Abstract
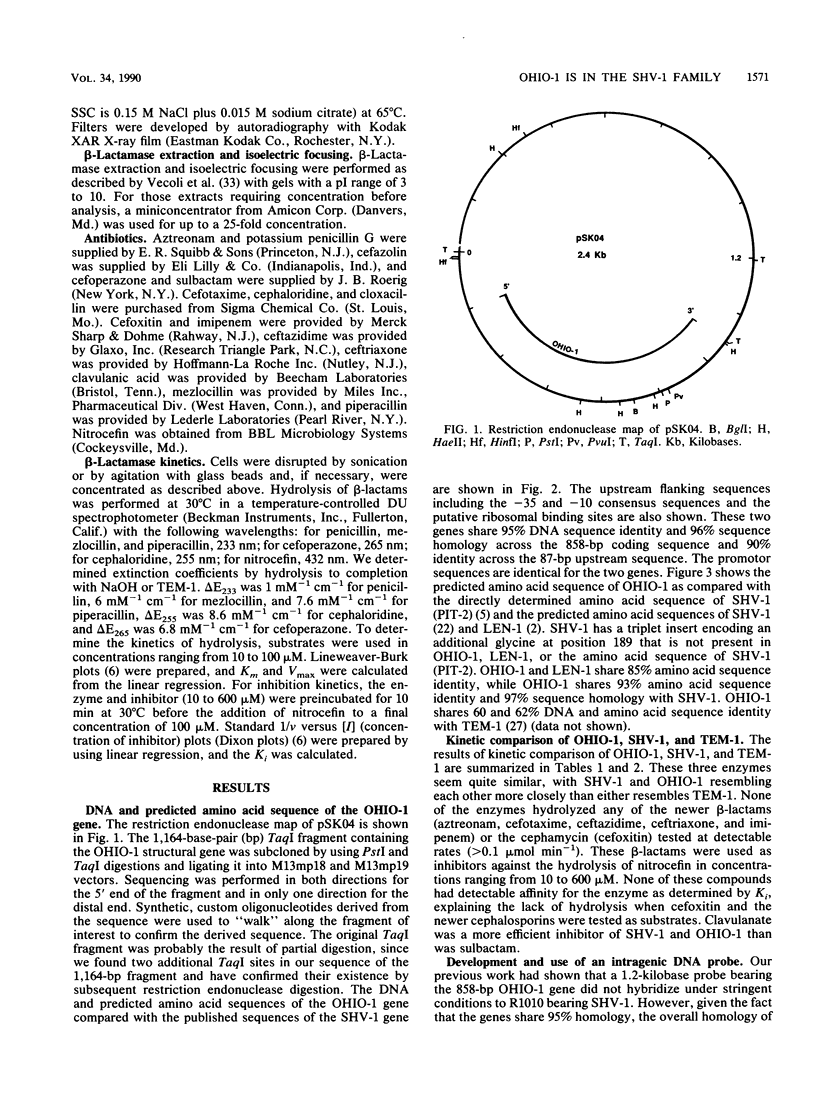
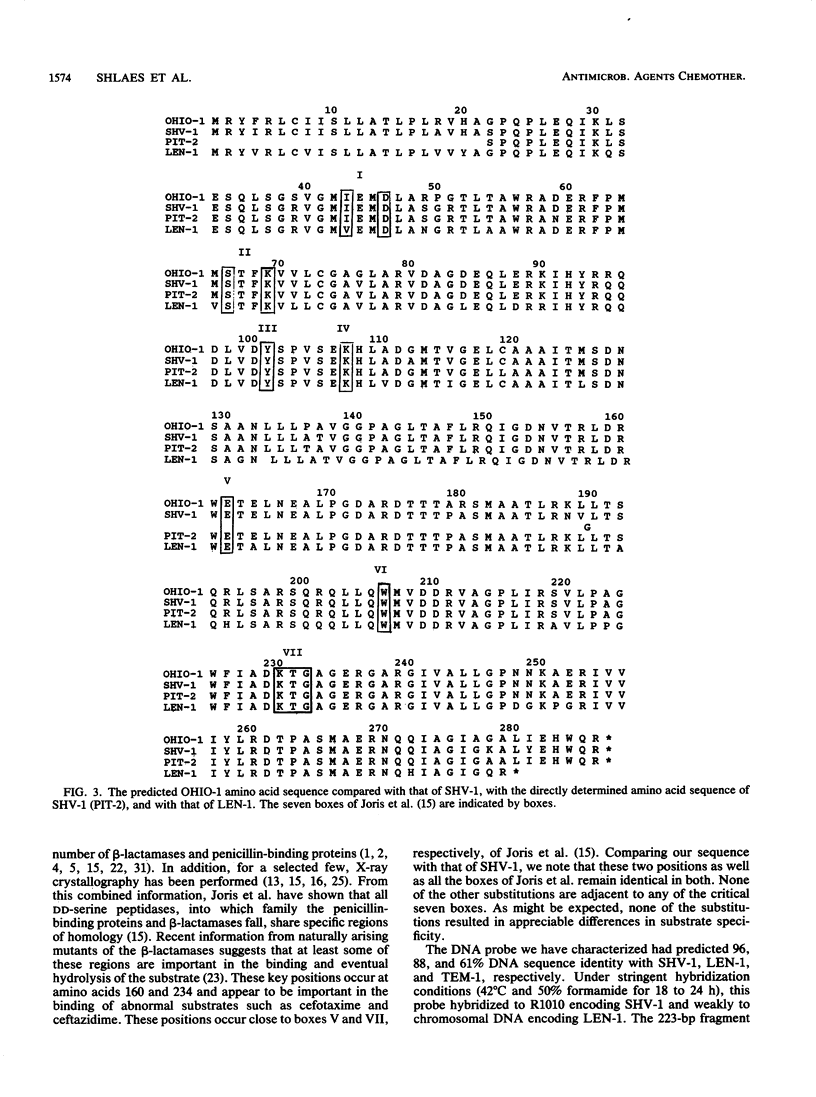
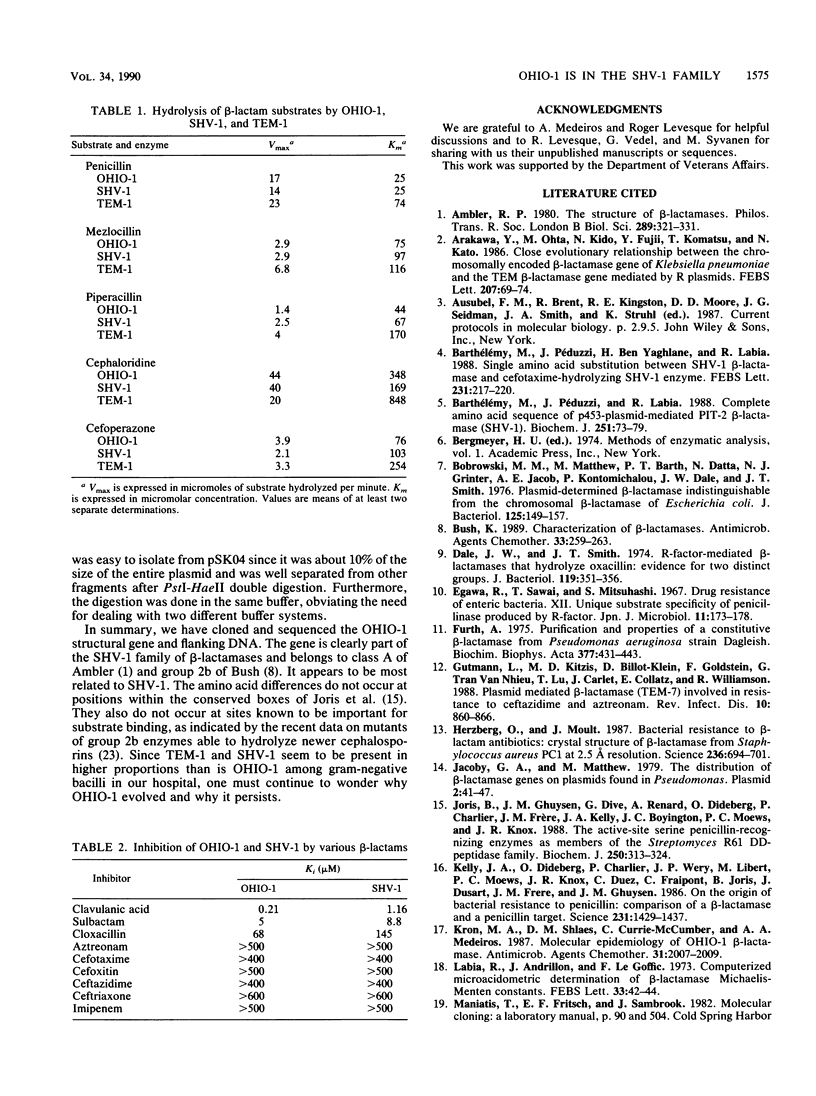
The OHIO-1 beta-lactamase gene was subcloned in a 1.16-kilobase TaqI fragment in the 2.4-kilobase chimeric plasmid pSK04. After directional subcloning into M13, the DNA sequence of this fragment was determined. The results showed an open reading frame of 858 base pairs (bp) encoding a protein of 286 amino acids. The structural gene showed 95, 87, and 60% DNA sequence identity with SHV-1, LEN-1, and TEM-1, respectively, and 93, 85, and 62% predicted amino acid sequence identity, respectively. The 87 bp upstream of the OHIO-1 structural gene had 96% identity with the upstream flanking sequence of SHV-1, including the -35 and -10 consensus sequences and the putative ribosomal binding site. A 223-bp DNA probe derived from a PstI-HaeII fragment in the C-terminal sequence of OHIO-1 had predicted 96, 88, and 61% sequence identity with SHV-1, LEN-1, and TEM-1, respectively. This probe hybridized to SHV-1 and poorly to LEN-1, but not to TEM-1 or a variety of other plasmid-mediated beta-lactamase genes, under stringent conditions. Screening of plasmid DNA derived from 40 ampicillin-resistant clinical isolates by Southern hybridization with the 223-bp probe uncovered no strains encoding OHIO-1. Isoelectric focusing of the same collection did identify two strains producing enzymes resembling SHV-1, however. We have also performed a kinetic comparison of OHIO-1, SHV-1, and TEM-1. OHIO-1 and SHV-1 were indistinguishable from each other but could be distinguished from TEM-1. These data clearly place OHIO-1 within the SHV-1 family of beta-lactamases.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Arakawa Y., Ohta M., Kido N., Fujii Y., Komatsu T., Kato N. Close evolutionary relationship between the chromosomally encoded beta-lactamase gene of Klebsiella pneumoniae and the TEM beta-lactamase gene mediated by R plasmids. FEBS Lett. 1986 Oct 20;207(1):69–74. doi: 10.1016/0014-5793(86)80014-x. [DOI] [PubMed] [Google Scholar]
