuo (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), two anonymous reviewers, and the co-editor for critical comments on the manuscript, Itsuro Takamure (Hokkaido University) for providing rice bc mutants, Tefu Qin (Institute of Wood Industry, Chinese Academy of Forestry) for assistance in measuring the contents of cellulose and lignin, and the MAFF DNA Bank at the National Institute of Agrobiological Resources (Tsukuba, Ibaraki, Japan) and the Clemson University Genomic Institute for providing restriction fragment length polymorphism probes, filters of BAC libraries, and BAC clones. This work was supported by grants from the State High-Tech Program (2001AA222021), the State Key Basic Research Program of China (G19990116 and G19990160), and the National Natural Science Foundation of China.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011775.
References
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., and Ye, Z.H. (2002). Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 14, 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N., and McCann, M. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 52–108.
- Chen, M., et al. (2002). An integrated physical and genetic map of the rice genome. Plant Cell 14, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff, M.O. (1979). Atlas of Protein Sequence and Structure, Vol. 5, Suppl. 3. (Washington, DC: National Biomedical Research Foundation).
- Eisenhaber, B., Bork, P., Yuan, Y., Loeffler, G., and Eisenhaber, F. (2000). Automated annotation of GPI anchor sites: Case study C. elegans. Trends Biol. Sci. 25, 340–341. [DOI] [PubMed] [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Hofte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12, 2409–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1993). PHYLIP: Phylogeny Inference Package, Version 3.5. (Seattle: University of Washington).
- Ferguson, M.A.J., et al. (1994). Glycosylphosphatidylinositol molecules of the parasite and the host. Parasitology 108, s45–s54. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hoebler, C., Barry, L.D., and Delort-Laval, J. (1989). Rapid hydrolysis of plant cell wall polysaccharides by gas-liquid chromatography. J. Agric. Food Chem. 37, 360–367. [Google Scholar]
- Jones, L., Ennos, A.R., and Turner, S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26, 205–216. [DOI] [PubMed] [Google Scholar]
- Kao, Y.Y., Harding, S.A., and Tsai, C. (2002). Differential expression of two distinct phenylalanine ammonia-lyase genes in condensed tannin-accumulating and lignifying cells of quaking aspen. Plant Physiol. 130, 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T. (1995). Report of committee on gene symbolization, nomenclature and linkage groups. Rice Genet News 12, 9–153. [Google Scholar]
- Kirk, T.K., and Obst, J.R. (1988). Lignin determination. Methods Enzymol. 161, 87–101. [Google Scholar]
- Kokubo, A., Kuraishi, S., and Sakurai, N. (1989). Culm strength of barley: Correlation among maximum bending stress, cell wall dimensions, and cellulose content. Plant Physiol. 91, 876–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo, A., Sakurai, N., Kuraishi, S., and Takabe, K. (1991). Culm brittleness of barley (Hordeum vulgare L.) mutants is caused by smaller number of cellulose molecules in cell wall. Plant Physiol. 97, 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E., and Newberg, L. (1987). Mapmaker: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. [DOI] [PubMed] [Google Scholar]
- Li, X., et al. (2003). Control of tillering in rice. Nature 422, 618–621. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Nickle, T.C., Meinke, D.W., Last, R.L., Conklin, P.L., and Somerville, C.R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a re-quirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98, 2262–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z., He, Y., Dai, Y., Liu, X., and Li, J. (2000). Deficiency in fatty acid synthase leads to premature cell death and dramatic alteration in plant morphology. Plant Cell 12, 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36. [DOI] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Hofte, H. (1998). A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., and Krogh, A. (1998). Prediction of signal peptides and signal anchors by a hidden Markov model. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB 6). (Menlo Park, CA: AAAI), pp. 122–130. [PubMed]
- Pagant, S., Bichet, A., Sugimoto, K., Lerouxel, O., Desprez, T., McCann, M., Lerouge, P., Vernhettes, S., and Hofte, H. (2002). KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14, 2001–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Peles, E., Nativ, M., Lustig, M., Grumet, M., Schilling, J., Matinez, R., Plowman, G.D., and Schlessinger, J. (1997). Identification of a novel conactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J. 16, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, Q., Li, Y.H., Zeng, D., Teng, S., Wang, Z., Li, X., Dong, Z., Dai, N., Sun, L., and Li, J. (2001). Isolation and genetic characterization of a fragile plant mutant rice (Oryza sativa L.). Chin. Sci. Bull. 46, 2082–2085. [Google Scholar]
- Ratcliffe, O.J., Riechmann, J.L., and Zhang, J. (2000). INTERFASCICULAR FIBERLESS1 is the same gene as revoluta. Plant Cell 12, 315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg, K.G., Ying, Y.S., Kamen, B.A., and Anderson, R.G. (1990). Cholesterol controls the clustering of the glycophospholipid-anchored membrane receptor for 5-methyltetrahydrofolate. J. Cell Biol. 111, 2931–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier, F., Schindelman, G., DeSalle, R., and Benfey, P.N. (2002). The COBRA family of putative GPI-anchored proteins in Arabidopsis, a new fellowship in expansion. Plant Physiol. 130, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Salamov, A.A., and Solovyev, V.V. (2000). Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C., Gilson, P., Oxley, D., Youl, J.J., and Bacic, A. (1998). GPI-anchors on arabinogalactan-proteins: Implications for signalling in plants. Trends Plant Sci. 3, 426–431. [Google Scholar]
- Sedbrook, J.C., Carroll, K.L., Hung, K.F., Masson, P.H., and Somerville, C.R. (2002). The Arabidopsis SKU5 gene encodes an extracellular glycosylphosphatidylinositol-anchored glycoprotein in- volved in directional root growth. Plant Cell 14, 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrier, D.J., Prime, T.A., and Dupree, P. (1999). Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Shi, H., Kim, Y.S., Guo, Y., Stevenson, B., and Zhu, J.K. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strivastava, L.M. (1966). Histochemical studies on lignin. Tappi 49, 173–183. [Google Scholar]
- Taylor, N.G., Laurie, S., and Turner, S.R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12, 2529–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis in the secondary cell wall. Plant Cell 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udenfriend, S., and Kodukula, K. (1995). How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu. Rev. Biochem. 64, 563–591. [DOI] [PubMed] [Google Scholar]
- Updegraff, D.M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32, 420–424. [DOI] [PubMed] [Google Scholar]
- Wadsworth, G.J., Redinbaugh, M.G., and Scandalios, J.G. (1988). A procedure for the small-scale isolation of plant RNA suitable for RNA blot analysis. Anal. Biochem. 172, 279–283. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., Morrison, W.H., and Ye, Z.H. (2002). A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14, 3101–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]