Abstract
Cyclophilins are peptidyl-prolyl cis-trans isomerases involved in catalyzing conformational changes and accelerating the rate of protein folding and refolding in several cellular systems. In the present study, we analyzed the expression pattern and intracellular distribution of the cellular isomerase cyclophilin A (CypA) during vaccinia virus (VV) infection. An impressive increase in CypA stability was observed, leading to a practically unchanged accumulation of CypA during infection, although its synthesis was completely inhibited at late times. By confocal microscopy, we observed that CypA went through an intense reorganization in the cell cytoplasm and colocalized with the virosomes late in infection. CypA relocation to viral factories required the synthesis of viral postreplicative proteins, and treatment of infected cells with cyclosporine (CsA) prevented CypA relocation, clearly excluding the virosomes from CypA staining. Immunoelectron microscopy of VV-infected cells showed that CypA was incorporated into VV particles during morphogenesis. Biochemical and electron microscopic assays with purified virions confirmed that CypA was encapsidated within the virus particle and localized specifically in the core. This work suggests that CypA may develop an important role in VV replication.
Vaccinia virus (VV) is the prototype member of the family Poxviridae. Virus gene expression is tightly regulated in a cascade fashion, and three distinct classes of viral genes are recognized: early, intermediate, and late. The sequential expression of VV genes is also temporally coordinated with virus genome replication. DNA synthesis requires the expression of early proteins and is strictly needed for the subsequent stages of intermediate and late gene expression (reviewed in reference 42). VV replication and progeny assembly take place exclusively in the cytoplasm of infected cells at discrete foci enriched in virus DNA and proteins termed viral factories or virosomes (13, 34).
The VV genome encodes most of the proteins required for nucleic acid metabolism and virion morphogenesis. Therefore, it is generally accepted that the VV life cycle occurs nearly independently of host cell functions (42). Nevertheless, many attempts to investigate the possible involvement of host proteins in VV replication have been made on the basis of biochemical and microscopic strategies. Cellular proteins have been implicated in VV postreplicative transcription, e.g., VITF-2, YY1, and heterogeneous ribonucleoproteins A2 and RBM3 (6, 46, 58). Several morphogenetic studies also revealed the role of microtubules, actin tails, and intermediate filaments in progeny assembly and release (11, 21, 29, 56). The molecular chaperones Hsp70 (33) and, more recently, Hsp90 were shown to be important to VV replication (32).
In a previous report, we have described the possible involvement of cyclophilins (Cyps), particularly CypA, in the VV replication cycle (16). Cyps are abundant and ubiquitously distributed proteins possessing peptidyl-prolyl cis-trans isomerase (PPIase) activity (19, 22, 49). They were discovered originally as specific ligands for the immunosuppressive drug cyclosporine (CsA) and then referred to as immunophilins (28). Cyp isomerase activity is inhibited upon binding to CsA (22, 49). The major Cyp isoform is a cytoplasmic 18-kDa protein termed CypA. The PPIase activity enables CypA to behave as a molecular chaperone and a catalyst of conformational changes in several cellular processes, assisting protein folding and accelerating the rate of polypeptide refolding (19, 49). Actually, in several cell systems, proteins belonging to the Cyp family have been associated with protein trafficking during secretion (9, 36, 50), mitochondrial physiology (45), formation of receptor heterocomplexes (49), RNA splicing (31), gene expression, and the cell cycle (1), among other processes. CypA was also reported to bind human immunodeficiency virus type 1 (HIV-1) Gag polyprotein by the CA domain (24, 39, 53). This interaction allows CypA to be incorporated into HIV-1 particles and is necessary for the production of infectious virions (8, 24).
We have previously shown that CsA inhibits VV replication in cell culture. Virus early protein synthesis is slightly affected, and viral DNA accumulation seems to be the primary aim and the rate-limiting step in VV growth in the presence of CsA. However, VV morphogenesis may be particularly targeted (15). CypA was suggested to be involved in the antiviral effect of CsA. The ability of different CsA analogs to block VV replication correlates with their CypA-binding activity, suggesting that CypA plays a role in the VV life cycle (16).