- Barthélémy M., Peduzzi J., Labia R. Complete amino acid sequence of p453-plasmid-mediated PIT-2 beta-lactamase (SHV-1). Biochem J. 1988 Apr 1;251(1):73–79. doi: 10.1042/bj2510073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélémy M., Péduzzi J., Ben Yaghlane H., Labia R. Single amino acid substitution between SHV-1 beta-lactamase and cefotaxime-hydrolyzing SHV-2 enzyme. FEBS Lett. 1988 Apr 11;231(1):217–220. doi: 10.1016/0014-5793(88)80734-8. [DOI] [PubMed] [Google Scholar]
- Bobrowski M. M., Matthew M., Barth P. T., Datta N., Grinter N. J., Jacob A. E., Kontomichalou P., Dale J. W., Smith J. T. Plasmid-determined beta-lactamase indistinguishable from the chromosomal beta-lactamase of Escherichia coli. J Bacteriol. 1976 Jan;125(1):149–157. doi: 10.1128/jb.125.1.149-157.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Characterization of beta-lactamases. Antimicrob Agents Chemother. 1989 Mar;33(3):259–263. doi: 10.1128/aac.33.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J. W., Smith J. T. R-factor-mediated beta-lactamases that hydrolyze oxacillin: evidence for two distinct groups. J Bacteriol. 1974 Aug;119(2):351–356. doi: 10.1128/jb.119.2.351-356.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth A. J. Purification and properties of a constitutive beta-lactamase from Pseudomonas aeruginosa strain Dalgleish. Biochim Biophys Acta. 1975 Feb 19;377(2):431–443. doi: 10.1016/0005-2744(75)90323-x. [DOI] [PubMed] [Google Scholar]
- Gutmann L., Kitzis M. D., Billot-Klein D., Goldstein F., Tran Van Nhieu G., Lu T., Carlet J., Collatz E., Williamson R. Plasmid-mediated beta-lactamase (TEM-7) involved in resistance to ceftazidime and aztreonam. Rev Infect Dis. 1988 Jul-Aug;10(4):860–866. doi: 10.1093/clinids/10.4.860. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 A resolution. Science. 1987 May 8;236(4802):694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A., Matthew M. The distribution of beta-lactamase genes on plasmids found in Pseudomonas. Plasmid. 