Abstract
The Mediator complex forms the bridge between transcriptional activators and RNA polymerase II. Mediator subunit Med1/TRAP220 is a key component of Mediator originally found to associate with nuclear hormone receptors. Med1 deficiency causes lethality at embryonic day 11.5 because of defects in heart and placenta development. Here we show that Med1-deficient 10.5 days postcoitum embryos are anemic but have normal numbers of hematopoietic progenitor cells. Med1-deficient progenitor cells have a defect in forming erythroid burst-forming units (BFU-E) and colony-forming units (CFU-E), but not in forming myeloid colonies. At the molecular level, we demonstrate that Med1 interacts physically with the erythroid master regulator GATA-1. In transcription assays, Med1 deficiency leads to a defect in GATA-1-mediated transactivation. In chromatin immunoprecipitation experiments, we find Mediator components at GATA-1-occupied enhancer sites. Thus, we conclude that Mediator subunit Med1 acts as a pivotal coactivator for GATA-1 in erythroid development.
Keywords: Mediator of transcriptional regulation, erythroid progenitors
Hematopoietic development is regulated in large part by nuclear proteins that activate or repress sets of genes characteristic of distinct lineages. Studies of gene expression have formed the conceptual framework for investigating the transcriptional control of erythropoiesis (1, 2). Among erythroid transcription factors, GATA-1 is of particular interest, because it is essential for erythropoiesis (3). The Mediator complex is the key coactivator for many transcription factors (reviewed in refs. 4–8). The interaction of activators with Mediator stimulates preinitiation complex assembly on promoter DNA. Mediator (TRAP/SMCC/DRIP/CRSP/NAT/ARC/ PC2) is a large (22- to 28-subunit) protein complex that binds RNA polymerase II (polII) and controls transcription from class II genes. It serves as an interface for regulatory factors and the polII. Substantial evidence argues that Mediator activates transcription, at least in part, by direct interactions with DNA-binding transcriptional activators bound at upstream promoter elements and enhancers with polII and one or more of the general initiation factors bound at the core promoter. Notably, different Mediator subunits are targets for interactions with the transcriptional activation domains of different DNA-binding transcriptional activators (4, 7, 9). As a consequence, it is of great interest to identify subunits that serve as contact sites for the large collection of DNA-binding transcriptional activators. Prominent mammalian activators that interact with Mediator are the hormone receptors. For example, Med1/TRAP220 was shown to be required for the transcription of nuclear hormone receptor and PPARγ target genes but not for p53 or VP16 activated transcription (10, 11). In another study, gene targeting of Med23 revealed an essential role in MAP kinase signaling through Elk-1, as well as for activation by the adenoviral E1A protein (12).
Med1/TRAP220 is the main Mediator subunit target for nuclear receptors such as the thyroid hormone, estrogen, vitamin D, and retinoic acid receptors (6). These receptors bind to and strongly rely on Mediator for their function. Recently, it was shown that Med1 exists predominantly in a Mediator subpopulation enriched in polII (13). Previously, it was suggested that Med1 interacts not only with hormone receptors but also with GATA factors (14). These data are supported by in vitro studies with Pit-1, GATA-2, and Med1 at the TSHβ promoter (15). Med1 deficiency leads to death at embryonic day 11.5. Although Med1 is not required for cell division, Med1 is critical for embryonic development of placental and cardiac development (10, 14, 16).
Generation of mature red blood cells is a tightly regulated, multistep process. Erythroid cells derive from hematopoietic stem cells (Fig. 1B). The first specific progenitors to be formed are referred to as erythroid burst-forming units (BFU-Es) (17, 18). More committed erythroid precursor cells give rise to erythroid colony-forming units (CFU-Es) and can be prospectively isolated (19). Later on in erythroid differentiation, erythroblasts extrude their nuclei and become reticulocytes, which further loose specific organelles to generate the fully mature circulating erythrocytes.
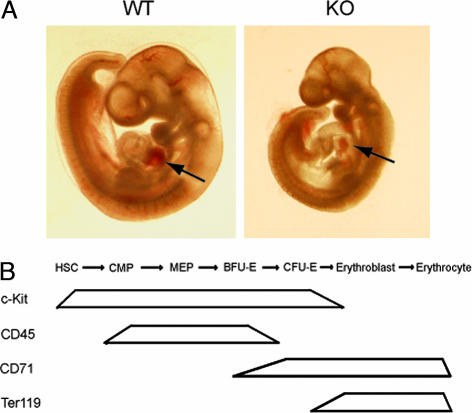
Fig. 1.
