Abstract
Toll-like receptors (TLRs)-2 and -4 are important proteins in innate immunity, recognizing microbial products and eliciting host defense responses. Both use the adapter proteins MyD88 and MyD88 adapter-like (Mal) to activate signaling pathways. Here we report that Mal but not MyD88 interacts with caspase-1, the enzyme that processes the precursors of the proinflammatory cytokines IL-1β and IL-18. The interaction was found in a yeast two-hybrid screen and was confirmed by reciprocal GST pull-downs and coimmunoprecipitation of endogenous proteins. We were unable to implicate Mal in regulating caspase-1 activation. However, we found that Mal was cleaved by caspase-1 and that inhibition of caspase-1 activity blocked TLR2- and TLR4-mediated NF-κB and p38 MAP kinase activation but not IL-1 or TLR7 signaling, which are Mal independent. These responses, and the induction of TNF, were also attenuated in caspase-1-deficient cells. Finally, unlike wild-type Mal, a mutant Mal, which was not cleaved by caspase-1, was unable to signal and acted as a dominant negative inhibitor of TLR2 and TLR4 signaling. Our study therefore reveals a role for caspase-1 in the regulation of TLR2 and TLR4 signaling pathways via an effect on Mal. This functional interaction reveals an important aspect of the coordination between TLRs and caspase-1 during the innate response to pathogens.
Keywords: signaling, Toll-like receptor
Toll-like receptors (TLRs) are important activators of the host innate immune response. Ten TLRs have been discovered in humans, and all are characterized as having extracellular leonine-rich repeat domains and an intracellular Toll/IL-1 receptor (TIR) domain (1). Upon ligand activation of the TLRs, cytosolic TIR domain-containing adapter proteins are recruited (1). Four signaling adaptor proteins have been identified, myeloid differentiation factor 88 (MyD88), MyD88 adaptor-like (Mal), TIR domain-containing adaptor inducing IFN-β (Trif), and Trif-related adaptor molecule (TRAM), and although the TLRs have similar signal transduction pathways, there is specificity with regard to their adaptor usage (2). MyD88 is recruited by all TLRs except TLR3 (3) and leads activation of the transcription factor NF-κB (2). Mal is required for signaling by the LPS receptor TLR4 and the bacterial lipoprotein receptor TLR2 (4), and Mal acts as a bridging adapter for MyD88 recruitment (1). Trif mediates TLR3 and TLR4 signaling and is involved in the activation of another transcription factor, IRF3 (5). Finally, TRAM mediates TLR4 signaling exclusively (5), acting as a bridging adapter to recruit Trif to the TLR4 complex.
Caspase-1 plays a key role in inflammatory responses by cleaving pro-IL-1β, pro-IL-18, and probably pro-IL-33 into their bioactive forms (6, 7). Caspase-1 occurs in multiprotein complexes called inflammasomes (8), three of which have been characterized to date (9). One of these inflammasomes comprises the Nod-like receptor (NLR) protein Nalp1 in a complex with caspase-1, caspase-5, and the adapter protein apoptosis-associated speck-like protein (ASC) (10). The second inflammasome contains two other NLRs, Nalp3 and Nalp2, along with caspase-1, ASC, and Cardinal (10). The Nalp3-containing inflammasome is activated by bacterial RNA, certain bacterial toxins and ATP, and uric acid crystals (9, 11, 12). The third inflammasome contains another NLR termed Ipaf, and caspase-1 is activated by bacterial flagellin (13, 14). ASC has a particularly important role in caspase-1 activation because ASC-deficient macrophages are unable to activate caspase-1 in response to a number of stimuli (15). In all cases so far, TLR ligands are required to prime inflammasomes for activation, although the mechanism is not known.
In an effort to discover proteins that interact with Mal, we carried out a yeast two-hybrid screen and identified caspase-1 as a Mal-interacting protein. Further, signaling by TLR2 and TLR4, but not TLR7 or IL-1, is impaired in caspase-1-deficient cells, and cleavage of Mal by caspase-1 is required for Mal to signal. We have therefore revealed an additional function for caspase-1 in the regulation of TLR2 and TLR4 signaling and provided an important functional link between TLRs and NLRs in inflammasomes.
Results
Mal Interacts with Caspase-1.
