Abstract
We report the latest release (version 3.0) of the CATH protein domain database (http://www.cathdb.info). There has been a 20% increase in the number of structural domains classified in CATH, up to 86 151 domains. Release 3.0 comprises 1110 fold groups and 2147 homologous superfamilies. To cope with the increases in diverse structural homologues being determined by the structural genomics initiatives, more sensitive methods have been developed for identifying boundaries in multi-domain proteins and for recognising homologues. The CATH classification update is now being driven by an integrated pipeline that links these automated procedures with validation steps, that have been made easier by the provision of information rich web pages summarising comparison scores and relevant links to external sites for each domain being classified. An analysis of the population of domains in the CATH hierarchy and several domain characteristics are presented for version 3.0. We also report an update of the CATH Dictionary of homologous structures (CATH-DHS) which now contains multiple structural alignments, consensus information and functional annotations for 1459 well populated superfamilies in CATH. CATH is directly linked to the Gene3D database which is a projection of CATH structural data onto ∼2 million sequences in completed genomes and UniProt.
INTRODUCTION
The numbers of new structures being deposited in the Protein Data Bank (PDB) continues to grow at a considerable rate. In addition, structures being targeted by world wide structural genomics initiatives are more likely to be novel or only very remotely related to domains previously classified in CATH (1,2). Only 2% of structures currently solved by conventional crystallography or NMR are likely to adopt novel folds (see Figures 1 and 2). A higher proportion of new folds are expected to be solved by structural genomics structures; indeed a recent study has already showed that to be the case (1,2). Although the influx of more diverse structures and subsequent analysis will inform our understanding of how domains evolve, it has resulted in increasing lags between the numbers of structures being deposited and classified in CATH. In response to this situation we have significantly improved our automated and manual protocols for domain boundary assignment and homologue recognition.
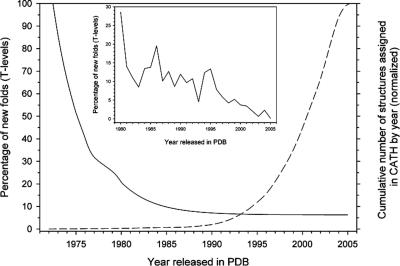
Figure 1.
Annual decrease in the percentage of new structures classified in CATH which are observed to possess a novel fold. The raw data for years 1972–2005 was fit to a single exponential equation by nonlinear regression using Sigma Plot (SPSS, Version 9.0) and the fit is shown as a solid black line. The inset shows a close-up of the raw data for new topologies over the years 1980–2005. For comparison, the numbers of structural domains solved each year and deposited in the PDB and classified in CATH is depicted in the dashed line.
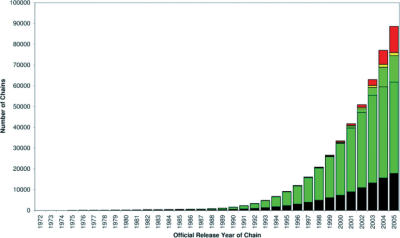
Figure 2.
Annual proportion of protein structures deposited in the PDB which are classified in CATH, rejected or pending classification. The colour scheme reflects different categories of PDB chains. Black: not accepted by the CATH criteria; Red: unprocessed chains; Dark green: cumulative count of all chains processed in CATH release 2.6. Light green: cumulative count of all chains processed in CATH release 3.0.
Significant changes have been implemented in the CATH classification protocol to achieve a more highly automated system. A seamless flow of structures between the constituent programs has been achieved by building a pipeline which integrates web services for each major comparison stage in the classification (see Figure 3). Secondly, completely automatic decisions are now being made for new protein chains with close relatives already assigned in the CATH database. There are two situations that preclude the CATH update process from being fully automated. We rely on expert manual curation for particularly challenging protein domain boundary assignments (DomChop stage) and also for classifications of remote folds and homologues (HomCheck stage). These two manual stages will remain an integral part of the system (Figure 3).
