Abstract
The 52 000 MW Ro/SS-A (Ro52) protein is a major target of autoantibodies in autoimmune conditions such as systemic lupus erythematosus and Sjögren's syndrome. Recent genomic and bioinformatic studies have shown that Ro52 belongs to a large family of related RING/Bbox/coiled-coil (RBCC) tripartite motif proteins sharing overall domain structure and 40–50% identity at the amino acid level. Ro52 also has a B30.2 domain at the C-terminus. Using the human genome draft sequence, the genomic organization of the Ro52 gene on human chromosome 11p15.5 has been deduced and related to the protein domain structure. We show that the steady-state levels of Ro52 mRNA are normally very low but are induced by cell activation with interferon-γ. In transient transfection of HeLa cells, epitope-tagged Ro52 protein was localized to unidentified membrane proximal rod-like structures. Using in vitro coupled transcription/translation followed by immunoprecipitation, the autoimmune response to Ro52 protein was investigated and two distinct interactions were resolved. The Ro52 C-terminal B30.2 domain interacts with human immunoglobulin independently of antibody specificities. Sera derived from patients with Sjögren's syndrome and systemic lupus erythematosus, in addition, contained specific autoantibodies directed towards the rest of the Ro52 molecule. The majority of these autoimmune sera also immunoprecipitated the Ro52-related molecule RNF15. A possible role for Ro52 protein in alterations of plasma membranes during cellular activation or apoptosis is discussed.
Introduction
Autoimmune rheumatic diseases such as systemic lupus erythematosus and Sjögren's syndrome are characterized by the presence of autoantibodies to diverse cellular constituents, including double-stranded DNA, histones and Ro/SS-A and Ro/SS-B proteins. The role of Ro/SS-A (Ro52 and Ro60) and Ro/SS-B (La) proteins in the development and pathophysiology of systemic autoimmune conditions is a paradigm for understanding the normal mechanisms of B/T-cell tolerance to self antigen and development of autoimmunity.1 It is unclear why the immune response targets these particular autoantigens and whether this autoimmune response is the result or cause of the underlying immune pathology. Enzyme-linked immunosorbent assay (ELISA) methods using recombinant 52 000 MW Ro/SS-A antigen are commonly used diagnostically for the detection of Ro52 autoantibodies in these diseases.2 Autoantibodies to 52 000 MW Ro/SS-A (Ro52) are found in >70% of sera from patients with primary Sjögren's syndrome, 30% in systemic lupus erythematosus, as well as in 10% of rheumatoid arthritis and most congenital heart block patients. The presence of these antibodies is often related to disease severity, lymphopenia, photosensitive dermatitis and, possibly, to pulmonary and renal disease, suggesting that they have an immunopathological role.3
Antibody responses to Ro and La induced in animal models, using recombinant protein as immmunogens, are consistent with the concept of epitope-determinant spreading. Immunization with Ro60 for example leads not only to anti-Ro60 antibodies, but also to anti-Ro52 and anti-La antibodies.4–6 The involvement of chaperone molecules such as the endoplasmic-reticulum-resident grp78, hsp70 and calreticulin have also been discussed in relation to the determinant spreading phenomenon in Ro52- and Ro60-immunized animals.7–9
One suggestion to account for these observations concerning antibody epitope spreading, is that the immune responses to Ro52, Ro60 and La are linked because the molecules themselves co-localize or are physically associated in vivo as part of Ro ribonucleoprotein complexes (RoRNPs). Other evidence shows that Ro52 is not associated directly with RoRNPs10 and that Ro52 is normally resident in the cytoplasm.11,12 Both nuclear and cytoplasmic locations have been detected for RoRNPs and Ro6013 whereas La is more consistently localized to the nucleus. These discrepancies regarding the cellular localization of Ro proteins remain to be resolved.14 Evidence that cross-reacting determinants between these proteins are conformational,15 suggests that neither direct physical association nor amino acid similarity is required to account for intermolecular determinant spreading.
The specific targeting by the immune system of apparently unrelated intracellular components of diverse subcellular location, has been explained more recently by the observation that autoantigens are clustered into distinct populations of blebs at the surface of apoptotic cells. These surface structures may constitute an important immunogenic target in autoimmune disease.16,17 Under normal conditions, autoantigens are not associated but become clustered and concentrated in apoptotic blebs. Stress-related cell surface expression of Ro52, and alterations in the distribution of Ro and La have been detected in normal cultured keratinocytes subjected to ultraviolet irradiation or heat-shock, both in vitro models of apoptosis. This has been related to the light-sensitivity exhibited by some patients with systemic lupus erythematosus and Sjögren's syndrome.18,19
The function of Ro52 is not known but the molecule contains distinct zinc finger and leucine zipper motifs, suggesting a possible role in binding to DNA/RNA.20,21 Zinc-binding capabilities have been investigated22 and epitope-mapping studies using synthetic peptides and recombinant antigen have shown immuno-dominant epitopes localizing predominantly towards the N-terminal zinc finger domains.23 Variation in the specificity of autoantibodies directed against different parts of the Ro52 protein have been detected in different patient sera and associations with particular human leucocyte antigen (HLA) class I and class II haplotypes have been made.24
The gene for human Ro52 has been previously mapped to chromosome 11.3 More recent bioinformatic and genomic approaches have identified Ro52 as belonging to a large family of related proteins, sharing overall domain structure and approximately 40% amino acid identity. These molecules have been variously termed acid finger (AFP), ret or RING finger-like (RFP-like) or more recently tripartite motif proteins. In the human genome, more than 50 distinct proteins consisting of the RING/B-box/coiled-coil (RBCC) triple motif structure like Ro52, have been documented in sequence databases to date. A B30.2 domain, originally identified as a single exon mapping within the human major histocompatibility complex region of chromosome 625 is found at the C-terminus of Ro52, in a subset of other tripartite motif proteins, and in a number of cell surface structures related to butyrophilin/B7 co-stimulatory receptors.26 In the light of these observations, we initiated a molecular investigation of Ro52, the expression of the gene, the structure and intracellular localization of the protein and how these may be related to the autoimmune response directed towards the molecule.
