Abstract
Hereditary spastic paraplegia (HSP) is a progressive upper-motor neurodegenerative disease. The eighth HSP locus, SPG8, is on chromosome 8p24.13. The three families previously linked to the SPG8 locus present with relatively severe, pure spastic paraplegia. We have identified three mutations in the KIAA0196 gene in six families that map to the SPG8 locus. One mutation, V626F, segregated in three large North American families with European ancestry and in one British family. An L619F mutation was found in a Brazilian family. The third mutation, N471D, was identified in a smaller family of European origin and lies in a spectrin domain. None of these mutations were identified in 500 control individuals. Both the L619 and V626 residues are strictly conserved across species and likely have a notable effect on the structure of the protein product strumpellin. Rescue studies with human mRNA injected in zebrafish treated with morpholino oligonucleotides to knock down the endogenous protein showed that mutations at these two residues impaired the normal function of the KIAA0196 gene. However, the function of the 1,159-aa strumpellin protein is relatively unknown. The identification and characterization of the KIAA0196 gene will enable further insight into the pathogenesis of HSP.
Hereditary spastic paraplegia (HSP) has a worldwide prevalence of 1–18 in 100,0001–3 and is characterized by central-motor-system deficits leading to lower-limb spastic paraperesis.4–6 This is due to a “dying back” phenomenon whereby upper motor neurons degenerate progressively, commencing with the longest axons.7,8 HSP can be classified into pure and complicated forms.5 In pure HSP, lower-limb spasticity is the only major symptom. Alternatively, in complicated HSP, this spasticity can be accompanied by other neurological or nonneurological symptoms, such as ataxia, dementia, mental retardation, deafness, epilepsy, ichthyosis, retinopathy, ocular neuropathy, and extrapyramidal disturbances.5,9 There is clinical heterogeneity within families, where age at onset and severity can differ markedly; between families that map to the same locus; and certainly between families that map to separate loci. This heterogeneity complicates genotype-phenotype correlations for HSP.
HSP is also extremely genetically heterogeneous. From >30 loci mapped (SPG1–33), 11 genes have been identified. This disease can be transmitted in a dominant (13 loci), a recessive (15 loci), or an X-linked manner (4 loci).9–11 By far, the most common locus for the disease is SPG4 (MIM 604277), with mutations in the microtubule-severing protein spastin accounting for ∼40% of dominant HSP cases.12,13
Families that map to SPG8 are considered to have one of the more aggressive subtypes of HSP, with disease onset occurring for patients as early as their 20s or 30s. It was first identified in a white family as a 6.2-cM region between markers D8S1804 and D8S1774.14 The family had 15 patients affected with spasticity, hyperreflexia, extensor plantar reflexes, lower-limb weakness, decreased vibration sensation, and limited muscle wasting. The candidate region was further reduced to 3.4 cM because of a lower recombinant in a second family, which narrows the interval between markers D8S1804 and D8S1179.15 This family, as well as a third Brazilian family linked to SPG8, also presented with pure adult-onset HSP.16 For two of the families, a muscle biopsy was performed14,16; however, no gross histological or histochemical abnormalities were observed. Ragged red fibers have been observed in muscle biopsies of patients with HSP who have paraplegin mutations.17
In the present study, we identified four additional families that are linked to the SPG8 locus. Genes were screened in an expanded candidate SPG8 locus defined by these four families, along with the British and Brazilian families described above.15,16 This led to the identification of three point mutations in the KIAA0196 gene encoding the strumpellin protein product.
Material and Methods
Subjects
Protocols were approved by the Ethics Committee of the Centre Hospitalier de l’Université de Montréal. Patients gave informed consent, after which patient information and blood was collected. DNA was extracted from peripheral blood through use of standard protocols.
Genotyping and Locus Exclusion
PCR-amplified fragments incorporating α-35S–2-deoxyadenosine 5-triphosphate were resolved on 6% denaturing polyacrylamide gels. Alleles were run alongside an M13mp18 sequence ladder and were scored on the basis of allele sizes and frequencies from the Fondation Jean Dausset-CEPH database. LOD-score calculations and multipoint analysis were performed using the MLINK program of the LINKMAP software package.18
Mutation Screening
The 28 exons of KIAA0196 were screened by automated sequencing, including at least 50 bp of each intronic region. Primers were designed using the PrimerSelect program (Lasergene) and were synthesized by Invitrogen Canada. Primer sequences and amplification conditions for each exon are listed in table 1.
Table 1. .
Primers and Amplification Conditions for KIAA0196[Note]
| Exon | Primer(5′→3′) |
| KIAA0196×1 (forward) | GCCAAGAGTGTTAATCTAGCAAAGTC |
| KIAA0196×1(reverse) | TTCATGGTTCCCAGAGAAAACACG |
| KIAA0196×2 (forward) | TCTGCTTTAAGTTTGGGATGTCTA |
| KIAA0196×2 (reverse) | TTAAGATGACCAGTGCCACAGGTA |
| KIAA0196×3 (forward) | AATATCAAACTGTGGCCCTAAATC |
| KIAA0196×3 (reverse) | TACACCGAGGAGGCTCATAACTTC |
| KIAA0196×4 (forward) | CATCCCAGCCATCTGTCCTGATAC |
| KIAA0196×4 (reverse) | ACATACACTGCATTTTACCGACAGC |
| KIAA0196×5 (forward) | AATGGAATTCTACTTTATTGGACT |