Med1-deficient embryos are pale (anemic). (A) Med1-deficient (KO) but not wild type (WT) are pale, in particular in the heart region (see arrows). (B) Expression of markers during erythroid development; HSC, hematopoietic stem cell; CMP, common myeloid progenitor; MEP, megakaryocyte-erythrocyte progenitor. Expression of markers during erythroid development is shown.
Erythrocyte differentiation is among the best-studied model systems used to define the mechanisms responsible for lineage commitment, cellular differentiation, and tissue-specific gene expression (reviewed in refs. 1, 2, and 20). The lineage-restricted transcription factor GATA-1 orchestrates several key aspects of erythroid development. Binding sites for GATA-1 are found in the regulatory regions of virtually all erythroid-cell-specific genes (21). Mice deficient for GATA-1 succumb to fatal anemia because of an inability of erythroid precursor cells to survive and mature (22, 23). Efforts to understand GATA-1 functions have identified a number of protein interactions with transcription factors or cofactors such as TAL-1, EKLF, PU.1, Sp1, and FOG-1 (reviewed in refs. 1 and 20). GATA-1 is also reported to associate with chromatin remodeling proteins, including the CBP/p300 histone acetyltransferase and the SWI/SNF chromatin remodeling complex (24, 25). GATA-1 is able to act not only as an activator but also as a repressor (26, 27). Recently, a corepressor complex that can associate with GATA-1 by FOG-1 has been identified (27, 28).
This study describes the role of the Mediator subunit Med1/TRAP220 in erythropoiesis. Med1 deficiency leads to a defect in formation of erythroid colonies in methylcellulose assays. Med1 physically interacts with master regulator GATA-1 and GATA-1-dependent transcription is impaired in the absence of Med1. In addition, Mediator components are found in vivo at GATA-1-occupied enhancer sites. Therefore, we conclude that the Mediator complex, in particular subunit Med1, plays a pivotal role in GATA-1-mediated transcription.
Results
Med1/TRAP220 Day 10.5 Embryos Are Pale but Have a Normal Number of c-Kit+ Progenitor Cells.
Previous studies have shown that ablation of Mediator subunit Med1 leads to embryonic death at day 11.5, which is caused by defects in placental and cardiac development (10, 16, 29). Here, we show that at day 10.5, Med1-deficient embryos are phenotypical pale compared with wild-type embryos, especially visible in the heart region (Fig. 1A). That prompted us to examine erythrocyte development in detail (Fig. 1B). To exclude that this is due to a lack of stem cells in Med1-deficient embryos, we performed FACS analysis with the stem/progenitor marker c-Kit using yolk sac, aorta-gonads mesonephros (AGM) and fetal blood cells of 10.5 days postcoitum (dpc) embryos. In all three locations, c-Kit+ progenitor cells were found in equal numbers, comparing both wild-type and Med1-deficient embryos (Fig. 2B and data not shown). In addition Med1 mRNA expression level remains constant during erythropoiesis as checked by RT-PCR (data not shown).
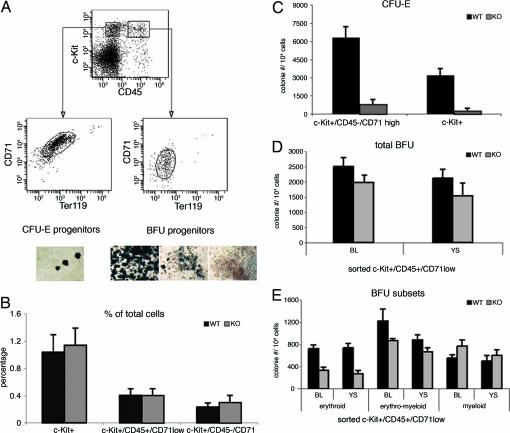
Fig. 2.