Materials and methods
Plasmids, sequencing and sequence analysis
UniGene Cluster Hs.1042 for Sjögren's syndrome antigen A1 (52 000 MW, ribonucleoprotein autoantigen Ro/SS-A) contains 46 cDNAs, including 5′ and 3′ sequences from IMAGE:593027 with an insert size of 2·0 kilobases. By sequence assembly (DNAStar, Madison, WI) and comparison to full length 52 000 MW A Ro/SS-A gene sequence (GenBank M62800), we identified this clone as representing the full-length open reading frame for 52 000 MW Ro/SS-A. The clone was obtained from the Resource Centre, Human Genome Mapping Project, Hinxton http://www.hgmp.mrc.ac.uk. A similar approach was used in order to clone digitally full-length cDNA for RNF15 (IMAGE:1130436).
The following primers were used for PCR reactions, expression constructs and in sequencing: Ro52 forward, aagatatctcagcagcacgcttgac from codon 3; Ro52 reverse, ttctcgagttaatagtcagtggatccttg containing a stop codon; Ro52 sequence 1, ggtatgtgcccagtctcgg; Ro52 sequence 2, tccagagtgaaagtgctgg; Ro52 forward 2, aagatatcgagaagctccaggtggc (from exon 2); Ro52 reverse 2, ttctcgagttatgcacatgtcctcagcatc (from exon 5); RNF15 forward, aagatatctcaaccaccagca; and RNF15 reverse, ttctcgagttagtcacctgggggaggcag.
Forward primers contained EcoRV sites and reverse primers contained XhoI sites and stop codons. All primers were obtained from MWGBiotech (Ebersberg, Germany). Standard polymerase chain reaction (PCR) conditions were used except with the addition of Expand High Fidelity Taq polymerase (Roche, Lewes, UK). Cloning of PCR products was carried out using a ZeroBlunt TOPO-PCR cloning kit (Invitrogen, Groningen, Netherlands). Plasmid DNA was purified using a standard alkaline lysis method and sequenced using vector and original PCR primers with ABI fluorecent dye terminator technology. Reactions were run and analysed on an ABI 377 sequencer (Applied Biosystems, Foster City, CA). Inserts were subcloned into pcDNA3HisB producing N-terminal fusions with the anti-Xpress™ antibody epitope (Invitrogen). Sequence data were analysed using Seqman software (DNAstar).
Amino acid comparisons were carried out using GCG Pileup on Unix running on TIN at the Human Genome Mapping Project, Hinxton, Cambridgeshire, UK and Boxshade at http://www.ch.embnet.org/software/BOX. Amino acid sequences were obtained at GenBank (HTTP://www.ncbi.nlm.nih.gov) using accession numbers: 52 000 MW Ro/SS-A autoantigen; Sjögren's syndrome antigen A1 (NP_003132); TRIM6 (AF220030); RNF 21 (interferon-responsive finger protein or TRIM34, NP_067629); ring finger protein 15 (RNF15) Ro/SS-A ribonucleoprotein homologue (RoRet, NP_006346.1); RFP (NP_006501.1).
Coiled-coil domains were predicted using coils at http://www.ch.embnet.org/software/COILS. Genomic structures were deduced using BLAST running at NCBI using the 52 000 MW Ro/SS-A cDNA vs. the homo sapiens chromosome 11 working draft sequence NT_009151.3|Hs11–9308 (latest update 16 April 2001).
Tissue culture and reverse transcription PCR
Cell lines were obtained from the European Collection of Cell Cultures (ECACC, Salisbury, UK) and grown in RPMI-1640 medium (Gibco Life Technologies Ltd., Paisley, UK) containing 10% fetal calf serum, l-glutamine (2 mm) and penicillin/streptomycin (100 U) (Gibco). Recombinant interferon-γ (IFN-γ; R & D Systems, Minneapolis, MN) was used for time–course activation at 100 U/ml in tissue culture medium for the indicated time periods. Messenger RNA (mRNA) was prepared from cell lines using an mRNA purification kit (Amersham, Little Chalfont, UK). Reverse transcription was carried out using 1 µg mRNA and Moloney murine leukaemia virus reverse transcriptase and other reagents provided in the Prostar First Strand reverse transcription (RT)-PCR kit (Stratagene, Amsterdam, Netherlands). Standard PCR conditions were used except with the addition of Expand High Fidelity Taq polymerase (Roche). RT-PCR products were resolved by electrophoresis in 1% agarose gels containing ethidium bromide. Analysis of band intensities was carried out using NIH Image 1.62 software. Primers used for amplification of Ro52 cDNA were Ro52 forward and Ro52 reverse. The control primer sequences were: DMα forward, gagcatgatcacattcctgccg; DMα reverse, ggaaatgtgccatccttctgaa; GAPDH forward, accacagtccatgccatcac; and GAPDH reverse, tccaccaccctgttgctgta.
Fluorescence and confocal microscopy
Transient transfections (Lipofectamine, Gibco-BRL) of Chinese hamster ovary (CHO) and HeLa cells growing on coverslips were fixed with 3% paraformaldehyde and rendered permeable with 0·1% Triton X-100. Alternatively, cells were fixed and rendered permeable using ice-cold 50% acetone/methanol. Primary (anti-Xpress monoclonal antibody, Invitrogen) and secondary [rabbit anti-mouse fluorescein isothiocyanate (FITC); Sigma, Poole, UK] antibodies were applied sequentially in 3% bovine serum albumin/phosphate-buffered saline at various dilutions. For double staining, sheep anti-mouse FITC and donkey anti-rabbit Texas red were used (Amersham). The antibody to the β-subunit of AP-2 has been described previously.27 A fluorescence microscope (Leica, Solms, Germany), fitted with standard fluorescein or rhodamine filters with a 100 × oil immersion objective lens was used. Confocal images were collected with a Leica TCS SP system equipped with a 63 × Leitz Plan-Apo objective (NA 1.4) at a resolution of 1024 × 1024 pixels and using a zoom of 2.0. Adobe Photoshop software (Adobe Systems, San Jose, CA) was used for image processing.
Patient sera
All patient sera were obtained with the approval of the Local Research Ethics Committee. Patients with systemic lupus erythematosus each met four or more of the revised classification criteria of the American College of Rheumatology28. Patients with Sjögren's syndrome met four or more of the preliminary criteria for classification proposed by the European Community29. Sera from patients with systemic lupus erythematosus (n = 25) and primary Sjögren's syndrome (n = 20) were studied. Anti-Ro52 antibodies in patient sera were detected using ELISA (Axis-Shield Diagnostics, Dundee, UK). A total of 14 patient sera were subsequently analysed by immunoprecipitation.
