Abstract
In vivo studies have previously shown that Saccharomyces cerevisiae ribosomal protein (RP) gene expression is controlled by the transcription factor repressor activator protein 1 (Rap1p) in a TFIID-dependent fashion. Here we have tested the hypothesis that yeast TFIID serves as a coactivator for RP gene transcription by directly interacting with Rap1p. We have found that purified recombinant Rap1p specifically interacts with purified TFIID in pull-down assays, and we have mapped the domains of Rap1p and subunits of TFIID responsible. In vitro transcription of a UASRAP1 enhancer-driven reporter gene requires both Rap1p and TFIID and is independent of the Fhl1p-Ifh1p coregulator. UASRAP1 enhancer-driven transactivation in extracts depleted of both Rap1p and TFIID is efficiently rescued by addition of physiological amounts of these two purified factors but not TATA-binding protein. We conclude that Rap1p and TFIID directly interact and that this interaction contributes importantly to RP gene transcription.
Eukaryotic mRNA gene transcription is controlled by the action of modular enhancer-binding transactivators, proteins composed of separable DNA binding domains (DBD) and activation domains (AD). DNA-bound transactivators functionally interact with the mRNA gene transcription machinery, the so-called general transcription factors (GTFs) TFIIA, -B, -D, -E, -F, and -H plus RNA polymerase II (RNAP II), to stimulate formation and/or function of the RNAP II preinitiation complex (PIC) (67). Activators also collaborate with one or more factors termed coactivators, proteins that serve as receptors for the transfactor-AD activation signal. Coactivators link transactivator-enhancer DNA binding to the PIC (43) and can be divided into several classes: those that are chromatin active, the mediator complex, and the individual components of the mRNA gene transcription machinery itself.
One of the first and likely rate-limiting steps in mRNA gene transcription is the binding of TFIID to the promoter (36, 40, 47). TFIID is a multisubunit assembly composed of 15 evolutionarily conserved (86) subunits, the TATA-binding protein (TBP) and 14 TBP-associated factors (TAFs) (72). This GTF displays high-affinity, sequence-specific promoter DNA binding activity. Two classes of yeast mRNA-encoding genes have been defined with respect to TFIID TAF function; the first, which is TAF dependent (TAFdep), requires TAF function for transcription, while the second and smaller group is TAF independent (TAFind) (29, 31, 45, 58, 81, 92). Both types of genes require TBP for wild-type (WT) levels of transcription, but in the case of the TAFind genes, TBP appears to be recruited via mechanisms distinct from TFIID (4, 80).
When mRNA-encoding genes are monitored for TBP and TAF occupancy by chromatin immunoprecipitation (ChIP) assay, TAFdep genes exhibit higher TAF/TBP occupancy ratios than TAFind genes (39, 45). Three models for TAF function have been proposed (22, 23): TAFs mediate core promoter recognition; TAFs provide essential catalytic activities that modulate PIC formation and/or function; and finally, TAFs may serve as coactivators, by providing a direct molecular bridge between enhancer-bound transactivators and the other components of the mRNA gene transcription machinery. These functions of TAFs likely operate in gene-specific transfactor-directed ways, and potentially multiple TAF modalities could be utilized on a single transcription unit.
Ribosomal protein genes (RPGs) are TAF dependent and require TAF function in vivo for normal levels of transcription (45, 58, 81, 92). TFIID associates with RPG promoters, resulting in high Taf1p/TBP ChIP occupancy ratios. RPGs often contain TATA-less promoters, a feature that may contribute to their TAFdep nature (9, 53); TFIID functions primarily at TATA-less promoters (3). An essential regulatory element of the RPGs is the upstream activating sequence (UAS) or enhancer. Nearly all of the 137 RP gene enhancers contain multiple binding sites for repressor activator protein 1 (Rap1p) (42, 93), and most of these UASRAP1 elements appear to be bound by Rap1p in vivo (48). Mutation of UASRAP1 sites disrupts Rap1p binding in vitro and in vivo, concomitantly decreasing Taf1p and TBP occupancy at the promoter. In the case of the RPGs, this leads to a significant drop in RPG transcription. These and other observations have led to the hypothesis that the recruitment of TFIID to the RPGs is driven by Rap1p (46, 53), though the molecular mechanism by which this occurs is unknown.
Rap1p, encoded by a single-copy essential gene in yeasts, is an abundant (ca. 104 molecules/cell) DNA-binding protein that plays multiple roles in vivo. It is both a key transcriptional activator of many coregulated genes (83) and a repressor that silences transcription at HML, HMR, and telomeres (41). Additionally, Rap1p contributes to telomere length modulation (59) and maintenance of recombination hot spots (11) and can provide barrier function by preventing the spread of silenced chromatin (5, 17). Most Rap1p binding sites are devoid of nucleosomes (93). Mapping studies have identified Rap1p DNA bending, BRCT, DBD, toxicity, noncanonical AD, and silencing domains (60) (Fig. 1A).
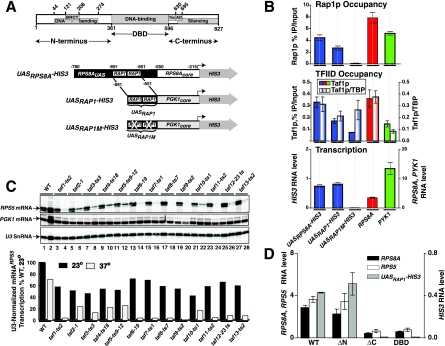
FIG. 1.
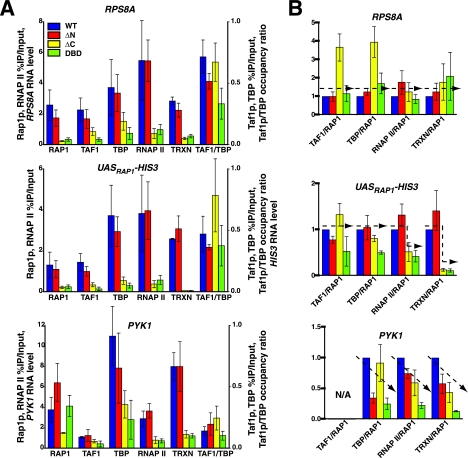
Rap1p drives UASRAP1-directed transcription in vivo. (A) Rap1p domain structure and reporter genes. The diagram illustrates the localization of Rap1p domains (top) and the various reporter genes used in this study (bottom). (B) Requirement for Rap1p binding for both TFIID recruitment and transcription in vivo. Rap1p occupancy (top), Taf1p occupancy and Taf1p/TBP occupancy ratio (middle), and in vivo transcription (bottom) of three reporter genes (UASRPS8A-HIS3, UASRAP1-HIS3, and UASRAP1M-HIS3) and two control genes (TAFdep RPS8A and TAFind PYK1) are shown. Occupancy was calculated as percent enrichment of a gene segment in the IP relative to the input. Background occupancy was estimated as enrichment of the same proteins on the POLI gene and was subtracted. Transcription was assessed following reverse transcription of the mRNAs by quantitative real-time PCR. Error bars indicate standard deviations. (C) Dependence of RP, but not PGK, gene transcription on TFIID-Tafp function. Cells carrying the indicated TAFts alleles were cultured at 23°C to mid-log phase and split, and half of the culture was incubated at 23oC while the other half was shifted to 37oC. Total RNA was extracted after 2 h and purified, and then PGK, RPS5, and U3 RNA levels were scored by primer extension. Extension products were gel fractionated, and the sequencing gel was exposed to an imaging screen for quantitation (the scanned screen image is shown [top]; odd-numbered lanes, 23oC cultures; even-numbered lanes, 37oC cultures). The RPS5 RNA level was normalized to U3 RNA and graphed as percent of WT at 23oC. (D) Effect of deletion of Rap1p domains on Rap1p-driven transcription in vivo. Steady-state mRNA levels for the three indicated genes (RPS8A, RPS5, and the UASRAP1-HIS3 reporter) in the strains expressing the four indicated variants of Rap1p shown were determined by Q-RT-PCR analyses as for panel B.
Rap1p plays a critical role in yeast physiology by activating the expression of the most vigorously transcribed mRNA-encoding genes, such as those encoding ribosomal proteins, ribosome biogenesis (RiBi) regulon proteins (33, 90), translation factors, and glycolytic enzymes, all of which are required for rapid cell proliferation (69, 93). It has been estimated that Rap1p drives nearly 40% of all RNA polymerase II-mediated transcription events (48). Rap1p utilizes different mechanisms of activation for each of these coordinated, complex regulatory events. For example, while Rap1p is required for transcription of both RP- and glycolytic enzyme-encoding genes, it collaborates with transcription factors Gcr1p and Gcr2p only on the TAFind glycolytic enzyme-encoding genes (10). By contrast, ribosomal protein, but not glycolytic enzyme, gene transcription is dependent on the H4-histone acetylase Esa1p (66). DNA-bound Rap1p thus selectively interacts with a variety of cofactors, in a context-dependent manner, for the purpose of activating unique sets of genes. Rap1p also functions with several other proteins that modulate RPG and RiBi regulon gene regulation, transcription factors Fhl1p, Ifh1p, Crf1p, Sfp1p, and Hmo1p (15, 26, 33, 49, 50, 70, 77, 91, 99). These factors are thought to integrate and transduce growth regulatory signals to the RP/RiBi genes, and together with Rap1p they modulate expression of the many genes important for ribosome production, cell division, and growth. Interestingly, Rap1p occupancy on UASRAP1 sites in vivo does not appear to vary significantly with transcription rate, suggesting either that non-Rap1p coregulators turn RP gene transcription on and off or that modification of Rap1p and/or other factors controls transcription levels of these genes (see references 63 and 69 for recent reviews). The choice of Rap1p interaction partner may be directed by Rap1p conformation or other UASRAP1-determined features, which in turn may be dictated either by the exact sequence of the UASRAP1 site bound or by neighboring DNA sequences that provide docking sites for other transfactors (26, 63, 69, 99). It has been proposed that these multiple protein-protein interactions may occur on the nuclear matrix (8, 55, 78). Though the details of the many regulatory interactions described above remain to be elucidated, it is clear that both Rap1p and TFIID play central roles in the process (46, 53). For all of these reasons, RPGs are among the most attractive genes in yeast for analyzing the molecular mechanisms by which TFIID might function as a transcriptional coactivator.
The goal of the studies described here was to analyze the mechanism behind the previously characterized functional interaction between Rap1p and TFIID on the ribosomal protein genes. We tested the hypothesis that TFIID plays a key coactivator role on RPGs by making direct, specific interactions with Rap1p. We have found that Rap1p and TFIID do indeed directly interact, and we mapped Rap1p and TFIID domains that contribute importantly to their interaction. These data represent the first demonstration of direct and specific interactions between a purified yeast transactivator and the purified yeast TFIID holocomplex, and they set the stage for the dissection of the coactivator function(s) of the multisubunit TFIID TBP-TAF complex.
MATERIALS AND METHODS
Yeast strains.