CFU-E and BFU colony-forming potential and morphology of embryonic 10.5-dpc erythroid progenitor populations. (A) Day 10.5 embryonic blood, AGM or yolk sac was analyzed with the markers c-Kit, CD45, Ter119, and CD71. C-Kit+/CD45−/Ter119−/CD71high give rise to CFU-Es, and c-Kit+/CD45+/Ter119−/CD71low cells give rise to BFU colonies. Embryonic precursor cells from A were sorted and plated into methylcellulose for colony formation of either CFU-E or BFU. Colonies were stained with benzidine in order to look at the morphology and to measure the potential for BFU and CFU-E (A Lower). For BFU colonies, three different colony types were identified: erythroid, blue; erythroid/myeloid, mixed; myeloid, white. (B) Med1-deficient embryos (10.5 dpc) have the same amount of c-Kit+ progenitor cells and progenitors that give rise to BFU (CD45+/CD71low) and CFU-E colonies (CD45−/CD71high). (C) Sorted Med1-deficient progenitors (c-Kit+/CD45−/Ter119−/CD71high or simply c-Kit+) yield 10-fold less CFU-E colonies compared with wild type. (D) Total BFU numbers of sorted Med1-deficient progenitors cells are only slightly reduced compared with wild type. (E) Sorted Med1-deficient progenitors (c-Kit+/CD45+/Ter119−/CD71low) show a 2- to 3-fold reduction in forming BFU-E colonies. The levels of erythroid/myeloid and myeloid colonies are normal.
Analysis of Fetal Day 10.5 Precursor Cells by Flow Cytometry and Methylcellulose Assays.
To investigate erythroid development in Med1-deficient embryos, we first established a FACS scheme to analyze and sort erythroid progenitor cells for functional methylcellulose assays. For the FACS-scheme erythroid-specific marker TER119 and transferrin receptor (CD71) were used, as described for definitive erythropoiesis (30). To identify and sort progenitors from day 10.5 embryos, we used in addition to Ter119 and CD71 stem cell factor receptor c-Kit and panleukocyte marker CD45, which was found to be present also in the stem cell compartment (31). An expression profile of these markers during erythroid development is depicted in Fig. 1C. In the fetal blood of day 10.5-dpc embryos, there are ≈1% of c-Kit+ progenitor cells (Fig. 2A), which can be separated further in CD45+ and CD45- fractions. The sorted c-Kit+/CD45+ cells, staining negative for Ter119 and low for CD71, gave rise predominantly to BFU colonies and not CFU-Es [supporting information (SI) Fig. 6]. The c-Kit+/CD45- population, low for Ter119 and high for CD71, gave rise to CFU-Es but scarcely to BFU colonies (SI Fig. 6). Using this scheme, we performed single-cell sorts into 96-well plates of the different cell populations mentioned above and assayed them for colony formation. Although the precursor cells are present in both Med1-deficient and wild-type embryos (Fig. 2 B and D), we observed a 10× reduction of CFU-E formation in the Med1-deficient cells (Fig. 2C). For the BFU colonies, formation of myeloid and erythromyeloid colonies was normal, whereas formation of BFU-Es was 2- to 3-fold reduced (Fig. 2E). The absolute numbers of colonies formed from wild type with Med1-deficient progenitors can be seen in SI Table 1. As an aside, we note that the efficiency of colony formation using embryonic c-Kit+ cells is markedly higher than using cells from bone marrow (data not shown).
Med1 Physically Interacts with Erythroid Master Regulator GATA-1.
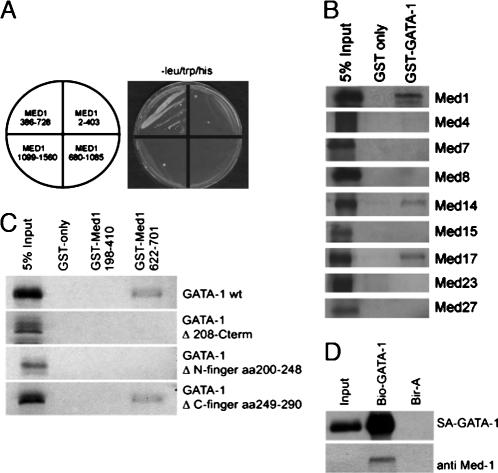
Next, we sought to investigate the molecular basis for the defective erythropoiesis in Med1-deficient embryos. Med1 is part of the Mediator complex that has been shown to interact with ligand-bound hormone receptor (reviewed in refs. 4 and 6). Recently, two groups described an interaction between GATA factors and Med1 (14, 32). Because GATA-1 is a master regulator in erythropoiesis, we investigated whether GATA-1 and Med1 can physically interact. To address this question, we used the yeast two-hybrid assay with four different regions of Med1 (amino acids 2–403, 386–728, 680-1085, and 1099–1560). Med1 fragments were fused to the Gal4 DNA-binding domain (bait), and GATA-1 was fused to the Gal4-activation domain (prey). Only amino acids 386–728 of Med1 were able to interact with GATA-1 (Fig. 3A). We verified these results using a GST-pulldown assays (SI Fig. 7). In vitro-transcribed and -translated GATA-1 was offered to interact with different regions of Med1 fused to GST and immobilized on glutathione beads. We could narrow down the interaction region of Med1 to amino acids 622–701 (SI Fig. 7A). In a modified GST-pulldown assay offering GST-GATA-1 and either HeLa nuclear extract (SI Fig. 7B) or affinity-purified Mediator only (SI Fig. 7C), endogenous Mediator binds to GATA-1. In further GST-pulldown assays using GST-GATA-1 and different in vitro synthesized Mediator subunits, not only Med1 but also Med17/TRAP80 and Med14/Rgr1/TRAP170 was found to interact with GATA-1 (Fig. 3B). In contrast, Med4, Med7, Med8, Med15, Med23, and Med27 did not interact with GST-GATA-1. These results suggest that GATA-1 contacts not only one but at least three different Mediator subunits. GATA-1 harbors two zinc-finger domains; the C-terminal zinc finger is required for the binding to DNA, and the N-terminal zinc finger known to interact with FOG-1 and GATA-1 itself. Using GATA-1 mutants (delta N-terminal zinc finger, delta C-terminal zinc finger, and the C-terminal truncation lacking both the N- and C-terminal zinc fingers), we could show that the N- but not the C-terminal zinc finger of GATA-1 is required for interaction with Med1 (Fig. 3C).