In vitro transcription/translation and immunoprecipitation (IP)
In vitro transcription/translation (TnT) was carried out using rabbit reticulocyte lysate (TnT kit, Promega, Southampton, UK). Briefly, 1 µg of plasmid DNA was mixed with rabbit reticulocyte lysate and 35S methionine/cysteine (4 µl of 1000 Ci/mmol Promix, Amersham) in a total volume of 30 µl and incubated for 90 min at 30°. The integrity and purity of the resulting 35S-labelled protein was checked using standard 12% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) gels. Immunoprecipitation of TnT products (3 µl) was carried out using human serum (4 µl) or anti-Xpress antibody (2 µg) in a total volume of 300 µl RIPA buffer (50 mm Tris pH 7·4, 150 mm NaCl, 0·5% nonidet P-40). Incubation was for 1 hr at 4°, with mixing. Immune complexes were precipitated by the addition of 25% (w/v) protein G agarose (20 µl, Sigma) and continued incubation at 4° for 1 hr, followed by three washes by centrifugation using RIPA buffer. TnT/IP products were resuspended finally in SDS–PAGE loading buffer and heat denatured (95° for 2 min). Following electrophoresis, SDS–PAGE gels were fixed (25% propan-2-ol, 10% methanol), immersed in Amplify (Amersham) for 30 min each, then dried under a vacuum at 80°. Dried gels were exposed to X-ray film for 1–5 days.
For Western blotting, protein lysates from transfected cells made in RIPA buffer were separated in 12% SDS–PAGE gels. Proteins were transferred to a PVDF membrane and probed with antiXpress antibody using standard procedures.
Results
Ro52-related molecules, protein domain structure and initial characterization
A BLASTp analysis using the complete amino acid sequence of Ro52 identifies over 50 sequences producing significant alignment scores. Among the most related molecules in the human genome are TRIM6 with 42% amino acid identity, RNF 21 (interferon-responsive finger protein or TRIM34)30 and RNF15 (RoRet) both 40% identical and ret finger protein (RFP) at 35% identity. Figure 1(a) shows an amino acid alignment of Ro52 with TRIM6, RNF15 and RFP. Significant stretches of amino acid identity extend over the whole length of the molecules. Similarity of Ro52 to butyrophilin and related BTN molecules on the other hand is restricted to the C-terminal B30.2 domain.
Figure 1.
(a) Amino acid alignment of Ro52 protein with other representatives of the tripartite motif family TRIM6, RNF15 (RoRet) and RFP. Alignments were produced using Pileup followed by Boxshade with amino acid sequences obtained using accession numbers listed in the Materials and Methods section. Numbers and vertical lines above the Ro52 sequence indicate protein domain boundaries deduced from the exon structure of the gene. This implies that the related molecules shown have similar structures at the genomic level.(b) Relationship of the genomic structure of the Ro52 gene, deduced from the human genome draft sequence, to the protein domain structure. Separate exons encode separate domains of the protein. Amino acid sequences conforming to the RING and B-box motifs found encoded by the complex single first exon are shown.
By interrogation of the human genome draft sequence using the Ro52 cDNA, we show that the Ro52 gene covers six coding exons on human chromosome 11p15.5.31 The genomic structure of the gene may be related to the protein domain structure as shown (Fig. 1b). The first coding exon (exon 1) consists of two distinct cysteine-rich motifs suggestive of zinc binding. The first conforms to the C3HC4 RING finger32,33 and the second is a B-box.34 The RING domain is found more widely in other proteins where it is not associated with a B-box, such as the eponymous RING1 (RNF1), breast cancer type 1 (BRCA1) and V(D)J recombination activating gene 1 (RAG1). The B-box domains however, are only found in the context of the RBCC motif.34,35 In the Ro52 gene the RING and B-box domains are encoded by a single exon. This association of the RING domain with the B-box as a complex single exon in the manner exemplified by Ro52 may have evolved by duplication and/or fusion of a more simple exon encoding a zinc-binding domain. It is unclear whether this arrangement is found in other tripartite molecules, particularly those with two B-box domains, or in other organisms. This single exon may represent a recent pairing of domains in evolutionary terms.36
Exons 2 and 3 encode leucine-rich zipper-like motifs predicted to form the predominantly α-helical coiled-coil domains. Coiled-coil domains are also found in other proteins involved in both protein and DNA interactions37 and not only in the context of the RBCC motif. Exons 4 and 5 encode amino acid sequences with no obvious functional motifs. Exon 4 is very short, encoding eight amino acids, and exon 5 shows the most diversity in terms of amino acid sequence between the related molecules illustrated here (Fig. 1). Exon 6 encodes the so-called B30.2 or RFP-like domain. The B30.2 domain is of unknown function but again is found at the C-terminus of a large number of tripartite motif proteins, and on a set of cell surface receptor molecules related to butyrophilin/B7.25,26
There are no obvious protein targeting motifs in the Ro52 amino acid sequence and hydrophobicity plots do not reveal any stretches of potential membrane-spanning hydrophobic residues. The theoretical molecular weight of Ro52 is 54 000, with one potential N-linked glycosylation site positioned in the C-terminal B30.2 domain. Other related tripartite motif proteins illustrated here have similar theoretical molecular weights.
For initial characterization of Ro52 protein, we used in vitro coupled TnT with rabbit reticulocyte lysate and Western blotting of transfected cells. The full-length Ro52 cDNA was cloned into pcDNA3.1HisB vector, which contains the human CMV promoter and T7 promoter priming site together with an N-terminal anti-Xpress antibody epitope. Radiolabelled Ro52 TnT product migrated as a single band in SDS–PAGE protein gels, consistent with a calculated theoretical molecular weight of 58 000 for recombinant Ro52 protein, including the N-terminal tag (not shown). The use of reducing or non-reducing conditions did not alter the size of the TnT product. The addition of canine pancreatic microsomal membranes, used in the study of initial post-translational modification, also had no effect on the size of Ro52 protein. The single potential N-linked glycosylation site identified in the Ro52 sequence was apparently not modified, although N-linked glycosylation was detected in control protein.
In Western blotting experiments of lysates produced from cells transfected with the Ro52 expression construct using the antiXpress antibody, a single band consistent with the theoretical molecular weight of 58 000 for recombinant Ro52 was detected (Fig. 2). Again, use of reducing or non-reducing conditions did not alter the size of the protein. Using these systems therefore, we did not detect any post-translational modifications of Ro52.
Figure 2.
Western blot analysis of lysates from untransfected cells (lane 1) and cells transfected with the Ro52 expression construct (lane 2) probed with antiXpress monoclonal antibody. A single band consistent with the theoretical molecular weight of 58 000 for recombinant Ro52 was detected in transfected cells. Western blotting did not identify any post-translational modifications of the Ro52 molecule.