| KIAA0196×5 (reverse) | CTCAAAAGGTTTTAAAAGGTTCTACC |
| KIAA0196×6 (forward) | TGGGCTTTGGAAAAACTGATGTGTCT |
| KIAA0196×6 (reverse) | AAGTTTACCTAAGTGATGTTATGTCC |
| KIAA0196×7 (forward) | CAAAAAGCAACGTTAATAGGTGTAA |
| KIAA0196×7 (reverse) | ATCATTGCATTAAATTATCTAAGTG |
| KIAA0196×8 (forward) | TTAATCACAGCCAGAACTAGGATGTAG |
| KIAA0196×8 (reverse) | GACAGGGGAGAGCTTTTCAGGTATGCT |
| KIAA0196×9 (forward) | TGGCACTCCATGTCAGATTCAACTGT |
| KIAA0196×9 (reverse) | ATGTCTATATTCCCCATTAGG |
| KIAA0196×10 (forward) | CAGGGTCAATGTTAATTTATAGTGTA |
| KIAA0196×10 (reverse) | AGATGGAGGCCAACTGTGACTCTC |
| KIAA0196×11 (forward) | TGCTCCAGGCATTTTTGTCG |
| KIAA0196×11 (reverse) | GAACAGACTGCTGGGTGGGTCATA |
| KIAA0196×12 and 13 (forward) | ATGAGCACCATAGAGTCCATTCAGG |
| KIAA0196×12 and 13 (reverse) | ATTATGCTCTCGTGGAAAAACTGCTA |
| KIAA0196×14 (forward) | CTTTTTGAAACAAGAAACAGATATACC |
| KIAA0196×14 (reverse) | GGCAAGTAAAAACATCTGTACATCCAC |
| KIAA0196×15 (forward) | TTTGCAGCATTTTTAGAAGGATTAGC |
| KIAA0196×15 (reverse) | TTCCCCTGAGAATACTGAGGCGAACA |
| KIAA0196×16 (forward) | GGAGGCCAGGGAAGGGGAGGTTACC |
| KIAA0196×16 (reverse) | GGAATGTCAAACAGCCAGATGATGT |
| KIAA0196×17 (forward) | ACTTTGCTGAAATAAACAGAGTCC |
| KIAA0196×17 (reverse) | GTAAGGTCTTGTTCGCGATAGGTT |
| KIAA0196×18 (forward) | AGAACGAATAGTTGACAATCTACTC |
| KIAA0196×18 (reverse) | TGAGGTTTGGGATGTGTACTCTAA |
| KIAA0196×19 (forward) | AATTATATGGAAAAGGGATAACTAGGT |
| KIAA0196×19 (reverse) | TAAAGGGTCAGAATATGAGTTGACAAG |
| KIAA0196×20 (forward) | TTGGTGCCGCATGTCCTGTTGAGTC |
| KIAA0196×20 (reverse) | AAGTCTTATCTTCCCAAGTTGAAAC |
| KIAA0196×21 and 22 (forward) | CCCAGCCTCTGTTCTGCATAGCAT |
| KIAA0196×21 and 22 (reverse) | AAGAACAGATCCAGAAACGGAGAT |
| KIAA0196×23 (forward) | AAGGCCCAGTGAAGAATTGTCATC |
| KIAA0196×23 (reverse) | CTGAAGAAACTGGGGTGCGTAGAT |
| KIAA0196×24 (forward) | CTGAGGCTTGAAAAGATTACATCAC |
| KIAA0196×24 (reverse) | CTTCCCCTTTGTCATGAGCTTTCAC |
| KIAA0196×25 (forward) | TCCCACACTCCCCCTATATTCACCTC |
| KIAA0196×25 (reverse) | AGAAAAGATCTCATATCCGACATAGG |
| KIAA0196×26 (forward) | GACCCCTGGAATGCCCTACCAATC |
| KIAA0196×26 (reverse) | CTGGCAGGGTGACTAAGGATGGAC |
| KIAA0196×27 (forward) | GATAGATAGCAGGGATCGTGTTGT |
| KIAA0196×27 (reverse) | AGGCATCTACTGTGAACGACCTAT |
| KIAA0196×28 (forward) | AAAGGGGCTGTTTCAAGGAGTCG |
| KIAA0196×28 (reverse) | AGTTTTTGAATCATAAGCGAGACG |
Note.— PCR was performed using 50 ng DNA, 20 pmol of each primer, 10× buffer, 0.25 nM deoxyribonucleotide triphosphate, and 0.15 μl of Taq (Qiagen). Initial denaturation for 5 min at 94°C was followed by 30 cycles of 30-s denaturation at 94°C, 30 s annealing at 55°C (for all exons except 15 and 26), and 45-s elongation at 72°C. A final extension at 72°C was performed for 7 min. For exon 15, a 50°C annealing temperature was used, and, for exon 26, 10 cycles of a touchdown reaction were performed from 68°C to 63°C, followed by 25 cycles at 63°C.
Variants were first tested in 12 control individuals by sequencing, followed by allele-specific oligomerization (ASO).19,20 In brief, 4 μl of PCR product was hybridized onto Hybond-N+ Nylon membranes (Amersham Biosciences) by use of a dot-blot apparatus. P32-labeled probes specific to the mutation or normal sequence were hybridized and then visualized on autoradiographic film after overnight exposure. ASO primers for exon 11 are 5′-ACTAGAAAACCTTCAAGCT-3′ (normal) and 5′-ACTAGAAGACCTTCAAGCT-3′ (mutated). For exon 14, ASO primers of 5′-GGAGAGTTGGTATC-3′ (normal) and 5′-GGAGAGTTCGTATC-3′ (mutated) were used. Exon 15 ASO primers were 5′-CACTGAAGGTTTTG-3′ (normal) and 5′-CACTGAAGTTTTTG-3′ (mutated).
Protein-Sequence Alignment
Cluster analysis was performed using the Probcons (v. 1.09) program. Proteins from aligned species included Homo sapiens (Q12768), Canis familiaris (GenBank accession number XP_532327), Pan troglodytes (GenBank accession number XP_519952), Drosophila melanogaster (GenBank accession number CG12272), Caenorhabditis elegans (GenBank accession number CE13235), Xenopus tropicalis (GenBank accession number MGC89323), Rattus norvegicus (GenBank accession number XP_343250), Danio rerio (GenBank accession number BC045490), Gallus gallus (GenBank accession number XP_418441), Dictyostelium discoideum (GenBank accession number EAL63144), and Mus musculus (GenBank accession number NP_705776.2).
Homology Modeling
The size of the strumpellin protein (1,159 aa) made it prohibitive to obtain a template for the entire protein. Instead, 200 aa (amino acids 501–725) around the two mutations were selected and inputted in the Phyre program version 2.0 (Phyre Protein Fold Recognition Server). The template with the highest score was selected—namely, 1dn1b from the Neuronal-Sec1 syntaxin 1a complex. The SwissProt database viewer (v. 3.7)21 was used to visualize the model, with concentration on the α-helix in which the two mutations lie and on a second α-helix in nearby 3D space. Peptides incorporating one or the other identified point mutation were visualized in the same manner.
Expression Studies
Northern-blot and RT-PCR analyses
The KIAA0196 cDNA pBluescript clone was kindly provided by the Kazusa DNA Research Institute. A 1-kb probe specific to the C-terminal region of strumpellin was generated by digesting the KIAA0196 pBluescript vector with XhoI and NotI. Thirty micrograms of total RNA per sample was loaded. RNA was extracted from various regions of the brain of a control individual. An RT-PCR was performed using Moloney murine leukemia virus–reverse transcriptase (Invitrogen). Primers in exons 10 (forward) and 15 (reverse) of KIAA0196 were used. Glyceraldehyde-3-phosphate dehydrogenase cDNA was amplified as a control.