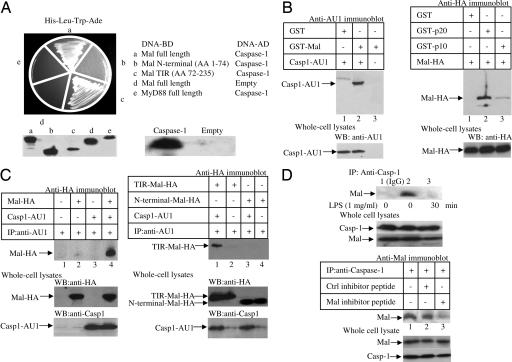
By using a yeast two-hybrid-based screening assay with full-length Mal as a bait and a target splenocyte cDNA library, we identified a total of 20 Mal-positive unique interacting clones. The most interesting encoded a portion of caspase-1 spanning the p20 and p10 subunits, which, in the active enzyme, comprise the catalytic domain. When tested in a bait/interactor format, Mal was shown to interact with caspase-1 (Fig. 1A, Upper Left, segment a). The N-terminal domain of Mal did not interact with caspase-1 (segment b), but the TIR domain clearly interacted (segment c). MyD88 did not interact with caspase-1 (segment e). Fig. 1B Left confirms the interaction whereby GST-Mal isolated caspase-1 from lysates prepared from HEK293 cells transfected with AU1-tagged caspase-1 (lane 2). A GST fusion of the p20 subunit of caspase-1 isolated Mal from HEK293 cells transfected with HA-tagged Mal (Fig. 1B Right, lane 2). The p10 subunit also isolated Mal but to a lesser extent (lane 3), indicating that Mal interacts largely with the p20 subunit. Moreover, AU1-tagged caspase-1 coimmunoprecipitated with HA-Mal (Fig. 1C Left, lane 4), and HA-Mal-TIR coimmunoprecipitated with AU1-tagged caspase-1 (Fig. 1C Right, lane 1). Also, using lysates prepared from THP1 cells, we found that endogenous Mal coimmunoprecipitated with endogenous caspase-1 (Fig. 1D Upper, lane 2). This result was not due to Mal nonspecifically associating with the beads (Fig. 1D Upper, lane 1). Treatment of THP-1 cells with LPS for 30 min disrupted the association between endogenous caspase-1 and Mal (Fig. 1D Upper, lane 3). Also, the endogenous Mal-caspase-1 complex was disrupted by the Mal inhibitor peptide, which disrupts interactions mediated by the TIR domain of Mal (16), but not a control peptide (Fig. 1D Lower, compare lanes 2 and 3), confirming the importance of the TIR domain of Mal for the interaction with caspase-1.
Fig. 1.
Mal interacts with caspase-1. (A) Yeast two-hybrid analysis of Mal, Mal variants, and MyD88 with caspase-1. Immunoblots indicate the presence of respective proteins. (B and C) Interaction of GST-Mal (B Left) or GST-p20 or -p10 (B Right) with lysates from HEK293 cells transfected with either caspase-1-AU1 (B Left) or HA-Mal (B Right) and blotted with relevant antibodies. For immunoprecipitations, HEK293 cells were transfected with plasmids encoding Mal-HA or caspase-1-AU1 (C Left) or encoding TIR-Mal-HA, N-terminal Mal-HA, or caspase-1-AU1 (C Right). (D) THP-1 cells were treated with LPS (top blot) for the indicated times or the Mal inhibitor peptide (20 μM) or a control peptide (20 μM) for 1 h (bottom blot). After immunoprecipitation of the complexes with an IgG control antibody (top blot, lane 1) or an anti-caspase-1 antibody (top blot, lanes 2 and 3, and third blot from the bottom, lanes 1–3), and immunoblotting was performed using an anti-Mal antibody.
Mal Is Not Required for Caspase-1 Activation.
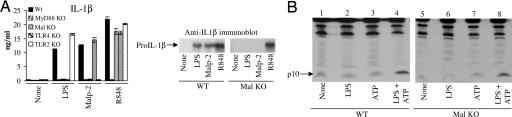
We next analyzed the production of mature IL-1β in Mal-deficient macrophages in response to TLR ligands. Fig. 2A Left shows that wild-type macrophages produced IL-1β in response to LPS, the TLR2 ligand Malp-2, and the TLR7/8 ligand R848. None of the ligands induced IL-1β in MyD88-deficient cells, and the LPS and Malp-2 response was abolished in TLR4- and TLR2-deficient cells, respectively. IL-1β production was also significantly impaired in Mal-deficient macrophages treated with LPS or Malp2, as shown by an ELISA on cell supernatants (Fig. 2A Left) and upon Western blotting of pro-IL-1β whole-cell lysates (Right), the latter also being the case in MyD88-deficient cells (data not shown). The inability of LPS and Malp-2 to induce IL-1β production in Mal-deficient macrophages is therefore likely to be due to effects on expression rather than processing. We next tested more directly whether Mal was required for caspase-1 activation by examining caspase-1 processing. The processed p10 subunit of caspase-1 could be detected in bone marrow-derived and peritoneal macrophages (data not shown) from both wild-type (Fig. 2B, lane 4) and Mal-deficient (Fig. 2B, lane 8) mice.
Fig. 2.
Caspase-1 activation does not require Mal. (A) Wild-type, MyD88-, Mal-, TLR4-, and TLR2-deficient peritoneal macrophages were treated with LPS (100 ng/ml), Malp-2 (0.01 nM), or R848 (0.01 nM) for 16 h followed by ATP (5 mM) for 20 min. IL-1β production was then assessed by ELISA (Left) and Western blot analysis (Right). (B) Wild-type and Mal-deficient bone marrow-derived macrophages were treated with medium or LPS (100 ng/ml) for 16 h followed by ATP (5 mM for 20 min). Lysates were immunoblotted with a caspase-1 anti-p10 antibody. Results shown are representative of three separate experiments.
TLR2 and TLR4 Signaling Is Impaired in Caspase-1-Deficient Cells.