Ro52 transcript levels are up-regulated by IFN-γ
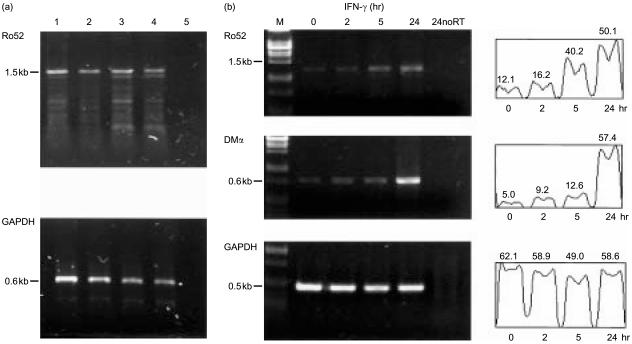
Using RT-PCR, we analysed mRNA derived from a number of tissue culture cell lines to look for expression of the Ro52 transcript. Ro52 mRNA was detectable only at low levels in all cell lines tested, including HT1080, HeLa, HepG2, 293, Jurkat, Molt4, Raji and undifferentiated THP-1 and U937 cells. Figure 3(a) shows typical results from RT-PCR analysis for Ro52 transcripts from mRNA derived from Raji, THP-1, HL60 and HeLa. After treatment with IFN-γ, we observed that steady-state levels of Ro52 mRNA were rapidly induced (Fig. 3b). Up-regulation of Ro52 transcript levels was consistently visible after 5 hr of IFN-γ treatment of HeLa cells, rising to a maximum at 24 hr (Fig. 3b, top panel). Quantification of band intensities showed that the induction by IFN-γ represented a fourfold increase in Ro52 mRNA after 24 hr exposure. As a positive control for transcript induction by IFN-γ, we analysed class II HLA-DMα transcripts by RT-PCR. The increase in HLA-DM transcript levels was visible after 24 hr IFN-γ treatment (Fig. 3b, middle panel) and this correlated with increased cell surface expression of HLA class II molecules, determined by fluorescence-activated cell sorting (FACs) analysis (data not shown). The abundant and constitutive expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts were used as a loading control.
Figure 3.
(a) RT-PCR analysis of cell lines for Ro52 transcript expression: mRNA derived from Raji (lane 1), THP-1 (lane 2), HL-60 (lane 3), HeLa (lane 4), no RT template (lane 5).(b) Up-regulation of Ro52 transcripts by IFN-γ: mRNA isolated from HeLa cells treated for the times indicated, with 100 U/ml IFN-γ, were analysed for Ro52 expression. Up-regulation of Ro52 transcripts was detected after 5 hr of IFN-γ treatment. Analysis for HLA-DMα transcripts was used as a positive control for IFN-γ treatment and GAPDH to account for template input. Band intensity profile scans (expressed in arbitrary units) for each transcript analysed are shown. M=λ PstI DNA marker. 24noRT represents the no reverse transcriptase enzyme control using mRNA template from cells treated for 24 hr with IFN-γ.
Analysis of autoimmune and normal sera by immunoprecipitation
In order to investigate the immune response to Ro52, immunoprecipitation experiments with autoimmune sera were carried out using TnT products (TnT/IP). We investigated the specificity of human sera derived from patients with autoimmune conditions to the different domains of the Ro52 molecule and whether other tripartite motif molecules showing high amino acid identity may be recognized. For these experiments we used DNA contructs containing the full-length Ro52 cDNA, and constructs with deletions of either exon 1 (ΔE1, RING/Bbox domain) or exon 6 (ΔE6, B30·2 domain) as well as a construct of full-length RNF15, all in the pcDNA3 vector.
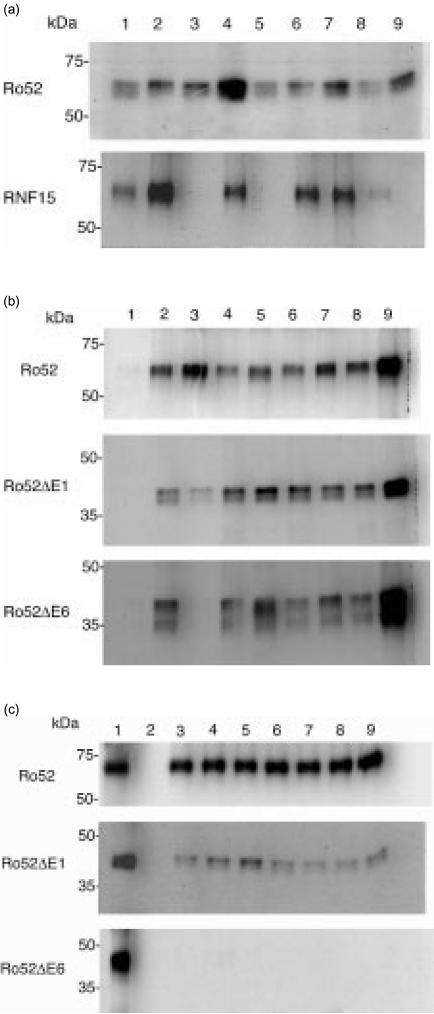
Figure 4(a) shows TnT/IP analysis of Ro52 and the Ro52-related protein RNF15, immunoprecipitated with sera from patients with Sjögren's syndrome. All patient sera precipitated Ro52 protein (Fig. 4a, top panel, lanes 2–9). There was wide variation between the strength of the resulting signal, indicating differences in the relative affinities of different sera to Ro52 protein, or differences in concentration of interacting antibodies. The same sera (SS patients 2–9) tested against RNF15 (Fig. 4a bottom, lanes 2–9), appeared to show cross-reactivity and again variation in signal intensity which did not necessarily correlate with that found in the Ro52 experiment. For example, serum 2 interacted strongly with RNF15 but relatively weakly with Ro52. Patient sera 3, 5 and 9 showed no interaction with RNF15. Precipitation using monoclonal anti-Xpress was used as a positive control in each case (lane 1).
Figure 4.