Constructs
Each mutation was introduced into the KIAA0196 pBluescript clone by site-directed mutagenesis through use of the primers 5′-CTGGAGAGTTCGTATCCTATGTG-3′ for the exon 14 variant and 5′-CCTATGTGAGAAAATTTTTGCAGATC-3′ for the exon 15 variant, along with primers of their complementary sequence. Wild-type and mutant KIAA0196 cDNAs were cloned, upstream of Myc and His tags, into a pCS2 vector and were transcribed in vitro by use of the SP6 mMESSAGE mMachine kit (Ambion) for zebrafish studies. The protein expression from each of these constructs was validated after their transient expression in cell (HeLa) culture and subsequent western-blot analysis with an anti-Myc antibody. A band at the appropriate height (∼134 kDa for strumpellin) was observed.
Zebrafish Knockdown Studies
Morpholino injections
Wild-type zebrafish were raised and mated as described elsewhere.22 Antisense morpholinos (AMOs) were purchased from Genetools. The morpholino sequences were designed against the zebrafish strumpellin ortholog BC045490. The oligonucleotide CTCTGCCAGAAAATCAC(CAT)GATG (KIAA MO) binds to the ATG of the KIAA0196 gene, which prevents its translation, and CTCTcCCAcAAAATgAg(CAT)cATG (CTL MO) is a 5-bp mismatch control. AMO injections were performed as described elsewhere, at a concentration of 0.8 mM.23 The rescue injections were performed as mentioned above, with morpholino and mRNA concentrations of 0.8 mM and 50 ng/μl, respectively.
Immunohistochemistry
Standard protocols were used for immunohistochemistry.22 In brief, 3-d-old embryos were fixed in 4% paraformaldehyde, were washed, and were blocked at room temperature. Primary antibody (anti-acetylated tubulin; 1:50 [Sigma]) was added overnight. After extensive washing, the embryos were incubated with the fluorescently labeled secondary antibody Alexa 568 (Molecular Probes). Imaging was performed on an UltraView LCI confocal microscope (Perkin Elmer) with use of Methamorph Imaging software (Universal Imaging). The statistical significance between the different conditions was calculated using a χ2 test.
Results
Clinical Information and Family Details
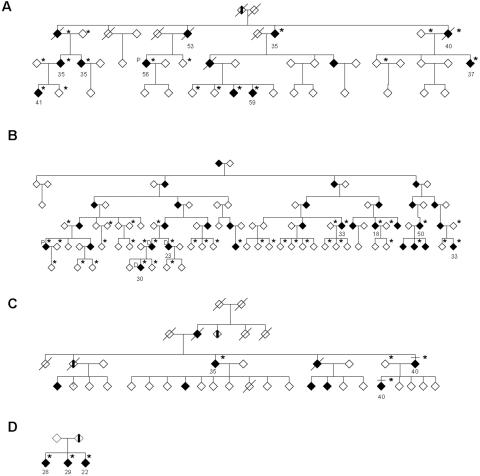
Family FSP24 with the SPG8 mutation is from British Columbia. It is composed of 13 members affected with a spastic gait and lower-limb stiffness (fig. 1A); genetic information is available for 10 of them. Symptoms were first observed in individuals between ages 35 and 53 years. Intrafamilial phenotypic heterogeneity exists, as shown by the symptoms presented and the range of disease severity in patients. Deep-tendon reflexes were brisk or increased, and decreased vibration sensation was also noted in three patients. Occasional bladder-control problems were also observed. Walking aids were required for some individuals, whereas one is confined to a wheelchair. Together, these features are consistent with a pure, uncomplicated HSP similar to that described for other families linked to the SPG8 locus. Family FSP29 is of European descent and resides in the United States. There are 31 affected individuals in the family, and genetic information is available for 10 of them (fig. 1B). Age at onset was quite variable, with symptom onset ranging in patients from their 20s to their 60s. The family was negative for mutations in the spastin gene.
Figure 1. .
Pedigrees for families with KIAA0196 mutations. A, Family FSP24. B, Family FSP29. C, Family FSP34. D, Family FSP91. Blackened boxes represent affected individuals, and a diagonal line through the symbol means the individual is deceased. A vertical blackened bar indicates an individual with an unconfirmed phenotype. Sex of each individual has been masked to preserve confidentiality. Individuals marked “P” represent proximal recombinants; “D” represents the distal recombinant. An asterisk (*) indicates that DNA and clinical information have been collected for the particular individual. The age at onset of affected individuals is listed below each symbol, although this information is not available for each patient. All studied affected patients are heterozygous for a c.1956C→T mutation (pedigrees A, B, and C) or a c.A1491G mutation (pedigree D) in KIAA0196.
Linkage Analysis
Two large families that map to the SPG8 locus were identified. In family FSP24, seven markers spanning the candidate region from markers D8S586 to D8S1128 were genotyped in the 18 individuals studied (fig. 1A). A disease haplotype segregated with the disease in all 10 affected individuals (table 2). A recombination event occurred in one individual (fig. 1A) between markers D8S586 and D8S1804, which defined the proximal border of the locus in this family. A lower recombinant was neither identified nor searched for, since the haplotype extended beyond the limits of the SPG8 locus. The maximum LOD score for this family was 3.43 at θ=0, by use of CEPH allele frequencies for the marker D8S1804, along with a maximum multipoint of 4.20 at marker D8S1799.
Table 2. .
Haplotype Comparison between SPG8-Linked Families[Note]
| Alleles Number for Family |
||||
| Marker | Position (Mb) |
FSP24 | FSP29 | FSP34 |
| D8S586 | 121.2 | 1 | 11 | 11 |
| D8S1804 | 124.8 | 5 | 3 | 3 |
| D8S1832 | 125.4 | 2 | 2 | Not typed |
| D8S1179 | 125.9 | 3 | 9 | 9 |
| D8S1774 | 127.5 | 3 | 5 | 4 |
| D8S1128 | 128.5 | 7 | 5 | 1 |
Note.— Flanking markers in the candidate region are D8S1832 and D8S1774 for family FSP29. The KIAA0196 L619F mutation was at position 126.1 Mb for all three families. The allele for rs2293890 (126.4 Mb) was G for family FSP24 and was C for both families FSP29 and FSP34.