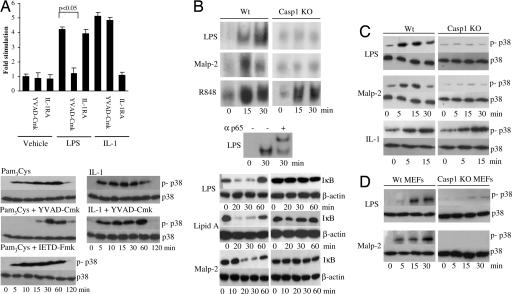
We next tested whether caspase-1 was required for Mal to signal. As shown in Fig. 3A, preincubation of cells with the caspase-1 inhibitor YVAD-Cmk blocked the activation of NF-κB by LPS. This blockage was unlikely a result of IL-1 mediating the effect of these stimuli because pretreatment with the IL-1 receptor antagonist had no effect. The cells are also unresponsive to IL-18. As also shown in Fig. 3A, the inhibition did not block the effect of IL-1 on NF-κB. We also examined p38 MAP kinase activation. We tested Pam3Cys here because unlike LPS, it only uses the adapters MyD88 and Mal (4, 16, 17). Pretreatment of THP-1 cells with the caspase-1 inhibitor blocked the early phase activation of p38 MAP kinase (Fig. 3A Lower, compare top panel with middle panel), whereas a caspase-8 inhibitor was without effect (Fig. 3A Lower). The caspase-1 inhibitor did not affect IL-1-mediated activation of p38 MAP kinase (Fig. 3A Lower, compare upper and lower panels on the right side of the figure). Moreover, LPS and Malp-2 stimulated caspase-1-deficient peritoneal macrophages displayed an impairment in NF-κB activation when compared with the TLR 7/8 ligand R848, as judged in an EMSA (Fig. 3B, top three panels) or by measuring IκB degradation (Fig. 3B, bottom three blots). Similarly, p38 MAP kinase activation was impaired in caspase-1-deficient macrophages and murine embryonic fibroblasts treated with LPS or Malp2 (Fig. 3 C and D) but not IL-1 (Fig. 3C). In addition, TNF-α levels were reduced in caspase-1-deficient macrophages treated with LPS and Pam3Cys (Table 1), whereas TNF-α levels were normal after stimulation with R848. Complete impairment of TNF-α production was not evident in caspase-1-deficient cells because noncleaved Mal may still retain its structural role as a bridging adaptor between TLR2/4 and MyD88, thereby facilitating impaired signaling through MyD88. These data indicate a role for caspase-1 in TLR-2 and TLR-4 signaling but not in TLR 7/8 or IL-1 signaling, the difference between these sets of receptors being Mal utilization.
Fig. 3.
Caspase-1 is required for Mal to signal. (A) (Upper) U373 cells were transfected with a 5x NF-κB reporter gene plasmid. Cells were left untreated or pretreated with YVAD-Cmk (100 μM) or IL-1 receptor antagonist (1 μg/ml) for 1 h. Thereafter, cells were untreated or incubated with LPS (1 μg/ml) or IL-1 (100 ng/ml) for 6 h. Shown is the mean relative stimulation of luciferase activity ± SD for a representative experiment from three separate experiments. (Lower) THP-1 cells were left untreated or pretreated with YVAD-Cmk (100 μM) or IETD-Fmk (50 μM) for 1 h followed by treatment with Pam3Cys (1 μg/ml) or IL-1 (1 μg/ml) for 0–120 min. Activation of p38 was analyzed using an anti-phospho-p38-specific antibody. (B) (Top) Time course of NF-κB activation in wild-type and caspase-1-deficient peritoneal macrophages stimulated with LPS (10 ng/ml), Malp-2 (10 nM), and R848 (10 μM) as detected by EMSA. (Middle) Supershift assay was performed by using an anti-p65 antibody for 1 h before analysis by EMSA. Protein:DNA complexes are shown. (Bottom) Wild-type and caspase-1-deficient murine embryonic fibroblast were treated with LPS (100 ng/ml), lipid A (100 ng/ml), or Malp-2 (10 nM) as indicated, followed by immunoblot analysis of the cell lysates with antibodies directed against IκBα or β-actin. (C) Time course of p38 activation in wild-type and caspase-1-deficient peritoneal macrophages stimulated with LPS (100 ng/ml), Malp-2 (10 nM), and IL-1 (1 μg/ml) analyzed by immunoblotting with phospho-p38-specific antibodies. Total p38 levels are also shown. (D) Time course of p38 activation in wild-type and caspase-1-deficient peritoneal murine embryonic fibroblasts stimulated with LPS (100 ng/ml) or Malp-2 analyzed by immunoblotting with phospho-p38-specific antibodies. Total p38 levels are also shown.
Table 1.
TNF-α secretion from wild-type and caspase-1 KO macrophages
| Stimulus | TNFα concentration, pg/ml |
|
|---|---|---|
| Wild type | Caspase-1 KO | |
| None | 110 ± 5 | 98 ± 32 |
| LPS | 3,108 ± 24 | 2,160 ± 14 |
| Pam3Cys | 1,906 ± 7 | 848 ± 48 |
| R848 | 1,213 ± 22 | 1,320 ± 51 |
Wild-type or caspase-1-deficient peritoneal macrophages (0.5 × 106/well) were treated with LPS (10 ng/ml), Pam3Cys (100 ng/ml), or R848 (10 μM) for 16 h. Cytokine production was then assessed by ELISA. Results shown are mean ± SD. A similar result was obtained in another independent experiment.