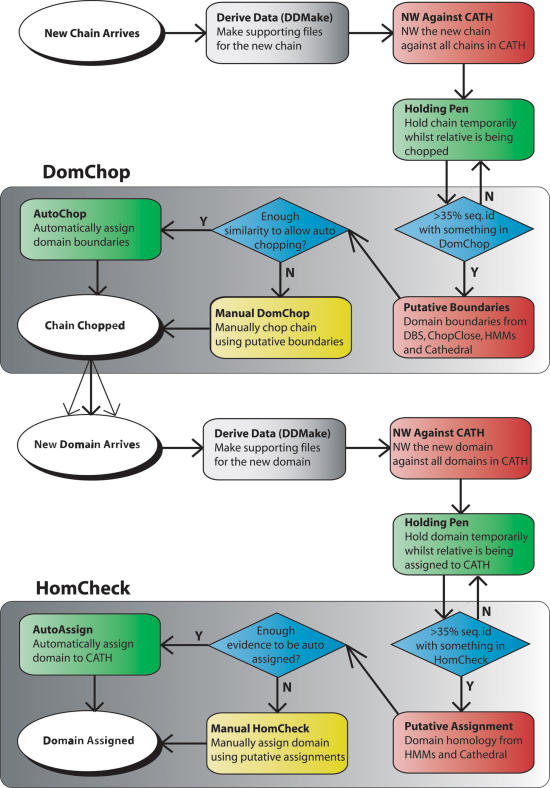
Figure 3.
Flow diagram of the CATH classification pipeline. This schematic illustrates the processes involved in classifying newly determined structures in CATH. The CATH update protocol workflow from new chain to assigned domain is split into two main processes; DomChop where chains are divided into domains and HomCheck where domains are classified into homologous families. Grey boxes denote production of meta-data, red denotes algorithms, blue denotes workflow decision, yellow denotes manual process. Definition of abbreviations and terms are as follows: NW, Needleman–Wunsch (23) sequence alignment algorithm; HMM, hidden Markov model (11); ChopClose, program which determines domain boundaries based on sequence identity with domains in CATH (Lewis T.E. et al. unpublished); DomChop, manual validation of domain boundary assignment; HomCheck, manual validation of homology assignment; CATHEDRAL (4), structure comparison program.
In this paper we report our ongoing development of the automated procedures. These critical new features should better enable CATH to keep pace with the PDB (3) and facilitate its development. Key statistics on the domain structure populations and characteristics are also presented.
A REVISED CATH CLASSIFICATION HIERARCHY: CATHSOLID
In order to provide information on the sequence diversity between superfamily members, we have introduced additional levels into the CATH hierarchy. The CATH hierarchal classification scheme now consists of nine levels. Class is derived from secondary structure content and Architecture describes the gross orientation of secondary structures, independent of connectivity. The Topology level clusters structures into fold groups according to their topological connections and numbers of secondary structures. The Homologous superfamilies cluster proteins with highly similar structures, sequences and/or functions (4,5). The new extension of the CATH classification system now includes five ‘SOLID’ sequence levels. S, O, L, I further divides domains within the H-level using multi-linkage clustering based on similarities in sequence identity (35, 60, 95 and 100%) (see Table 1). The D-level acts as a counter within the I-level and is appended to the classification hierarchy to ensure that every domain in CATH has a unique CATHsolid identification code (see Table 1). Specific details on the nature of the SOLID-levels can be found in the ‘General Information’ section of the CATH website, http://www.cathdb.info. CATH only includes experimentally determined protein structures with a 4 Å resolution or better, 40 residues in length or longer and having 70% or more side chains resolved.
Table 1.
CATH version 3.0 statistics
| C | A | T | H | S | O | L | I | D |
|---|---|---|---|---|---|---|---|---|
| Mainly alpha | 5 | 316 | 674 | 1877 | 2280 | 2862 | 5207 | 18 271 |
| Mainly beta | 20 | 195 | 428 | 1843 | 2436 | 3653 | 6152 | 23 482 |
| Alpha–beta | 14 | 506 | 941 | 3956 | 5103 | 6239 | 12 184 | 43 025 |
| Few 2° structures | 1 | 93 | 104 | 165 | 197 | 267 | 387 | 1373 |
| Total | 40 | 1110 | 2147 | 7841 | 10 016 | 13 021 | 23 930 | 86 151 |
DOMAIN BOUNDARY ASSIGNMENTS
We have further improved our automated domain boundary prediction method—CATHEDRAL (4). This is used to search a newly determined multi-domain structure against a library of representative structures from different fold groups in the CATH database to recognise constituent domains. CATHEDRAL performs an initial rapid secondary structure comparison between structures using graph theory to identify putative fold matches which are then more carefully aligned using a slower, more accurate, dynamic programming method. A new scoring scheme has been implemented which combines information on the class of the domains being compared, their sizes, similarity in the structural environments and the number of equivalent residues. A support vector machine is used to combine the different scores and select the best fold match for each putative domain in a new multi-domain protein.