We reconstructed several of the SPY-ADE strains of Mencia et al. (53). These strains express an HA3-tagged variant of Taf1p and various UAS-HIS3 reporter genes (see Table 1 and Fig. 1A for construct details); reporter genes were sequenced prior to integration. We introduced two additional genetic modifications into these yeast strains. First, a UASGAL-controlled WT RAP1 allele, derived from strain SCR101 (21), was integrated into the RAP1 locus. Second, these strains were transformed with pRS415 expressing Myc5-tagged forms of Rap1p (WT [amino acids {aa} 1 to 827], ΔN [aa 361 to 827], ΔC [aa 1 to 596], or DBD [aa 361 to 596]) controlled by the normal RAP1 enhancer-promoter (i.e., RAP1 position −437 to +3 [21]). These modifications of the SPY-ADE strains allowed us to switch from expression of the chromosomal WT RAP1 copy to expression of to the plasmid-borne RAP1 variants by changing the carbohydrate source in the growth medium from galactose to glucose.
TABLE 1.
Yeast strains
| Strain | Genotype |
|---|---|
| SAR229-MYC5WT | MATaade2::hisG leu2::PET56 met17-Δ0 trp1-63 ura3-Δ0 HA3TAF1 UASRPS8A(229)-RPS8ACORE-HIS3::TRP1::DED1 URA3::UASGAL1RPL25core-RAP1(pRS415-UASRAP1-MYC5-RAP1) |
| SAR41-MYC5WT | MATaade2::hisG leu2::PET56 met17-Δ0 trp1-63 ura3-Δ0 HA3TAF1 UASRPS8A(41)-RPS8ACORE-HIS3::TRP1::DED1 URA3::UASGAL1RPL25core-RAP1 (pRS415-UASRAP1-MYC5-RAP1ΔN) |
| SAR41M-MYC5WT | MATaade2::hisG leu2::PET56 met17-Δ0 trp1-63 ura3-Δ0 HA3TAF1 UASRPS8A(41M)-RPS8ACORE-HIS3::TRP1::DED1 URA3::UASGAL1RPL25core-RAP1(pRS415-UASRAP1-MYC5-RAP1) |
| SAR41-MYC5ΔN | MATaade2::hisG leu2::PET56 met17-Δ0 trp1-63 ura3-Δ0 HA3TAF1 UASRPS8A(41)-RPS8ACORE-HIS3::TRP1::DED1 URA3::UASGAL1RPL25core-RAP1(pRS415-UASRAP1-MYC5-RAP1ΔN) |
| SAR41-MYC5ΔC | MATaade2::hisG leu2::PET56 met17-Δ0 trp1-63 ura3-Δ0 HA3TAF1 UASRPS8A(41)-RPS8ACORE-HIS3::TRP1::DED1 URA3::UASGAL1RPL25core-RAP1(pRS415-UASRAP1-MYC5-RAP1ΔC) |
| SAR41-MYC5DBD | MATaade2::hisG leu2::PET56 met17-Δ0 trp1-63 ura3-Δ0 HA3TAF1 UASRPS8A(41)-RPS8ACORE-HIS3::TRP1::DED1 URA3::UASGAL1RPL25core-RAP1(pRS415-UASRAP1-MYC5-RAP1DBD) |
| YSLS18 | MATα ura3-52 lys2-801amberade2-101ochre taf1Δ::TRP1 his3-Δ200 leu2-Δ1(pRS313-HA1TAF130) |
| YPH252 | MATα ura3-52 lys2-801amberade2-101ochre trp1-Δ1 his3-Δ200 leu2-Δ1 |
| DR35 | W303α FHL1Δ::HIS3 IFH1 Δ::G418r |
| W303α | MATα ura3-1/ura3-1 his3-11,15/his3-11,15 ade2-1/ade2-1 leu2-3,112/leu2-3,112 trp1-1/trp1-1 can1-100/can1-100 |
| YLSTAF1 | MATaTAF1-TAP4.5-KANr leu2 trp1 ura3-52 prb1-1122 pep4-3 pre1-451 gal2 |
Protein expression and purification.
RAP1 sequences encoding Rap1p WT (aa 1 to 827), ΔN (aa 361 to 827), ΔC (aa 1 to 596), DBD (aa 361 to 596), and ΔDBD (aa 1 to 361 fused to Rap1p aa 596 to 827) were amplified by PCR, introducing EcoRI and XhoI restriction endonuclease cleavage sites at the N-terminal and C-terminal ends of amplified open reading frame (ORF) DNA fragments. After restriction endonuclease digestion, these fragments were cloned into EcoRI-XhoI-digested pET28+ and introduced into Escherichia coli BL21(DE3), and the proteins were expressed following IPTG (isopropyl-β-d-thiogalactopyranoside) addition. The resulting His6-tagged recombinant proteins were purified with Ni-nitrilotriacetic acid-agarose and either HiTrap SP HP (Rap1WT, Rap1ΔN, Rap1ΔC, and Rap1DBD) or HiTrap Q HP (Rap1ΔDBD) ion-exchange chromatography. Proteins were eluted from the ion exchange columns by using a 150 to 1,000 mM NaCl gradient in a buffer containing 20 mM HEPES (pH 7.6), 10% (vol/vol) glycerol, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM benzamidine, and 0.001% (vol/vol) Nonidet P-40. Recombinant His6-Taf4p, His6-Taf5p, and His6-Taf12p were purified as described previously (73). Glutathione S-transferase (GST)-Taf12p fusion proteins were generated by PCR by inserting BamHI (upstream) and SalI (downstream) sites flanking the TAF12 ORF-carrying segments in all PCR primers. After digestion with BamHI and SalI, the resulting fragments were cloned into a similarly digested pET27b derivative expression plasmid. This Kanr-marked plasmid has had the DNA sequence encoding its normal pelB periplasmic targeting sequence replaced with a DNA fragment expressing, in frame, His6-GST and a protease 3C cleavage sequence upstream of the TAF12 ORF insertion site. Plasmids were introduced into E. coli and induced with IPTG, and GST-fusion proteins were purified by glutathione agarose affinity chromatography. TAP-tagged Taf1p TFIID was purified from yeast strain YLSTAF1 by immunoglobulin G (IgG)-Sepharose, calmodulin-agarose, and Mono-S fast protein liquid chromatographies (Bio-Rad Uno-S1) (unpublished data). TFIID was >95% pure as assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Pull-down assays.
Yeast strain YSLS18 (74), which contains a single hemagglutinin (HA) epitope at the N terminus of Taf1p, was used for the preparation of a Bio-Rex 70 1 M fraction containing partially purified TFIID; as a control, an untagged Bio-Rex 70 1 M fraction was prepared from strain W303α. TFIID from 1.2 ml of each 1 M Bio-Rex 70 fraction (12.5 mg total protein) was incubated with 100 μl anti-HA-protein A-Sepharose beads (2.5 μg monoclonal antibody [MAb] 12CA5 IgG/μl beads) as described previously (72). After overnight binding at 4°C, beads were washed five times with 1.5 ml buffer B200 (20 mM HEPES [pH 7.6], 10% [vol/vol] glycerol, 200 mM potassium acetate, 1 mM dithiothreitol, 0.1 mM PMSF, 1 mM benzamidine, and 0.002% [vol/vol] Nonidet P-40), and then 15-μl aliquots of beads were distributed to individual 70-μl (final volume) binding reaction mixtures in B200. Bovine serum albumin (BSA) (Sigma) was added to a final concentration of 0.5 mg/ml prior to addition of a 3× molar excess of Rap1p (2.25 fmol) to TFIID (0.75 fmol). Binding was allowed to proceed for 30 min at room temperature. The beads were washed five times with 600 μl buffer B200, and then TFIID, along with the TFIID-bound proteins, was eluted with 50 μl of 2 mg/ml of HA peptide in B200. Aliquots of the input and HA-peptide eluate were fractionated by SDS-PAGE, blotted to polyvinylidene difluoride (PVDF) membranes, and probed with polyclonal antibodies against Rap1p and Taf13p; antibody-antigen complexes were detected by chemiluminescence (73, 74).
Determination of Kd,app values for the interaction of Rap1p and TFIID.
The apparent Kd(Kd,app) was calculated using the formula Kd,app = [TFIID][Rap1p]/[TFIID-Rap1p]. The concentration of the TFIID-Rap1p complex was determined by calculating the fraction of total Rap1p bound to TFIID {percent binding = 100[PD(Rap1p)/IN(Rap1p)], where PD is the pull-down fraction and IN is the input fraction) corrected for the efficiency of elution of TFIID from the protein A-MAb 12CA5 IgG beads. The amounts of TFIID and Rap1p in the input, beads, and pull-down fractions were determined by quantitative immunoblotting using dilution series of known quantities of purified TFIID and Rap1p electrophoresed in parallel lanes of SDS-polyacrylamide gels, polyclonal anti-Tafp, and anti-anti-Rap1p IgGs. Quantitation was performed using a Bio-Rad Chemi-Doc imager and Bio-Rad QuantityOne software to measure chemiluminescence signals from Western blots. The average Kd,app values determined in three independent experiments are presented.
In vitro transcription and primer extension analysis of RNAs.
Woontner whole-cell extracts (WCEs) (95) were prepared, antibody depleted, and assayed by primer extension as described previously (35, 72, 79). mRNAHIS3 transcribed from the UASRAP1-HIS3 reporter was scored using 32P-labeled primer ATCGCAATCTGAATCTTGGTTTCATTTGTAATACGC (specific activity, ∼6,700 cpm/fmol; 25 fmol primer/reaction); extension produced a cDNA of 107 nucleotides. Extension products were detected with X-ray film and quantitated via imaging (Kodak K-screen; Bio-Rad Molecular Imager FX and Bio-Rad QuantityOne software) and corrected for recovery by quantitation of a 32P-labeled 330-nucleotide internal recovery control DNA added to each reaction mixture with the transcription stop mix. Quantitative immunoblotting using known amounts of purified His6-Rap1p, His6-TBP, and TFIID was used to determine the content of these proteins in the WCE utilized for the experiments of Fig. 2; ∼270 fmol Rap1p/μl WCE, 125 fmol TBP/μl WCE, ∼32 fmol TFIID/μl WCE, and 4 μl (or 4-μl equivalents) of WCE were used for these in vitro transcription assays.
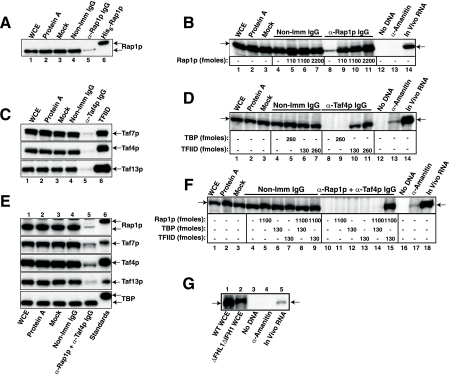
FIG. 2.