Mal Is Cleaved by Caspase-1.
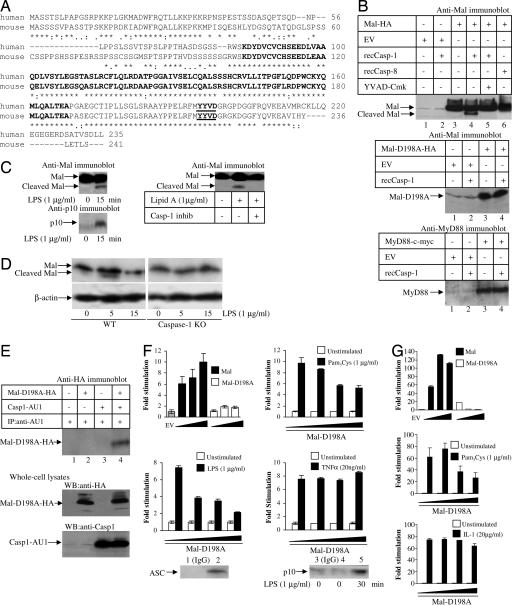
Human and mouse Mal have a putative caspase-1 cleavage (18) site (Fig. 4A). As shown in Fig. 4B Top, treatment of Mal with active recombinant caspase-1 led to the appearance of an additional form of Mal, 4 kDa less than the major form (lane 4), consistent with cleavage of Mal by caspase-1 at the putative caspase-1 cleavage site, and this effect was blocked by YVAD-Cmk, a caspase-1 inhibitor (lane 5). Mal was not cleaved by caspase-8 (lane 6) despite caspase-8 activity being confirmed in vitro as judged by its ability to cleave the fluorogenic substrate Ac-IETD-AMC (data not shown). In addition, mass spectrometry confirmed that the 21-kDa fragment generated by caspase-1 cleavage lacked the portion of Mal C-terminal to D198 and thus established the identity of the cleaved Mal fragment as detected by Western blot analysis (data not shown). When the caspase-1 cleavage site in Mal was mutated, generating Mal-D198A, no cleavage occurred (second panel, lane 4) and MyD88 was also not cleaved by caspase-1 (Fig. 4B Bottom, lane 4). Examination of endogenous Mal in THP-1 cells showed it to be cleaved after 15-min treatment of cells with either LPS (Fig. 4C Left) or Lipid A (Fig. 4C Right). Mal cleavage correlated with the appearance of the p10 subunit of caspase-1 in the cell lysates (Fig. 4C Left Lower), indicative of concomitant caspase-1 activation. Mal cleavage was blocked by a caspase-1 inhibitor (Fig. 4C Right, lane 3). THP-1 cells display caspase-1 activation in response to LPS alone and thus do not require costimulation with ATP (19, 20). In mouse macrophages, treatment with LPS led to the appearance of an additional form of Mal that was 2.5 kDa smaller in molecular mass than the major form of Mal (Fig. 4D Left), consistent with cleavage of murine Mal by caspase-1 at the putative caspase-1 cleavage site, and Mal was unaffected in LPS-treated caspase-1-deficient macrophages (Fig. 4D Right). These data therefore demonstrate that Mal is cleaved by caspase-1.
Fig. 4.
Mal is a substrate for caspase-1. (A) ClustalW alignment of the amino acid sequence of human (top rows) and mouse (lower rows) Mal with the TIR domain in bold and the proposed caspase-1 cleavage site underlined and in bold. (B) for Mal cleavage assay, whole-cell lysates generated from HEK293 cells transfected with a plasmid encoding Mal-HA (Top), Mal-D198A-HA (Middle), or MyD88-cMyc (Bottom) were treated with active recombinant caspase-1 or caspase-8 (Top) in the presence/absence of the caspase-1 inhibitor, YVAD-Cmk. Gels were blotted as indicated. (C) THP1 cells was screened for Mal cleavage following LPS (Left Upper) or Lipid A (± caspase-1 inhibitor; Right Upper) treatment by immunoblotting with an anti-Mal antibody. Cell lysates were also immunoblotted with an antibody against the p10 subunit of caspase-1 (Left Lower). (D) Wild-type and caspase-1 peritoneal macrophages were screened for Mal cleavage after LPS treatment by immunoblotting with an anti-Mal antibody or an anti-β-actin antibody as a loading control. (E) HEK293 cells were transfected with plasmids encoding Mal-D198A-HA or caspase-1-AU1. Immunoprecipitated caspase-1-AU1 was probed for the presence of Mal by immunoblotting. (F) HEK293 cells were transfected with a 5x NF-κB reporter gene plasmid and cotransfected with plasmids encoding empty vector (EV), Mal (1, 40, and 80 ng), or Mal-D198A (1, 40, and 80 ng) (Top Left). HEK293-TLR2 (Top Right), HEK293-TLR4 (Bottom Left), or HEK 293 cells (Bottom Right) were transfected with Mal-D198A (0, 1, 40, and 80 ng) for 24 h. Cells were left untreated or treated with Pam3Cys, LPS, or TNF-α for 6 h. In addition, ASC and the p10 subunit of caspase-1 were immunoprecipitated from untreated (lanes 1–4) or LPS-treated (1 μg/ml, 30 min, lane 5) HEK293-TLR4 cells using an anti-human ASC antibody (lane 2; Genentech) or an anti-p10 antibody (lanes 4 and 5) with an IgG antibody serving as a control (lanes 1 and 3). (G) HEK293 cells were transfected with an IL-8 reporter gene plasmid and cotransfected with plasmids encoding empty vector (EV), Mal (1, 40, and 80 ng), or Mal-D198A (1, 40, or 80 ng) (Top). HEK293-TLR2 (Middle) or HEK293 R1 cells (Bottom) were transfected with Mal-D198A (0, 1, 40, or 80 ng) for 24 h. Cells were left untreated or treated with Pam3Cys or IL-1 for 6 h. For all luciferase assays, mean relative stimulation of luciferase activity ± SD from triplicate determinations for a representative experiment from three separate experiments is shown.