The same seven markers tested in family FSP24 were genotyped for family FSP29. A disease haplotype was established for all 10 studied affected individuals; it included many informative recombination events. The proximal recombinant occurred between markers D8S1799 and D8S1832 in three affected individuals (fig. 1B), and the distal recombinant was between markers D8S1774 and D8S1128 for another affected individual (fig. 1B). This yielded a candidate interval of 3.15 Mb. The maximum LOD score for this family was 5.62 (θ=0) for marker D8S1179 when CEPH allele frequencies were used. Multipoint analysis was also conducted for this family in this region, which yielded a maximum LOD score of 6.73, 0.5 cM centromeric to the D8S1128 marker.
Gene Screening
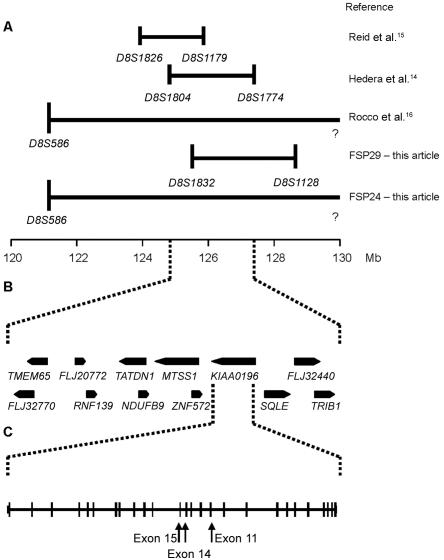
The previously published SPG8 locus spanned 3.4 cM (1.04 Mb) between markers D8S1804 and D8S1179 on chromosome 8q23-8q24. We screened nine known genes surrounding this candidate region, as annotated in the University of California–Santa Cruz Genome Browser (UCSC) May 2004 update, along with many clustered ESTs and mRNAs that aligned to the locus, without detecting a mutation. Therefore, we opted to redefine the candidate region on the basis of the critical interval determined by an upper recombinant in our FSP29 family at marker D8S1832, and a lower recombinant at D8S1774 was based on published data (fig. 2A).14 This increased the size of the region to 5.43 cM (3.15 Mb), a region that contains three additional known genes (fig. 2B). These additional genes were screened, and three mutations were identified in the KIAA0196 gene (fig. 2C).
Figure 2. .
Region spanning the SPG8 locus. A, Markers defining the borders of each described family with the SPG8 mutation and the scaled marker positions on chromosome 8q24.13. B, Candidate region used to search for the SPG8 gene between markers D8S1804 and D8S1774. Genes in the region are shown in their observed orientation. C, The 28-exon KIAA0196 gene, drawn to scale, with the location of three mutations in exons 11, 14, and 15 highlighted.
Mutation Analysis
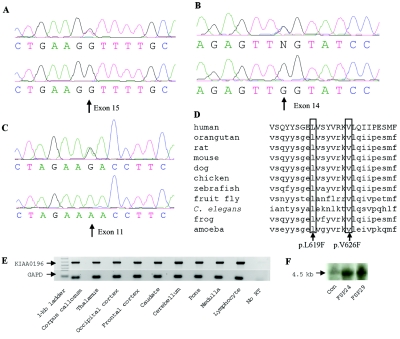
A valine→phenylalanine mutation was identified in amino acid 626 for families FSP24 and FSP29 (p.V626F) (fig. 3A). All studied affected individuals from each family were screened and were positive for this mutation. The same mutation was also found to segregate in a British family.15 This G→T nucleotide change is at position 1956 of the mRNA (GenBank accession number NM_014846.2). A total of 500 ethnically matched control individuals (400 from North America and 100 from CEPH) were negative for this mutation, on screening by a combination of ASO and sequencing. No unaffected members or spouse control individuals in any family were positive for the mutations.
Figure 3. .
Mutation analysis of the KIAA0196 gene. A–C, Sequence trace of a patient with HSP above the sequence trace of a control individual. Exon 15 (A), 14 (B), and 11 (C) heterozygous point mutations are indicated. D, Multiple-sequence alignment for strumpellin homologues surrounding the two coding changes (boxed). The Probcons (v.1.09) program was used for cluster analysis. E, RT-PCR of multiple brain regions performed using a KIAA0196-specific probe. F, Northern blot of the KIAA0196 transcript performed using 30 μg of total RNA and a 1-kb C-terminal probe.
A second mutation was identified in the Brazilian family16 in exon 14, a G→C transition at position 1937 of the mRNA (fig. 3B). This leucine→phenylalanine change (p.L619F) is only 7 aa away from the V626F mutation. It was not found, with use of ASO, in 500 controls.
The KIAA0196 gene was screened in probands from 24 additional dominant HSP–affected families that are negative for mutations in both spastin and atlastin, resulting in the identification of two more families with missense mutations in the KIAA0196 gene. Thus, the frequency of mutations in our SPG3A- and SPG4-negative autosomal dominant cohort is ∼8% (2 of 24). FSP34 has the same p.V626F change in its three affected studied family members. This family is originally from Great Britain and resides in Canada (fig. 1C). Haplotype analysis of this family with markers D8S1804, D8S1179, D8S1774, and D8S1128 indicated that there is allele sharing between this family and family FSP29, which suggests an ancestral haplotype (table 2). An additional mutation was found in three affected siblings of another North American family of European origin, family FSP91 (fig. 1D). This c.A1491G transition results in an asparagine→aspartate amino acid change (p.N471D) and is not present in the 500 controls tested (fig. 3C).
Mutated amino acids at positions 619 and 626 are strictly conserved across all 11 examined species, from human to the social amoeba, D. discoideum (fig. 3D). Indeed, the entire region surrounding these two mutations appears to be functionally relevant for the protein, since 73 consecutive aa (amino acids 576–649) are 100% identical in the human, dog, chicken, mouse, rat, and orangutan. Despite this high level of conservation, this region is an unknown domain, on the basis of searches of the National Center for Biotechnology Information (NCBI) Conserved Domain Database, NCBI BLAST, and the Sanger Institute’s Pfam database. Position 471 is conserved across all species except D. melanogaster (with a glutamine residue) and X. tropicalis (with a histidine residue).
The exon 15 mutation is in the very first nucleotide of the exon, which leads to the speculation that the splicing of this exon might be compromised in our study families. Splice-site prediction programs, including NetGene2, suggested that the strength of the splice-site acceptor may be reduced by 33% in the mutant form. However, both normal and mutant alleles were observed in cDNA analysis, with use of several pairs of primers, of patient lymphoblasts. The KIAA0196 gene was expressed ubiquitously, including all regions of the brain that were examined by RT-PCR (fig. 3E). There were no alternative splice isoforms detected in control brain samples or patient whole-blood samples by RT-PCR and northern-blot analysis (fig. 3E and 3F). For the full KIAA0196 gene, all spliced ESTs and mRNAs from the UCSC browser, May 2004 draft, were analyzed for potential alternative splice products. One alternative first exon often appears; however, of the 356 entries, only 2 (AK223628 and DA202680) contain exons that are skipped. Thus, overall, the gene is not frequently spliced, and the two spliced entries may represent spurious transcripts.