1979 Jan;2(1):41–47. doi: 10.1016/0147-619x(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Joris B., Ghuysen J. M., Dive G., Renard A., Dideberg O., Charlier P., Frère J. M., Kelly J. A., Boyington J. C., Moews P. C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988 Mar 1;250(2):313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. A., Dideberg O., Charlier P., Wery J. P., Libert M., Moews P. C., Knox J. R., Duez C., Fraipont C., Joris B. On the origin of bacterial resistance to penicillin: comparison of a beta-lactamase and a penicillin target. Science. 1986 Mar 21;231(4744):1429–1431. doi: 10.1126/science.3082007. [DOI] [PubMed] [Google Scholar]
- Kron M. A., Shlaes D. M., Currie-McCumber C., Medeiros A. A. Molecular epidemiology of OHIO-1 beta-lactamase. Antimicrob Agents Chemother. 1987 Dec;31(12):2007–2009. doi: 10.1128/aac.31.12.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labia R., Andrillon J., Le Goffic F. Computerized microacidimetric determination of beta lactamase Michaelis-Menten constants. FEBS Lett. 1973 Jun 15;33(1):42–44. doi: 10.1016/0014-5793(73)80154-1. [DOI] [PubMed] [Google Scholar]
- Matthew M., Sykes R. B. Properties of the beta-lactamase specified by the Pseudomonas plasmid RPL11. J Bacteriol. 1977 Oct;132(1):341–345. doi: 10.1128/jb.132.1.341-345.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros A. A., Cohenford M., Jacoby G. A. Five novel plasmid-determined beta-lactamases. Antimicrob Agents Chemother. 1985 May;27(5):715–719. doi: 10.1128/aac.27.5.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier J., Levesque R. C. Cloning of SHV-2, OHIO-1, and OXA-6 beta-lactamases and cloning and sequencing of SHV-1 beta-lactamase. Antimicrob Agents Chemother. 1990 Aug;34(8):1577–1583. doi: 10.1128/aac.34.8.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A., Labia R., Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989 Aug;33(8):1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin L. G., Medeiros A. A., Yolken R. H., Moxon E. R. Ampicillin treatment failure of apparently beta-lactamase-negative Haemophilus influenzae type b meningitis due to novel beta-lactamase. Lancet. 1981 Nov 7;2(8254):1008–1010. doi: 10.1016/s0140-6736(81)91214-9. [DOI] [PubMed] [Google Scholar]
