Abstract
Hematopoietic stem cells are resistant to HIV-1 infection. Here, we report a novel mechanism by which the cyclin-dependent kinase inhibitor (CKI) p21Waf1/Cip1/Sdi1 (p21), a known regulator of stem cell pool size, restricts HIV-1 infection of primitive hematopoietic cells. Modifying p21 expression altered HIV-1 infection prior to changes in cell cycling and was selective for p21 since silencing the related CKIs, p27Kip1 and p18INK4C, had no effect on HIV-1. We show that p21 blocked viral infection by complexing with HIV-1 integrase and aborting chromosomal integration. A closely related lentivirus with a distinct integrase, SIVmac-251, and the other cell-intrinsic inhibitors of HIV-1, Trim5α, PML, Murr1, and IFN-α, were unaffected by p21. Therefore, p21 is an endogenous cellular component in stem cells that provides a unique molecular barrier to HIV-1 infection and may explain how these cells remain an uninfected “sanctuary” in HIV disease.
Introduction
HSCs are a self-replenishing source of all blood and immune cells. In contrast to its more differentiated offspring in either the myeloid or lymphoid lineages, which are highly susceptible to infection by HIV-1, the HSC is naturally resistant despite the presence of CD4 and functional CXCR4 (1–6). We sought to determine whether molecular regulators of stem cell function participated in restricting HIV-1.
A characteristic distinguishing feature of adult stem cells is their relative cell cycle quiescence. Cyclin-dependent kinase (CDK) inhibitors (CKIs), p21Waf1/Cip1/Sdi1 (p21) and p18INK4C (p18) serve as G1 checkpoint regulators and play important roles in stem cell physiology (7, 8). These proteins affect size and self-renewal capacity of the HSC pool in vivo (7, 8). One of the regulators, p21, is known to be highly expressed in stem cells but not in amplifying or progenitor cells descendent from stem cells (7, 9–11). In this report, we show that p21 plays an indispensable role in maintaining the intrinsic cellular defense against HIV-1 infection in HSCs. Of note, the effect of p21 on HIV susceptibility was not due to an effect on cell cycle entry and was independent of the known mediators of HIV-1 resistance, tripartite motif protein 5α (Trim5α), promyelocytic leukemia protein (PML), copper metabolism domain containing 1 (Murr1), and IFN-α (12–14). Rather, it was associated with restricting viral DNA integration into the host cell genome.
Results
p21-restricted HIV-1 replication in primitive hematopoietic cells.
HSCs and progenitor cells are CD34+ cells and represent only 0.5–3% of mononucleosis cells in human bone marrow. To assess the role of p21 in HIV-1 replication in these cells, we first knocked down p21 expression with 2 types of siRNA, an in vitro–synthesized siRNA and an in vivo plasmid–transcribed short hairpin RNA (shRNA) (15). Both forms of siRNA suppressed p21 mRNA expression in cells in the range of 62%–98% (Figure 1A), as measured by real-time RT-PCR, which detected as few as 10 copies of target mRNA in a sample of 50 ng total cellular RNA (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI28971DS1). Western blot analysis confirmed that p21 protein expression was decreased in p21 siRNA–treated cells but not in control cells (Figure 1B).
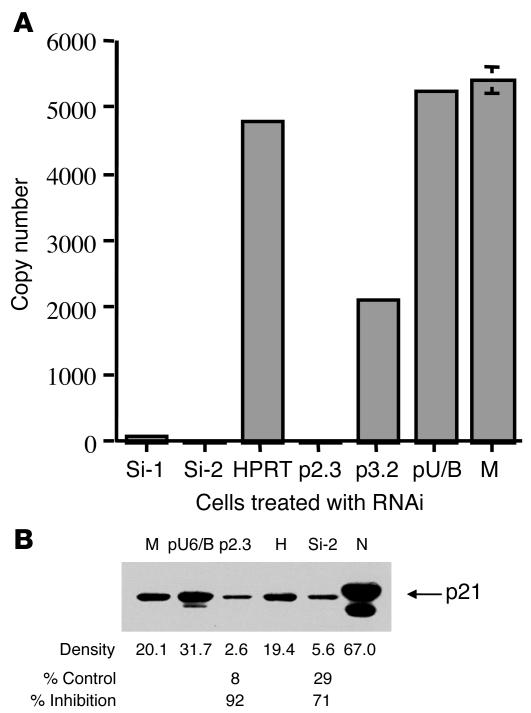
Figure 1. Inhibition of p21 expression by RNAi.
(A) Inhibition of p21 mRNA expression by RNAi. p21 mRNA levels were decreased after transfection of chemically synthesized siRNA (Si-1, Si-2) or cell-expressed shRNA (p2.3, p3.2) specific for p21. The decrease of p21 mRNA did not occur in control cells treated with siRNA specific for hypoxanthine phosphoribosyl transferase (HPRT) or vector only (pU/B) or mock-treated cells (M) (or a mutated form of Si-2, shown in ref. 15). RNA copy number in 50 ng of total cellular RNA was detected by TaqMan real-time RT-PCR assay (Supplemental Figure 1) performed in quadruplicate. Error bars are not visible when the SD is less than 2.0%. (B) Inhibition of p21 expression with p21 siRNA examined by Western blot. The same cell aliquots used in A were examined for p21 protein expression after RNAi treatment. M, p21 protein detected in cells mock treated with siRNA; pU6/B, treated with vector only; p2.3, treated with p21 shRNA; H, treated with HPRT siRNA; and Si-2, treated with p21 siRNA. N, nuclear extract of p21 protein served as a positive control. *Percentage of control was calculated by comparing the densitometer readings of p2.3 with pU6/8 (8%) and Si-2 with H (29%). Results are representative of 3 independent experiments.
It has been shown by us and others that HSCs and progenitor cells resist HIV-1 infection (1–3). Treatment of bone marrow CD34+ cells with p21 siRNA resulted in a marked increase in productive HIV-1 infection. At 14 days after infection, HIV-1 Gag p24 levels were 50-fold higher in p21 siRNA–treated cells than in cells that were either mock treated or treated with control siRNA (Figure 2A).
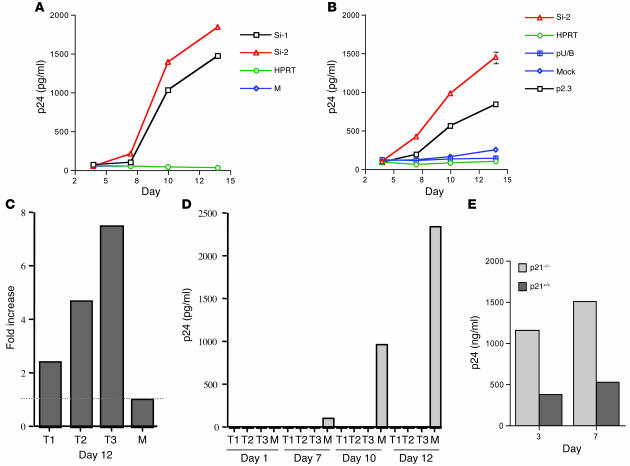
Figure 2. p21 restricted HIV-1 replication in cells.