Rap1p and TFIID are both necessary for UASRAP1-driven WCE in vitro transcription. (A) Efficacy and specificity of Rap1p immunodepletion. WCEs were treated as indicated: no treatment (WCE), with protein A beads alone (Protein A), subjected to incubation but with no additions (Mock), with nonimmune rabbit IgG bound to protein A beads (Non-Imm IgG), or with anti-Rap1p IgG bound to protein A beads (α-Rap1p IgG). Equivalent amounts of each of these WCEs, along with 2 ng purified recombinant His6-Rap1p as a positive control, were subjected to SDS-PAGE and blotted, and the blots were probed with anti-Rap1p antibody (Rap1p and His6-Rap1p are indicated by arrows). (B) Transcription of UASRAP1-HIS3 is Rap1p dependent and rescued in Rap1p-depleted WCE by readdition of purified recombinant Rap1p. The control (lanes 1 to 7) and immunodepleted (lanes 8 to 11) WCEs of panel A, with either no additions (lanes 1, 2, 3, 4, 8, 12, and 13) or the addition of the indicated amounts of purified recombinant Rap1p were used for in vitro transcription experiments with a plasmid carrying the UASRAP1-HIS3 reporter gene as a template (Fig. 1A). Transcription of the UASRAP1-HIS3 reporter gene was monitored by primer extension analysis using an mRNAHIS3-complementary probe. Primer extension of specific in vitro transcription products (mRNAHIS3; arrow, left) comigrated with the extension product produced from authentic, in vivo mRNAHIS3 produced from the integrated UASRAP1-HIS3 reporter gene (arrow, right, lane 14). Additional control reactions (lanes 12 and 13) used untreated WCE and were performed in the absence of template DNA (lane 12) or in the presence of 10 μg/ml α-amanitin (lane 13). (C) Efficacy and specificity of TFIID immunodepletion. WCE was treated as for panel A except that anti-Taf4p IgG was used as the specific antibody. Equivalent amounts of each of these WCEs, along with 2.5 ng purified TFIID as a positive control, were subjected to SDS-PAGE and blotted, and the blots were probed with antibodies recognizing the indicated proteins (Taf7p, Taf4p, and Taf13p). (D) Transcription of UASRAP1-HIS3 is TFIID dependent and rescued in TFIID-depleted WCE by readdition of purified TFIID but not by addition of TBP. Transcription experiments were conducted with control and anti-Taf4p IgG/TFIID-immunodepleted WCEs, as shown, in the presence and absence of the indicated amounts of TBP or TFIID. All WCEs, controls, depletions, and transcription assays were as for panels A and B. (E) Efficacy and specificity of TFIID/Rap1p double immunodepletion. WCE was treated as for panels A and C except that both bead-bound anti-Taf4p IgG and anti-Rap1p were used. Equivalent amounts of each of the resulting WCEs, along with 0.8 ng purified TFIID, 2 ng His6-Rap1p, and 0.8 ng His6-TBP, were run in parallel as positive controls. Proteins were subjected to SDS-PAGE and blotted, and the blots were probed with antibodies recognizing the indicated proteins (Rap1p, Taf7p, Taf4p, Taf13p, and TBP). (F) TFIID and Rap1p, but not TBP, can efficiently rescue specific transcription in doubly depleted WCE. Transcription experiments were conducted with control and anti-Taf4p/anti-Rap1p IgG doubly immunodepleted WCEs, as shown, in the presence and absence of the indicated amounts of Rap1p, TBP, or TFIID. All controls, depletions, and transcription assays were as described above. (G) The UASRAP1-driven HIS3 reporter gene is efficiently transcribed in the absence of the Fhl1p-Ifh1p coactivator complex. WCEs were prepared in parallel from WT (YPH252) and ΔFHL1 ΔIFH doubly deleted yeast (DR35). Equivalent amounts of protein from these two extracts (WT WCE and ΔFHL1/ΔIFH1 WCE) were tested for in vitro transcription of the UASRAP1-driven HIS3 reporter gene. Controls were as for panels B, D, and F. Transcription was quantitated by imaging as for Fig. 1C.
Electrophoretic mobility shift Rap1p DNA binding assays.
Purified recombinant Rap1p proteins (20, 40, 80, 100, 200, 400, 600, 100, 2,000, and 4,000 fmol) were incubated with 20 fmol 32P-labeled double-stranded DNA (6,400 cpm/pmol) containing the Rap1p consensus binding site (boldface) ATATACACCCATACATTGA in 20 μl of 20 mM HEPES (pH 7.6), 10% vol/vol glycerol, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 1 mM PMSF, 25 μg/ml BSA, 2.5 μg/ml poly(dG-dC), and 2.5 mM β-mercaptoethanol for 30 min room temperature. Reaction products were fractionated on 6% polyacrylamide Tris-borate-EDTA gels, dried, and exposed to X-ray film and imaging screens for quantitation.
Chromatin immunoprecipitation assay.
Yeast strains were grown to approximately 2 units of optical density at 600 nm/ml on synthetic complete medium (82) with 2% galactose without leucine and uracil (SC-LU) at 30°C. Cells were collected, washed with H2O, and used to inoculate SC-LU cultures containing 2% glucose. Cells were then grown to log phase (1 unit of optical density at 600 nm/ml) and cross-linked with 1% (vol/vol) formaldehyde (30 min room temperature). Cross-linking was terminated by the addition of glycine (125 mM final concentration; 5 min). Cells were harvested by filtration, resuspended in immunoprecipitation (IP) buffer (50 mM HEPES [pH 7.6], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin), and disrupted by 10 1-min pulses in a 96-well Midi-beater (Bio-Spec Products, Bartlesville, OK). Chromatin was purified and sheared as described by Kuras and Struhl (40). IPs were performed for 18 h at 4°C with 1 mg of protein extract and the following antibodies: 30 μg anti-TBP, 10 μg anti-RNAPII C-terminal domain MAb (8WG16), 60 μg anti-HA MAb (12CA5), 30 μg anti-myc MAb (9E10), and 30 μg rabbit IgG (Sigma) as a negative control. All IgGs were cross-linked to protein A-Sepharose beads (Sigma). The amount of UAS or core promoter fragments from the RPS8A, RPS5, PYK1, or HIS3 reporter genes in input and IP samples were determined by quantitative real-time PCR (Q-RT-PCR) amplification using the IQ SYBR green Supermix and iCycler system (both from Bio-Rad Laboratories). Chromatin enrichment for each gene was calculated as 100(IP/input). Background, estimated for the POL1 ORF in each IP, was subtracted. Each value represents the average from at least three independent experiments. PCR oligonucleotides used were RPS5-UAS (CAGCCTTGAGTTCTCAAATTTGC and AAAGAATTCTCCTTCCCCGTAGC), RPS5-Core (GGCCAACTTCTACGCTCACGTTAG and CGGTGTCAGACATCTTTGGAATGGTC), RPS5-ORF (GGTCCAAGAGAAGACACCACCAGAG and CGGTTAGACTTGGCAACACGTTCC), RPS8A-UAS (GGGACAAGGATAGGTGAACATTTG and GTGTAAGGGTGTTAAAGAGGGTGTATGG), RPS8A-Core (CACACCAGGACAAAAAGTATGTGCT and AAAGAACGTACAAAAGTTGCGGAA), RPS8A-ORF (AAAGATCCGCTACCGGTGCCAAG and TCTTGGAGATACCTTCAGAAGCCC), PYK1-UAS (CGCCCTGGTCAAACTTCAGAAC and GAGATACAGACATCACACGCCATAG), PYK1-Core (TGGTAAATGAATGCTTGTGATGTCTT and TTGTTTTGATTGGTGTCTTGTAAATAGAAA), PYK1-ORF (CCAACCTCCACCACCGAAAC and GGGCTTCAACATCATCAGTCCA), REPORTERRPS8A-UAS (GCTGAACTGGTGTCCATGTCGC and GTGTAAGGGTGTTAAAGAGGGTGTATGG), REPORTERRPS8A-Core (GCTTGAGGGCTTTCTCTTACGC and GGGCTTTCTGCTCTGTCATCTTTG), REPORTERRAP1UAS (CTGTCGTAACCTTCAGTTCATCG and CCCTTGGTTAGAAGAAAAGAGTGTG), REPORTERRAP1.Core (CACACTCTTTTCTTCTAACCAAGGGG and TGGGAAGATCGAGTGCTCTAT), HIS3-ORF (TAACGTCCACACAGGTATAGGGTTT and AGCCTTGAACGCACTCTCACTAC), and POL1-ORF (TGCACCAGTTAATTCTAAAAAGGCA and AAAACACCCTGATCCACCTCTGAA).
RNA analysis.
Total RNA was prepared from equal numbers of yeast cells by using hot phenol (38). One microgram of in vitro-synthesized RNA from the E. coli ycxA gene (88) was added prior to RNA extraction and used as a recovery control. DNase I-treated total RNA was used for RT-PCR assessment of RPS8A, RPS5, PYK1, and HIS3 gene expression. Reverse transcription was performed with the Bio-Rad iScript cDNA synthesis kit; cDNAs produced were quantitated by Q-RT-PCR. The number of transcripts was estimated by comparison to a standard curve generated with known amounts of yeast genomic DNA amplified in parallel during the PCR. Alternatively, specific RNAs were measured by primer extension analyses as described previously (33).
Protein blotting assays.
Purified TFIID and recombinant Tafps were fractionated on 4 to 12% polyacrylamide gradient denaturing gels (Invitrogen or Bio-Rad). Following electrophoresis, gels were equilibrated for 10 min in transfer buffer (30 mM Bicine, 25 mM bis-Tris, 1 mM EDTA, 60 μM chlorobutanol, 20% [vol/vol] methanol) and then electrotransferred to a PVDF membrane (Immobilon-P, 0.45 μm; Millipore) for 2 h at 12 V. Blots were blocked in binding buffer (20 mM HEPES KOH [pH 7.6], 75 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 0.05% NP-40, 1 mM dithiothreitol, 1% BSA [Sigma, A-2153, lot 98H1450]) for 1.5 h at 4oC, followed by incubation at 4°C overnight in binding buffer containing 7.3 nM recombinant Rap1p (∼120 μl/cm2). Blots were then washed at room temperature (five times for 15 min; 100 ml/wash) in binding buffer without BSA. Tafp-bound Rap1p was detected by incubation for 60 min at room temperature with affinity-purified anti-Rap1p IgG (1:30,000) in Tris-buffered saline-0.1% Tween 20 plus 1% nonfat dry milk. Primary antibody was removed by washing (four times for 10 min; 100 ml/wash) in Tris-buffered saline-0.05% Tween 20, and antibody-antigen complexes were detected using horseradish peroxidase-conjugated goat anti-rabbit IgG(Fc) (Pierce) and Lumi Light (Roche) reagent. Binding-competition assays were performed as described above except a 5- or 10-fold mole excess of either Taf12p or human PDE5A (100 kDa and pI 5.67 [versus Rap1p, 92.4 kDa and pI 4.68]) was added to the binding reaction mixtures. Binding was qualitatively visualized with X-ray film and quantitatively detected with a Bio-Rad Chemi-Doc imager and Bio-Rad QuantityOne software.
RESULTS
Rap1p collaborates with TFIID in vivo to drive UASRAP1-directed transcription.