Benchmarking against a set of 964 ‘difficult’ multi-domain chains, whose 1593 constituent domains were remotely related to folds in CATH (<35% sequence identity) and originated from 245 distinct fold groups and 462 superfamilies, showed that 90% of domains within these chains could be assigned to the correct fold group and for 78% of them, the domains boundaries were within ±15 residues of boundaries assigned by careful manual validation. Larger variations in domain boundaries are often due to the fact that in many families significant structural variation can occur during evolution so that distant relatives vary considerably in size. If no close relative has been classified in CATH, it is likely that the only CATHEDRAL match will be to a relative with significant structural embellishments thus making it harder to determine the correct boundaries.
Since domain boundary assignment of remote homologues is one of the most time consuming stages in the classification we combine multiple information for each new structure on a web page to guide manual curation. Pages display scores from a range of algorithms which include structure based methods: CATHEDRAL (4), SSAP (6,7), DETECTIVE (8), PUU (9), DOMAK (10), sequence based methods such as hidden Markov Models (HMMs) (11) and relevant literature. These pages are now viewable for information on putative boundaries for new multi-domain structures currently being classified in CATH (e.g. http://www.cathdb.info/cgi-bin/cath/Chain.pl?chain_id=2g3aA).
For protein chains which are closely related to chains that are already chopped in CATH, an automated protocol has been developed (ChopClose). ChopClose identifies any previously chopped chains that have sufficiently high sequence identity and overlap with the query chain. Using SSAP (6), the query is aligned against each of these chains in turn and in each case the domain boundaries are inherited across the alignment. The process of inheriting the boundaries often requires some adjustments to be made to account for insertions, deletions or unresolved residues. If the inheritance from one of the chains meets various criteria (SSAP score ≥80, sequence identity >80%, RMSD ≤6.0 Å, longest end extension ≤10 residues etc) then the resulting boundaries are used to chop the chain automatically—AutoChop. For cases where ChopClose's best result does not meet all the criteria for automatic chopping, it is provided as support information for a manual domain boundary assignment. Refer to Figure 3 for the location of AutoChop/ChopClose within the CATH update protocol.
NEW HOMOLOGUE RECOGNITION METHODS
We have assessed a number of HMM based protocols for improving homologue recognition. A new protocol (Samosa), exploiting models built using multiple structure alignments to improve accuracy, gives some improvements in sensitivity (4–5%). However, a protocol exploiting an 8-fold expanded HMM library based on sequence relatives of structural domains, gives an increase of nearly 10% in sensitivity (12). In addition, HMM–HMM-based approaches have been implemented using the PRC protocol of Madera and co-workers (http://supfam.org/PRC/). These allow recognition of extremely remote homologues some of which are not easily detected by the structure comparison methods (discussed further below). HMM based database scans developed for the CATH classification protocol are collectively referred to as HMMscan below.
For some very remotely related homologues, confidence in an assignment can be improved by combining information from multiple prediction methods. We have investigated the benefits of using machine learning methods to do this automatically. A neural network was trained using a dataset of 14 000 diverse homologues (<35% sequence identity) and 14 000 non-homologous pairs with data from different homologue comparison methods including structure comparison (CATHEDRAL, SSAP), sequence comparison (HMM–HMM), and information on functional similarity. The latter was obtained by comparing EC classification codes between close relatives of the distant homologues and using a semantic similarity scoring scheme for comparing GO terms, based on a method developed by Lord et al. (13). On a separate validation set of 14 000 homologous pairs and 14 000 non-homologous pairs 97% of the homologues can be recognised at an error rate of <4%.
NEW UPDATE PROTOCOL
Automatic methods
Previously, CATH data was generated using a group of independent programs and flat files. Over the past two years we have developed an update protocol for CATH that is driven by a suite of programs with a central library and a PostgreSQL database system. A classification pipeline has been established which links in a completely automated fashion the different programs that analyse the sequences and structures of both protein chains and domains. The CATH update protocol can essentially be divided into two parts, domain boundary assignment and domain homology classification (see Figure 3). The aim of the protocol is to minimise manual assignment and provide as much support as possible when manual validation is necessary.