KIAA0196 Profile
The KIAA0196 gene spans 59.7-kb pairs of genomic DNA, is 28 exons long, and codes for a protein of 1,159 aa (strumpellin). The European Bioinformatics Institute’s InterProScan program predicts a spectrin-repeat–containing domain in amino acids 434–518. Thus, the mutation at position 471 may abrogate the binding of the spectrin domain with other spectrin-repeat–containing proteins. In examination of the secondary structure by use of PSIPRED,24 74% of the protein is considered to be α-helical. The program further predicts an α-helix in the protein from amino acids 606–644, which encompasses the two other mutations that have been identified.
Homology Modeling
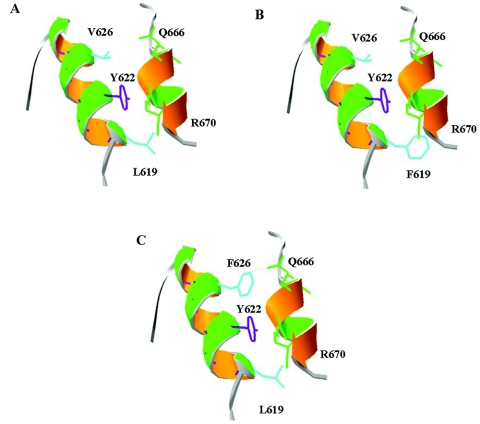
Given the high proportion of KIAA0196 considered to be α-helical, it is not surprising that the optimal homology-modeling candidates are similar in secondary-structure composition. This is true for 1dn1b, a stat-like t-SNARE protein neuronal-Sec1 syntaxin 1a complex. This is the most appropriate model for strumpellin, according to the Phyre program (Phyre Protein Fold Recognition Server). The two mutated residues lie within an α-helix from amino acids 619–628 that is in close 3D proximity to another α-helix from residues 665–670 (fig. 4A). A mutation in either Val-626 or Leu-619 to a phenylalanine residue would appear to have significant structural implications, given the change in bulkiness between the residues. In addition, Tyr-622 points in the same direction from the α-helix residue. To have two amino acids with aromatic rings in such a physical proximity could force apart the alignment of the two α-helices or induce alterations in the α-helix backbone. The N471D mutation was identified well after the two other mutations and so was not tested in homology modeling or in subsequent zebrafish-rescue experiments.
Figure 4. .
Three-dimensional modeling of strumpellin, with d1dn1b as a template, through use of SwissProt database viewer. Two helices from the 1,159-aa protein are shown, including amino acids 614–634 in one α-helix and amino acids 662–672 from a nearby α-helix in the antiparallel direction. A, Residues L619 and V626, in the same orientation in an α-helix opposite a second helix in an antiparallel direction. Only residue side chains that are closest in physical space are shown. B, The L619F mutation adds a bulky phenylalanine side group that likely exceeds the space available between the two α-helices. C, V626F mutation. The ε carbon of the F626 aromatic ring impinges on Q666 and may force apart the two α-helices.
Zebrafish-Rescue Experiments
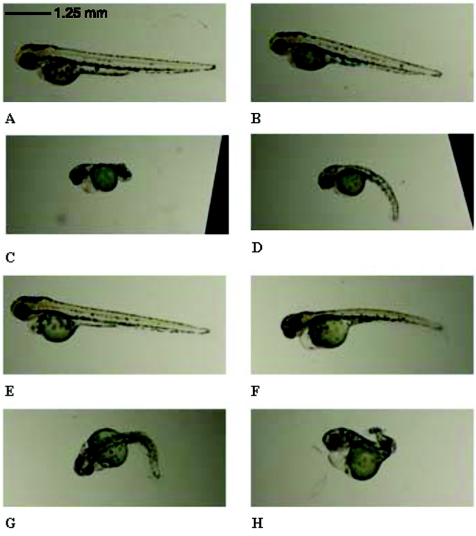
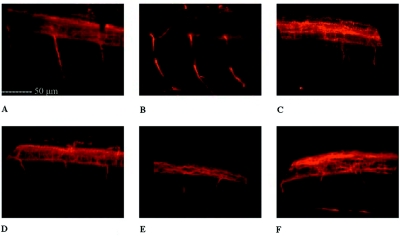
To validate the functional phenotype of the SPG8 mutations in vivo, we developed a zebrafish model. Morpholino oligonucleotide knockdown of the KIAA0196 protein ortholog in zebrafish (KIAA MO) resulted in an enlarged heart cavity, along with a curly-tail phenotype that severely impaired the ability of the fish to swim properly. The overall phenotype ranged in severity and was classified in three major groups: normal, slightly curly, and severely curly. This phenotype was clearly visible after dechorionating by 1 d post fertilization (dpf). At 3 dpf, wild-type zebrafish are ∼5 mm long, with a straight tail (fig. 5A). Fish injected with a mismatch-control morpholino (CTL MO) were initially used to titer a KIAA MO–specific nontoxic injection dose (fig. 5B). Injection of the KIAA MO resulted in 66 (37%) of 178 fish with a severely curly tail and 50 (28%) of 178 fish with a slightly curly tail (table 3 and fig. 5C and 5D). The KIAA MO fish had a significantly different distribution of phenotypic groups compared with those with CTL MO injections (P<.001). When wild-type human KIAA0196 mRNA was coinjected with KIAA MO, the curly-tail phenotype was rescued to levels comparable to CTL MO injections (P=.51) (fig. 5E and 5F). This suggests that, in zebrafish, human KIAA0196 mRNA can compensate for the loss of endogenous zebrafish mRNA. Conversely, coinjection of human KIAA0196 mRNA incorporating either the exon 14 or exon 15 mutation failed to significantly rescue the phenotype (fig. 5G and 5H). Injection of mutant exon 14 or exon 15 mRNA alone (without morpholinos) did not lead to a curly-tail phenotype or influence lethality in zebrafish, which suggests that the two mutations do not exert a dominant negative effect. Approximately 200 embryos were injected per experimental condition (table 3). The difference in distribution between KIAA MO injection alone and KIAA MO coinjection with wild-type mRNA was significant (P<.001). Similarly, coinjection of wild-type mRNA versus either exon 14 or exon 15 mutant mRNA was significantly different, with a P value <.001. There was no statistical difference between the coinjection of the exon 14 mutant and the exon 15 mutant (P=.10). On histochemical analysis of the embryos by use of an antiacetylated tubulin stain for growing axons, we found that the motor neurons in the spinal cord did not develop normally (fig. 6). Motor-neuron axons in fish injected with KIAA MO alone or with the mutant mRNAs were shorter and showed abnormal branching. The structure of interneurons in the spinal cord was also different. The absence of the KIAA0196 gene or mutations in this gene during early development thus seemed to hamper axonal outgrowth.