Promoter-enhancer swap and mutagenesis experiments have demonstrated that the RAP1-containing UASRPG is required for RP gene transcription activation and that the RAP1 elements of this enhancer are sufficient to induce TAF dependency upon a TAFind core promoter (45, 46, 53, 87). We confirmed these findings by reconstructing and analyzing several chimeric HIS3 reporter genes (Fig. 1A) characterized previously by Mencia et al. (53). The first reporter gene, designated UASRPS8A-HIS3, contains a 229-bp RPS8A enhancer fragment fused to the RPS8A core promoter, driving transcription of the HIS3 gene. The other two chimeras contain either WT or doubly mutant versions of a 41-bp subfragment of the RPS8A UAS composed of just two Rap1p binding sites; the tandem RAP1 sites are fused to the PGK core promoter and drive transcription of the HIS3 reporter. These two minimal enhancer-reporter genes are termed UASRAP1-HIS3 and UASRAP1M-HIS3, respectively. All reporter genes were integrated at the HIS3 locus.
We scored transcription and occupancy of Rap1p, Taf1p, and TBP on the HIS3 reporter genes and, in parallel, on two control Rap1p-driven genes, the TAFdep RPS8A gene and the TAFind PYK1 gene. Protein occupancy was measured by ChIP and Q-RT-PCR. As expected (53) all genes carrying intact RAP1 sites exhibit high-level occupancy of Rap1p (compare Rap1p occupancy on UASRAP1M with Rap1p occupancy with all other genes tested [Fig. 1B, top]). Rap1p occupancy on the UASRPS8A and UASRAP1 reporters, although significant, was approximately half that of the authentic RPS8A gene (Fig. 1B, top panel), presumably because these two genes both lack some of the positive-acting cis elements present in the intact gene; this is also likely the explanation for why transcription of these genes is 5- to 10-fold lower than that of RPS8A (Fig. 1B, bottom panel). TFIID and TBP occupancy was high and tracked with Rap1p on the UASRPS8A-HIS3, UASRAP1-HIS3, and RPS8A genes (Fig. 1B, middle panel). By contrast, mutation of both RAP1 sites in the UASRAP1M-HIS3 reporter gene resulted in a drop in Rap1p, Taf1p, and TBP occupancy and transcription. Nonetheless, the Taf1p/TBP occupancy ratio remained high on this RAP1-mutated reporter, indicating that TFIID recruitment still occurs on the cis-linked promoter, although at a reduced level. This result is consistent with the idea that it is the binding of Rap1p to the UASRAP1 enhancer of these genes that drives TFIID recruitment and PIC formation.
To further support this conclusion we analyzed specific mRNA gene transcription in yeast strains carrying mutant alleles of either the Rap1p-encoding (deletion mutations) or Tafp-encoding (temperature conditional mutations) genes. For the TFIID functional tests, cells were grown at permissive and nonpermissive temperatures, total RNA extracted, and both mRNAPGK and mRNARPS5 quantitated by primer extension. Transcription of the RP but not the TAFind-PGK gene was significantly reduced in all the 13 tested TAFts strains when cells were shifted to the nonpermissive temperature (Fig. 1C; compare RPS5 mRNA and PGK signals). Similarly, transcription of RPS5, RPS8A, and the UASRAP1-HIS3 reporter gene were all dramatically reduced in strains expressing certain RAP1 truncation mutations (Fig. 1D; compare ΔC and DBD with WT). Together these data (Fig. 1B to D) show that Rap1p binding to UASRAP1, TFIID recruitment, TFIID function, PIC formation, and RP gene transcription are all interdependent, results that are consistent with the idea that TFIID serves as coactivator for Rap1p on the RPGs.
In vitro transcription of the UASRAP1-driven reporter gene is both Rap1p and TFIID dependent.
We next asked whether we could recapitulate the UASRAP1/Rap1p/TFIID dependency seen in vivo with a WCE in vitro transcription system. WCE was efficiently and specifically depleted of either Rap1p (Fig. 2A, compare lanes 1 to 4 with lane 5), or TFIID (Fig. 2C, compare lanes 1 to 4 with lane 5) when incubated with anti-Rap1p IgG or with anti-Taf4p IgG coupled to protein A beads. Approximately 90% of Rap1p and TFIID was removed by antibody depletion. Activated transcription dropped to basal levels when either Rap1p (Fig. 2B, compare lanes 1 to 4 with lane 8) or TFIID (Fig. 2D, compare lanes 1 to 4 with lane 8) was removed. However, activated transcription could be specifically and efficiently rescued by readdition of either purified Rap1p in the Rap1p-depleted WCE (Fig. 2B, lanes 9 to 11) or purified TFIID in the case of TFIID-depleted WCE (Fig. 2D, lanes 10 and 11) when physiological levels of these proteins were added back to the two depleted WCEs. Importantly, TBP addition was ineffective at transcription rescue in the TFIID-depleted WCE (Fig. 2D, lane 9 versus lanes 10 and 11).
To explore the WCE transcription system in more depth, we performed two additional experiments. In the first, we immunologically depleted the WCE of both Rap1p and TFIID and then tested for transcription reconstitution, while in the second, we generated WCE from a yeast strain carrying null mutations of FHL1 and IFH1, genes that encode the potent heterodimeric DNA binding transactivator of RP gene transcription, Ifh1p-Fhl1p (50, 70, 77, 91). These two experiments more rigorously tested the in vitro collaborative involvement of TFIID and Rap1p and also probed the possible role of the Ifh1p-Fhl1p coactivator in Rap1p- and TFIID-dependent transactivation of the UASRAP1-driven reporter gene.
When WCE was doubly depleted of Rap1p and TFIID (Fig. 2E, compare control [lanes 1 to 4] with doubly depleted WCE [lane 5]), readdition of both proteins was required for efficient reconstitution of UASRAP1-activated transcription (Fig. 2F, lane 15 versus lanes 10, 11, and 13). Importantly, as with the singly TFIID-depleted WCE (Fig. 2D), TBP could not collaborate with Rap1p to restore transcription (Fig. 2F, compare lane 14, +Rap1 and TBP, with lane 15, Rap1p and TFIID), suggesting that Rap1p and TFIID might physically interact through Rap1p-Tafp interactions. Finally, the UASRAP1-HIS3 reporter was efficiently transcribed in WCE derived from yeast strain DR35 (70), cells that carry null mutations in ΔFHL1 and ΔIFH1 and hence are genetically depleted of the Fhl1p-Ifh1p complex (Fig. 2G, compare lanes 1 and 2) (transcript levels produced in the WCE prepared from the Δfhl Δifh cells were 87% of those in the WT WCE). Together these data argue that the in vitro transcription profiles we observe result from positive, potentially direct interactions between Rap1p and TFIID.
Rap1p interacts directly with TFIID.
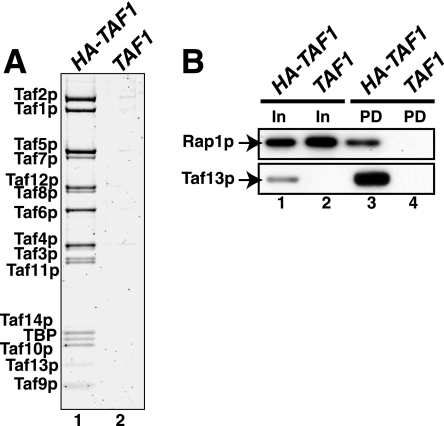
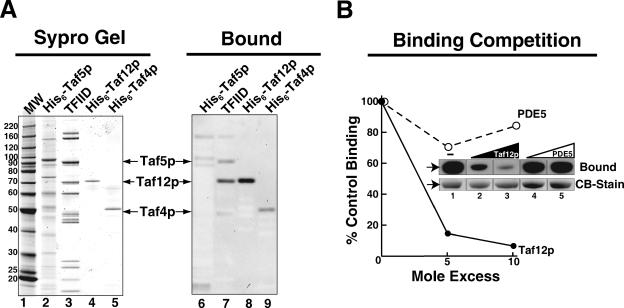
Having documented important in vivo and in vitro functional interactions between Rap1p and TFIID, we next asked whether these two proteins physically interact. To test this hypothesis, we performed a TFIID-Rap1p pull-down experiment; this experiment both enabled us to test for direct interaction and allowed us to estimate the relative affinity of such interaction. In this assay TFIID was specifically loaded onto anti-HA-coated protein A beads by incubation with the Bio-Rex fractions derived from either HA-TAF1 or TAF1 cells (see purity and specificity of TFIID loading in Fig. 3A, lanes 1 and 2). Purified recombinant Rap1p was added to both sets of beads (Fig. 3B, lanes 1 and 2), and then TFIID, along with TFIID-bound proteins, was eluted from the beads with HA-peptide. The TFIID and Rap1p contents of the control (TAF1) (Fig. 3B, lane 4) and TFIID (HA-TAF1) (Fig. 3B, lane 3) pull-down samples were measured by immunoblotting. The results of this assay show that Rap1p interacts directly with TFIID and that binding appears rather tight (we estimate the apparent binding affinity, Kd,app, for the interaction between Rap1p and TFIID to be ∼300 nM). This experiment was repeated using several different preparations of similarly purified TFIID or with TFIID purified through an additional Mono-S fast protein liquid chromatographic step (68), with equivalent results. We conclude from the experiment of Fig. 3 that Rap1p binds TFIID directly, specifically, and with high affinity.
FIG. 3.
Direct, high-affinity binding of Rap1p with TFIID. (A) Specificity of loading of HA1-Taf1p-tagged TFIID beads for pull-down assay. Equivalent volumes of TFIID purified from HA-tagged Bio-Rex 70 1 M fraction (HA-TAF1) and untagged control Bio-Rex 70 1 M fraction (TAF1) were fractionated by SDS-PAGE and stained with Sypro Ruby, and TFIID subunits were visualized using a Bio-Rad FX fluorescence scanner; the image is shown. (B) Purified recombinant Rap1p interacts with TFIID. Anti-HA MAb beads were specifically loaded with TFIID (In) (A). The resulting control and TFIID-containing beads were washed extensively and incubated with a threefold mole excess (relative to TFIID) of purified E. coli-expressed Rap1p. Rap1p specifically TFIID bound was eluted with HA-peptide and detected by immunoblotting with anti-Rap1p IgG (pull-down [PD]); recovery of TFIID in the eluate was scored by probing for Taf13p.
Mapping the Rap1p domains mediating TFIID binding.