(a) Investigating the autoimmune response to Ro52 using in vitro transcription/translation followed by immunoprecipitation (TnT/IP). Sera derived from patients with autoimmune Sjögren's syndrome were used to immunoprecipitate Ro52 and the Ro52-related molecule, RNF15. All sera immunoprecipitated Ro52 (top panel, lanes 2–9). Immunoprecipitation of RNF15 was observed in some cases (bottom panel, lane 2–9). Lane 1 in each panel represents immunoprecipitation using the anti-Xpress mouse monoclonal. (b) TnT/IP using domain deletion constructs of Ro52. Sera from six patients with autoimmune systemic lupus erythematosus were used to immunoprecipitate full-length Ro52 (top panel), ΔE1 (middle) and ΔE6 (bottom) material (lanes 4–9 in each case). Lane 1 in each panel, mouse monoclonal (9E10); lane 2, anti-Xpress antibody; lane 3, normal human serum. (c) TnT/IP of Ro52 with normal human sera. Sera from seven normal laboratory volunteers (lanes 3–9 in each panel) were used to immunoprecipitate full-length Ro52 (top), ΔE1 (middle) but not ΔE6 (bottom) material. Lane 1, anti-Xpress antibody; lane 2, normal mouse serum.
Based on our analysis of the Ro52 genomic and protein domain structures, we made domain deletion constructs in order to investigate the autoantibody response to the different domains of Ro52 (Fig. 4b). A further group of six systemic lupus erythematosus patient sera were used. All of these sera were able to immunoprecipitate full-length Ro52 protein (Fig. 4b top, lanes 4–9). Interactions with Ro52 protein missing exon 1 (Fig. 4b, middle) and with the ΔE6 material (Fig. 4b, bottom) were also detected. As with the previous experiment, there was some variation in signal intensity with different sera and doublet bands were visible when precipitating Ro52-domain deletion protein. Lane 1 in each panel represents a non-precipitating isotype-matched mouse monoclonal (9E10) and lane 2 shows use of the anti-Xpress monoclonal antibody. This result suggested that, by this methodology, no particular domain of the Ro52 protein was targeted specifically by autoantibodies.
We used normal human serum in the experiments shown in Fig. 4(b) as a control. To our surprise, this serum precipitated full-length Ro52 and Ro52ΔE1 protein but not the ΔE6 material (Fig. 4b, lane 3 in each case). This suggested that normal serum interacted with the B30.2 domain encoded by exon 6, although the weak signal obtained with the ΔE1 material vs. the full-length Ro52 possibly indicated a more complex interaction. We therefore carried out a similar experiment using human sera collected from seven laboratory volunteers. Figure 4(c) shows the result of the analysis of TnT/IP using full-length Ro52 and domain deletion constructs with normal human sera (lanes 3–9). All sera immunoprecipitated full-length Ro52 (Fig. 4c, top) and the Ro52ΔE1 protein (Fig. 4c, middle), but not the ΔE6 material (Fig. 4c, bottom). Little variation in signal intensity was detected in these experiments using normal human serum, although full-length Ro52 protein appeared to be precipitated more efficiently than the ΔE1 material (compare lanes 3–9 with lane 1, showing immunoprecipitation using the anti-Xpress antibody). Lane 2 in Fig. 4(c) shows the negative result obtained using normal mouse serum.
Taken together, these results show that there are two interactions of sera and Ro52 antigen. First, the interaction of normal human immunoglobulin with the C-terminal B30.2 domain. This interaction is species specific since mouse immunoglobulin did not immunoprecipitate human Ro52. Second, specific autoantibodies in sera from patients with Sjögren's syndrome or systemic lupus erythematosus, which are directed towards the rest of the Ro52 molecule.
Intracellular localization of Ro52 by fluorescence microscopy
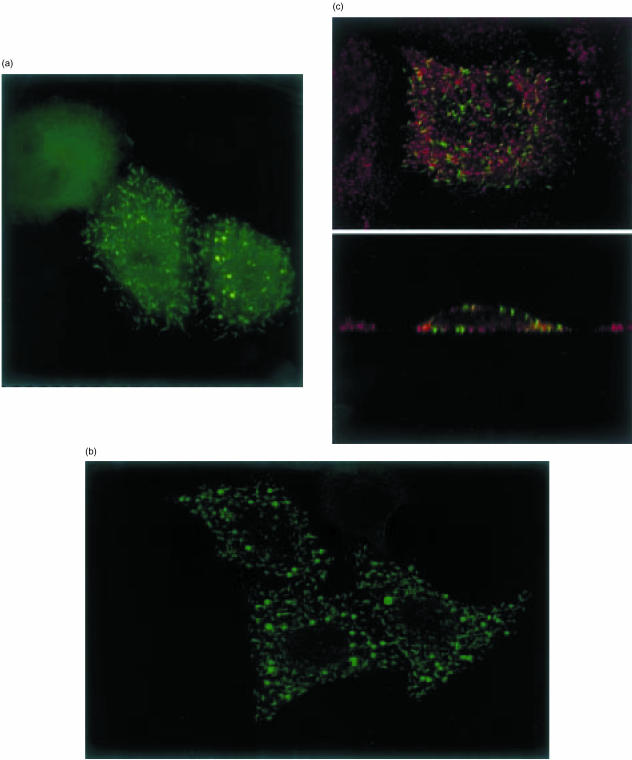
We transiently transfected HeLa and CHO cells with a Ro52 construct epitope-tagged at the N-terminus to determine the intracellular distribution of the protein by indirect immunofluorescence microscopy. The tag is an eight-amino-acid peptide which is unlikely to interfere with the localization of Ro52 and can be detected using the commercial anti-Xpress antibody. Figure 5 shows typical examples of the staining patterns observed in HeLa cells 48 hr after transfection. The most prominent pattern was the labelling of short rod-like structures of strikingly uniform morphology (Fig. 5a). In addition, spots of intense fluorescence were detected, which appeared vesicular in structure (Fig. 5b). Side views (zx sections) taken from the same cells showed that the short rod-like structures were typically located directly underneath the plasma membrane. This was especially evident in cells that had been double labelled with antibodies to the adaptor protein complex 2 (AP-2), which is localized to coated pits at the plasma membrane (Fig. 5c). Compared to this complex, Ro52 was present in spatially distinct and much larger aggregates that extended from the plasma membrane towards the inside of the cells. Ro52 was not exposed on the cell surface, since staining was observed only in cells that had been rendered permeable. In a minority of cells only diffuse staining throughout the cytoplasm was seen (Fig. 5a). In the remaining cells, both diffuse cytosolic staining and strongly labelled rod-like or round structures were visible to varying degrees, suggesting that these cells were in a transitional state from a cytosolic to a membrane-proximal location of the Ro52 protein. In this case, the rod-like and round structures were further inside the cells, suggesting that the transition involved at least two steps: formation of larger Ro52-containing aggregates and translocation of aggregates to the plasma membrane. We cannot rule out the possibility that Ro52 was associated with intracellular membranes; however, the staining was not typical for any previously identified compartment in the secretory or endocytic pathway and no overlap was found with known markers, such as the lysosome-associated membrane protein 1 (LAMP-1), early endosomal antigen 1 (EEA1) (not shown), or adaptor protein 2 of clathrin-coated pits (AP-2). In all cases, Ro52 was excluded from the nucleus. Similar results were obtained in CHO cells (not shown). The distribution of Ro52 was the same in cells that were fixed with paraformaldehyde and permeabilized with detergent or cells treated with acetone/methanol, showing that they were not simply due to fixation artefacts. Staining of transfected cells with isotype-matched control antibody was negative. Our theory that the rod-like structures in close proximity to the plasma membrane were the typical final distribution in this system was substantiated by the fact that none of these structures were seen at earlier times after transfection. This indicated that the localization to these structures was a concentration- and time-dependent phenomenon. Changes in the growth conditions, such as treatment with IFN-γ, did not affect the distribution of Ro52.