Processing of both parts of the classification protocol are similar, requiring related meta-data and the triggering of the same automated algorithms. Methods include pairwise sequence similarity comparisons and scans by other homologue detection or fold recognition algorithms such as HMMscan and CATHEDRAL that provide data for either manual or automated assignment. Many of the automated steps in the protocol have been established as a web service and the pipeline integrates both automated steps together with ‘holding stages’ in which domains are held prior to processing and await the completion of manual validation of predictions (see below).
Web pages to support manual validation
For each manual stage (domain boundary assignment—DomChop and homologue recognition—HomCheck) in the classification we have developed a suite of web pages bringing together all available meta-data from prediction algorithms (e.g. DBS, CATHEDRAL, HMMscan for DomChop, CATHEDRAL, HMMscan, for HomCheck) and information from the literature and from other family classifications with relevant data (e.g. Pfam). For each protein or domain shown on the pages, information on the statistical significance of matches is presented. The web pages will shortly be made viewable and will provide interim data on protein chains and domains not fully classified in CATH for biologists interested in any entries pending classification.
OVERVIEW OF THE CURRENT RELEASE (VERSION 3.0)
Assigning domain boundaries and relationships between protein structures is computationally challenging. Since the last CATH release version (2.6), the number of domains in the CATH database has increased by 20% in version 3.0 and now totals 86 151. This is a more than 10-fold increase in the number of domains classified in CATH since its creation. Improvements in automation and also in the web based resources used to aid manual validation, have allowed us to increase the proportion of hard-to-classify structures processed in CATH and this is reflected in a significant increase in the proportion of new folds in the database—now more than 1000. The detailed breakdown of numbers of domains in the nine CATH levels is given in Table 1. We conducted an analysis of the domains in version 3.0 and have derived statistics for several fundamental features:
Percentage of new topologies
An analysis of the percentage of new folds arising since the early 1970s to the present age is shown in Figure 1. The numbers of new folds has been decreasing over time with respect to the number of new structures being deposited and it can be seen that currently approximately 2% of new structures classified in CATH are observed to be novel folds. For comparison the number of domain structures solved over time is also graphically represented in Figure 1.
Number of domains within a protein chain
Integral to the construction of the CATH database is designating domain boundaries. We conducted an analysis of the number of chains versus number of domains in a chain. It is interesting to note that 64% of all protein structures currently solved and classified in CATH are single domain chains (data not shown). The next most prevalent are two domain chains (27%) and following this we find that the number of chains containing three or more domains rapidly decreases. The average size of the single domain chains is 159 residues in length.
The CATH Dictionary of Homologous Superfamilies (CATH-DHS)
The CATH-DHS has also been recently updated. Data on structural similarity and superfamily variability is presented as a significant update to the Dictionary of Homologous Superfamilies (DHS) web-resource (14). The DHS also provides functional annotations of domains within each H-level (superfamily) in CATH v 2.5.1.
For each superfamily, pair-wise structural similarity scores between relatives, measured by SSAP, are presented. The DHS now contains 3307 multiple structural alignments for 1459 superfamilies. For each superfamily, multiple alignments are generated for all the relatives and also for subgroups of structurally similar relatives and sequence similar relatives. Alignments are performed using the residue-based CORA algorithm (15) and presented both as CORAPLOTS (14) and in the form of a 2DSEC diagram (16), alongside co-ordinate data of the superposed structures in PDB format. Sequence representations of the alignments are available to download in FASTA format. In the CORAPLOT images of the multiple alignment, residues in each domain are coloured according to ligand binding and residue type. EquivSEC plots are also shown that describe the variability in orientation and packing between equivalent secondary structures (16).
To identify sequence relatives for CATH superfamilies, sequences from UniProt (17) were scanned against HMMs of all CATH domains (12). Homologous sequences were identified as those hits with an E-value < 0.01 and a 60% residue overlap with the CATH domain. This protocol recognised over one million domain sequences in UniProt which could be integrated in the CATH-DHS. The harvested sequences in each superfamily were compared against other relatives by BLAST (18) to determine the pair-wise sequence identity, and then clustered at appropriate levels of sequence identity (35 and 95%) using multi-linkage clustering. Information and links to other functional databases ENZYME (19), GO (Gene Ontology Consortium, 2000), KEGG (20), COG (21), SWISSPROT (22) are also included by BLASTing the sequences from each superfamily against sequences provided by these resources. Only 95% sequence identity hits, with an 80% residue overlap which were used to annotate sequences.