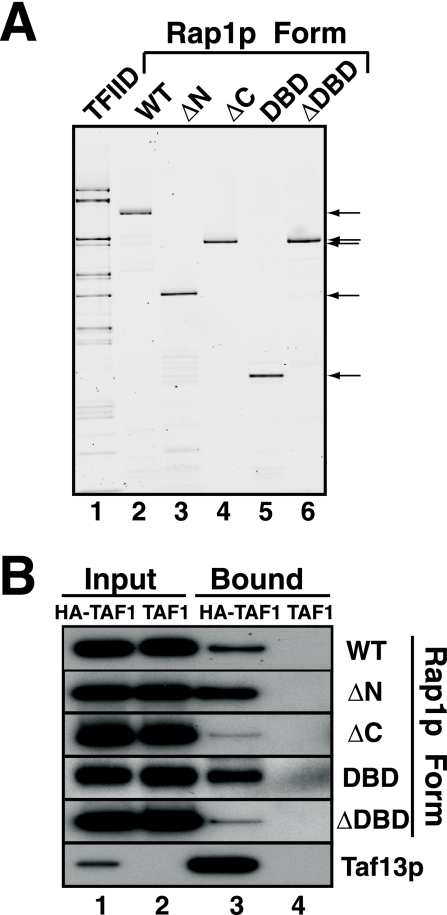
Having shown that Rap1p binds directly to TFIID, we next mapped the domain(s) of Rap1p responsible by using in vitro (TFIID pull-down) and in vivo (ChIP) assays. The purity of the reagents used for the pull-down studies is illustrated in Fig. 4A, where purified TFIID and five E. coli-expressed variants of Rap1p (WT, ΔN, ΔC, ΔDBD, and DBD; see Fig. 1A for Rap1p domain organization) were fractionated by SDS-PAGE and visualized with Sypro Ruby. The various purified recombinant proteins were analyzed by gel shift for in vitro DNA binding activity and in vitro transactivation potential by using Rap1p-depleted WCE; all but the ΔDBD variant were active in these two functional tests (Table 2). The TFIID-Rap1p pull-down assay demonstrated that all Rap1p variants bound to TFIID but with various affinities: ΔN ∼ DBD > WT ≫ ΔC ∼ ΔDBD (Fig. 4B). Quantitation of these and similar experiments (not shown; see Materials and Methods for details) allowed us to calculate Rap1p-TFIID binding affinities, relative to WT Rap1p, for the interaction of these five forms of Rap1p with TFIID as follows: ΔN and DBD variants, 2.1; WT, 1.0; ΔDBD, 0.2; and ΔC, 0.16.
FIG. 4.
Mapping of Rap1p domains responsible for Rap1p-TFIID complex formation. (A) Purity of TFIID and Rap1p variants. Purified TFIID (∼2,000 ng) and ∼100 ng of the five forms of purified Rap1p (WT, ΔN, ΔC, DBD, and ΔDBD) indicated (arrows) were fractionated by SDS-PAGE and detected by Sypro Ruby staining. (B) Binding of Rap1p variants to TFIID. The five forms of Rap1p were tested for TFIID binding as detailed for the experiment of Fig. 3B; TFIID-Rap1p binding was quantitated as detailed in Materials and Methods.
TABLE 2.
Relative activities of WT and truncated forms of Rap1p
| Rap1p form | [Rap1p] in vivoa | UASRAP1 DNA binding in vitrob | Transactivation potential in vitroc | Rap1p-TFIID (Kd)d | Predicted transcriptione |
|---|---|---|---|---|---|
| WT | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| ΔN | 0.35 | 1.00 | 1.80 | 2.14 | 1.35 |
| ΔC | 0.90 | 1.00 | 0.20 | 0.17 | 0.03 |
| DBD | 0.22 | 0.50 | 0.30 | 2.14 | 0.07 |
In vivo steady-state Rap1p concentration determined by quantitative immunoblotting.
Relative DNA binding affinity determined by gel mobility shift assay using a 32P-labeled 19-mer probe, ATATACACCCATACATTGA (the RAP1 site is in boldface).
Rap1p transactivation potential determined by in vitro transcription assay using anti-Rap1p IgG-depleted WCE. Assays performed as for Fig. 2B.
Relative Rap1p-TFIID binding values determined by pull-down assay; data are taken from the experiment of Fig. 4 (see text).
Product of multiplying the values in the previous four columns.
To provide an in vivo correlate of these data, we next performed ChIP assays using yeast cells expressing the WT, ΔN, ΔC, and DBD forms of Rap1p. Because the ΔDBD form of Rap1p fails to bind DNA, either in vitro or in vivo, it is unable to support viability and thus could not be analyzed in these in vivo assays (19). For this set of ChIP experiments, we constructed pseudodiploid yeast strains that express both a chromosomal WT RAP1 allele under control of the UASGAL enhancer and a plasmid-borne Myc5-tagged version of either the WT, ΔN, ΔC, or DBD RAP1 allele, all under the control of the normal chromosomal RAP1 enhancer-promoter sequences. These pseudodiploid yeast cells were grown in galactose, conditions where transcription of the plasmid-borne RAP1 enhancer-promoter-controlled Myc5-tagged RAP1 alleles was autorepressed (20). After shifting cells to glucose medium to repress the chromosomal UASGAL-RAP1 allele and activate transcription of the Myc5-RAP1 alleles, cells were grown for seven doublings to allow for the complete degradation of the untagged WT Rap1p (data not shown). Aliquots of the cultures were then removed for determination of Myc5-Rap1p protein and mRNA levels, while the remainder was processed for ChIP.
Cells expressing all the variants as the sole source of Rap1p were viable; cells expressing Rap1p-ΔN grew like the WT, while strains expressing Rap1p-ΔC or Rap1p-DBD alone grew very slowly (19). Moreover, the strains expressing Rap1p-ΔC and Rap1p-DBD both exhibited total protein and total RNA levels that were reduced two- to threefold compared to WT levels. We conclude that these dramatic effects on growth, RNA synthesis, and total protein synthesis are a direct consequence of decreased Rap1p function, because steady-state levels of these two truncated Rap1p proteins are similar to those for the WT and ΔN Rap1p variants (Table 2).
In order to assess the effects of the truncation mutations on Rap1p function in vivo, we performed ChIP on the yeast strains expressing the WT, ΔN, ΔC, and DBD forms of Rap1p. ChIP immunoprecipitates were probed by Q-RT-PCR to measure Rap1p, TFIID, TBP, and RNAP II occupancy on RPS8A, PYK1, and UASRAP1-HIS3. Presented in Fig. 5A are occupancy values for each protein as well as normalized transcription levels for each gene in the four different strains. Full-length Rap1p efficiently bound the RAP1 enhancer sites in all three genes (Fig. 5A), and deletion of Rap1p N-terminal sequences did not change this pattern of occupancy on any gene (Fig. 5A). By contrast, deletion of the C terminus of the protein (i.e., in either the ΔC- or DBD-alone Rap1p variant) dramatically reduced Rap1p binding to the RPS8A and UASRAP1-HIS3 enhancers (Fig. 5A, top and middle panels). Surprisingly, though, on the TAFind PYK1 gene we found that while ΔC-Rap1p bound with reduced efficiency, the DBD variant actually bound the UASPYK1 RAP1 sites as well as full-length WT Rap1p, suggesting the possibility of potential Rap1p intramolecular regulatory interactions. However, in spite of the high Rap1p occupancy levels of the Rap1p DBD on the PYK1 enhancer, occupancy of TBP and RNAP II on this gene, and indeed on all genes tested, was lower than for the full-length and ΔN-Rap1p proteins, indicating that the Rap1p DBD is deficient in promoting PIC formation and transcription of all three genes (Fig. 5A).
FIG. 5.
Contribution of Rap1p domains to PIC formation and transcription in vivo. (A) Effect of Rap1p truncations on ChIP occupancy and transcription. ChIP assays were used to determine the in vivo occupancy of Rap1p and Taf1p/TFIID, TBP, and RNAP II on RPS8A, PYK1, and the UASRAP1-HIS3 reporter gene. Occupancy was calculated as percent enrichment in the IP over the input. Transcription (TRXN) was assessed by real time Q-RT-PCR as for Fig. 1D. Error bars indicate standard deviations. B, Assessment of the relative efficiencies of Rap1p variants in the promotion of PIC formation and transcription. Data for Taf1p, TBP, and RNAP II occupancy and transcription from panel A were normalized to Rap1p occupancy and plotted relative to WT Rap1p set to a value of 1.0; data trends are indicated by dashed lines (see text).
Importantly, even though overall Taf1p and TBP occupancies are reduced in strains expressing the ΔC and DBD Rap1p variants, Taf1p/TBP occupancy ratios on the two TAFdep genes, RPS8A and UASRAP1-HIS3, remained high, indicating that all four forms of Rap1p are indeed capable of recruiting TFIID to these two TAFdep promoters, albeit with apparent reduced efficiency (but see below). Lastly, as expected, we observe low Taf1p occupancy and a correspondingly low Taf1p/TBP ratio on the TAFind PYK1 gene (Fig. 5A, bottom panel).
Next, we wanted to more directly assess whether either the efficiency of TFIID and RNAP II recruitment or transcription of these genes was affected by the deletion of Rap1p domains. In order to measure this we normalized the values of the different parameters (i.e., Taf1p, TBP, and RNAP II occupancy and transcription) to the amount of Rap1p variant DNA occupancy and then expressed these values relative to that for the WT full-length Rap1p set as 1.0. When these data was plotted (Fig. 5B), three general patterns (dashed lines) were observed. First, with the RPS8A gene, within error (except for the ΔC variant), the normalized efficacies of TFIID and RNAP II occupancy and transcription were roughly equal to (or slightly higher than) WT for all Rap1p forms tested (Fig. 5B, top panel). This result indicates that each Rap1p variant, once UASRPS8A enhancer bound, is at least as efficient as full-length Rap1p at recruitment of the PIC and activation of transcription on this gene. These results might reflect compensation by other cis/trans elements of the intact RPS8A enhancer. Second, in the case of the UASRAP1-HIS3 reporter gene, the normalized efficiency of recruitment of TFIID and RNAP II and transcription, were approximately equal in the case of the WT and ΔN-Rap1p forms, but both values were lower for the strains expressing Rap1p-ΔC and Rap1p-DBD (Fig. 5B, middle panel). These data are consistent with our in vitro Rap1p-TFIID interaction data (Fig. 4B) that show that all Rap1p truncations can bind to TFIID. Third, with the TAFind PYK1 gene, TBP and RNAP II occupancy and transcription decreased linearly for the ΔN, ΔC, and DBD forms of Rap1p (Fig. 5B, bottom panel), suggesting that on this TAFind gene, different functions of Rap1p are utilized during PIC formation and activation of transcription.
Finally we considered several factors, such as relative affinity for UASRAP1, in vivo protein levels, in vitro transactivation potential, and TFIID binding affinity, in order to calculate a predicted value for the expression of the Rap1p-driven TFIID-dependent UASRAP1 reporter gene (Table 2, predicted transcription). These predictions were made under the assumption that Rap1p occupancy on the UASRAP1 enhancer was the dominant driver of PIC formation and transcription. Interestingly, the unique pattern of occupancy and transcription of the UASRAP1-HIS3 reporter gene seen in vivo (Fig. 5) was very close to that predicted (Table 2), consistent with our hypothesis that on this model chimeric reporter gene the Rap1p-TFIID activator-coactivator module does indeed drive transcription.
Mapping the TFIID subunits responsible for binding Rap1p.
Protein blotting was used to globally assess the binding of Rap1p to all TFIID subunits in a single experiment. Duplicate samples of purified TFIID, partially purified recombinant His6-tagged Taf5p, and highly purified recombinant His6-Taf12p and His6-Taf4p were fractionated by SDS-PAGE. Half of the gel was stained directly to indicate the mobility of the respective proteins (Fig. 6A, left panel), and the other identical half was blotted to a PVDF membrane and incubated with Rap1p and the resulting Rap1p-Tafp complexes were detected using anti-Rap1p IgG (Fig. 6A, right panel); Rap1p bound preferentially to Taf12p. We determined that this binding was dose dependent, was saturable (not shown), and specifically competed with excess Taf12p (Fig. 6B; compare Taf12p and PDE5 competition curves). Moreover, the “specific activity” of Rap1p binding to Taf12p binding was roughly equivalent to that for both TFIID-Taf12p and E. coli-expressed Taf12p. Together these data suggest that the binding of Rap1p to TFIID is dominated by interactions between Rap1p and the Taf12p subunits of TFIID, though Rap1p also clearly binds, with slightly reduced affinity, to Taf4p and Taf5p.