(A) HIV-1 replication in p21 siRNA–treated CD34+ primary cells. HIV-1 p24 levels were detected at days 4, 7, 10, and 14 after HIV-1 infection. Cells were treated with chemically synthesized p21 siRNA (Si-1, Si-2) or control siRNA (HPRT) or were mock treated. Culture supernatants collected at each time point were analyzed by HIV-1 p24 antigen assay. HIV-1 growth kinetics in mock-treated cells was indistinguishable from that in HPRT siRNA–treated cells. Each point represents the p24 level from 2 separate experiments run in duplicate. Values represent means ± SD (0.2–4.7). (B) HIV-1 replication in p21 siRNA–treated CMK cells. HIV-1 p24 levels were detected at days 4, 7, 10, and 14 after viral infection. Cells were treated with chemically synthesized p21 siRNA (Si-2), control siRNA, p21 shRNA (p2.3), or vector without shRNA (pU/B) or were mock treated. Each point represents the p24 level from 2 separate experiments run in duplicate. (C and D) TPA-induced p21 expression inhibited HIV-1 replication in CMK cells. Cells were treated with TPA for 1 day (T1), 2 days, or 3 days or were mock treated before HIV-1 infection. (C) p21 mRNA levels examined at day 12 compared with mock-treated control. (D) HIV-1 p24 levels determined at days 1, 7, 10, and 12. Each bar represents the p24 level from 2 separate experiments run in duplicate. Values represent means ± SD (0.3–2.6). (E) HIV-1 p24 expression from transduced p21–/– and p21+/+ fibroblasts. The p24 level in culture supernatant was determined at days 3 and 7 after transduction with a single cycle of HIV-1NLX.Luc(R–) infection. Results are from a representative experiment, and samples run in triplicate. Values represent means ± SD (0.07–0.3).
To assess the knockdown of p21 in cells more tractable to in-depth study, we examined HIV-1 replication in CMK cells, a p53-deficient human megakaryoblastic cell type with known ability to modulate p21 (16). CMK cells were derived from megakaryoblasts, which are precursor cells of megakaryocytes and platelets (17). Treating these cells with p21 siRNA increased HIV-1 replication up to 14-fold in comparison with that in controls (Figure 2B). Further, expression of p21 in CMK cells can be stimulated by 12-O-tetradecanoylphorbol-13-acetate (TPA) or retinoic acid without affecting the levels of other CKIs, such as p27Kip1, p16INK4A, p15INK4B, and p18 (16). To determine whether stimulation of p21 expression could inhibit HIV-1 replication, we treated CMK cells with TPA for varying intervals prior to infection with a high dose of HIV-1 (104 50% tissue culture infective dose (TCID50)/106 cells). The expression of p21 mRNA was increased in a dose-responsive manner after TPA treatment, and HIV-1 replication was blocked in all TPA-treated cells. This was noted in cells treated with TPA for as few as 24 hours. In contrast, mock-treated cells, which had low levels of p21 expression, had high levels of HIV-1 replication (Figure 2, C and D).
Further evaluation of the role for p21 in HIV infection was performed using cells that had been engineered to be deficient in p21. Brown et al. generated a normal diploid human embryonic lung fibroblast line that was then rendered p21–/– by homologous recombination (18). The parent and p21-deficient cells were then transduced with HIV-1NLX.Luc(R–) (a pseudotype of HIV-1 that only replicates a single round in cells) at 25 reverse transcriptase unit (RTU)/cell, and production of HIV-1 p24 was monitored in the cell supernatant. Cells deficient in p21 produced approximately 3-fold greater p24, consistent with an inhibition on HIV-1 production imposed by p21 (Figure 2E).
p21-restricted viral DNA integration into the host cell genome.
To elucidate the mechanism by which p21 affects HIV-1 susceptibility in primary hematopoietic cells, we examined the different replication steps in the HIV-1 life cycle. First, we examined cell-surface expression of the HIV-1 coreceptors CD4 and CXCR4. There was no detectable change in the amount of each receptor between cells treated by p21 siRNA and control siRNA as determined by flow cytometric analysis (Supplemental Figure 2).
Soon after entry, viral reverse transcriptase converts genomic RNA into linear double-strand cDNA, and viral integrase integrates the viral cDNA into a host cell chromosome (19). To determine whether p21 inhibited reverse transcription or integration, we examined the levels of total HIV-1 cDNA, 2–long terminal repeat (2-LTR) circles, and integration by real-time PCR assays (Supplemental Figure 3) in 2 independent infection systems. First, we used vesicular stomatitis virus — glycoprotein (VSV-G)–pseudotyped HIV-1 to infect CMK cells in a single-round infection after siRNA treatment. From the same infectious dose, viral cDNA was detected in p21 siRNA– and control siRNA–treated cells at comparable levels (Figure 3A). However, 2-LTR circles, markers of aborted integration (20, 21), were significantly increased in control siRNA–treated cells in contrast with p21 siRNA–treated cells (Figure 3B). At 24 hours after infection, a high copy number (4.3 ± 0.11 × 103) of HIV-1 DNA was detected in the genomes of p21 siRNA–treated cells, but only a marginal level (7.9 ± 0.03 × 10) of integrated HIV-1 DNA was detected in control siRNA–treated cells (Figure 3C). Note the inverse relationship between viral 2-LTR circles and integrated DNA in p21 siRNA– and control siRNA–treated cells (Figure 3, B and C). The single-cycle HIV-1 growth experiments demonstrated that p21 prevented viral DNA integration and enhanced 2-LTR circles in cells, which is a marker of aborted integration.
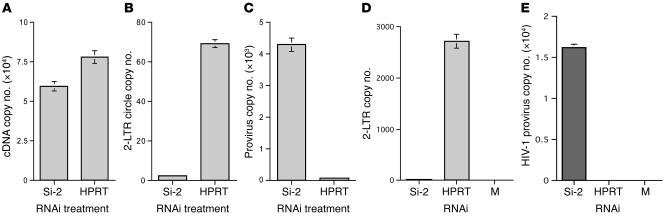
Figure 3. Knockdown of p21 increased HIV-1 integration.
(A–C) HIV-1 DNA kinetics in p21 siRNA–treated CMK cells. Cells were infected by VSV-G–pseudotyped HIV-1 after siRNA treatment. (A) Total levels of HIV-1 cDNA were determined by real-time PCR at 7 hours after infection. (B) At 20 hours after infection, more 2-LTR circles were detected in p21 siRNA–treated cells than in control siRNA–treated cells. (C) At 24 hours after infection, p21 siRNA–treated cells had a significantly increased amount of integrated viral DNA in comparison with control cells. Each sample (500 ng) was tested in triplicate (47–49). Results are representative of 3 separate experiments. (D and E) Detection of HIV-1 2-LTR circle and provirus levels in CD34+ primary cells using quantitative PCR. (D) HIV-1 2-LTR circles were detected at 16 hours after infection. (E) HIV-1 integration was increased significantly in p21 siRNA–treated cells in contrast with control siRNA– or mock-treated cells. Each bar represents the mean from 4 individual tests, with each sample run in triplicate. Each sample was tested in an equal amount of total cellular DNA from 6 × 103 CD34+ primary cells.