Fig. 3.
Mediator interacts with GATA-1 directly. (A) Yeast two-hybrid assay using different regions of Med1 as bait (fused to the Gal4 DNA-binding domain) and GATA-1 as prey (fused to Gal4 activation domain); only Med1 region 386–728 drives the expression of the two different reporter genes. (B) GST-pulldown assay with GATA-1 as bait (fused to GST) and in vitro transcribed/translated Mediator subunits; GATA-1interacts with Med1, Med14, and Med17. (C) GST-pulldown assay using different Med1 regions (fused to GST) and different in vitro transcribed/translated GATA-1 mutants; the GATA-1 N-terminal- but not the C-terminal zinc finger is required for interaction with Med1. (D) Biotinylation-tagging approach using in vivo biotin-tagged GATA-1 in mouse erythroid leukemia cells. Med1 (and bio-GATA-1) are precipitated with streptavidin beads only in cells expressing biotin-tagged GATA-1 (lane 2) but not in control cells expressing only BirA (lane 3).
To investigate the association of endogenous Mediator with GATA-1 in vivo, we used the biotinylation tagging approach in erythroid cells (ref. 40; Fig. 3D). Input and pulled-down proteins were analyzed by Western blot for the presence of Med1, GATA-1 (positive control), and DEK (negative control). Only GATA-1 and Med1 but not DEK are present in the pulled-down fraction (Fig. 3D, lane 2, and data not shown).
Mediator Subunit Med1 Is Required for Transcriptional Activation by GATA-1.
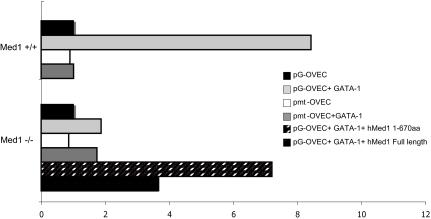
We next tested whether the Mediator subunit is required for GATA-1 transcriptional activation at endogenous promoters and in a GATA-1 reporter assay (Fig. 4). A real-time PCR-based reporter assay was used with the reporter plasmid pG-OVEC containing four copies of an optimal GATA-1-binding site (34) transfected into mouse embryonic fibroblasts (MEFs) together with (or without) GATA-1. A mutated plasmid (pmt-OVEC), to which GATA-1 can no longer bind, was used as a negative control (35). The GATA-responsive promoter was activated only if GATA-1 was cotransfected in both Med1+/+ (10-fold) and but not in Med1−/− MEFs (Fig. 4). Importantly, GATA-1 transactivation function in Med1−/− MEFs was rescued by expression of either the full length human Med1 or the Med1 amino acids 1–670. The expression of the rabbit β-globin expression was increased up to 4-fold. The mutated pmtOVEC control plasmid was not activated in either of the different MEFs. Using the same reporter assay as in Fig. 4, different GATA-1 constructs were tested. The deletion of the N-terminal zinc finger, shown to interact with Med1, leads to a complete abrogation of rabbit β-globin expression (SI Fig. 8A). The same is true for the C-terminal zinc-finger deletion mutant or C-terminal truncation mutant (Delta 208-end) lacking both N- and C-terminal zinc fingers.
Fig. 4.