Figure 5.
Representative results of confocal imaging of epitope tagged Ro52 protein in transfected HeLa cells. Defined rod-like structures were visible in a proportion of transfected cells (a) Localization to intensely staining round structures and exclusion from the nucleus was detected (b) Rod-like structures localized to a membrane proximal position extending towards the inside of cells by zx imaging (c) No co-localization was detected using antibodies directed towards known cellular components LAMP-1, EEA1 or AP-2. Red fluorescence in (c) represents detection of endogenous AP-2.
In order to identify which domain of the Ro52 protein was responsible for intracellular localization, we used domain deletion constructs. Deletions of either exon 1 (RING/Bbox) or exon 6 (B30.2) both gave a diffuse cytoplasmic staining pattern (data not shown). From this data it was not possible to identify a single Ro52 protein domain as being responsible for the intracellular targeting.
Discussion
We initiated a molecular investigation of Ro52 in order to study the autoimmune response to the protein. Ro52 is related by significant stretches of amino acid identity and overall domain structure to a large number of RBCC or tripartite motif proteins, which are encoded throughout the human genome. Transcript expression, intracellular localization, and the autoimmune response to the Ro52 molecule have been analysed, assuming that studies of Ro52 may be applicable to the other members of the tripartite motif protein family. In addition, we were intrigued by the presence of the B30.2 domain at the C- terminus of Ro52, and on a set of cell surface glycoproteins related to butyrophilin/B7 co-stimulatory receptors, a subset of the immunoglobulin gene superfamily.26
Very low steady-state levels of full-length transcripts of Ro52 were found in all tissue culture cell lines examined, and these were induced rapidly in HeLa cells by cellular activation with IFN-γ. A fourfold increase in Ro52 mRNA levels was detected after 24 hr exposure of cells to this cytokine, a potent modulator of immune responses. Ro52 has previously been included in a list of genes whose expression is modulated in human fibrosarcoma HT1080 cells by IFN-α, IFN-β and IFN-γ, identified using microarray technology.38 Confocal immunofluorescence microscopy of transfected HeLa cells, using an epitope attached to the N-terminus of Ro52 protein, revealed localization to membrane proximal structures. Disrupting the integrity of Ro52 protein using deletion constructs abrogated this localized immunofluorescence, suggesting that a complex protein conformation of the molecule, involving association with other cellular components, was involved. The localization did not appear to involve post-translational modification of Ro52 since this was not detected by Western blot analysis of transfected cells. Membrane changes accompanying cellular activation by IFN-γ result in increased recruitment of cell surface receptors, for instance HLA class II molecules for antigen presentation. Increased levels of Ro52 transcripts after IFN-γ treatment of HeLa cells together with a membrane proximal location for the protein may imply a role for Ro52 in these processes.
We investigated the autoimmune response to Ro52 and the related molecule RNF15. Using TnT/IP experiments we have shown that sera from about 60% of patients with autoimmune Sjögren's syndrome immunoprecipitated RNF15. This may represent cross-affinity of anti-Ro52 antibodies to the RNF15 molecule, which shares significant stretches of amino acid identity with Ro52. This observation could be extended to other Ro52-related molecules, for example RFP, TRIM6 and RNF21. Alternatively, these data may indicate the existence of a separate population of anti-RNF15 antibodies in patient sera. These may represent a confounding factor in diagnosis and may point to an involvement of Ro52-related molecules in the pathology of autoimmune rheumatic disease.
We also investigated whether different domains of the Ro52 molecule were preferentially targeted by the autoimmune response using domain deletion constructs. We first noted that all patient sera immunoprecipitated Ro52 protein, whether the full-length molecule was used or constructs missing either exon 1 or exon 6. This suggested to us initially that there was no specific targeting of autoantibodies to the different domains of the protein. However, in subsequent experiments using normal sera, we showed that these were also able to immunoprecipitate the Ro52 protein, except when exon 6, encoding the B30.2 domain, was deleted. Since we used protein G agarose to precipitate the immune complexes, this suggested a specific interaction of human immunoglobulin with the B30.2 domain of Ro52. This interpretation is consistent with the observations of Yang et al. who showed interaction of Ro52 with human immunoglobulin G, identified initially using a yeast two-hybrid approach39 and confirmed by immunopreciptation using bacterially expressed Ro52.40 At this stage however, we could not exclude the possibility that another serum component mediated the immunoprecipitation of Ro52 by immunoglobulin. Differences in signal intensities obtained with different protein constructs precipitated with the normal human serum suggested a more complex interaction than with the B30.2 domain alone, possibly including the RING/B-box domain. In our experiments, sera from patients with Sjögren's syndrome were still able to precipitate Ro52ΔE6 material, showing that these sera contained specific antibodies directed against the rest of the Ro52 molecule. Mouse serum did not interact with human Ro52 in these experiments showing that a highly specific interaction occurs between human immunoglobulin and human Ro52 protein. Whether other Ro52-related molecules with C-terminal B30.2 domains interact with immunoglobulin remains to be determined.