We then examined wild-type HIV-1 integration in primary CD34+ cells and CMK cells. At 16 hours after infection, cells treated with control siRNA had significantly higher levels of 2-LTR circles than cells treated with p21 siRNA (2,721.8 ± 142.6 copies versus 21.7 ± 1.46 copies/6 × 103 cells) (Figure 3D). Examining the HIV-1 provirus at 18 days after infection revealed that cells treated with p21 siRNA had significantly increased levels of viral integrated DNA in the cell genome in contrast with the controls. An average of 2.9 integrated copies per cell was detected in p21 siRNA–treated cells in contrast with no copies detected per cell in control siRNA–treated cells (Figure 3E). In CMK cells, an average of 2.4 integrated copies per cell were detected in cells treated with p21 siRNA in contrast with 0.07 copies per cell in cells treated with control siRNA (data not shown). Taking these data together, knocking down p21 expression dramatically increased the levels of viral integration in both primary primitive CD34+ cells and in the CMK cell line. The high level of 2-LTR circles detected in cells that expressed p21 represented the end products of aborted HIV-1 integration. A high level of 2-LTR circles has been previously seen in cells infected with integrase mutant viruses, which specifically lose the ability to integrate, or in cells treated with specific inhibitors that inhibit the function of HIV-1 integrase (20, 21).
HIV-1 preintegration complexes (PICs) are the functional unit required for integration of the retroviral genome into the host genome. To determine whether p21 interacts with the viral integration machinery, PICs were isolated from control and siRNA-treated CMK cells, and interaction of p21 with PIC components was determined by coimmunoprecipitation–Western blot assays. The viral matrix protein is a PIC component (22, 23), and anti-matrix antibodies coimmunoprecipitated integrase from the initial cytoplasmic extracts (Figure 4A) as well as from PICs purified by spin column chromatography (Figure 4B). Similarly, anti-p21 monoclonal antibody coimmunoprecipitated integrase from starting and purified PICs (Figure 4, A and B). In addition, p21 siRNA, but not the control siRNA, aborted coimmunoprecipitation of integrase from purified PIC samples (Figure 4B). Further, p21 was not detected in PICs derived from H9 cells, a T cell line highly susceptible to HIV-1IIIB (the original HIV-1 clone isolated from an AIDS patient) infection (Figure 4C). Combined, these data demonstrate that p21 in primitive primary cells or CMK cells interacted with the HIV-1 integration machinery and that the presence of p21 in these complexes was associated with an inability of viral DNA to integrate into cellular DNA.
Figure 4. p21 was present in HIV-1 PIC.
(A–C) Detection of p21 in HIV-1 PICs. CMK (restrictive) and H9 (permissive) cells were infected with HIV-1IIIB and cytoplasmic PICs isolated at 7 hours after infection. Viral matrix protein is a component of PICs (22, 23). (A) Crude cytoplasmic PICs were isolated from CMK cells. Antibodies against p21 or HIV-1 Gag protein matrix (MA) (50) recovered similar levels of integrase (IN) (lanes 1 and 2). Purified integrase served as a marker (lane 3). Mock-infected cell control is shown in lane 4. (B) Analyses following PIC purification by spin column chromatography. Antibodies against p21 coimmunoprecipitated p21 with integrase in CMK cells (lane 4). Antibodies against p21 failed to recover IN when CMK cells were treated with p21 siRNA (lane 6) but not when cells were treated with mutated p21-siRNA (lane 7) (15). Isotype controls IgG3k (for MA) and IgG1 (for p21) failed to recover IN (lanes 2 and 8). Purified integrase served as a marker (lane 1). (C) Anti-MA antibodies coimmunoprecipitated p21 in PICs made from CMK cells (lane 1) but not from H9 cells (lane 2). p21-containing nuclear extract served as a marker (lane 5).
p21-restricted HIV-1 infection before cell cycling.
To address the relationship between cell cycle and viral replication in p21 siRNA–treated cells, we analyzed cell cycle status by flow cytometry. Cells were stained with Hoechst 33342 for DNA and pyronin Y (PY) for RNA content at 10 hours and 42 hours after siRNA treatment. Up to the time of HIV-1 infection (42 hours after siRNA treatment), there was no significant change in cycle status between cells treated with p21 siRNA and with control siRNA in either primitive CD34+ cells (Figure 5A) or in CMK cells (Figure 5B). Since this analysis provides only a snapshot of cell cycle status, we conducted a BrdU incorporation assay, which provided a more sensitive and more temporally integrated analysis. At 42 hours after silencing, there was no difference in BrdU incorporation detected between cells treated with p21 siRNA and with control siRNA (Figure 5, C and D). It was only after 68 hours following siRNA treatment that we did note more cells incorporating BrdU in p21 siRNA–treated cells than in control cells (Supplemental Figure 4). In our system, this was 26 hours after viral infection. This late-occurring change in BrdU incorporation may well be derived from the virus interaction with host cells, either by synthesis of viral DNA in cells or virus-induced further downregulation of p21. Studies have shown that HIV-1 DNA integration occurs before cell cycling, such as in HIV-1 infection of quiescent T lymphocytes and nondividing cells (22–26). We found that HIV-1 infection ultimately downregulated p21 expression in infected cells that were without RNA interference (RNAi) treatment (Supplemental Figure 5). Others report that active HIV-1 infection of T cells is associated with a reduced expression of p21 (27, 28).
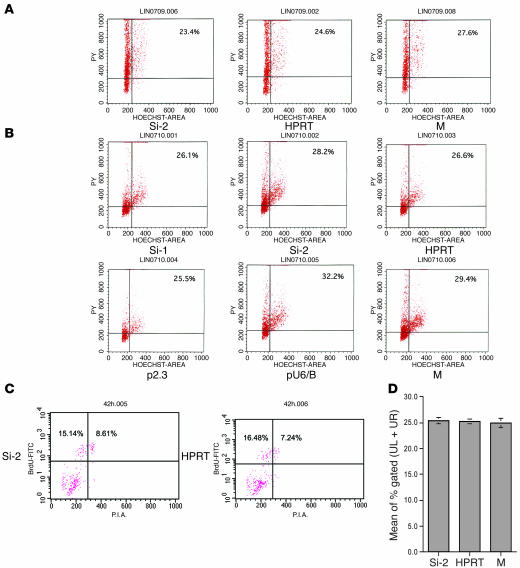
Figure 5. p21 restricted HIV-1 infection before cell cycling.