Mediator subunit Med1 is required for the transcriptional activation by GATA-1. Real-time PCR transcription assays in transfected MEFs. Cotransfection of β-Gal was used to normalize transfection efficiency. The x axis represents the relative fold activation of the rabbit globin reporter gene. GATA-1 activates the transcription of wild-type pG-OVEC in Med1+/+ MEFs but not in Med1−/− MEFs. Either full length or amino acids 1–670 of human Med1 (hMed1) are able to reconstitute transcriptional activation to ≈50% of wild-type level. Mutated minimal promoter transcription reporter plasmid [pOVEC (pmt-OVEC)], containing mutated GATA-1-binding sites shows no activation in the different MEFs.
To look at endogenous expression of GATA-1 target genes in Med1-deficient embryos, we sorted c-Kit+ progenitor cells from the fetal blood of day 10.5 embryos (SI Fig. 8B). As expected, the Med1 transcript is not present in the Med1-deficient embryos, whereas hypoxanthine phosphoribosyltransferase and c-Kit transcript levels are comparable with wild-type level. GATA-1 is known to positively autoregulate its own expression during erythroid differentiation (33). Therefore, we observe not only a severe reduction of GATA-1 target genes βH1 globin and β-major globin but also a strong reduction in GATA-1 expression level.
Mediator Colocalizes with GATA-1 Regulatory Sequences in Vivo.
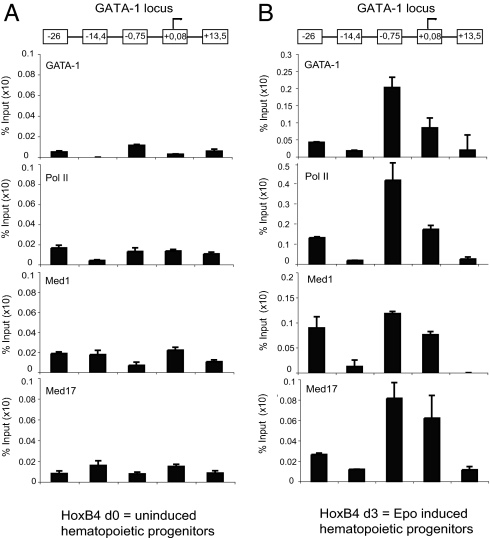
To investigate whether the (activating) Med1-GATA-1 interaction correlates with colocalization in vivo, ChIP experiments were performed (Fig. 5). First, we used bone marrow progenitor cells immortalized by transduction with HoxB4 (D.v.E., unpublished work) that were either noninduced (Fig. 5A and SI Fig. 9B) or induced with erythropoietin for 3 days (Fig. 5B and SI Fig. 9B). Preliminary FACS and RT-PCR experiments suggest that these cells correspond to early committed erythroid progenitor cells. GATA-1 is specifically found to bind at enhancer elements −26 and −0.75 kb with respect to the start of transcription in the GATA-1 locus. Both polII and Mediator (subunits Med1 and Med17) are found to be present to −0.75 and +0.08 kb but not at control sequences −14.4 and +13.5 kb. Furthermore, in SI Fig. 9A, similar ChIP experiments were performed by using erythroid cells isolated from the spleen of phenylhydrazine-treated mice (36). Erythroid (Ter119+) cells were first enriched by depleting all other cell types with a mix of lineage markers. Strikingly, the overall ChIP profile of GATA-1, Med components, and polII looks similar to the profile obtained in erythropoietin-induced progenitor cells (Fig. 5B). Similarly, ChIP experiments looking at the GATA-1 target gene β-globin show that Mediator subunits Med1 and Med17 as well as polII occupancy correlate with the presence of GATA-1 (SI Fig. 9B).
Fig. 5.
ChIP experiments reveal Mediator and polII recruitment to GATA-1 enhancer elements in vivo. ChIP analysis was performed with hematopoietic progenitor cells either not induced (A) or induced with erythropoietin for 3 days (B; both images show the GATA-1 locus). Mediator subunits Med1, Med17, and polII are found only in committed erythroid precursors (B) at GATA-1 enhancer elements −0.75, +0.08, and −26 kb but not to −14.4 and +13.5 kb. The y axis represents the specific enrichment for each given antibody, in which the value of serum control was removed. Antibodies used were GATA-1 (N6); Med1/TRAP220 (M255), Med17/CRSP77 (G17), and Pol II, H-224 (all from Santa Cruz Biotechnology).