FIG. 6.
Mapping of TFIID subunits capable of binding Rap1p. (A) Protein blotting test of Rap1p-Tafp interaction. Purified TFIID and recombinant His6-tagged Tafps were fractionated by SDS-PAGE. The gel was loaded with molecular weight (MW) standards (lane 1), ∼2,000 ng purified TFIID (lanes 3 and 7), ∼250 ng E. coli-expressed His6-Taf5p (lanes 2 and 6), ∼200 ng His6-Taf12p (lanes 4 and 8), and ∼100 ng E. coli-expressed His6-Taf4p (lanes 5 and 9). One part of the gel (lanes 1 to 5) was excised and stained directly with Sypro Ruby (Sypro Gel) to indicate the mobility of TFIID subunits and recombinant Tafps, while the other identical part (lanes 6 to 9) was blotted to a PVDF membrane and probed with Rap1p. Tafp-bound Rap1p was detected by immunoblotting with anti-Rap1p IgG (Bound). (B) Specificity of Rap1p binding to Taf12p. Equal amounts of E. coli-expressed His6-Taf12p (250 ng) were fractionated by SDS-PAGE and blotted to PVDF, and individual lanes were excised from the blot and incubated with Rap1p in combination with 5- or 10-fold mole excess of either E. coli-expressed His6-Taf12p (inset lanes 2 and 3) or purified bovine phosphodiesterase 5 (PDE5) (inset lanes 4 and 5). No exogenous competitor was added to Rap1p (inset lane 1) to determine 100%, control binding to Taf12p. PVDF-bound Taf12p was detected by staining the membrane slices with Coomassie blue (CB-Stain), while Rap1p-Taf12p complex was detected by immunoblotting with anti-Rap1p IgG (Bound) as for panel A. Rap1p binding was quantitated and graphed relative to binding in the absence of competitor. Averages of results from two replicate assays are plotted.
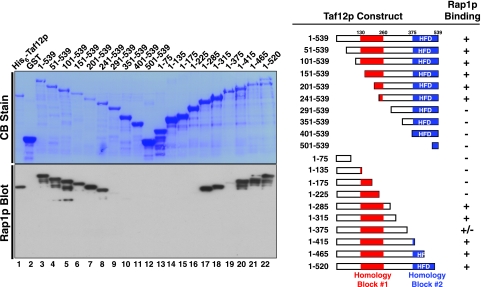
In order to determine which region of Taf12p interacts with Rap1p, we generated a systematic family of N- and C-terminal truncation variants of Taf12p. These deletion mutants were cloned and expressed as GST fusion proteins in E. coli (Fig. 7, top left panel). We assessed the binding of Rap1p to this family of Taf12p proteins by protein blotting (Fig. 7, bottom left panel). Rap1p bound to all Taf12p variants containing the C-terminal residues of conserved homology block 1 (Fig. 7, right panel), indicating that this domain of Taf12p contributes importantly to the interaction with Rap1p.
FIG. 7.
Mapping of the Rap1p binding domain of Taf12p. Serial truncations from the N or C terminus of Taf12p were expressed as GST fusion proteins and purified by glutathione-agarose chromatography. Proteins were fractionated on two identical SDS-polyacrylamide gels and blotted to PVDF membranes; one blot was directly stained with Coomassie blue (CB Stain), while the other blot was probed with Rap1p, and Taf12p-bound Rap1p was detected with anti-Rap1p IgG (Rap1p Blot). A schematic of Taf12p indicating the locations of two conserved amino acid sequence blocks, block 1 (highly conserved among Saccharomyces cerevisiae sensu stricto strains) and block 2 (the histone fold domain [HFD]), are indicated, as is a summary of the binding data (right).
DISCUSSION
In this report we describe our efforts to test the hypothesis that TFIID serves as coactivator on ribosomal protein-encoding genes by making direct, specific interactions with the multifunctional DNA binding transcription factor Rap1p. We found that these two proteins do interact, both in vivo and in vitro. This is the first demonstration of direct transfactor binding to the yeast TFIID holocomplex, and it sets the stage for a detailed dissection of the molecular mechanism of action of the coactivator function of TFIID.
Rap1p and TFIID functionally interact to drive transcription of authentic RP genes and a chimeric UASRAP1-driven reporter gene in vivo.
Using mutant forms of Rap1p, TFIID-Tafp subunits, and reconstructed RAP1+ and RAP1− versions of a minimal UASRAP1-driven HIS3 reporter gene, we confirm and extend the observations of others (46, 53, 87) by demonstrating a biochemical and genetic requirement for Rap1p, UASRAP1, and TFIID function for efficient RP gene transcription in vivo (Fig. 1). Most importantly, these data indicate that the simple chimeric UASRAP1-HIS3 reporter gene accurately recapitulates the critical features of authentic Rap1p-controlled RP gene regulation noted above (i.e., dependence/recruitment on Rap1p, RAP1, and TFIID) and therefore could appropriately be utilized for in vitro studies designed to probe the molecular mechanisms of the functional in vivo interplay between Rap1p and TFIID.
Rap1p and TFIID functionally interact to drive UASRAP1-directed gene transcription in vitro.
We were able to reproduce the efficient and accurate RNAP II-catalyzed transcription of the UASRAP1-driven chimeric HIS3 reporter gene in vitro with a WCE transcription system (Fig. 2). The UASRAP1 enhancer used to drive HIS3 reporter gene transcription in vitro consists only of 41 bp of the RPS8A enhancer, CTTTACATCCATACACCCTCTTTAACACCCTTACACTTTTA, and is comprised of just two 14-bp high-affinity Rap1p binding sites (boldface) separated by 6 bp of DNA and flanked by 3 bp upstream and 4 bp downstream. This fact minimizes the possibility that other known RP gene transcription factors, such as Fhl1p-Ifh1p, Hmo1p, or Sfp1p, might contribute to our transcription results. It is important to emphasize then that due to the minimal constitution of this chimeric gene, both our in vivo (Fig. 1 and 5) and in vitro (Fig. 2) transcription experiments that utilize the UASRAP1-HIS3 gene are reporting interactions between just Rap1p and the transcription machinery.
Specific transcription of the UASRAP1-driven reporter gene in vitro depends upon both Rap1p and TFIID, since immunodepletion of either protein dramatically decreases specific transcription to basal, non-Rap1p-activated levels. Importantly, UASRAP1-directed gene transcription could be rescued in the depleted extracts by the addition of the appropriate purified protein, and in the case of the TFIID-depleted WCE, TBP was unable to reconstitute Rap1p-activated transcription. Doubly depleted extracts (i.e., without both Rap1p and TFIID) required the readdition of both purified proteins for rescue of UASRAP1-driven reporter gene transcription. The Fhl1p-Ifh1p coactivator complex was not required for efficient transcription in our in vitro system (Fig. 2).
We draw a number of conclusions from these in vitro transcription experiments that corroborate and complement our in vivo studies. First, the reconstitution results indicate that the component that was antibody depleted was the targeted molecule (i.e., Rap1p or TFIID) and not other RPG transcription factors. Second, the purified proteins added to the depleted WCEs were highly active, since Rap1p and TFIID were added back to approximate their levels in the initial WCE and at these levels efficiently reconstitute activated transcription. Third, the fact that TFIID, but not TBP, reconstitutes Rap1p-driven transactivation of transcription argues that Rap1p interacts with a Tafp(s) within the context of the TFIID holocomplex. This idea is underscored by the fact that while ≥90% of TFIID Tafps are immunodepleted from treated WCE, ≤15% of WCE TBP was removed by anti-Taf4p IgG (72). Fourth, TFIID and Rap1p appear to be in excess in the WCE, because addition of these purified proteins to control WCE has no dramatic stimulatory effect upon transcription. Fifth, efficient Rap1p-driven transactivation does not obligatorily require nuclear matrix tethering. Sixth, functional Rap1p-TFIID interaction can occur independent of nucleosomes and/or chromatin, since naked DNA was used as the template in all reactions. Finally, these data argue that if posttranslational modifications (PTMs) are mandatory for complementing activity, such PTMs must be catalyzed by WCE-endogenous activities, because Rap1p was purified from E. coli.
Rap1p-TFIID functional collaboration is mediated by direct, specific, high-affinity protein-protein interactions between transactivator and TFIID holocomplex.
We found that Rap1p bound directly and with high affinity (Kd,app in the nanomolar range) to purified TFIID (Fig. 3B). PTMs are not required for the interaction of pure Rap1p with pure TFIID. Given the probable minimal 10 μM intranuclear concentrations of both Rap1p and TFIID (i.e., roughly 5 to 10,000 molecules of each in 20 × 10−15 liter) and our estimate of a binding constant for Rap1p-TFIID complex formation the nanomolar range, these two proteins could readily bind in vivo.
TFIID pull-down and ChIP studies with Rap1p variants support the idea that specific Rap1p domains directly and specifically recruit TFIID to RP and UASRAP1-HIS3 genes (Fig. 4 and 5), a proposition supported by the results of Rap1p-Tafp protein blotting experiments (Fig. 6). These data indicate that primary contacts between TFIID and Rap1p are mediated through a C-terminal domain (aa 597 to 827) and DBD (aa 361 to 596) of Rap1p and a domain within the N-terminal half of the Taf12p subunit of TFIID (Fig. 7). N-terminal Taf12p sequences have been shown by others to contribute to transcriptional readout in several other in vivo contexts (27, 56, 96). In addition to the tight binding of Rap1p to Taf12p, we reproducibly detect somewhat lower-affinity binding of Rap1p to Taf4p and Taf5p. It is notable that when all the parameters vis-à-vis Rap1p and TFIID are taken into account (Table 2), we are able to make a reasonable prediction as to UASRAP1-HIS3 reporter gene transcriptional readout (Fig. 5B and Table 2), a result consistent with the hypothesis that the TFIID-Rap1p interactions identified here are physiologically relevant and likely contribute significantly to activating ribosomal protein gene transcription in vivo.
The Taf4p-Taf12p heterodimer: a nexus for transfactor-TFIID interactions?
It is interesting that we observe Rap1p-Taf12p and Rap1p-Taf4p interactions in our direct protein binding assays (Fig. 6 and 7); transfactor-Taf12p and transfactor-Taf4p interactions have been observed previously (16, 25, 27, 28, 37, 65). Indeed, to date, transfactor-Taf4p interactions are arguably the most frequently observed interactions between DNA binding transfactors and TFIID. Factors Sp1, CREB, RAR, HP1, CBF, RanBPM, simian virus 40 small t antigen, NFAT, AhR, and c-Jun have all been reported to interact with Taf4p (1, 6, 7, 13, 18, 19, 28, 32, 34, 54, 68, 71, 89, 94). Given these numerous Taf4p-Taf12p-transfactor interactions, it will be interesting to dissect the molecular details of Rap1p-Tafp binding. For example, is there a single domain, or multiple domains, within the Taf4p or Taf12p subunits that can bind the transfactor? Are there multiple pathways by which transfactor-Tafp interaction results in transactivation?