(A) Cell cycle status of CD34+ primary cells at 42 hours after siRNA treatment. Cells treated with p21 siRNA and control siRNAs were examined by Hoechst (DNA dye) and PY (RNA dye) staining. Cell cycle status was examined at both 10 hours (not shown) and 42 hours after siRNA treatment. No significant changes in the cell cycle status were observed at both time points among CD34+ primary cells that were treated with p21 siRNA or control siRNA or those that were mock treated. (B) Cell cycle status of CMK cells at 42 hours after siRNA treatment. No significant changes were observed in the cell cycle status in megakaryoblasts treated with p21 siRNA (Si-1, Si-2), control siRNA, p21 shRNA, or vector only or those that were mock treated. (C and D) Cell cycle status determined by BrdU incorporation assay. (C) Cell cycle status of CD34+ primary cells was examined at both 10 hours (not shown) and 42 hours after siRNA treatment. No significant changes were observed between cells treated with p21 siRNA and cells treated with control siRNA. Cells incorporating BrdU (UL + UR) consisted of 23.75% and 23.72% p21 siRNA– or control siRNA–treated cells, respectively. (D) Mean values of cells incorporating BrdU in each condition from 4 independent assays are shown.
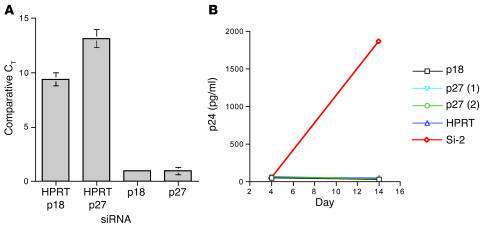
To determine whether other CKIs affect HIV-1 infection, we examined the effects of silencing p27Kip1 (p27, another member of the CIP/KIP sub-family) and p18 (a member of the INK4 subfamily) expression on HIV-1 replication in CD34+ cells. Neither p27 nor p18 silencing increased HIV-1 replication in CD34+ cells (Figure 6, A and B).
Figure 6. Silencing of p27 and p18 had no effect on HIV-1.
(A and B) Silencing p18 or p27 did not increase HIV-1 replication. (A) The effect of siRNA specific for p18 or p27 was determined in CD34+ cells using a real-time TaqMan RT-PCR assay by comparing CT values from cells treated with siRNA specific for p18 or p27 to cells treated with control siRNA. Samples run in quintuplicate were normalized to endogenous GAPDH levels. (B) HIV-1 replication in CD34+ cells treated with siRNA specific for p18 or p27 was determined at days 4, 7, 10, and 14 after HIV-1 infection (shown at days 4 and 14) by p24 antigen assays. No active replication was detected in cells treated with p18, p27, or control siRNA in contrast with cells treated with p21 siRNA. Values represent means ± SD (0.22–2.1).
Inhibition of HIV-1 infection is specific and independent of other restriction factors.
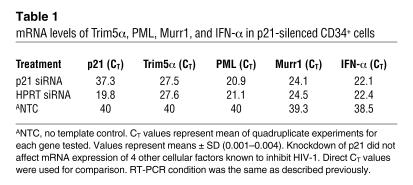
To determine whether expression of other host genes known to inhibit HIV-1 could account for the differences seen in susceptibility after silencing p21, we examined expression of Trim5α, PML, Murr1, and IFN-α by using real-time RT-PCR assays. In cells with or without silencing of p21, we could detect no changes in mRNA expression of any of these cellular virus inhibitors (Table 1). The role of p21 in inhibiting HIV-1 replication in primitive cells, therefore, is not mediated by increased expression of 4 other known modulators of innate cell resistance to HIV-1.
Table 1 .
mRNA levels of Trim5α, PML, Murr1, and IFN-α in p21-silenced CD34+ cells
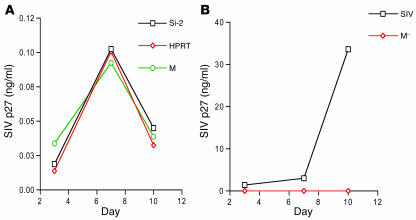
To assess whether the effect of p21 on HIV-1 replication was virus specific, we studied the infectivity of SIVmac-251, a dual tropic (T- and M-tropic) lentivirus related to HIV-1 (29–31), in both p21 siRNA–treated primary cells and in CMK cells. A low level of viral antigen p27 was detected (0.103 ± 0.001 ng/ml) in culture supernatant of CD34+ cells at day 7, but this dropped to background levels by day 10 (<0.05 ng/ml; Figure 7A). In contrast, the more mature bone marrow CD34– primary cells from the same donors were actively infected by SIVmac-251 (SIV capsid protein p27 level >3.92 ng/ml at day 7; Figure 7B). Silencing of p21 expression, therefore, did not induce SIV to replicate in primary CD34+ cells and CMK cells, in marked contrast to what was observed with HIV-1.
Figure 7. Silencing of p21 had no effect on SIVmac-251.
(A and B) SIVmac-251 infection of CD34+ cells and CD34– cells. (A) Silencing of p21 expression in CD34+ cells did not induce SIVmac-251 replication. SIV antigen p27 was detected from CD34+ cells treated with p21 siRNA or control siRNA or cells that were mock treated (SIV infection only). (B) SIVmac-251 replicated in CD34– cells of the same donor. SIV antigen p27 was detected in CD34– cells infected with virus (SIV) but not in mock-infected CD34– cells (M–). Assays were run in duplicate. Values represent means ± SD (0.01–0.66).
Discussion
p21 was previously shown to regulate stem cell function and megakaryocyte differentiation (7, 10, 16). In this study, we demonstrated that p21 modulated HIV-1 integration and viral replication in primary primitive hematopoietic cells independently of a direct effect on cell cycling or other known mediators of HIV-1 resistance. The single-cycle infection with VSV-G–pseudotyped HIV-1 demonstrated that p21 functions at the level of preventing viral DNA integration and enhancing the amount of 2-LTR circles in cells, a measure of abortive integration. This cell-autonomous effect of p21 on HIV-1 infection was associated with p21 coimmunoprecipitating with the viral preintegration complex. A role for p21 in other chromatin-modifying phenomena has been documented by others. Cellular DNA repair systems play essential roles in the completion of retroviral DNA integration (32, 33), and one component in DNA repair and replication is proliferating cell nuclear antigen (PCNA) (34, 35). p21 appears to directly bind PCNA and inhibit PCNA-dependent DNA repair and replication in a cyclin/CDK-independent manner (34, 35). p21 is also known to interact with human cytosin-5 DNA methyltransferase–PCNA complex, which regulates DNA stability in the absence of a cyclin/CDK (36). Whether either of these interactions is related to the role of p21 in viral integration we have defined remains to be determined, but p21 as a participant in protein complexes modifying chromatin is well described.
p21 has pleiotropic effects that are highly cell-type specific, and repression of p21 does not always increase cell cycling, as might be predicted from its function as a CKI. The absence of p21 increases the cycling of hematopoietic and neural stem cells (7, 37) but results in endothelial–cell cycle arrest (38, 39). Similar cell-type–specific effects of p21 may also apply to its effect on HIV-1. Vazquez et al. reported that HIV-1 infection induced p21 expression and that suppression of p21 with 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), a potent inducer of heme oxygenase-1 and Nrf2/ARE signaling (40, 41), repressed HIV-1 replication in macrophages (42). The mechanism of this p21-induced HIV-1 replication in macrophages was not further explored. The role of p21 in the study of Vazquez et al. is different from that reported in other cell types. Clark and colleagues noted that productive high-titer infection of HIV-1 in T lymphocytes was associated with a loss of p21, not an increase (27, 28). Therefore, p21 appears to have cell-type–specific effects that may be contradictory, depending on the host cell type. T lymphocytes and stem cells appear to have a similar profile of p21 effect on HIV-1 that is distinct from that of macrophages, and it is tempting to speculate that it may be due to the distinct ability of these cells to undergo self-renewing cell divisions compared with the terminally differentiated, low-proliferating macrophage.