Intact Mal Does Not Activate NF-κB.
We next carried out experiments on Mal-D198A, which cannot be degraded by caspase-1. Mal-D198A can still interact with caspase-1 (Fig. 4E, lane 4), but as shown in Fig. 4B, it is not a substrate. Whereas Mal overexpression activated NF-κB, Mal-D198A was inactive (Fig. 4F). Mal-D198A acted as a dominant-negative inhibitor toward both LPS and Pam3Cys stimuli (Fig. 4F) but had no effect on TNF-α signaling (Fig. 4F). Using an IL-8 promoter-dependent reporter gene, we show that Mal-D198A does not induce this promoter and that induction of the IL-8 promoter by Pam3Cys was blocked by Mal-D198A (Fig. 4G), whereas Mal-D198A did not affect induction by IL-1 (Fig. 4G). Moreover, HEK293 cells contain ASC, a component of TLR-mediated caspase-1 activity (15) (Fig. 4F), and caspase-1 is activated by treatment of these cells with LPS (Fig. 4F, lane 5). The mutant Mal was therefore inactive most likely because it was not cleaved by caspase-1. Taken together, our data therefore strongly suggest that cleavage of Mal by caspase-1 is required for Mal to mediate NF-κB activation.
Discussion
Our study provides a clear, specific aspect to Mal that distinguishes it from the other adapters in TLR signaling (4, 5, 21, 22), in that it is cleaved by caspase-1. Whereas Mal was not required for caspase-1 activation, caspase-1 appeared to be required for the ability of Mal to activate NF-κB. TLR2 and TLR4 signaling was blocked by a caspase-1 inhibitor and was attenuated in caspase-1-deficient macrophages and murine embryonic fibroblasts. All responses tested were affected. Moreover, IL-1 and TLR7 signaling remained normal in these cells. Because the only known difference for these signals between TLR2 and TLR4 on the one hand and IL-1RI and TLR7 on the other is a role for Mal, this result provides circumstantial evidence that caspase-1 was targeting Mal for activation. We then demonstrated that Mal is a substrate for caspase-1 and that a mutant form of Mal, which cannot be cleaved, was unable to activate NF-κB. Finally, we demonstrated that this mutant form of Mal could still interact with caspase-1 but when overexpressed acted as a dominant negative inhibitor of TLR2 and TLR4 signaling. We were therefore able to conclude that Mal is a substrate for caspase-1 and must undergo cleavage to be active. The effect of caspase-1 on Mal is unique among the TIR adapters. We found that MyD88 was not a substrate, and Trif and Tram are unlikely to be regulated by caspase-1 because similar to MyD88 they lack a caspase-1 cleavage site in their primary structures. We have therefore revealed a mechanism for Mal activation and a function for caspase-1 in the regulation of TLR2 and TLR4 signaling.
The first question that arises is how caspase-1 might be activated during TLR2 and TLR4 activation to allow for Mal processing. Caspase-1 has been shown to occur in different protein complexes termed inflammasomes (8), of which three have been described to date (9). How caspase-1 is activated in these inflammasomes is poorly understood, but in all cases TLR ligands such as LPS appear to be required for priming of the inflammasome for subsequent activation by agents such as ATP (15, 23). There is, however, disagreement in the literature concerning this priming event and whether TLR activation alone is sufficient to activate caspase-1. A recent study has shown that ultrapure LPS alone can activate caspase-1 in murine macrophages (11) or in certain cell lines such as THP-1 (8). It is possible that the issue here is magnitude; TLR stimulation alone will activate caspase-1, but this response requires amplification by endogenous factors such as uric acid or ATP, which acts via Nalp3 (16). Given that Mal requires cleavage by caspase-1 to be active, it is therefore possible that during infection, bacterial products will be sensed by TLRs but will also lead to the activation of caspase-1 in inflammasomes, an effect amplified by the generation of endogenous ligands. Caspase-1 will then process Mal into its active form, amplifying signaling by TLR2 and TLR4.
Regarding the TLR adapters, caspase-1 activation by LPS has been shown to be normal in MyD88-, Trif- (24), and, from our study, Mal-deficient macrophages. The caspase-1 assay relies on cells primed with LPS or Pam3Cys and then treated with ATP (15). The response measured is the appearance of the p10 or p20 subunits of caspase-1, which are indicative of autoprocessing. We cannot fully rule out a role for Mal in caspase-1 function, however, because the precise role of caspase-1 processing for its activity is uncertain. Although caspase-1 activation was unaffected in Mal-deficient cells, we found that the production of IL-1β and IL-18 was impaired, identifying these cytokines as being on the MyD88/Mal-dependent pathway. This effect was due to Mal having a role in the induction of pro-IL-1β.