Whilst this work was being completed, an analysis of 37 tripartite motif proteins (termed TRIM) was presented.41 Using green fluorescent protein tagged constructs, Reymond et al. showed localization of different members of this family of molecules to a variety of intracellular locations, none of which could be identified using counter staining with antibodies to known cellular components. Results for Ro52 (called TRIM21, and available at web site http://www.tigem.it/TRIM) were similar to those presented here. These authors concluded that tripartite motif (TRIM) proteins are localized to novel compartments within cells.41 The tripartite motif molecules do not contain recognized cell-sorting motifs, suggesting that interactions with other cellular components, particularly of the cytoskeleton, may be involved in the unique localizations observed. A role for tripartite motif proteins and other proteins containing RING domains as intracellular molecular scaffolds has been suggested35 and evidence for a general biochemical role for RING domains as E3 ubiquitin protein ligases has been presented.42,43
Further experiments will be directed towards investigating any potential function for the novel molecular interaction of Ro52 and human immunoglobulin in vivo. Yang et al.39 have shown that the interaction is isotype specific and have proposed a role in the recognition and removal by the immune system of cells undergoing apoptosis. This is suggested to occur via initial cell surface exposure of the Ro52 molecule in apoptotic cells followed by binding of immunoglobulin and recruitment of macrophages. Re-localization of Ro52 protein to the cell surface has been reported in cells undergoing apoptosis. This process has not been directly visualized, hampered by uncertainty over the intracellular localization of Ro52 protein. Our observation that Ro52 localizes to distinct membrane proximal structures offers a means by which this potential re-localization may be viewed directly in apoptotic cells, and a means by which the mechanisms involved may be examined. The observation presented here and elsewhere39 concerning the interaction of Ro52 with human immunoglobulin, suggests a direct functional role in apoptosis. Cell surface exposure of Ro52 and recruitment of immunoglobulin molecules would effectively signal the apoptotic status of the cell. This may represent a general cellular mechanism for opsonization and removal of apoptotic cells by macrophages, by binding of immunoglobulin to cell surfaces via the exposed B30.2 domain.
Given the results presented here, the use of ELISA methods to determine autoantibody status to Ro52 in sera from patients with systemic autoimmune conditions must be questioned. Possible confounding effects of direct interaction of the protein with human immunoglobulin have not previously been addressed. We have shown that the TnT/IP approach is a highly sensitive and specific method for the characterization of interactions of human sera with the Ro52 protein, particularly with the use of domain deletion constructs. A more thorough investigation of patient sera in the light of these results may help in diagnosis and subclassification of this group of autoimmune conditions.
Acknowledgments
We thank Dr M. S. Robinson for providing us with antibody, Professors P. Sissons and A. C. Minson for access to the Leica Confocal Microscope and Professor Paul Luzio and Dr Howard Davidson for helpful discussion.
References
- 1.Gordon T, Topfer F, Keech C, Reynolds P, Chen W, Rischmueller M, McCluskey J. How does autoimmunity to La and Ro initiate and spread? Autoimmunity. 1994;18:87–92. doi: 10.3109/08916939409007981. [DOI] [PubMed] [Google Scholar]
- 2.McCauliffe DP, Wang L, Satoh M, Reeves WH, Small D. Recombinant 52 kDa Ro (SSA) ELISA detects autoantibodies in Sjogren's syndrome sera that go undetected by conventional serologic assays. J Rheumatol. 1997;24:860–6. [PubMed] [Google Scholar]
- 3.Frank MB, Itoh K, Fujisaku A, Pontarotti P, Mattei MG, Neas BR. The mapping of the human 52-kD Ro/SSA autoantigen gene to human chromosome 11, and its polymorphisms. Am J Hum Genet. 1993;52:183–91. [PMC free article] [PubMed] [Google Scholar]
- 4.Keech CL, Gordon TP, McCluskey J. The immune response to 52-kDa Ro and 60-kDa Ro is linked in experimental autoimmunity. J Immunol. 1996;157:3694–9. [PubMed] [Google Scholar]
- 5.Tseng CE, Chan EK, Miranda E, Gross M, Di Donato F, Buyon JP. The 52-kd protein as a target of intermolecular spreading of the immune response to components of the SS-A/Ro-SS-B/La complex. Arthritis Rheum. 1997;40:936–44. doi: 10.1002/art.1780400523. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita G, Keech CL, Sontheimer RD, Purcell A, McCluskey J, Gordon TP. Spreading of the immune response from 52-kDa Ro and 60-kDa Ro to calreticulin in experimental autoimmunity. Lupus. 1998;7:7–11. doi: 10.1191/096120398678919606. [DOI] [PubMed] [Google Scholar]
- 7.Topfer F, Gordon T, McCluskey J. Intra- and intermolecular spreading of autoimmunity involving the nuclear self-antigens La (SS-B) and Ro (SS-A) Proc Natl Acad Sci USA. 1995;92:875–9. doi: 10.1073/pnas.92.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCluskey J, Farris AD, Keech CL, Purcell AW, Rischmueller M, Kinoshita G, Reynolds P, Gordon TP. Determinant spreading: lessons from animal models and human disease. Immunol Rev. 1998;164:209–29. doi: 10.1111/j.1600-065x.1998.tb01222.x. [DOI] [PubMed] [Google Scholar]
- 9.Kinoshita G, Purcell AW, Keech CL, Farris AD, McCluskey J, Gordon TP. Molecular chaperones are targets of autoimmunity in Ro (SS-A) immune mice. Clin Exp Immunol. 1999;115:268–74. doi: 10.1046/j.1365-2249.1999.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boire G, Gendron M, Monast N, Bastin B, Menard H. Purification of antigenically intact Ro ribonucleoproteins; biochemical and immunological evidence that the 52-kD protein is not a Ro protein. Clin Exp Immunol. 1995;100:489–98. doi: 10.1111/j.1365-2249.1995.tb03728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keech CL, Gordon TP, McCluskey J. Cytoplasmic accumulation of the 52 kDa Ro/SS-A nuclear autoantigen in transfected cell lines. J Autoimmun. 1995;8:699–712. doi: 10.1006/jaut.1995.0052. [DOI] [PubMed] [Google Scholar]
- 12.Pourmand N, Blange I, Ringertz N, Pettersson I. Intracellular localisation of the Ro 52kD auto-antigen in HeLa cells visualised with green fluorescent protein chimeras. Autoimmunity. 1998;28:225–33. doi: 10.3109/08916939808995370. [DOI] [PubMed] [Google Scholar]