We have previously shown that silencing p21 increases HIV-1–derived lentivector transduction in primary primitive hematopoietic cells (15). Although the in vitro transduction efficiency was only 3- to 4-fold higher in p21-silenced cells than in p21–mock-silenced cells, the in vivo transduction efficiency was magnified to 7- to 8-fold, possibly due to the transduction of primitive hematopoietic cells. Further examination of transduced cells showed that the p21-silenced cells had a higher number of integrated lentivectors than p21–mock-silenced cells. Therefore, our previous study documented a potential therapeutic application in modulating p21 levels to improve gene therapy vector transduction. In this study, we demonstrate a mechanism by which p21 may restrict lentiviral infection and define it as a molecule that protects HSCs from infection by pathogenic HIV-1. The basis for this protection was unexpectedly not directly due to cell cycle changes, but rather blocking HIV-1 integration. These results define what we believe to be a novel protective mechanism against HIV-1 that is endogenous to stem cells and distinct from other virus-resistance mechanisms defined to date. The enhanced HIV replication observed in the p21–/– fibroblasts provides additional confirmation that p21 expression inhibits HIV replication in an entirely different cell type. A definitive confirmation of the role of p21 expression such as would be achieved by adding it back to a p21-null human HSC must await development of such a cell line. Why p21 functions distinctly among CKIs in restricting HIV-1 is an intriguing and still-to-be-answered question. p21 has been called a universal CKI and appears to be capable of interacting with essentially all of the CDK complexes (43, 44) while CKIs such as p18, p27, or p57 selectively interact with specific CDK complexes. Our studies report an additional unique function for p21. They indicate that this cell cycle checkpoint protein not only maintains HSC homeostasis but uniquely protects the stem cell genome from viral DNA invasion.
Methods
Cells and viruses.
Human CD34+ primary cells were obtained from bone marrow of healthy adult volunteers via a protocol approved by the Institutional Review Board of Massachusetts General Hospital. Mononuclear cells from bone marrow were selected with Ficoll/Paque PLUS (Amersham) density gradient centrifugation. CD34+ cells were enriched by immunomagnetic selection according to the manufacturer’s instructions (Miltenyi Biotec) with 96% purity. Human megakaryoblastic leukemia cells (CMKs) were grown in RPMI with 20% FBS, and cells in log growth were used in experiments (16). p21+/+ cells were normal diploid human lung embryonic fibroblasts (LF-1 strain), and the p21–/– (HO 7.2-1; HO, homozygous derivative) subclone was obtained by targeted homologous recombination (18). Both cell lines were generous gifts from J. Sedivy (Brown University, Providence, Rhode Island, USA). The viral pool of HIV-1IIIB was harvested from H9/HTLV-IIIB cell culture (NIH AIDS Research & Reference Reagent Program), and viral stocks were titrated with the TCID50 assay in H9 and in human peripheral blood mononuclear cells (45). HIV-1NLX.Luc(R–) pseudotyped with the VSV-G envelope was prepared as previously described (15, 46). In brief, HIV-1NLX.Luc(R–) has the backbone of pHIvec2.GFP but with a luciferase gene inserted upstream of GFP (15). Three package plasmids, pCMV.G, pCMV.E, and pCMV.R (15), were used to make the pseudotyped virus, and the viral titer was determined by the reverse transcription assay.
HIV-1 infection and detection, lentivector transduction.
At 42 hours after siRNA treatment, 1 × 106 cells were infected with 1 × 103 TCID50 of HIV-1IIIB. Cell number and viability after siRNA treatment were determined by trypan blue exclusion. Viruses and cells were incubated at 37°C for 2 hours, and the unabsorbed viruses were washed out with PBS. Cells were cultured at 37°C, and the culture supernatant or cells were harvested at indicated time points. Beckman Coulter p24 antigen kinetics assay was used to detect HIV-1 levels in culture supernatant. Cells were transduced with HIV-1NLX.Luc(R–) at 45 RTU/cell in integration experiments and 25 RTU/cell in p24 assays. Transduction was conducted 12–14 hours after cell seeding, with a spin transduction method (15).
SIV infection and detection.
SIVmac-251 was a gift from K. Reimann (Beth Israel Deaconess Medical Center, Boston, Massachusetts, USA) (29). Infection was performed as described previously (29). Beckman Coulter SIV p27 antigen ELISA assay was used to detect SIV replication in the supernatant of cell cultures.
Alu-LTR PCR assay.
The Alu-LTR (with Alu denoting the Alu repeat in the human genome) PCR assay (47–49) was used to determine levels of HIV-1 DNA integration (Supplemental Figure 3). We adapted this assay with some modification. In brief, a nested PCR assay was carried out with 2 sets of Alu-LTR primers and a probe. The sequences of the primers for first step PCR were the same as that of Alu forward (MH535) and Alu reverse (SB704) (47). The PCR was performed with Taq polymerase (Invitrogen) and a DNA thermal cycler (PerkinElmer). Viral DNA was detected by using 500 ng of cellular genomic DNA and subjected to denaturation at 94°C for 4 minutes, then 20 cycles of denaturation at 94°C for 0.5 minutes, annealing at 55°C for 0.5 minutes, and extension at 72°C for 3 minutes, with a last cycle extension at 72°C for 7 minutes. Following the initial PCR, a real-time PCR was performed by using an aliquot equivalent to 1/20 of the 20-cycle PCR product with the second-round Alu-LTR primers and TaqMan probe. The sequences of the second set of primer and probe are as follows: Alu forward (3′ of LTR), 5′GGTAACTAGAGATCCCTCAGACCCT-3′; Alu reverse (5′ of Alu), 5′-gcgtgagccaccgc-3′; and Alu probe, 5′-TTAGTCAGTGTGGAAAATCTCTAGCAGGCCG-3′. The copy numbers of integrated viral DNA in samples were derived from IS cell line (a gift from U. O’Doherty, University of Pennsylvania, Philadelphia, Pennsylvania, USA) (49) adjusted with the known copy number of SM2 plasmid spiked with H9 cellular DNA (47, 48). The gel-purified genomic DNA of OM10.1 cells was used as a reference standard (48). Each sample was tested in triplicate. The copy number of viral DNA in samples was calculated directly from the result spreadsheet, which was converted by the threshold cycle numbers (CT value ≤ 36 with FAM, 6-carboxyfluorescein) (47, 48) versus the standard curves with Sequence Detector version 1.7 software (Applied Biosystems).