Our results also provide a previously unsuspected function for caspase-1 activity. Pro-IL-1β, pro-IL-18, and possibly pro-IL-33 are the only previously described physiological substrates for caspase-1 (6, 18, 25). We can now add Mal to this list. Our findings may explain the profound phenotype of caspase-1-deficient mice, which, unlike IL-1β and IL-18 knockout (KO) mice, and as recently shown in IL-1β/IL-18 double KO mice (26), are completely resistant to the effects of LPS (27–29) and are also highly susceptible to infection with pathogens such as Escherichia coli (30, 31). Similar to our study, others have shown defects in LPS responses in caspase-1-deficient macrophages such as induction of TNF, IL-6, and IL-1α (20, 29, 31). TNF production in response to LPS was not affected by a neutralizing antibody to IL-18 or in IL-1β-deficient mice (32, 33). Our data however provide an explanation because the defect in TLR4 signaling in caspase-1-deficient mice is likely to be because Mal is not cleaved.
Similar to our study, two other recent studies have found proteins that interact with caspase-1. Chae et al. (34) have shown that the protein Pyrin interacts directly with caspase-1 and interferes with its activation. Sarkar et al. (20) have shown that Receptor Interacting Protein-2 (RIP2) interacts with caspase-1 and that this interaction is somehow required for NF-κB activation, although direct evidence for this effect was not provided. Similar to our data, NF-κB activation by LPS was attenuated in caspase-1-deficient macrophages and was inhibited by a catalytically inactive form of caspase-1. This effect was interpreted as being due to impairment in RIP2 activation. Our results provide an additional explanation because this response may be due to the lack of Mal cleavage in caspase-1-deficient cells and somehow RIP2 may also be required for this process. An important outstanding question concerns the reason why Mal is cleaved by caspase-1. It is possible that an inhibitory domain is released from Mal. Alternatively, an active moiety from Mal is released. These possibilities should be examined. Ultimately, Mal is fully degraded in a SOCS-1-dependent manner (35).
In conclusion, our findings demonstrate a role for caspase-1 in inflammation beyond its ability to process pro-IL-18 or pro-IL-1β, implicating it in the regulation of many proinflammatory genes. Thus, Mal has a specific property among the TLR adapter proteins in that it requires cleavage by caspase-1 to elicit the downstream signal NF-κB. These findings therefore reveal an important mechanism for the coordinated activation of host defense responses by TLRs and NLR-containing inflammasomes (9).
Materials and Methods
Biological Reagents and Cell Culture.
Thioglycollate was from REMEL Inc. (Lenexa, KS). Highly purified protein-free LPS derived from E. coli strain 011:B4 was used in all treatments. Synthetic Malp-2 was from the EMC Microcollection (Tuebingen, Germany). Pam3Cys, human recombinant caspase-1 and caspase-8, YVAD-Cmk, IETD-Fmk, Mal inhibitor, and control peptides were from Calbiochem (San Diego, CA). pcDNA3.1-caspase-1-AU1 was a gift from Seamus Martin (Trinity College, Dublin). Antibodies to caspase-1 (A-19) and p10 subunit (M-20) were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-Mal antibody was raised against full-length human Mal (Clone R4; Trinity College, Dublin).
Sources of Macrophages.
Mal/TIRAP KO, MyD88 KO, TLR4 KO, and TLR2 KO mice were constructed as described (4, 36, 37). MyD88 KO mice were backbred to C57BL/6 for 15 generations, and Mal/TIRAP KO mice were on a mixed C57BL/6 and 129 background. Caspase-1 KO and wild-type mice were supplied by W. Wong (BASF Bioresearch Corporation, Worcester, MA). All mice were confirmed as being homozygous mutants by PCR genotyping of DNA. All of the animal protocols used in this study were approved by the Institutional Animal Care and Use Committee at the University of Massachusetts Medical School (Worcester, MA) and in accordance with Animals (Scientific Procedures) Act of 1986, United Kingdom.
Yeast Two-Hybrid Analysis.
Full-length Mal, TIR domain of Mal, N-terminal domain of Mal, and full-length MyD88 were subcloned into pGBKT7 downstream of the Gal4 DNA-binding domain. pGBKT7:full-length Mal was used as a bait to screen a cDNA library prepared from human splenocytes expressed in the yeast strain AH109 essentially as described by the manufacturer (Matchmaker Gal4 Two-Hybrid System 3; Clontech, Palo Alto, CA). The combinations tested on His-Leu-Trp-Ade indicate protein–protein interactions.
Reporter Assays.