Discussion
Because Med1 deficiency leads to a lack of erythroid precursors, we investigated whether Mediator subunit Med1 can directly interact with master regulator GATA-1 (Fig. 3). In yeast two-hybrid and GST-pulldown assays, Med1 and GATA-1 interaction was clearly demonstrated and mapped. Interestingly, not only Med1 but also Med14 and Med17 interact with GATA-1. Other groups reported similar findings for activator–Mediator interactions, such as hormone receptor (Med1, Med14, and Med24), VP16 (Med25 and Med17), p53 (Med1 and Med17), and STAT2 (Med14 and Med17; summarized in ref. 4). Therefore, it is a common theme that one activator contacts multiple Mediator subunits. Strikingly, often the same Mediator subunits (Med1, Med14, and Med17) are described as the Mediator interaction surface. It is likely that these three subunits are positioned on the surface of Mediator and have evolved to form physical contacts with diverse set of transcriptional regulators. Because Med1 interacts with the same region in GATA-1 like FOG-1 and GATA-1 itself (reviewed in ref. 20), it will be interesting to find out whether Mediator binding can compete for the binding of these factors to GATA-1.
Using a reporter assay in MEFs, Med1 is clearly an essential coactivator for GATA-1 (Fig. 4). Interestingly, the dose of Med1 seems to be limiting, because heterozygous MEFs display only half the amount of transcriptional activation compared with wild-type MEFs (data not shown). In the Med1 rescue experiments, the N-terminal part of Med1 is surprisingly more effective than full length Med1 in rescuing transcriptional activation. The explanation for this difference might be because of the higher expression level of the N-terminal part (data not shown).
ChIP experiments were performed to verify whether the physical interaction between Med1 and GATA-1 also takes place in vivo (Fig. 5 and SI Fig. 9A). GATA-1 is able to stimulate its own GATA-1 promoter activity (33); the GATA-1-binding sites could be systematically mapped on the GATA-1 gene locus (37). We confirmed the described GATA recruitment to GATA-1 gene enhancer elements and found a clear correlation with the presence of Mediator subunits Med1 and Med17, as well as polII. Occupancy of GATA-1, Mediator, and polII is observed at element −0.75 and +0.08 kb close to the promoter and, to a lesser extent, far upstream at −26 kb.
Preliminary data indicate that Mediator is also found at the GATA-2 promoter where GATA-1 is known to repress transcription (26). The putative dual role of Mediator in activation and repression needs further investigation. Furthermore, our data reveal recruitment of polII to an enhancer far away from the core promoter. Similarly, it has been demonstrated that polII is recruited to the β-globin locus control region, and that this occupancy depends at least in part on the presence of GATA-1 (38). Further support for polII occupancy at enhancers has been described for the androgen-specific gene PSA and for the TCR-β locus, both supported by in vivo footprinting and ChIP experiments (reviewed in ref. 39).
We have identified a specific defect in the erythropoiesis of Mediator subunit Med1-deficient embryos at the CFU-E and also the BFU-E stage. We demonstrate that Mediator can physically interact with erythroid master regulator GATA-1 and is localized at the GATA-1 enhancer sites in erythroid cells by using ChIP experiments. In Med1-deficient cells, GATA-1-dependent transcriptional activation is defective in both reporter assays and endogenous GATA-1 target genes. Therefore, we conclude that Mediator subunit Med1 is crucial for erythropoiesis because it acts as a cofactor for master regulator GATA-1. In the future, conditional Med1-deficient mice will be instrumental in analyzing all hematopoietic lineages.
Materials and Methods
Animals and Tissue Preparation.
Med1/TRAP220 heterozygous deficient mice were generated by Roeder and colleagues (10). Embryos at 10.5 dpc, staged by somite counting, were dissected and genotyped by PCR. Embryonic yolk sac and AGM were isolated and digested with 0,1% collagenase D and DNase I in PBS/5% FCS at room temperature over 2 h. The cell suspension was filtered and stored on ice until further use. Embryonic blood was collected over 1 h by bleeding the headless embryo in PBS/5% FCS on ice.
Flow Cytometry Analysis and Cell Sorting.
Isolated YS, AGM, and embryonic blood cells were blocked with 10 μg of rat IgG antibody in PBS/5% FCS (Jackson ImmunoResearch, West Grove, PA) over 15 min on ice. Cells were washed once with PBS/FCS and stained with antibodies against c-Kit, CD45, CD71, and Ter119 (PharMingen, San Diego, CA) in PBS/FCS over 1 h on ice. Flow cytometry analysis and cell sorting were performed by using BD FACS Aria (Beckton Dickinson, Franklin Lakes, NJ). A single-cell sort of c-Kit+/CD45+/CD71−/Ter119− and c-Kit+/CD45−/CD71+/Ter119− cells into 96-well plates containing methylcellulose and growth factors was performed to investigate colony-forming ability.