2-LTR circle PCR assays.
The 2-LTR circle PCR assays were performed as previously described (47, 48). The 2-LTR standard was a generous gift from F. Bushman (University of Pennsylvania, Philadelphia, Pennsylvania, USA). PCR assay was performed as described, and the primers and probes were the same as described (47, 48). Viral 2-LTR circles were detected from 500 ng cellular genomic DNA in quintuplicate.
PIC isolation.
CMK and H9 cells in log growth were infected by HIV-1IIIB at 8,000 TCID50/106 (CMK) or 2,000 TCID50/106 cells (H9), respectively. At 7 hours after infection, cells were harvested and washed twice in buffer K (20 mM HEPES, pH 7.6, 5 mM MgCl2, 150 mM KCl, 1 mM dithiothreitol, and 20 μg/ml aprotinin) and lysed in 1 ml of buffer K-0.025% (w/v) digitonin. Crude cytoplasmic extract cleared by brief centrifugation was treated with RNase A (0.1 mg/ml) for 30 minutes at room temperature. To partially purify PICs, 2 ml of lysate was passed through a BSA-coated Sepharose CL-4B (Amersham Biosciences) spin column (12 ml) equilibrated in buffer K-0.025% digitonin. Both the crude cytoplasmic extract and the column eluate were used for immunoprecipitation.
Coimmunoprecipitation–Western blot assay.
p21 protein in PIC samples was detected following immunoprecipitation with anti-matrix antibody (mAb 3H7, a gift from A. Engelman, Dana-Farber Cancer Institute, Boston, Massachusetts, USA) or anti-p21 antibody (mAb SC-187; Santa Cruz Biotechnology Inc.) (15). HIV-1 Gag protein matrix (MA) is a PIC component, and mAb 3H7 has been previously used to immunoprecipitate PICs (50–52). Antibody (10 μl) was prebound to 15 μl protein A/G-Agarose (Santa Cruz Biotechnology Inc.) in 100 μl of buffer K for 1 hour at 4°C, and 200 μl of CL-4B column purified cytoplasmic PICs was added. The mixture was rocked for 4 hours at 4°C and washed, and the immune complexes were examined in 12% Tris-Glycine gel (Invitrogen). Integrase and p21 protein were detected by using mAb 8E5 (anti-integrase) and SC-187, respectively.
Western blot analysis of p21 after RNAi treatment was performed as previously described (15). In brief, each sample containing the same amount of lysate protein, determined with the BCA Protein Assay (Pierce Biotechnology), was separated on 16% SDS-PAGE (Invitrogen). Following transfer, the membrane was blocked with 5% milk and detected with mAb SC-187, a mouse anti-human p21 monoclonal antibody. The second antibody was an HRP-labeled rabbit anti-mouse IgG (Jackson ImmunoResearch Laboratories Inc.). The image was obtained by immediately exposing the membrane to film for from 30 seconds to 10 minutes. Hewlett-Packard ScanJet IIcx was used to scan the gel photographs, and density of each band was measured and analyzed with ImageQuant (version 1.3; Molecular Dynamics) software.
Cell culture and TPA assay.
CD34+ primary cells were cultured in StemSpan medium (StemCell Technologies Inc.) supplemented with 300 ng/ml SCF, Flt3 ligand, and 60 ng/ml IL-3 (R&D Systems) at 37°C under 5% CO2 (15, 53). The cell culture supernatants after viral infection were collected at indicated time points and replaced with the same amount of fresh medium. CMK cells after HIV-1 infection were cultured in RPMI 1640 medium containing 10% FBS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were cultured at 37°C under 5% CO2. In TPA experiments, cells were cultured in the presence or absence of 10 nM of TPA for 1, 2, and 3 days. Cell number and viability were determined with the trypan blue exclusion assay, and cells were washed with PBS before being infected with HIV-1IIIB at 1 × 104 TCID50/106 cells.
Real-time quantitative RT-PCR.
Real-time quantitative RT-PCR was performed as previously described (10, 15). In brief, the sequences of the primer set for detection of human p27 and p18 mRNA were as follows: p27 forward, 5′-CGGTGGACCACGAAGAGTTAA-3′; p27 reverse, 5′-ggctcgcctcttccatgtc-3′; and p27 probe, 5′-(FAM)CCGGGACTTGGAGAAGCACTGCA-3′(black hole quenchers [BHQ]; Integrated DNA Technologies); p18 forward, 5′-CCCCACAAAACCGGGAA-3′; p18 reverse, 5′-GCTGTAGGCACTCATTGAGCTG-3′; and p18 probe, 5′-(FAM)AAGGAAAGGACAGCGGCGGCA-3′(BHQ). The sequences of the primer set for detection of human Trim5α, PML, Murr1, and INF-α mRNA were as follows: Trim5α forward, 5′-ACCGTGGTCACCACAGGTT-3′; Trim5α reverse, 5′-TGCCTGGAGCTTCACTTGGTA-3′; and Trim5α probe, 5′-(FAM)CCCACAGAGGAGGTTGCCCAGGAG-3′(BHQ); PML forward, 5′-CCGCCCTGGATAACGTCTTT-3′; PML reverse, 5′-CCA CAATCTGCCGGTACACC-3′; and PML probe, 5′-(FAM)TCGAGAGTCTGCAGCGGCGC-3′(BHQ); Murr1 forward, 5′-CAAAAATCCGTGAGAGCCTCA-3′; Murr1 reverse, 5′-CAGCTCAGGCCCCGAAG-3′; and Murr1 probe, 5′-(FAM)AACCAGAGCCGCTGGAATAGCGG-3′(BHQ); IFN-α forward, 5′-CCTCGCCCTTTGCTTTACTG-3′; IFN-α reverse, 5′-GCCCAGAGAGCAGCTTGACT-3′; and IFN-α probe, 5′-(FAM)TGGTCCTGGTGGTGCTCAGCTGC-3′(BHQ). Human cellular GAPDH mRNA was used as the endogenous control in each RT-PCR reaction, and the primers and probes to detect GAPDH mRNA were from the TaqMan GAPDH Control Reagent kit (Applied Biosystems). Total cellular RNA from cells was isolated with the RNeasy kit (QIAGEN) and purified with DNase following the RNeasy clean-up protocol (QIAGEN). An equal amount of purified RNA (50 ng or 100 ng, dependent on each assay) in quadruplicate from each sample was examined in each experiment. A standard curve of the amplicon being measured was run in duplicate with 1 to 1 × 1010 copies in each assay, with 2 no-template controls. The amplicons from each reaction were analyzed using Sequence Detector version 1.7 software. The RNA copy number in each sample was converted according to the standard curve and the CT value of the amplicons in each sample (Supplemental Figure 1).
siRNAs and transfection of siRNA into cells.