Figure 5. .
Zebrafish knockdown and rescue of KIAA0196 function. A, Gross morphological features of normal wild-type zebrafish, depicted at 3 dpf. B, Injection of a 5-bp mismatch morpholino (CTL MO), which results in no obvious disease phenotype. C and D, KIAA MO–injected fish with a severely curly tail (C) or with a slightly curly tail (D). Their heart cavities are also enlarged, which is commonly seen in injected fish. E and F, Fish, injected with both KIAA MO and normal human KIAA0196 mRNA, with partially developed curly tail (F) or no effect at all (E), depending on the injected quantity. G and H, Disease phenotype not alleviated when the KIAA MO is injected with the mutant forms (×14 [panel G] and ×15 [panel H]) of the human mRNA. These fish resemble the KIAA MO fish (C and D).
Table 3. .
Phenotype Profile from Zebrafish Morpholino Oligonucleotide Knockdown
| Percentage (No.) of Zebrafish |
|||||
| Condition | Normal | Slightly Curly Tail | Severely Curly Tail | Dead | Total |
| KIAA0196 morpholino | 19.1 (34) | 28.1 (50) | 37.1 (66) | 15.7 (28) | 178 |
| Control morpholino | 56.1 (83) | 24.3 (36) | 7.4 (11) | 12.2 (18) | 148 |
| Wild-type rescue | 63.2 (127) | 19.4 (39) | 8.0 (16) | 9.5 (9) | 201 |
| Mutant ×14 rescue | 16.0 (32) | 37.0 (74) | 36.0 (72) | 11.0 (22) | 200 |
| Mutant ×15 rescue | 13.2 (29) | 37.4 (82) | 30.1 (66) | 19.2 (42) | 219 |
Figure 6. .
Immunohistochemical analysis of zebrafish with the KIAA0196 knockdown phenotype. A, Motor neurons in the ventral roots of wild-type zebrafish, segmented and oriented at 3 dpf. The spinal cord consists of the cell bodies of motor neurons and interneuron bundles. The picture was taken near the gut of the fish. B, Mismatch control has a motor-neuron distribution similar to the wild type. C, E, and F, Zebrafish injected with KIAA MO (C) and fish coinjected with mutant mRNA (×14 [panel E] and ×15 [panel F]), showing shorter, branching motor neurons that are not oriented. D, Wild-type KIAA0196 mRNA coinjections with KIAA MO, which partially rescue the motor-neuron phenotype. The axons are longer and oriented.
Discussion
HSP is one of the most genetically heterogeneous diseases, caused by mutations in at least 31 different genes. This means that >0.1% of genes in the human genome can be mutated and result in one predominant neurological outcome: the degeneration of upper-motor-neuron axons. Four families in this study share the same p.V626F mutation, which suggests that the altered nucleotide is a mutation hotspot. For one of these families,15 the KIAA0196 gene lies just beyond its distal candidate boundary. However, the flanking marker likely represents either a marker mutation or genotyping error in this family, since the p.V626F mutation is present in the supposed recombinant as well as all other affected individuals.
An SPG8 mutation causes a pure form of HSP with relatively little interfamilial variability in phenotype. Interestingly, two missense mutations were identified in highly conserved amino acids in a predicted α-helix. The helix consists of a heptameric repeat, with hydrophobic residues aligning in inaccessible regions at the center of the helix. The hydrophobic lysine and valine amino acids are 7 aa apart in the protein sequence; thus, it is expected that they would be buried in the helix, close in 3D space (fig. 5A). When replaced by a bulky phenylalanine residue at either position, the stability of the α-helix could very well be disrupted.
The one known domain in strumpellin is a spectrin repeat that consists of three α-helices of a characteristic length wrapped in a left-handed coiled coil.25 These spectrin repeats appear in the spectrin/dystrophin/α-actinin family. The spectrin proteins have multiple copies (15–20) of this repeat, which can then form multimers in the cell. Spectrin also associates with the cell membrane via spectrin repeats in the ankyrin protein. Likewise, four spectrin repeats are found in α-actinin beside two N-terminal calponin homology domains that anchor the complex to actin.26 This effectively connects the cell membrane with the actin-cytoskeletal network. The stability and structure of this network also provide appropriate routes for intracellular vesicular transport, a mechanism already linked to other mutated HSP genes. Proteins with three or fewer spectrin repeats can be considered to have transient association with the spectrin network. The single repeat in strumpellin is more likely to be involved in docking with one of the cytoskeletal spectrin repeats, which could help in protein localization or signal transduction. It will be interesting to determine with which protein(s) strumpellin interacts through its spectrin domain and, particularly, how the mutation identified at the core of this domain influences this potential interaction.
Proteins with a spectrin repeat have been identified in other neurological disorders—most notably, dystrophin, mutated in myotonic dystrophy (MIM 300377).27 The repeat also has been found in a form of cerebellar ataxia (MIM 117210).28 β-III spectrin itself is found to be mutated in spinocerebellar ataxia 5.29 Whereas none of the genes mutated in HSP have a spectrin domain, L1CAM (SPG1) has an indirect association.9,30 L1CAM is a single-pass transmembrane protein with a glycosylated extracellular component that facilitates the outgrowth and migration of neurons in the corticospinal tract. The intracellular C-terminus, however, binds to the spectrin-repeat–containing protein ankyrin that links the cell membrane to intracellular spectrin. Thus, strumpellin, with its spectrin domain, may also be involved in this process.
The only detail known so far about the human KIAA0196 gene is that it has previously been implicated in prostate cancer.31 An increase in gene-copy number was assayed by real-time quantitative PCR and FISH, which determined >10-fold overexpression of the gene in PC-3 prostate cancer lines and in approximately one-third of advanced prostate cancers examined.31 How this relates to a spastic paraplegia phenotype is not clear.