What is the mechanism by which Rap1p-TFIID interaction results in RP gene activation?
Examination of our earlier electron microscopy (EM)-based TFIID subunit mapping data (44) shows that Taf12p, which heterodimerizes with Taf4p (64, 85) and is present in two copies/TFIID molecule (72), localizes to both B and C lobes of the TFIID holocomplex. Interestingly, Taf5p (either its N terminus in lobe C or its C terminus in lobe B) also colocalizes with the Taf12p-Taf4p heterodimer. Thus, our observation that Rap1p binds tightly to Taf12p, and somewhat more weakly to both Taf4p and Taf5p, is likely to be physiologically important. Since TFIID contains two molecules of Taf4p-Taf12p per holocomplex and most RPGs contain two binding sites for Rap1p, it is possible that these two UASRAP1-bound Rap1p molecules simultaneously interact with both Taf12p subunits of TFIID, resulting in more efficient interaction with the TFIID coactivator. Such multiple Rap1p-Taf12p/TFIID interactions could contribute to the substantial transcription rates of this gene family.
How might the direct binding of Rap1p to TFIID-Taf12p stimulate PIC formation and transcription? The first potential mechanism we considered was that Rap1p-TFIID cooperative interactions would stimulate binding to DNA, as has been observed in the case of the interaction of two Drosophila transfactors with Drosophila TFIID (75, 76). However, under the conditions tested, numerous attempts to reproducibly detect such cooperative interactions between Rap1p and TFIID were unsuccessful (not shown). Consequently, we hypothesize that the interaction of DNA-bound Rap1p with TFIID either induces a specific conformational change in TFIID or, alternatively, results in the covalent modification of these or other proteins comprising the PIC.
Nogales, Tjian, and colleagues have used EM to show that VP-16 and SREBP activators, as well as the RNAP II CTD, can induce distinct conformational changes in the CRSP mediator complex (61, 84). Moreover, very recently these same workers utilized cryo-EM methods to demonstrate that human TFIID exhibits conformational heterogeneity, and they have suggested that the different forms of TFIID observed could be involved in integrating multiple gene regulatory cues from DNA-bound transfactors (24); such may be the case for yeast TFIID and Rap1p.
TFIID either contains intrinsic catalytic activities/functional domains or associates with proteins known to contain such activities (i.e., acetyltransferase, bromodomains, kinase, and ubiquitin ligase/protease) (2, 12, 14, 51, 52, 57, 62, 97, 98). Thus, it is possible that Rap1p-TFIID-DNA ternary complex formation could trigger covalent modification of either Rap1p, TFIID, or other components of the basal transcription machinery (30), resulting in (more) efficient PIC formation and/or PIC function and leading ultimately to enhanced rates of RPG transcription. Of course, some combination of both a triggered conformational change(s) and a triggered covalent modification(s) could be operative in the Rap1p-TFIID system. Finally, it should be noted that both models are consistent with the fact that Rap1p occupancy does not appear to dramatically change over a wide range of RP gene transcription rates (70, 77, 91). Additional biochemical, genetic, and EM studies that utilize mutant forms of Rap1p and TFIID (i.e., variant forms of Taf12p, Taf5p, and/or Taf4p) will be required to definitively distinguish which, if any, of these models are correct.
Summary and perspectives.
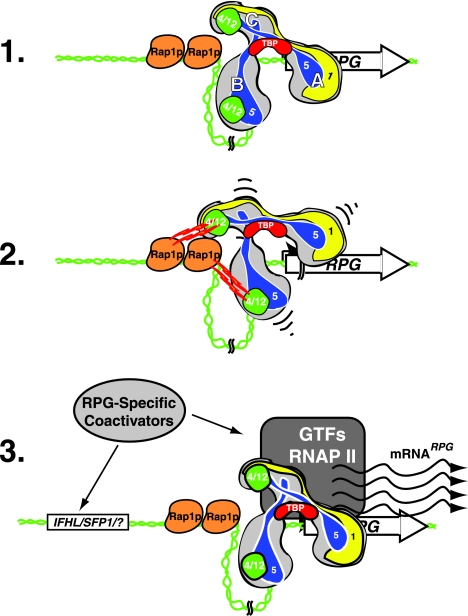
Together, our results are consistent with a model for RP gene activation wherein UASRAP1-bound-Rap1p directly interacts with the Taf12p subunit(s) of TFIID (Fig. 8). This interaction in turn leads to more efficient PIC formation (or function) and hence RP gene transcription. How TFIID is recruited and what signals/molecules trigger productive/activating interactions between the two proteins remain to be elucidated. Our future work will be aimed at studying the molecular details of the various protein-protein interactions described here. Equally important will be to undertake experiments to illuminate how the Rap1p-TFIID activator-coactivator module interacts with the other known RPG regulatory proteins such as NuA4, Fhl1p-Ifh1p, Sfp1p, and Hmo1p, molecules that play essential roles in RP gene regulation. Finally, from a more global point of view, the ideas and approaches outlined here should prove useful in formulating testable hypotheses regarding how TFIID mechanistically contributes to the regulation of transcription of the other 90% of yeast mRNA-encoding genes that are also TFIID dependent (3).
FIG. 8.
Model depicting Rap1p-TFIID interaction in RP gene transcription. Line 1, hypothetical RP gene with TFIID bound to the core promoter and two molecules of Rap1p bound to tandem RPG UASRAP1 enhancer sites. Illustrated within the trilobed (lobes A, B, and C) TFIID holocomplex (44) are the locations of TBP (red), Taf1p (yellow), and the Rap1p-interacting Tafps identified here: the two heterodimers of Taf4p-Taf12p (green) and the two molecules of Taf5p (blue). Line 2, interactions between the Rap1p DBD and C-terminal domain and the Taf12p/Taf4p and Taf5p subunits of TFIID (jagged red/yellow lines and small black curved lines). Line 3, the RP gene in the state of active transcription (wavy lines). Also shown are the additional known RP gene coregulators (i.e., Fhl1p-Ifh1p, Sfp1p, Hmo1p, Esa1p-NuA4, etc.) (light gray oval) and RNAP II plus GTFs (dark gray).
Acknowledgments
We thank our lab colleagues for freely sharing reagents, constructive criticism, and advice throughout the course of this work. We are grateful to Emmanuel Bessay, Allistair Chambers, Jackie Corbin, Ray Jacobson, Kevin Struhl, and Jon Warner for kindly supplying reagents and yeast strains used in this work.
The financial support of the NIH (grant GM52461) is gratefully acknowledged.
Footnotes
Published ahead of print on 30 October 2006.
REFERENCES
- 1.Asahara, H., B. Santoso, E. Guzman, K. Du, P. A. Cole, I. Davidson, and M. Montminy. 2001. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol. Cell. Biol. 21:7892-7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auty, R., H. Steen, L. C. Myers, J. Persinger, B. Bartholomew, S. P. Gygi, and S. Buratowski. 2004. Purification of active TFIID from Saccharomyces cerevisiae. Extensive promoter contacts and co-activator function. J. Biol. Chem. 279:49973-49981. [DOI] [PubMed] [Google Scholar]
- 3.Basehoar, A. D., S. J. Zanton, and B. F. Pugh. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699-709. [DOI] [PubMed] [Google Scholar]
- 4.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, X., M. Braunstein, G. J. Shei, and J. R. Broach. 1999. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc. Natl. Acad. Sci. USA 96:11934-11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunkhorst, A., M. Karlen, J. Shi, M. Mikolajczyk, M. A. Nelson, M. Metsis, and O. Hermanson. 2005. A specific role for the TFIID subunit TAF4 and RanBPM in neural progenitor differentiation. Mol. Cell Neurosci. 29:250-258. [DOI] [PubMed] [Google Scholar]
- 7.Brunkhorst, A., T. Neuman, A. Hall, E. Arenas, T. Bartfai, O. Hermanson, and M. Metsis. 2004. Novel isoforms of the TFIID subunit TAF4 modulate nuclear receptor-mediated transcriptional activity. Biochem. Biophys. Res. Commun. 325:574-579. [DOI] [PubMed] [Google Scholar]
- 8.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427-439. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, J. X., M. Floer, P. Ononaji, G. Bryant, and M. Ptashne. 2002. Responses of four yeast genes to changes in the transcriptional machinery are determined by their promoters. Curr. Biol. 12:1828-1832. [DOI] [PubMed] [Google Scholar]
- 10.Deminoff, S. J., and G. M. Santangelo. 2001. Rap1p requires Gcr1p and Gcr2p homodimers to activate ribosomal protein and glycolytic genes, respectively. Genetics 158:133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devlin, C., K. Tice-Baldwin, D. Shore, and K. T. Arndt. 1991. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol. Cell. Biol. 11:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dikstein, R., S. Ruppert, and R. Tjian. 1996. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell 84:781-790. [DOI] [PubMed] [Google Scholar]
- 13.Dunah, A. W., H. Jeong, A. Griffin, Y. M. Kim, D. G. Standaert, S. M. Hersch, M. M. Mouradian, A. B. Young, N. Tanese, and D. Krainc. 2002. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science 296:2238-2243. [DOI] [PubMed] [Google Scholar]
- 14.Durant, M., and B. F. Pugh. 2006. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:2791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fingerman, I., V. Nagaraj, D. Norris, and A. K. Vershon. 2003. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot. Cell 2:1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishburn, J., N. Mohibullah, and S. Hahn. 2005. Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell 18:369-378. [DOI] [PubMed] [Google Scholar]
- 17.Fourel, G., T. Miyake, P. A. Defossez, R. Li, and E. Gilson. 2002. General regulatory factors (GRFs) as genome partitioners. J. Biol. Chem. 277:41736-41743. [DOI] [PubMed] [Google Scholar]
- 18.Geles, K. G., R. N. Freiman, W. L. Liu, S. Zheng, E. Voronina, and R. Tjian. 2006. Cell-type-selective induction of c-jun by TAF4b directs ovarian-specific transcription networks. Proc. Natl. Acad. Sci. USA 103:2594-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill, G., E. Pascal, Z. H. Tseng, and R. Tjian. 1994. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA 91:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham, I. R., and A. Chambers. 1996. Rap1p is a negative regulator of the RAP1 gene. Curr. Genet. 30:93-100. [DOI] [PubMed] [Google Scholar]
- 21.Graham, I. R., R. A. Haw, K. G. Spink, K. A. Halden, and A. Chambers. 1999. In vivo analysis of functional regions within yeast Rap1p. Mol. Cell. Biol. 19:7481-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green, M. R. 2005. Eukaryotic transcription activation: right on target. Mol. Cell 18:399-402. [DOI] [PubMed] [Google Scholar]
- 23.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 24.Grob, P., M. J. Cruse, C. Inouye, M. Peris, P. A. Penczek, R. Tjian, and E. Nogales. 2006. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure 14:511-520. [DOI] [PubMed] [Google Scholar]
- 25.Hall, D. B., and K. Struhl. 2002. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J. Biol. Chem. 277:46043-46050. [DOI] [PubMed] [Google Scholar]
- 26.Hall, D. B., J. T. Wade, and K. Struhl. 2006. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamard, P. J., R. Dalbies-Tran, C. Hauss, I. Davidson, C. Kedinger, and B. Chatton. 2005. A functional interaction between ATF7 and TAF12 that is modulated by TAF4. Oncogene 24:3472-3483. [DOI] [PubMed] [Google Scholar]
- 28.Hiller, M., X. Chen, M. J. Pringle, M. Suchorolski, Y. Sancak, S. Viswanathan, B. Bolival, T. Y. Lin, S. Marino, and M. T. Fuller. 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131:5297-5308. [DOI] [PubMed] [Google Scholar]