- 13.Farris AD, Puvion-Dutilleul F, Puvion E, Harley JB, Lee LA. The ultrastructural localization of 60-kDa Ro protein and human cytoplasmic RNAs: association with novel electron-dense bodies. Proc Natl Acad Sci USA. 1997;94:3040–5. doi: 10.1073/pnas.94.7.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruijn GJ, Simons FH, van Venrooij WJ. Intracellular localization and nucleocytoplasmic transport of Ro RNP components. Eur J Cell Biol. 1997;74:123–32. [PubMed] [Google Scholar]
- 15.Deshmukh US, Lewis JE, Gaskin F, Dhakephalkar PK, Kannapell CC, Waters ST, Fu SM. Ro60 peptides induce antibodies to similar epitopes shared among lupus-related autoantigens. J Immunol. 2000;164:6655–61. doi: 10.4049/jimmunol.164.12.6655. [DOI] [PubMed] [Google Scholar]
- 16.Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes [see comments] J Exp Med. 1994;179:1317–30. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miranda-Carus ME, Askanase AD, Clancy RM, et al. Anti-SSA/Ro and anti-SSB/La autoantibodies bind the surface of apoptotic fetal cardiocytes and promote secretion of TNF-alpha by macrophages. J Immunol. 2000;165:5345–51. doi: 10.4049/jimmunol.165.9.5345. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa F, Itoh T, Wakita H, Yagi H, Tokura Y, Norris DA, Takigawa M. Keratinocytes from patients with lupus erythematosus show enhanced cytotoxicity to ultraviolet radiation and to antibody-mediated cytotoxicity. Clin Exp Immunol. 1999;118:164–70. doi: 10.1046/j.1365-2249.1999.01026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igarashi T, Itoh Y, Fukunaga Y, Yamamoto M. Stress-induced cell surface expression and antigenic alteration of the Ro/SSA autoantigen. Autoimmunity. 1995;22:33–42. doi: 10.3109/08916939508995297. [DOI] [PubMed] [Google Scholar]
- 20.Frank MB, McCubbin VR, Heldermon C. Expression and DNA binding of the human 52 kDa Ro/SSA autoantigen. Biochem J. 1995;305:359–62. doi: 10.1042/bj3050359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan EK, Hamel JC, Buyon JP, Tan EM. Molecular definition and sequence motifs of the 52-kD component of human SS-A/Ro autoantigen. J Clin Invest. 1991;87:68–76. doi: 10.1172/JCI115003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pourmand N, Pettersson I. The Zn2+ binding domain of the human Ro 52 kDa protein is a target for conformation-dependent autoantibodies. J Autoimmun. 1998;11:11–17. doi: 10.1006/jaut.1997.0171. 10.1006/jaut.1997.0171. [DOI] [PubMed] [Google Scholar]
- 23.Wahren-Herlenius M, Muller S, Isenberg D. Analysis of B-cell epitopes of the Ro/SS-A autoantigen. Immunol Today. 1999;20:234–40. doi: 10.1016/s0167-5699(99)01458-9. [DOI] [PubMed] [Google Scholar]
- 24.Ricchiuti V, Isenberg D, Muller S. HLA association of anti-Ro60 and anti-Ro52 antibodies in Sjogren's syndrome. J Autoimmun. 1994;7:611–21. doi: 10.1006/jaut.1994.1045. [DOI] [PubMed] [Google Scholar]
- 25.Henry J, Ribouchon MT, Offer C, Pontarotti P. B30.2-like domain proteins: a growing family. Biochem Biophys Res Commun. 1997;235:162–5. doi: 10.1006/bbrc.1997.6751. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes DA, Stammers M, Malcherek G, Beck S, Trowsdale J. The cluster of BTN genes in the extended major histocompatibility complex. Genomics. 2001;71:351–62. doi: 10.1006/geno.2000.6406. [DOI] [PubMed] [Google Scholar]
- 27.Page L, Robinson MS. Targeting signals and subunit interactions in coated vesicle adaptor. J Cell Biol. 1995;131:619–30. doi: 10.1083/jcb.131.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan E, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 29.Vitali C, Bombardieri S, Moutsopoulos HM, et al. Preliminary criteria for the classification of Sjögren’s syndrome. Results of a prospective concerted action supported by the European Community. Arthritis Rheum. 1993;36:340–7. doi: 10.1002/art.1780360309. [DOI] [PubMed] [Google Scholar]
- 30.Orimo A, Tominaga N, Yoshimura K, et al. Molecular cloning of ring finger protein 21 (RNF21) /Interferon-responsive finger protein (ifp1), which possesses two RING-B box-coiled coil domains in tandem. Genomics. 2000;69:143–9. doi: 10.1006/geno.2000.6318. 10.1006/geno.2000.6318. [DOI] [PubMed] [Google Scholar]
- 31.Tsugu H, Horowitz R, Gibson N, Frank MB. The location of a disease-associated polymorphism and genomic structure of the human 52-kDa Ro/SSA locus (SSA1) Genomics. 1994;24:541–8. doi: 10.1006/geno.1994.1664. 10.1006/geno.1994.1664. [DOI] [PubMed] [Google Scholar]
- 32.Freemont PS, Hanson IM, Trowsdale J. A novel cysteine-rich sequence motif. Cell. 1991;64:483–4. doi: 10.1016/0092-8674(91)90229-r. [DOI] [PubMed] [Google Scholar]
- 33.Lovering R, Hanson IM, Borden KL, et al. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc Natl Acad Sci USA. 1993;90:2112–16. doi: 10.1073/pnas.90.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borden KL. RING fingers and B-boxes: zinc-binding protein–protein interaction domains. Biochem Cell Biol. 1998;76:351–8. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- 35.Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–12. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 36.Borden KL, Freemont PS. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 37.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–82. [PubMed] [Google Scholar]
- 38.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–8. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Eversole T, Lee DJ, Sontheimer RD, Capra JD. Protein–protein interactions between native Ro52 and immunoglobulin G heavy chain. Scand J Immunol. 1999;49:620–8. doi: 10.1046/j.1365-3083.1999.00547.x. 10.1046/j.1365-3083.1999.00547.x. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y, Yang MW, Wang B, Weissler JC. Autoantigen Ro52 directly interacts with human IgG heavy chain in vivo in mammalian cells. Mol Immunol. 2000;37:591–602. doi: 10.1016/s0161-5890(00)00068-7. [DOI] [PubMed] [Google Scholar]
- 41.Reymond A, Meroni G, Fantozzi A, et al. The tripartite motif family identifies cell compartments. Embo J. 2001;20:2140–51. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freemont P. RING for destruction? Curr Biol. 2000;10:R84–7. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 43.Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–39. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]