Both synthetic siRNA duplexes and plasmid-derived shRNA were used in this study, and the sequences of siRNA and shRNA were as previously described (15). The transfection of siRNA and shRNA into cells was as previously described. In brief, siRNA was transfected into CD34+ cells by the siPORT method following manufacturer’s instructions (Ambion). In our assay system, siRNA can be efficiently transfected into cells by this method, but shRNA cannot. Electroporation was used to transfect shRNA into cells when sufficient numbers of cells were available (≥ 1.5 × 106 cells/assay condition).
Cell cycle analysis.
The siRNA-treated CD34+ cells and CMK cells were stained with CD34+ FITC (BD); this was followed by incubation with a DNA dye, Hoechst 33342 (Sigma-Aldrich), and an RNA dye, PY. The proportion of cells in G0 (PYlowHoechstlow) and in activated status (PYhighHoechsthigh) was measured from the CD34+ or CMK cells, representing quiescent and activated cells, at 10 hours and 42 hours after siRNA treatment, respectively.
BrdU incorporation assay.
The siRNA-treated CD34+ cells were incubated with BrdU in a final concentration of 10 μM. BrdU-pulsed cells were fixed in 70% ethanol at 20°C overnight and denatured by 2N HCl, 0.5% Triton X-100, for 30 minutes at room temperature followed by neutralization with borate buffer (pH 8.5). Treated cells were subjected to dual-color staining with anti-BrdU MoAb (BD) followed by goat anti-mouse FITC-conjugated and 5 μg/ml propidium iodide. Analyses were performed using the FACSCalibur flow cytometer and CellQuest (version 3.3) and ModFit (version 3.0) software (BD).
Supplementary Material
Acknowledgments
We thank Joseph Sodroski for insightful suggestions on this study; Alan Engelman for anti-HIV PIC antibodies and helpful suggestions in writing the manuscript; Chou-Wen Lin for helpful suggestions on the PIC assay; Nadia Carlesso and William Gordon for help on performing the BrdU experiments; David Dombkowski for performing FACS analysis; and Liying Ma for lab technical assistance. This work was supported by NIH grants K18 AI55313 (to J. Zhang), R01 HL044851 (to D.T. Scadden), and R01 HL71859 (to C.S. Crumpacker).
Footnotes
Nonstandard abbreviations used: Alu, Alu repeat in human genome; BHQ, black hole quenchers; CDK, cyclin-dependent kinase; CKI, CDK inhibitor; CT, threshold cycle; LTR, long terminal repeat; Murr1, copper metabolism domain containing 1; p18, p18INK4C; p21, p21Waf1/Cip1/Sdi1; PCNA, proliferating-cell nuclear antigen; PIC, HIV-1 preintegration complex; PML, promyelocytic leukemia protein; PY, pyronin Y; RNAi, RNA interference; RTU, reverse transcriptase unit; shRNA, short hairpin RNA; TCID50, 50% tissue culture infective dose; TPA, 12-O-tetradecanoylphorbol-13-acetate; TRIM5α, tripartite motif protein 5α; VSV-G, vesicular stomatitis virus — glycoprotein.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:473–481 (2007). doi:10.1172/JCI28971
David T. Scadden and Clyde S. Crumpacker contributed equally to this work.
See the related Commentary beginning on page 302.
References
- 1.Shen H., et al. Intrinsic human immunodeficiency virus type 1 resistance of hematopoietic stem cells despite coreceptor expression. J. Virol. 1999;73:728–737. doi: 10.1128/jvi.73.1.728-737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weichold F.F., et al. Neither human immunodeficiency virus-1 (HIV-1) nor HIV-2 infects most-primitive human hematopoietic stem cells as assessed in long-term bone marrow cultures. Blood. 1998;91:907–915. [PubMed] [Google Scholar]
- 3.Von Laer D., et al. CD34+ hematopoietic progenitor cells are not a major reservoir of the human immunodeficiency virus. Blood. 1990;76:1281–1286. [PubMed] [Google Scholar]
- 4.Lee B., Ratajczak J., Doms R.W., Gewirtz A.M., Ratajczak M.Z. Coreceptor/chemokine receptor expression on human hematopoietic cells: biological implications for human immunodeficiency virus-type 1 infection. Blood. 1999;93:1145–1156. [PubMed] [Google Scholar]
- 5.Louache F., Debili N., Marandin A., Coulombel L., Vainchenker W. Expression of CD4 by human hematopoietic progenitors. Blood. 1994;84:3344–3355. [PubMed] [Google Scholar]
- 6.Aiuti A., et al. Human CD34(+) cells express CXCR4 and its ligand stromal cell-derived factor-1. Implications for infection by T-cell tropic human immunodeficiency virus. Blood. 1999;94:62–73. [PubMed] [Google Scholar]
- 7.Cheng T., et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 8.Yuan Y., Shen H., Franklin D.S., Scadden D.T., Cheng T. In vivo self-renewing divisions of haematopoietic stem cells are increased in the absence of the early G1-phase inhibitor, p18INK4C. Nat. Cell Biol. 2004;6:436–442. doi: 10.1038/ncb1126. [DOI] [PubMed] [Google Scholar]
- 9.Steinman R.A. Cell cycle regulators and hematopoiesis. Oncogene. 2002;21:3403–3413. doi: 10.1038/sj.onc.1205325. [DOI] [PubMed] [Google Scholar]
- 10.Stier S., et al. Ex vivo targeting of p21Cip1/Waf1 permits relative expansion of human hematopoietic stem cells. Blood. 2003;102:1260–1266. doi: 10.1182/blood-2002-10-3053. [DOI] [PubMed] [Google Scholar]
- 11.Cheng T., Shen H., Rodrigues N., Stier S., Scadden D.T. Transforming growth factor beta 1 mediates cell-cycle arrest of primitive hematopoietic cells independent of p21(Cip1/Waf1) or p27(Kip1). Blood. 2001;98:3643–3649. doi: 10.1182/blood.v98.13.3643. [DOI] [PubMed] [Google Scholar]
- 12.Stremlau M., et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 13.Marcello A., et al. Recruitment of human cyclin T1 to nuclear bodies through direct interaction with the PML protein. EMBO J. 2003;22:2156–2166. doi: 10.1093/emboj/cdg205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganesh L., et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J.L., Attar E.C., Cohen K.S., Crumpacker C.S., Scadden D.T. Silencing p21Waf1/Cip1/Sdi1 expression increases gene transduction efficiency in primitive human hematopoietic cells. Gene Ther. . 2005;12:1444–1452. doi: 10.1038/sj.gt.3302544. [DOI] [PubMed] [Google Scholar]
- 16.Matsumura I., et al. Thrombopoietin-induced differentiation of a human megakaryoblastic leukemia cell line, CMK, involves transcriptional activation of p21(WAF1/Cip1) by STAT5. Mol. Cell. Biol. 1997;17:2933–2943. doi: 10.1128/mcb.17.5.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vainchenker W., Debili N., Mouthon M.A., Wendling F. Megakaryocytopoiesis: cellular aspects and regulation. Crit. Rev. Oncol. Hematol. 1995;20:165–192. doi: 10.1016/1040-8428(94)00159-q. [DOI] [PubMed] [Google Scholar]
- 18.Brown J.P., Wei W., Sedivy J.M. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 19. Coffin, J.M., Hughes, S.H., and Varmus, H.E. 1997. Retroviruses. Cold Spring Harbor Press. Cold Spring Harbor, New York, USA. 843 pp. [PubMed] [Google Scholar]