- 29.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 30.Imhof, A., X. J. Yang, V. V. Ogryzko, Y. Nakatani, A. P. Wolffe, and H. Ge. 1997. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 7:689-692. [DOI] [PubMed] [Google Scholar]
- 31.Irvin, J. D., and B. F. Pugh. 2006. Genome-wide transcriptional dependence on TAF1 functional domains. J. Biol. Chem. 281:6404-6412. [DOI] [PubMed] [Google Scholar]
- 32.Johannessen, M., P. A. Olsen, R. Sorensen, B. Johansen, O. M. Seternes, and U. Moens. 2003. A role of the TATA box and the general co-activator hTAF(II)130/135 in promoter-specific trans-activation by simian virus 40 small t antigen. J. Gen. Virol. 84:1887-1897. [DOI] [PubMed] [Google Scholar]
- 33.Jorgensen, P., I. Rupes, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, L. J., A. G. Seto, T. N. Nguyen, and J. A. Goodrich. 2001. Human Taf(II)130 is a coactivator for NFATp. Mol. Cell. Biol. 21:3503-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klebanow, E. R., D. Poon, S. Zhou, and P. A. Weil. 1996. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J. Biol. Chem. 271:13706-13715. [DOI] [PubMed] [Google Scholar]
- 36.Klein, C., and K. Struhl. 1994. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science 266:280-282. [DOI] [PubMed] [Google Scholar]
- 37.Klein, J., M. Nolden, S. L. Sanders, J. Kirchner, P. A. Weil, and K. Melcher. 2003. Use of a genetically introduced cross-linker to identify interaction sites of acidic activators within native transcription factor IID and SAGA. J. Biol. Chem. 278:6779-6786. [DOI] [PubMed] [Google Scholar]
- 38.Kohrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 39.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 40.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 41.Kyrion, G., K. Liu, C. Liu, and A. J. Lustig. 1993. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 7:1146-1159. [DOI] [PubMed] [Google Scholar]
- 42.Lascaris, R. F., W. H. Mager, and R. J. Planta. 1999. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics 15:267-277. [DOI] [PubMed] [Google Scholar]
- 43.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 44.Leurent, C., S. L. Sanders, M. A. Demeny, K. A. Garbett, C. Ruhlmann, P. A. Weil, L. Tora, and P. Schultz. 2004. Mapping key functional sites within yeast TFIID. EMBO J. 23:719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 46.Li, X. Y., S. R. Bhaumik, X. Zhu, L. Li, W. C. Shen, B. L. Dixit, and M. R. Green. 2002. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 12:1240-1244. [DOI] [PubMed] [Google Scholar]
- 47.Li, X. Y., A. Virbasius, X. Zhu, and M. R. Green. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 399:605-609. [DOI] [PubMed] [Google Scholar]
- 48.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 49.Marion, R. M., A. Regev, E. Segal, Y. Barash, D. Koller, N. Friedman, and E. K. O'Shea. 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101:14315-14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin, D. E., A. Soulard, and M. N. Hall. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119:969-979. [DOI] [PubMed] [Google Scholar]
- 51.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 52.Matangkasombut, O., and S. Buratowski. 2003. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 11:353-363. [DOI] [PubMed] [Google Scholar]
- 53.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 54.Mengus, G., M. May, L. Carre, P. Chambon, and I. Davidson. 1997. Human TAF(II)135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 11:1381-1395. [DOI] [PubMed] [Google Scholar]
- 55.Menon, B. B., N. J. Sarma, S. Pasula, S. J. Deminoff, K. A. Willis, K. E. Barbara, B. Andrews, and G. M. Santangelo. 2005. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 102:5749-5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michel, B., P. Komarnitsky, and S. Buratowski. 1998. Histone-like TAFs are essential for transcription in vivo. Mol. Cell 2:663-673. [DOI] [PubMed] [Google Scholar]
- 57.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 58.Moqtaderi, Z., Y. Bai, D. Poon, P. A. Weil, and K. Struhl. 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383:188-191. [DOI] [PubMed] [Google Scholar]
- 59.Moretti, P., K. Freeman, L. Coodly, and D. Shore. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8:2257-2269. [DOI] [PubMed] [Google Scholar]
- 60.Morse, R. H. 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16:51-53. [DOI] [PubMed] [Google Scholar]
- 61.Naar, A. M., D. J. Taatjes, W. Zhai, E. Nogales, and R. Tjian. 2002. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 63.Pina, B., J. Fernandez-Larrea, N. Garcia-Reyero, and F. Z. Idrissi. 2003. The different (sur)faces of Rap1p. Mol. Genet. Genomics 268:791-798. [DOI] [PubMed] [Google Scholar]
- 64.Reese, J. C., Z. Zhang, and H. Kurpad. 2000. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J. Biol. Chem. 275:17391-17398. [DOI] [PubMed] [Google Scholar]
- 65.Reeves, W. M., and S. Hahn. 2005. Targets of the Gal4 transcription activator in functional transcription complexes. Mol. Cell. Biol. 25:9092-9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 67.Roeder, R. G. 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579:909-915. [DOI] [PubMed] [Google Scholar]
- 68.Rojo-Niersbach, E., T. Furukawa, and N. Tanese. 1999. Genetic dissection of hTAF(II)130 defines a hydrophobic surface required for interaction with glutamine-rich activators. J. Biol. Chem. 274:33778-33784. [DOI] [PubMed] [Google Scholar]
- 69.Rudra, D., and J. R. Warner. 2004. What better measure than ribosome synthesis? Genes Dev. 18:2431-2436. [DOI] [PubMed] [Google Scholar]
- 70.Rudra, D., Y. Zhao, and J. R. Warner. 2005. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 24:533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saluja, D., M. F. Vassallo, and N. Tanese. 1998. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol. Cell. Biol. 18:5734-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanders, S. L., K. A. Garbett, and P. A. Weil. 2002. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol. 22:6000-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanders, S. L., E. R. Klebanow, and P. A. Weil. 1999. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274:18847-18850. [DOI] [PubMed] [Google Scholar]
- 74.Sanders, S. L., and P. A. Weil. 2000. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 275:13895-13900. [DOI] [PubMed] [Google Scholar]
- 75.Sauer, F., S. K. Hansen, and R. Tjian. 1995. DNA template and activator-coactivator requirements for transcriptional synergism by Drosophila bicoid. Science 270:1825-1828. [DOI] [PubMed] [Google Scholar]
- 76.Sauer, F., S. K. Hansen, and R. Tjian. 1995. Multiple TAFIIs directing synergistic activation of transcription. Science 270:1783-1788. [DOI] [PubMed] [Google Scholar]
- 77.Schawalder, S. B., M. Kabani, I. Howald, U. Choudhury, M. Werner, and D. Shore. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432:1058-1061. [DOI] [PubMed] [Google Scholar]
- 78.Schmid, M., G. Arib, C. Laemmli, J. Nishikawa, T. Durussel, and U. K. Laemmli. 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell 21:379-391. [DOI] [PubMed] [Google Scholar]
- 79.Schroeder, S. C., C. K. Wang, and P. A. Weil. 1994. Identification of the cis-acting DNA sequence elements regulating the transcription of the Saccharomyces cerevisiae gene encoding TBP, the TATA box binding protein. J. Biol. Chem. 269:28335-28346. [PubMed] [Google Scholar]
- 80.Sermwittayawong, D., and S. Tan. 2006. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J. 25:3791-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen, W. C., S. R. Bhaumik, H. C. Causton, I. Simon, X. Zhu, E. G. Jennings, T. H. Wang, R. A. Young, and M. R. Green. 2003. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 22:3395-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 83.Shore, D. 1994. RAP1: a protean regulator in yeast. Trends Genet. 10:408-412. [DOI] [PubMed] [Google Scholar]
- 84.Taatjes, D. J., A. M. Naar, F. Andel III, E. Nogales, and R. Tjian. 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295:1058-1062. [DOI] [PubMed] [Google Scholar]
- 85.Thuault, S., Y. G. Gangloff, J. Kirchner, S. Sanders, S. Werten, C. Romier, P. A. Weil, and I. Davidson. 2002. Functional analysis of the TFIID-specific yeast TAF4 (yTAF(II)48) reveals an unexpected organization of its histone-fold domain. J. Biol. Chem. 277:45510-45517. [DOI] [PubMed] [Google Scholar]
- 86.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 87.Tsukihashi, Y., M. Kawaichi, and T. Kokubo. 2001. Requirement for yeast TAF145 function in transcriptional activation of the RPS5 promoter that depends on both core promoter structure and upstream activating sequences. J. Biol. Chem. 276:25715-25726. [DOI] [PubMed] [Google Scholar]
- 88.van de Peppel, J., P. Kemmeren, H. van Bakel, M. Radonjic, D. van Leenen, and F. C. Holstege. 2003. Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep. 4:387-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vassallo, M. F., and N. Tanese. 2002. Isoform-specific interaction of HP1 with human TAFII130. Proc. Natl. Acad. Sci. USA 99:5919-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wade, C., K. A. Shea, R. V. Jensen, and M. A. McAlear. 2001. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell. Biol. 21:8638-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wade, J. T., D. B. Hall, and K. Struhl. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432:1054-1058. [DOI] [PubMed] [Google Scholar]
- 92.Walker, S. S., J. C. Reese, L. M. Apone, and M. R. Green. 1996. Transcription activation in cells lacking TAFIIS. Nature 383:185-188. [DOI] [PubMed] [Google Scholar]
- 93.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 94.Watt, K., T. J. Jess, S. M. Kelly, N. C. Price, and I. J. McEwan. 2005. Induced alpha-helix structure in the aryl hydrocarbon receptor transactivation domain modulates protein-protein interactions. Biochemistry 44:734-743. [DOI] [PubMed] [Google Scholar]
- 95.Woontner, M., P. A. Wade, J. Bonner, and J. A. Jaehning. 1991. Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol. Cell. Biol. 11:4555-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yatherajam, G., L. Zhang, S. M. Kraemer, and L. A. Stargell. 2003. Protein-protein interaction map for yeast TFIID. Nucleic Acids Res. 31:1252-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zanton, S. J., and B. F. Pugh. 2004. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc. Natl. Acad. Sci. USA 101:16843-16848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zanton, S. J., and B. F. Pugh. 2006. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 20:2250-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao, Y., K. B. McIntosh, D. Rudra, S. Schawalder, D. Shore, and J. R. Warner. 2006. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell. Biol. 26:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]