Methylcellulose Assays.
Cells obtained as described above were cultured in methylcellulose (M3334; Stem Cell Technologies, Vancouver, BC, Canada) to investigate CFU-E and BFU colony-forming capacity. CFU-E assays were supplemented with 2 units erythropoietin (Epo)/ml and incubated over 2 days at 37°C with 5% CO2. BFU assays were performed by adding 2 units of Epo/ml and IL-3 (supernatant) over 8 days at 37°C with 5% CO2. The cultures were stained with benzidine (12% acetic acid containing 0.4% benzidine and 0.3% H2O2) to indicate the amount of erythroid colonies.
GST-Pulldown and Biotinylation-Tagging Approaches.
GST pulldown assays were performed as described in ref. 41 (see also SI Text). Mouse erythroid leukemia cells expressing bacterial BirA biotin ligase as well as tagged GATA-1 were used for the purification as described (ref. 27; see also SI Text).
Transcription Assays.
MEFs (see SI Text) were transiently transfected with 10 μg of plasmid DNA in total by using the Amaxa (Cologne, Germany) MEF transfection kit. A CMV-β-Gal plasmid was included as transfection efficiency control. Plasmids used were: WT (pGOVEC) or mutated (pmtOVEC) plasmids containing a rabbit globin reporter (kindly provided by J. Strouboulis, Erasmus Medical Center, Rotterdam, The Netherlands); GATA-1 cDNA cloned into pcDNA 3.1; pcDNA-FLAG-human Med1 full length; pMSCV human Med1 amino acids 1–670 (kindly provided by K. Ge, The Rockefeller University); GATA-1 D amino acids 308-C term; GATA-1 D N-finger amino acids 200–248; and GATA-1 D C-finger amino acids 249–290 (kindly provided by G. Blobel, Pennsylvania School of Medicine, Philadelphia, PA). Transcription was assayed by real-time PCR with primers for exon 2 of the reporter gene.
ChIP Assays.
In the ChIP assays, we used either uninduced or HoxB4 cells (see below) induced with 3 units/ml EPO over 3 days. Chromatin preparation by using 1 × 107 of the indicated cells was done with 0.4% formaldehyde for 10 min at room temperature. Sonication to ≈500-bp fragments and immunoprecipitations was performed as described in the Upstate Biotechnology (Lake Placid, NY) protocol. For the GATA-1 ChIP, preclearing of the chromatin was done with Sepharose-G beads overnight at 4°C, and the GATA-1 protein-DNA complexes were immunoprecipitated for 3 h in an additional step using an AffiniPure rabbit anti-rat antibody (Jackson ImmunoResearch, West Grove, PA). Antibodies were used as follows: GATA-1, N6; TRAP220, M-255; TRAP80 (CRSP77), G17; and Pol II, H-224 (all from Santa Cruz Biotechnology, Santa Cruz, CA).
Cell Culture.
HoxB4 cells were cultured in SF-Medium [IMDM with l-glutamin and 25 mM Hepes supplemented with 2% FCS/300 μg/ml Primatone RL (Sigma, St. Louis, MO)/5 mg/ml insulin (Sigma)/1× Pen-Strep (Gibco, Carlsbad, CA)/1× MEM nonessential amino acids (Gibco)] containing 20 μg of stem cell factor/2% IL-6 and -3 final (supernatant). Induction of erythroid differentiation was done over 3 days with 3 units/ml EPO in IMDM/15% FCS/1× Pen-Strep/1% BSA.
Supplementary Material
Acknowledgments
We thank Dr. G. Terszowski (University of Ulm, Ulm, Germany) for help with the FACS-ARIA sorter; Dr. G. Blobel (Pennsylvania School of Medicine, Philadelphia, PA), Dr. J. W. Conaway (Stowers Institute, Kansas City, KS), Dr. J. Strouboulis (Erasmus Medical Center, Rotterdam, The Netherlands), and Dr. K. Ge (The Rockefeller University) for providing plasmids; Dr. T. Waldmann (Max Planck Institute, Freiburg, Germany) for the DEK antibody; and Dr. P. Vyas (John Radcliffe Hospital, Oxford, U.K.) for helpful discussions and for critically reading the manuscript. This work was supported by the German research foundation (DFG) Emmy–Noether Fellowship (BO-1639, to T.B.) and by funds from the Max Planck Society (to T.B.). Work done in the Roeder laboratory was supported by National Institutes of Health Grant DK071900.