HEK293, HEK293-TLR-4, HEK293-TLR2, and U373 cells (2 × 104 cells per well; 96-well plate) were transfected with 80 ng per well 5x NF-κB luciferase reporter gene plasmid and cotransfected with the expression vectors pcDNA3:Mal or pcDNA3:Mal-D198A by using GeneJuice essentially as described by the manufacturer (Novagen, Madison, WI). In all cases, 40 ng per well of phRL-TK reporter gene was cotransfected to normalize data for transfection efficiency. HEK293, HEK293-TLR2, and HEK293-R1 cells (2 × 104 cells per well; 96-well plate) were transfected with the 80 ng per well IL-8 promoter reporter plasmid (38) and cotransfected with the expression vectors pcDNA3:Mal-HA or pcDNA3:Mal-D198A-HA by using GeneJuice. In all cases, 40 ng per well of phRL-TK reporter gene was cotransfected to normalize data for transfection efficiency. After 24 h, reporter gene activity was measured as described (38). Data are expressed as the mean fold induction ± SD relative to control levels, for a representative experiment from a minimum of three separate experiments, each performed in triplicate.
Transfection, Coimmunoprecipitation, and GST Pull-Down Assays.
HEK293 cells (2 × 106 cells/10-cm dish) were transfected by using GeneJuice (Novagen) with the indicated plasmids where the total amount of DNA (4 μg per dish) was kept constant. Twenty-four h later, cells were lysed as described (38). The indicated antibodies (5 μg) were incubated with the cell lysates for 2 h, followed by the addition of 40 μl of 50% protein-G slurry for 1 h. The immune complexes were precipitated, washed, eluted by the addition of sample buffer, followed by SDS/PAGE and immunoblotting by using the indicated antibodies. For GST pull-down experiments, lysates prepared from HEK293 cells transfected with pcDNA3-caspase-1-AU1 or pcDNA3-Mal-HA were used in a GST pull-down assay whereby cell lysates were incubated for 2 h at 4°C with recombinant GST fusion protein coupled to glutathione-Sepharose. Complexes were washed three times in lysis buffer, separated by SDS/PAGE, and immunoblotted as indicated in the Fig. 1 legend.
Mal Cleavage Assay.
Lysates (600 μl per each 10-cm dish) prepared from HEK293 cells transfected (38) with the expression vectors pcDNA3-Mal-HA or pcDNA3-Mal-D198A-HA (3 μg of DNA per dish) were used as substrates in a Mal cleavage assay, as described for IL-1β (39) using human recombinant caspase-1 (2 μl per point) or caspase-8. Briefly, HEK293 cells overexpressing Mal, Mal-D198A, or MyD88 were lysed in 500 μl of low-stringency IP buffer (50 mM Hepes, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Nonidet P-40) for 15 min at 4°C followed by centrifugation at 800 × g for 5 min. Supernatants (20 μl of lysate per point) were incubated with 2 μl of recombinant caspase-1 (100 units) in assay buffer (100 mM Hepes, 10% sucrose, 10 mM DTT, 0.1% CHAPS, pH 7.5) for 1.5 h at 30°C. Lysates were subjected to SDS/PAGE and followed by immunoblot analysis.
EMSAs.
Murine peritoneal macrophages derived from wild-type and caspase-1-deficient mice were stimulated as indicated in the Fig. 3 legend, and their nuclear extracts were prepared as described. Nuclear extracts (4–8 μg of protein) were incubated for 30 min at 4°C with 10,000 counts per minute of double-stranded [γ-32P]ATP NF-κB oligonucleotide (5′-AGTTGAGGGGACTTTCCCAGGC-3′). Incubations were done in the presence of 2 μg of poly(dI:dC) as a nonspecific competitor and 10 mM Tris·HCl (pH 7.5) containing 100 mM NaCl, 1 mM EDTA, 5 mM DTT, 4% glycerol, and 100 μg/ml nuclease-free BSA. A supershift antibody specific for p65 (SC-8008X; Santa Cruz Biotechnology) was added to the nuclear extracts 1 h before hybridization with the oligonucleotide. DNA–protein complexes were resolved on native (5%) polyacrylamide gels that were subsequently dried and visualized by autoradiography.
Cytokine Analysis and Caspase-1 Processing Protocol.
Thioglycollate-elicited peritoneal macrophages (5 × 105 cells per well; 48-well plate) were stimulated with the following stimuli: LPS (100 ng/ml), Malp-2 (0.01 nM), and R848 (0.01 nM). For IL-1β release, peritoneal macrophages were then treated with 5 mM ATP for 20 min. The medium was then removed and replaced with 600 μl of fresh medium. After 3 h, these cell supernatants were removed and analyzed for IL-1β release according to the manufacturer's recommendations (R & D Systems, Minneapolis, MN). To measure caspase-1 processing, freshly made ATP (5 mM) in regular media was added for 20 min, and lysates were blotted for caspase-1 using the p10 antibody as described (15). A similar protocol was used for bone marrow-derived macrophages.
Immunoblotting for p38.
Cells were stimulated with ligand as described, and lysates were subjected to SDS/PAGE followed by immunoblot analysis with an anti-phospho-(Thr180/Tyr182) p38 MAPK or an antibody for total p38 (New England Biolabs, Ipswich, MA).
Data Analyses.
Statistical analysis was carried out by using the unpaired Student t test. P values of ≤0.05 were considered to indicate a statistically significant difference.