Analysis of other species has provided some insight into a potential function for KIAA0196. A 118-kDa homologue of the strumpellin protein was identified as part of a TATA-binding protein-related factor 2 (TRF2) complex in a Drosophila nuclear extract.32 Eighteen proteins were pulled down, along with TRF2, in this complex, including NURF and SWI, with functions for chromatin remodeling and transcription activation. TRF2 is selective for promoters lacking TATA or CAAT boxes. One protein of the complex is DREF, which binds to DRE elements common in controlling genes involved in cell-cycle regulation and cell proliferation.33,34
With little known about the function of KIAA0196, we decided to test the functionality of the missense changes with a zebrafish knockdown model. Interestingly, the KIAA MO–injected fish showed a severe tail phenotype characterized by abnormal motor-neuron outgrowth in the spinal cord. The knocked-down zebrafish resembles other mutants affecting midline development.35 Injection of wild-type human KIAA0196 mRNA concurrently with zebrafish KIAA MO knockdown rescued the phenotype to values not significantly different from a control morpholino injection. However, coinjecting human KIAA0196 mRNA containing either the exon 14 or the exon 15 mutation yielded a phenotype comparable to injection of KIAA MO alone. These experiments demonstrate the importance of the KIAA0196 gene in early development of zebrafish and suggest that the two missense mutations impair the normal function of strumpellin. Further characterization of the KIAA0196 knockdown phenotype is necessary to better understand the role of this gene in early fish development—more precisely, in motor-neuron outgrowth. This phenotype is remarkably similar to what was recently observed in SPG4 morpholino knockdown experiments in the zebrafish36: motor-neuron-axon outgrowth is impaired, and embryos also have a curly tail. This is another instance in which a dominant adult-onset disease in humans displays an embryonic phenotype in the zebrafish. It can be hypothesized that a complete knockdown of either SPG4 or SPG8 yields an embryonic phenotype, whereas one copy of the gene is enough in humans to remain at a subphenotypic state until adult stages. Importantly, in both instances, it is the motor neurons that are impaired.
The identification of KIAA0196 as the gene mutated in SPG8 adds another component to the various genes already identified for the disease. Two missense mutations in a conserved part of the protein have been identified, including one mutation common to four families. Additional work will aid in clarifying the function of the protein and how it relates to other proteins implicated in HSP and the overall disease pathogenesis. As more of the genes involved in HSP emerge, the responsible pathways and mechanisms of toxicity will be better understood. This will also help in elucidating the pathophysiology of a related disease—amyotrophic lateral sclerosis—in which the reduced life span of patients complicates the cloning of genes by linkage analysis.
Acknowledgments
We sincerely appreciate the cooperation of the families involved in this study. We thank Melanie Benard and Rosemary Rupps, for patient recruitment, and Daniel Rochefort, Pascale Hince, Julie Roussel, Liliane Karemera, Judith St-Onge, Kara Melmed, and James Lee, for expert technical assistance. P.N.V., P.A.D., and G.A.R. are supported by the Canadian Institutes of Health Research. E.R. is a Wellcome Trust advanced fellow in clinical science.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Conserved Domain Database, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- Fondation Jean Dausset–CEPH, http://www.cephb.fr/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for C. familiaris [accession number XP_532327], P. troglodytes [accession number XP_519952], D. melanogaster [accession number CG12272], C. elegans [accession number CE13235], X. tropicalis [accession number MGC89323], R. norvegicus [accession number XP_343250], D. rerio [accession number BC045490], G. gallus [accession number XP_418441], D. discoideum [accession number EAL63144], M. musculus [accession number NP_705776.2], and H. sapiens mRNA [accession number NM_014846.2])
- InterPro, http://www.ebi.ac.uk/interpro/ (for InterProScan from the European Bioinformatics Institute)
- NCBI BLAST, http://www.ncbi.nlm.nih.gov/blast/
- NetGene2, http://www.cbs.dtu.dk/services/NetGene2/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SPG4, myotonic dystrophy, and cerebellar ataxia)
- Pfam, http://www.sanger.ac.uk/Software/Pfam/
- Phyre Protein Fold Recognition Server, http://www.sbg.bio.ic.ac.uk/~phyre/
- Probcons, http://probcons.stanford.edu/
- UCSC Genome Browser, http://www.genome.ucsc.edu/
References
- 1.Silva MC, Coutinho P, Pinheiro CD, Neves JM, Serrano P (1997) Hereditary ataxias and spastic paraplegias: methodological aspects of a prevalence study in Portugal. J Clin Epidemiol 50:1377–1384 10.1016/S0895-4356(97)00202-3 [DOI] [PubMed] [Google Scholar]
- 2.Polo JM, Calleja J, Combarros O, Berciano J (1991) Hereditary ataxias and paraplegias in Cantabria, Spain: an epidemiological and clinical study. Brain 114:855–866 [DOI] [PubMed] [Google Scholar]
- 3.McMonagle P, Webb S, Hutchinson M (2002) The prevalence of “pure” autosomal dominant hereditary spastic paraparesis in the island of Ireland. J Neurol Neurosurg Psychiatry 72:43–46 10.1136/jnnp.72.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fink JK (1997) Advances in hereditary spastic paraplegia. Curr Opin Neurol 10:313–318 10.1097/00019052-199708000-00006 [DOI] [PubMed] [Google Scholar]
- 5.Harding AE (1983) Classification of the hereditary ataxias and paraplegias. Lancet 1:1151–1155 10.1016/S0140-6736(83)92879-9 [DOI] [PubMed] [Google Scholar]
- 6.Harding AE (1993) Hereditary spastic paraplegias. Semin Neurol 13:333–336 [DOI] [PubMed] [Google Scholar]
- 7.Behan WM, Maia M (1974) Strumpell’s familial spastic paraplegia: genetics and neuropathology. J Neurol Neurosurg Psychiatry 37:8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deluca GC, Ebers GC, Esiri MM (2004) The extent of axonal loss in the long tracts in hereditary spastic paraplegia. Neuropathol Appl Neurobiol 30:576–584 10.1111/j.1365-2990.2004.00587.x [DOI] [PubMed] [Google Scholar]