Abbreviations
- BFU
burst-forming units
- BFU-E
erythroid BFU
- CFU-E
erythroid colony-forming units
- MEF
mouse embryonic fibroblast
- polII
RNA polymerase II
- AGM
aorta-gonads mesonephros
- dpc
days postcoitum
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0604494103/DC1.
References
- 1.Cantor AB, Orkin SH. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 2.Perry C, Soreq H. Eur J Biochem. 2002;269:3607–3618. doi: 10.1046/j.1432-1033.2002.02999.x. [DOI] [PubMed] [Google Scholar]
- 3.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D'Agati V, Orkin SH, Costantini F. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 4.Blazek E, Mittler G, Meisterernst M. Chromosoma. 2005;113:399–408. doi: 10.1007/s00412-005-0329-5. [DOI] [PubMed] [Google Scholar]
- 5.Borggrefe T. Res Adv Biol Chem. 2004;2:9–20. [Google Scholar]
- 6.Ito M, Roeder RG. Trends Endocrinol Metab. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- 7.Kim YJ, Lis JT. Trends Biochem Sci. 2005;30:245–249. doi: 10.1016/j.tibs.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Kornberg RD. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Kuras L, Borggrefe T, Kornberg RD. Proc Natl Acad Sci USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 11.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 12.Stevens JL, Cantin GT, Wang G, Shevchenko A, Shevchenko A, Berk AJ. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG. Mol Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Crawford SE, Qi C, Misra P, Stellmach V, Rao MS, Engel JD, Zhu Y, Reddy JK. J Biol Chem. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 15.Urahama N, Ito M, Sada A, Yakushijin K, Yamamoto K, Okamura A, Minagawa K, Hato A, Chihara K, Roeder RG, Matsui T. Genes Cells. 2005;10:1127–1137. doi: 10.1111/j.1365-2443.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 16.Landles C, Chalk S, Steel JH, Rosewell I, Spencer-Dene B, Lalani el N, Parker MG. Mol Endocrinol. 2003;17:2418–2435. doi: 10.1210/me.2003-0097. [DOI] [PubMed] [Google Scholar]
- 17.Gregory CJ, Eaves AC. Blood. 1977;49:855–864. [PubMed] [Google Scholar]
- 18.Gregory CJ, Eaves AC. Blood. 1978;51:527–537. [PubMed] [Google Scholar]
- 19.Terszowski G, Waskow C, Conradt P, Lenze D, Koenigsmann J, Carstanjen D, Horak I, Rodewald HR. Blood. 2005;105:1937–1945. doi: 10.1182/blood-2004-09-3459. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. Mol Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orkin SH. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 22.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss MJ, Orkin SH. Proc Natl Acad Sci USA. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadam S, Emerson BM. Mol Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 26.Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J. EMBO J. 2005;24:2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA. EMBO J. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y, Qi C, Jia Y, Nye JS, Rao MS, Reddy JK. J Biol Chem. 2000;275:14779–14782. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
- 30.Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 31.Hole N, Graham GJ, Menzel U, Ansell JD. Blood. 1996;88:1266–1276. [PubMed] [Google Scholar]
- 32.Gordon DF, Tucker EA, Tundwal K, Hall H, Wood WM, Ridgway EC. Mol Endocrinol. 2006;20:1073–1089. doi: 10.1210/me.2005-0115. [DOI] [PubMed] [Google Scholar]
- 33.Tsai SF, Strauss E, Orkin SH. Genes Dev. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 34.Westin G, Gerster T, Muller MM, Schaffner G, Schaffner W. Nucleic Acids Res. 1987;15:6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whyatt DJ, deBoer E, Grosveld F. EMBO J. 1993;12:4993–5005. doi: 10.1002/j.1460-2075.1993.tb06193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krystal G. Exp Hematol. 1983;11:649–660. [PubMed] [Google Scholar]
- 37.Valverde-Garduno V, Guyot B, Anguita E, Hamlett I, Porcher C, Vyas P. Blood. 2004;104:3106–3116. doi: 10.1182/blood-2004-04-1333. [DOI] [PubMed] [Google Scholar]
- 38.Johnson KD, Grass JA, Park C, Im H, Choi K, Bresnick EH. Mol Cell Biol. 2003;23:6484–6493. doi: 10.1128/MCB.23.18.6484-6493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szutorisz H, Dillon N, Tora L. Trends Biochem Sci. 2005;30:593–599. doi: 10.1016/j.tibs.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 40.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis Proc Natl Acad Sci USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.