Acknowledgments
We thank Prof. Seamus Martin (Department of Genetics, Trinity College, Dublin) for helpful discussions, Kristen Halmen and Amit Roy for breeding and genotyping mice, Dr. Sanjeev Mariathasan (Genentech, South San Francisco, CA) for the generous gift of an anti-human ASC antibody and advice on the caspase-1 processing assay, Dr. Egil Lien for assistance with the isolation of peritoneal macrophages, and Dr. Cos Brikos for mass spectrometry advice. This work was supported by grants from Science Foundation Ireland (to L.A.J.O.), the Wellcome Trust (to K.A.F.), Health Research Board (to S.M.M.), and National Institutes of Health (to D.G.).
Abbreviations
- TLR
Toll-like receptor
- TIR
Toll/IL-1 receptor
- NLR
Nod-like receptor
- KO
knockout.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Miggin SM, O'Neill LA. J Leukoc Biol. 2006;80:220–226. doi: 10.1189/jlb.1105672. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Takeda K. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, Hoshino K, Takeuchi O, Kobayashi M, Fujita T, et al. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. J Exp Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, et al. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Creagh EM, Conroy H, Martin SJ. Immunol Rev. 2003;193:10–21. doi: 10.1034/j.1600-065x.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 8.Martinon F, Burns K, Tschopp J. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 9.Creagh EM, O'Neill LA. Trends Immunol. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Martinon F, Tschopp J. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, et al. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 12.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 13.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 14.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, et al. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 15.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 16.Horng T, Barton GM, Flavell RA, Medzhitov R. Nature. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. Nat Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 18.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, et al. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 19.Martinon F, Agostini L, Meylan E, Tschopp J. Curr Biol. 2004;14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 20.Sarkar A, Duncan M, Hart J, Hertlein E, Guttridge DC, Wewers MD. J Immunol. 2006;176:4979–4986. doi: 10.4049/jimmunol.176.8.4979. [DOI] [PubMed] [Google Scholar]
- 21.O'Neill LA, Fitzgerald KA, Bowie AG. Trends Immunol. 2003;24:286–290. doi: 10.1016/s1471-4906(03)00115-7. [DOI] [PubMed] [Google Scholar]
- 22.Oshiumi H, Sasai M, Shida K, Fujita T, Matsumoto M, Seya T. J Biol Chem. 2003;278:49751–49762. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- 23.Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M, Yaginuma K, Tsutsui H, Sagara J, Guan X, Seki E, Yasuda K, Akira S, Nakanishi K, Noda T, et al. Genes Cells. 2004;9:1055–1067. doi: 10.1111/j.1365-2443.2004.00789.x. [DOI] [PubMed] [Google Scholar]
- 25.Gu Y, Kuida K, Tsutsui H, Ku G, Hsiao K, Fleming MA, Hayashi N, Higashino K, Okamura H, Nakanishi K, et al. Science. 1997;275:206–209. doi: 10.1126/science.275.5297.206. [DOI] [PubMed] [Google Scholar]
- 26.Sarkar A, Hall MW, Exline M, Hart J, Knatz N, Gatson NT, Wewers MD. Am J Respir Crit Care Med. 2006;174:1003–1010. doi: 10.1164/rccm.200604-546OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng H, Fletcher D, Kozak W, Jiang M, Hofmann KJ, Conn CA, Soszynski D, Grabiec C, Trumbauer ME, Shaw A, et al. Immunity. 1995;3:9–19. doi: 10.1016/1074-7613(95)90154-x. [DOI] [PubMed] [Google Scholar]
- 28.Sakao Y, Takeda K, Tsutsui H, Kaisho T, Nomura F, Okamura H, Nakanishi K, Akira S. Int Immunol. 1999;11:471–480. doi: 10.1093/intimm/11.3.471. [DOI] [PubMed] [Google Scholar]
- 29.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, McDowell J, Paskind M, Rodman L, Salfeld J, et al. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 30.Joshi VD, Kalvakolanu DV, Hebel JR, Hasday JD, Cross AS. Infect Immun. 2002;70:6896–6903. doi: 10.1128/IAI.70.12.6896-6903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuida K, Lippke JA, Ku G, Harding MW, Livingston DJ, Su MS, Flavell RA. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 32.Netea MG, Fantuzzi G, Kullberg BJ, Stuyt RJ, Pulido EJ, McIntyre RC, Jr, Joosten LA, Van der Meer JW, Dinarello CA. J Immunol. 2000;164:2644–2649. doi: 10.4049/jimmunol.164.5.2644. [DOI] [PubMed] [Google Scholar]
- 33.Fantuzzi G, Dinarello CA. J Leukoc Biol. 1996;59:489–493. doi: 10.1002/jlb.59.4.489. [DOI] [PubMed] [Google Scholar]
- 34.Chae JJ, Wood G, Masters SL, Richard K, Park G, Smith BJ, Kastner DL. Proc Natl Acad Sci USA. 2006;103:9982–9987. doi: 10.1073/pnas.0602081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, Nicholson SE, Hilton DJ, O'Neill LA, Hertzog PJ. Nat Immunol. 2006;7:148–155. doi: 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- 36.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 37.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 38.Bowie A, Kiss-Toth E, Symons JA, Smith GL, Dower SK, O'Neill LA. Proc Natl Acad Sci USA. 2000;97:10162–10167. doi: 10.1073/pnas.160027697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thornberry NA. Methods Enzymol. 1994;244:615–631. doi: 10.1016/0076-6879(94)44045-x. [DOI] [PubMed] [Google Scholar]