- Samraoui B., Sutton B. J., Todd R. J., Artymiuk P. J., Waley S. G., Phillips D. C. Tertiary structural similarity between a class A beta-lactamase and a penicillin-sensitive D-alanyl carboxypeptidase-transpeptidase. 1986 Mar 27-Apr 2Nature. 320(6060):378–380. doi: 10.1038/320378a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlaes D. M., Currie-McCumber C. A., Lehman M. H. Introduction of a plasmid encoding the OHIO-1 beta-lactamase to an intermediate care ward by patient transfer. Infect Control Hosp Epidemiol. 1988 Jul;9(7):317–319. doi: 10.1086/645861. [DOI] [PubMed] [Google Scholar]
- Shlaes D. M., Medeiros A. A., Kron M. A., Currie-McCumber C., Papa E., Vartian C. V. Novel plasmid-mediated beta-lactamase in members of the family Enterobacteriaceae from Ohio. Antimicrob Agents Chemother. 1986 Aug;30(2):220–224. doi: 10.1128/aac.30.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi I., Tsukamoto K., Harada M., Sawai T. Carbenicillin-hydrolyzing penicillinases of Proteus mirabilis and the PSE-type penicillinases of Pseudomonas aeruginosa. Microbiol Immunol. 1983;27(12):995–1004. doi: 10.1111/j.1348-0421.1983.tb02934.x. [DOI] [PubMed] [Google Scholar]
- Vecoli C., Prevost F. E., Ververis J. J., Medeiros A. A., O'Leary G. P., Jr Comparison of polyacrylamide and agarose gel thin-layer isoelectric focusing for the characterization of beta-lactamases. Antimicrob Agents Chemother. 1983 Aug;24(2):186–189. doi: 10.1128/aac.24.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedel G., Paul G., Picard B., Philippon A. Biochemical, immunological and physicochemical comparisons between OHIO-1 and four SHV-type beta-lactamases. FEMS Microbiol Lett. 1989 Nov;53(1-2):5–9. doi: 10.1016/0378-1097(89)90357-1. [DOI] [PubMed] [Google Scholar]