Recent analysis of structural and functional divergence in highly populated CATH superfamilies (>5 structural relatives with <35% sequence identity) has been undertaken using data from the DHS. The 2DSEC algorithm was used to analyse multiple structural alignments of families and identify highly conserved structural cores and secondary structure embellishments or decorations to the common core. In some large superfamilies, extensive embellishments were observed outside the core, and although these secondary structure insertions were frequently discontinuous in the protein chain, they were often co-located in 3D space (16). In many cases, manual inspection revealed that the embellishment had aggregated to form a larger structural feature that was modifying the active site of the domain or creating new surfaces for domain or protein interactions. Data collected in the DHS clearly shows a relationship between structural divergence within a superfamily, sequence divergence of this superfamily amongst predicted domains in the genomes and the number of distinct functional groups that can be identified for the superfamily (see Figure 4).
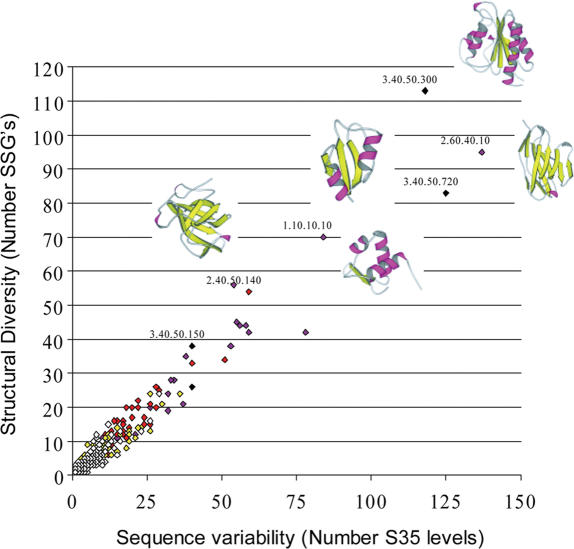
Figure 4.
Relationship between sequence variability, structural variability and functional diversity in CATH superfamilies. Structural variation in a CATH superfamily as measured by the number of diverse structural subgroups (SSAP score <80 between groups) is plotted against sequence diversity as measured by the number of sequence diverse subfamilies in the CATH-DHS (<35% sequence identity between groups). The colour of each point reflects the number of functions identified in that superfamily using GO as follows: white (0–25), yellow (26–50), red (51–100), maroon (101–200), black (200+).
LATERAL LINKS ACROSS THE CATH HIERARCHY TO CAPTURE EVOLUTIONARY DIVERGENCE AND EXPLORE THE STRUCTURAL CONTINUUM
Our analysis of structural divergence in CATH superfamilies (16) has revealed families where significant changes in the structures had occurred, in some cases 5-fold differences in the sizes of domains were identified and sometimes it was apparent that the ‘folds’ of these very diverse relatives had effectively changed. Therefore, in these superfamilies, more than one fold group can be identified, effectively breaking the hierarchical nature of the CATH classification which implies that each relative within a C.A.T.H. homologous superfamily should belong to the same C.A.T. fold group
In addition, an ‘all versus all’ HMM–HMM scan between all superfamily representatives revealed several cases of extremely remote homologues which had been classified into separate superfamilies and yet match with significant E-values. Structure comparison had failed to detect the relationship between these superfamilies because the structural divergence of the relatives was so extreme, sometimes constituting a change in architecture as well as fold group. In these cases homology was only suggested by the HMM-based scans and then manually validated by considering functional information and detailed evidence from literature. In order to capture information on these distant homologies, links have been created between the superfamilies both on our web pages and in the CATH database. The data can now be found as a link from the CATH homepage (http://www.cathdb.info).
In the near future, we also plan to provide web pages presenting cases of significant structural overlaps between superfamilies or fold groups. For these cases we are not currently able to find any additional evidence to support a distant evolutionary relationship and these examples highlight the recurrence of large structural motifs between some folds and the existence of a structural continuum in some regions of fold space.