- 9.Soderblom C, Blackstone C (2006) Traffic accidents: molecular genetic insights into the pathogenesis of the hereditary spastic paraplegias. Pharmacol Ther 109:42–56 10.1016/j.pharmthera.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 10.Klebe S, Azzedine H, Durr A, Bastien P, Bouslam N, Elleuch N, Forlani S, Charon C, Koenig M, Melki J, et al (2006) Autosomal recessive spastic paraplegia (SPG30) with mild ataxia and sensory neuropathy maps to chromosome 2q37.3. Brain 129:1456–1462 10.1093/brain/awl012 [DOI] [PubMed] [Google Scholar]
- 11.Zuchner S, Kail ME, Nance MA, Gaskell PC, Svenson IK, Marchuk DA, Pericak-Vance MA, Ashley-Koch AE (2006) A new locus for dominant hereditary spastic paraplegia maps to chromosome 2p12. Neurogenetics 7:127–129 10.1007/s10048-006-0029-1 [DOI] [PubMed] [Google Scholar]
- 12.Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine CS, Cruaud C, Durr A, Wincker P, et al (1999) Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet 23:296–303 10.1038/15472 [DOI] [PubMed] [Google Scholar]
- 13.Meijer IA, Hand CK, Cossette P, Figlewicz DA, Rouleau GA (2002) Spectrum of SPG4 mutations in a large collection of North American families with hereditary spastic paraplegia. Arch Neurol 59:281–286 10.1001/archneur.59.2.281 [DOI] [PubMed] [Google Scholar]
- 14.Hedera P, Rainier S, Alvarado D, Zhao X, Williamson J, Otterud B, Leppert M, Fink JK (1999) Novel locus for autosomal dominant hereditary spastic paraplegia, on chromosome 8q. Am J Hum Genet 64:563–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid E, Dearlove AM, Whiteford ML, Rhodes M, Rubinsztein DC (1999) Autosomal dominant spastic paraplegia: refined SPG8 locus and additional genetic heterogeneity. Neurology 53:1844–1849 [DOI] [PubMed] [Google Scholar]
- 16.Rocco P, Vainzof M, Froehner SC, Peters MF, Marie SK, Passos-Bueno MR, Zatz M (2000) Brazilian family with pure autosomal dominant spastic paraplegia maps to 8q: analysis of muscle beta 1 syntrophin. Am J Med Genet 92:122–127 [DOI] [PubMed] [Google Scholar]
- 17.Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, et al (1998) Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear encoded mitochondrial metalloprotease. Cell 93:973–983 10.1016/S0092-8674(00)81203-9 [DOI] [PubMed] [Google Scholar]
- 18.Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- 19.Bourgeois S, Labuda D (2004) Dynamic allele-specific oligonucleotide hybridization on solid support. Anal Biochem 324:309–311 10.1016/j.ab.2003.10.006 [DOI] [PubMed] [Google Scholar]
- 20.Labuda D, Krajinovic M, Richer C, Skoll A, Sinnett H, Yotova V, Sinnett D (1999) Rapid detection of CYP1A1, CYP2D6, and NAT variants by multiplex polymerase chain reaction and allele-specific oligonucleotide assay. Anal Biochem 275:84–92 10.1006/abio.1999.4293 [DOI] [PubMed] [Google Scholar]
- 21.Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- 22.Westerfield M (1995) The zebrafish book: a guide for laboratory use of zebrafish (Danio rerio). University of Oregon Press, Eugene [Google Scholar]
- 23.Nasevicius A, Ekker SC (2000) Effective targeted gene “knockdown” in zebrafish. Nat Genet 26:216–220 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- 24.Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- 25.Djinovic-Carugo K, Gautel M, Ylanne J, Young P (2002) The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett 513:119–123 10.1016/S0014-5793(01)03304-X [DOI] [PubMed] [Google Scholar]
- 26.Ylanne J, Scheffzek K, Young P, Saraste M (2001) Crystal structure of the α-actinin rod: four spectrin repeats forming a tight dimer. Cell Mol Biol Lett 6:234 [PubMed] [Google Scholar]
- 27.Koenig M, Monaco AP, Kunkel LM (1988) The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell 53:219–226 10.1016/0092-8674(88)90383-2 [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa K, Toru S, Tsunemi T, Li M, Kobayashi K, Yokota T, Amino T, Owada K, Fujigasaki H, Sakamoto M, et al (2005) An autosomal dominant cerebellar ataxia linked to chromosome 16q22.1 is associated with a single-nucleotide substitution in the 5′ untranslated region of the gene encoding a protein with spectrin repeat and Rho guanine-nucleotide exchange-factor domains. Am J Hum Genet 77:280–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeda Y, Dick KA, Weatherspoon MR, Gincel D, Armbrust KR, Dalton JC, Stevanin G, Durr A, Zuhlke C, Burk K, et al (2006) Spectrin mutations cause spinocerebellar ataxia type 5. Nat Genet 38:184–190 10.1038/ng1728 [DOI] [PubMed] [Google Scholar]
- 30.Jouet M, Rosenthal A, Armstrong G, MacFarlane J, Stevenson R, Paterson J, Metzenberg A, Ionasescu V, Temple K, Kenwrick S (1994) X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat Genet 7:402–407 10.1038/ng0794-402 [DOI] [PubMed] [Google Scholar]
- 31.Porkka KP, Tammela TL, Vessella RL, Visakorpi T (2004) RAD21 and KIAA0196 at 8q24 are amplified and overexpressed in prostate cancer. Genes Chromosomes Cancer 39:1–10 10.1002/gcc.10289 [DOI] [PubMed] [Google Scholar]
- 32.Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R (2002) TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420:439–445 10.1038/nature01167 [DOI] [PubMed] [Google Scholar]
- 33.Hirose F, Yamaguchi M, Handa H, Inomata Y, Matsukage A (1993) Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J Biol Chem 268:2092–2099 [PubMed] [Google Scholar]
- 34.Shimada M, Nakadai T, Tamura TA (2003) TATA-binding protein-like protein (TLP/TRF2/TLF) negatively regulates cell cycle progression and is required for the stress-mediated G(2) checkpoint. Mol Cell Biol 23:4107–4120 10.1128/MCB.23.12.4107-4120.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brand M, Heisenberg CP, Warga RM, Pelegri F, Karlstrom RO, Beuchle D, Picker A, Jiang YJ, Furutani-Seiki M, van Eeden FJ, et al (1996) Mutations affecting development of the midline and general body shape during zebrafish embryogenesis. Development 123:129–142 [DOI] [PubMed] [Google Scholar]
- 36.Wood JD, Landers JA, Bingley M, McDermott CJ, Thomas-McArthur V, Gleadall LJ, Shaw PJ, Cunliffe VT (2006) The microtubule-severing protein spastin is essential for axon outgrowth in the zebrafish embryo. Hum Mol Genet 15:2763–2771 10.1093/hmg/ddl212 [DOI] [PubMed] [Google Scholar]