- 20.Engelman A. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. . 1999;52:411–426. doi: 10.1016/s0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 21.Hazuda D.J., et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 22.Bukrinsky M.I., et al. A nuclear localization signal within HIV-I matrix protein that governs infection of nondividing cells. Nature. . 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Von Schwedler U., Kombluth R.S., Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. . 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G., Simm M., Potash M.J., Volsky D.J. Human immunodeficiency virus type 1 DNA synthesis, integration, and efficient viral replication in growth-arrested T cells. J. Virol. 1993;67:3969–3977. doi: 10.1128/jvi.67.7.3969-3977.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y., Zhang H., Siliciano J.D., Siliciano R.F. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J. Virol. 2005;79:2199–2210. doi: 10.1128/JVI.79.4.2199-2210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zack J.A. The role of the cell cycle in HIV-1 infection. Adv. Exp. Med. Biol. 1995;374:27–31. doi: 10.1007/978-1-4615-1995-9_3. [DOI] [PubMed] [Google Scholar]
- 27.Clark E., et al. Loss of G(1)/S checkpoint in human immunodeficiency virus type 1-infected cells is associated with a lack of cyclin-dependent kinase inhibitor p21/Waf1. J. Virol. 2000;74:5040–5052. doi: 10.1128/jvi.74.11.5040-5052.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D., et al. Inhibition of human immunodeficiency virus type 1 transcription by chemical cyclin-dependent kinase inhibitors. J. Virol. 2001;75:7266–7279. doi: 10.1128/JVI.75.16.7266-7279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz J.E., et al. Effect of CD8+ lymphocyte depletion on virus containment after simian immunodeficiency virus SIVmac251 challenge of live attenuated SIVmac239Delta3-vaccinated rhesus macaques. J. Virol. 2005;79:8131–8141. doi: 10.1128/JVI.79.13.8131-8141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoyama S. Molecular evolution of the human and simian immunodeficiency viruses. Mol. Biol. Evol. 1988;5:645–659. doi: 10.1093/oxfordjournals.molbev.a040520. [DOI] [PubMed] [Google Scholar]
- 31.Padow M., et al. Replication of chimeric human immunodeficiency virus type 1 (HIV-1) containing HIV-2 integrase (IN): naturally selected mutations in IN augment DNA synthesis. J. Virol. 2003;77:11050–11059. doi: 10.1128/JVI.77.20.11050-11059.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau A., Kanaar R., Jackson S.P., O’Connor M.J. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 2004;23:3421–3429. doi: 10.1038/sj.emboj.7600348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniel R., et al. Evidence that stable retroviral transduction and cell survival following DNA integration depend on components of the nonhomologous end joining repair pathway. J. Virol. 2004;78:8573–8581. doi: 10.1128/JVI.78.16.8573-8581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li R., Waga S., Hannon G.J., Beach D., Stillman B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994;371:534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- 35.Waga S., Hannon G.J., Beach D., Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 36.Chuang L.S., et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 37.Qiu J., et al. Regenerative response in ischemic brain restricted by p21cip1/waf1. J. Exp. Med. 2004;199:937–945. doi: 10.1084/jem.20031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaBaer J., et al. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 39.Noseda M., et al. Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Mol. Cell. Biol. 2004;24:8813–882. doi: 10.1128/MCB.24.20.8813-8822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stadheim T.A., Suh N., Ganju N., Sporn M.B., Eastman A. The novel triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) potently enhances apoptosis induced by tumor necrosis factor in human leukemia cells. J. Biol. Chem. 2002;277:16448–16455. doi: 10.1074/jbc.M108974200. [DOI] [PubMed] [Google Scholar]
- 41.Liby K., et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65:4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- 42.Vazquez N., et al. Human immunodeficiency virus type 1-induced macrophage gene expression includes the p21 gene, a target for viral regulation. J. Virol. 2005;79:4479–4491. doi: 10.1128/JVI.79.7.4479-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roninson I.B. Oncogenic functions of tumour suppressor p21(Waf1/Cip1/ Sdi1): association with cell senescence and tumour–promoting activities of stromal fibroblasts. Cancer Lett. 2002;179:1–14. doi: 10.1016/s0304-3835(01)00847-3. [DOI] [PubMed] [Google Scholar]
- 44.Harper J.W., et al. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. ACTG Virology Committee. 1994. ACTG Virology Manual for HIV laboratories. 3rd edition. F.B. Hollinger, editor. National Institute of Allergy and Infectious Diseases, NIH. Bethesda, Maryland, USA. http://aactg.s-3.com/labmanual.htm. [Google Scholar]
- 46.Lu R., Ghory H.Z., Engelman E. Genetic analyses of conserved residues in the carboxyl-terminal domain of human immunodeficiency virus type 1 integrase. J. Virol. 2005;79:10356–10368. doi: 10.1128/JVI.79.16.10356-10368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butler S.L., Hansen M.S., Bushman F.D. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J.L., Crumpacker C.S. Human immunodeficiency virus type 1 nucleocapsid protein nuclear localization mediates early viral mRNA expression. J. Virol. 2002;76:10444–10454. doi: 10.1128/JVI.76.20.10444-10454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Doherty U., Swiggard W.J., Jeyakumar D., McGain D., Malim M.H. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 2002;76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin C.W., Engelman A. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. . 2003;77:5030–5036. doi: 10.1128/JVI.77.8.5030-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niedrig N., et al. Epitope mapping of monoclonal antibodies against human immunodeficiency virus type 1 structural proteins using peptides. J. Virol. 1989;63:3525–3528. doi: 10.1128/jvi.63.8.3525-3528.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nilsen B.M., et al. Monoclonal antibodies against human immunodeficiency virus type 1 integrase: epitope mapping and differential effects on integrase activities in vitro. J. Virol. 1996;70:1580–1587. doi: 10.1128/jvi.70.3.1580-1587.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zandstra P.W., Lauffenburger D.A., Eaves C.J. A ligand-receptor signaling threshold model of stem cell differentiation control: a biologically conserved mechanism applicable to hematopoiesis. Blood. 2000;96:1215–1222. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.