ACCESSING CATH AND IMPROVEMENTS TO THE SERVER
The CATH database can be accessed at http://www.cathdb.info. The web interface may be browsed or alternatively searched with PDB codes or CATH domain identifiers. There is also a facility for keyword searches. With the version 3.0 release we now make the raw and processed data files available which include for example CATH domain PDB files, sequences, dssp files and they can be accessed through the CATH database main page. The Gene3D resource can be accessed through the CATH database or directly at http://www.cathdb.info/Gene3D. The DHS can be accessed through the CATH database or directly at http://www.cathdb.info/bsm/dhs.
Acknowledgments
We are grateful to Dr Janet Moloney, Dr Kanchan Phadwal, Dr Azara Janmohamed and Ms Elisabeth Rideal for valuable assistance with the domain boundary assignments and classification of domains in CATH (version 3.0). We acknowledge and thank the following for funding: L. Greene, I. Sillitoe and A. Cuff (MRC); M. Dibley (EU); T. Lewis (Wellcome Trust); C. Yeats (Biosapiens under the EU Framework Program 6); A. Reid and T. Dallman (BBSRC studentships); O. Redfern (EPSRC studentship); S. Addou (studentship from the Algerian government); R. Nambudiry (Argonne grant, USA). Funding to pay the Open Access publication charges for this article was provided by EU.
Conflict of interest statement. None declared.
REFERENCES
- 1.Todd A.E., Marsden R.L., Thornton J.M., Orengo C.A. Progress of structural genomics initiatives: an analysis of solved target structures. J. Mol. Biol. 2005;348:1235–1260. doi: 10.1016/j.jmb.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 2.Chandonia J.M., Brenner S.E. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 3.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearl F.M., Bennett C.F., Bray J.E., Harrison A.P., Martin N., Shepherd A., Sillitoe I., Thornton J., Orengo C.A. The CATH database: an extended protein family resource for structural and functional genomics. Nucleic Acids Res. 2003;31:452–455. doi: 10.1093/nar/gkg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearl F., Todd A., Sillitoe I., Dibley M., Redfern O., Lewis T., Bennett C., Marsden R., Grant A., Lee D., et al. The CATH domain structure database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:247–251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor W.R., Orengo C.A. Protein structure alignment. J. Mol. Biol. 1989;208:1–22. doi: 10.1016/0022-2836(89)90084-3. [DOI] [PubMed] [Google Scholar]
- 7.Orengo C.A., Taylor W.R. SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol. 1996;266:617–635. doi: 10.1016/s0076-6879(96)66038-8. [DOI] [PubMed] [Google Scholar]
- 8.Swindells M.B. A procedure for detecting structural domains in proteins. Protein Sci. 1995;4:103–12. doi: 10.1002/pro.5560040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holm L., Sander C. Parser for protein folding units. Proteins. 1994;19:256–268. doi: 10.1002/prot.340190309. [DOI] [PubMed] [Google Scholar]
- 10.Siddiqui A.S., Barton G.J. Continuous and discontinuous domains: an algorithm for the automatic generation of reliable protein domain definitions. Protein Sci. 1995;4:872–884. doi: 10.1002/pro.5560040507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karplus K., Barrett C., Hughey R. Hidden Markov models (HMMs) for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 12.Sillitoe I., Dibley M., Bray J., Addou S., Orengo C. Assessing strategies for improved superfamily recognition. Protein Sci. 2005;14:1800–1810. doi: 10.1110/ps.041056105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lord P.W., Stevens R.D., Brass A., Goble C.A. Semantic similarity measures as tools for exploring the gene ontology. Pac. Symp. Biocomput. 2003:601–612. doi: 10.1142/9789812776303_0056. [DOI] [PubMed] [Google Scholar]
- 14.Bray J.E., Todd A.E., Pearl F.M., Thornton J.M., Orengo C.A. The CATH Dictionary of Homologous Superfamilies (DHS): a consensus approach for identifying distant structural homologues. Protein Eng. 2000;13:153–165. doi: 10.1093/protein/13.3.153. [DOI] [PubMed] [Google Scholar]
- 15.Orengo C.A. CORA—topological fingerprints for protein structural families. Protein Sci. 1999;8:699–715. doi: 10.1110/ps.8.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves G.A., Dallman T.J., Redfern O.C., Akpor A., Orengo C.A. Structural diversity of domain superfamilies in the CATH database. J. Mol. Biol. 2006;360:725–41. doi: 10.1016/j.jmb.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 17.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Bairoch A., Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–8. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 22.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–53. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]