In the study described here, we further investigated the involvement of CypA in the VV replicative cycle. We show that CypA accumulation is not altered during VV replication in BSC-40 cells. Nevertheless, the intracellular distribution of CypA drastically changes with the progression of infection, leading to its full colocalization with viral factories. We also demonstrate that CypA is specifically incorporated into VV cores.
MATERIALS AND METHODS
Cells and viruses.
Monolayer cultures of BSC-40 cells were grown at 37°C in Dulbecco's modified Eagle medium (Gibco-BRL Laboratories) supplemented with 8% calf serum and 2% heat-inactivated fetal bovine serum as described previously (17). VV strain WR was propagated and titrated in BSC-40 cells as described elsewhere (17). VV mutant ts53 was kindly provided by R. Condit (University of Florida) and amplified in BSC-40 cells at 31°C (54).
Antibodies and CsA.
The polyclonal anti-CypA antibody was purchased from Affinity Bioreagents Inc., Golden, Colo., and Upstate Biotechnology, Lake Placid, N.Y. Monoclonal anti-α-tubulin (clone B-5-1-2) was purchased from Sigma Chemical Co., St Louis, Mo. Monoclonal anti-Hsp70 was a generous gift from M. A. Rebello (IPPMG-UFRJ, Rio de Janeiro, Brazil). The following antibodies against VV proteins were used: rabbit polyclonal antiserum raised against total VV structural proteins, which has been previously described (16); rabbit anti-F11L (18); monoclonal anti-A18R (a gift from R. Condit, University of Florida); polyclonal anti-D12L and anti-D1R (a gift from E. Niles, State University of New York, Buffalo); monoclonal anti-D8L (a gift from G. Smith, Imperial College, London, United Kingdom); rabbit anti-A12L (a gift from D. Hruby, Oregon State University); and polyclonal anti-H3L (a gift from B. Moss, National Institutes of Health).
CsA and the analog d-Ala(3-amino)8-cyclosporine (8′A-Cs) were a generous gift from Novartis Pharma AG, Basel, Switzerland. CsA was dissolved in 100% dimethyl sulfoxide and stored at −20°C as a 1,000×-concentrated stock solution. The analog 8′A-Cs was dissolved in 100% methanol (4 mg/ml) and stored at 4°C.
Preparation of mock- and VV-infected cell extracts.
Confluent monolayers of BSC-40 cells (6.5 × 106 cells) were infected with VV (multiplicity of infection [MOI] of 10) for 30 min at 37°C. The inoculum was then removed and replaced with complete medium (zero time of infection). At different times postinfection (p.i.), the monolayers were washed twice with phosphate-buffered saline (PBS), scraped from the dish, and centrifuged at 1,000 × g for 5 min at 4°C. The cells were then resuspended in lysis buffer [10 mM Tris-HCl, 150 mM NaCl, 0.5% Triton X-100, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (Calbiochem), 20 μg of leupeptin (Sigma) per ml (pH 7.5)] and placed on ice for 25 min. The postnuclear fraction was obtained by centrifugation at 7,200 × g for 15 min at 4°C. The supernatant was removed and stored at −70°C. Protein concentration was determined as previously described (3). For Western blot analysis, equal amounts of protein were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblotting.
For metabolic-labeling experiments, mock- or VV-infected cells were pulse-labeled for 1 h at 37°C with 100 μCi of [35S]methionine (Amersham/Pharmacia) per ml in methionine-free medium at the indicated times before extract preparation. For pulse-chase assays, mock- or VV-infected monolayers were [35S]methionine labeled as described above at 2 h p.i. (pulse). After this period, the cells were washed twice in PBS containing a 20-fold excess of unlabeled methionine and incubated with fresh medium supplemented with an excess of unlabeled methionine for the indicated times (chase). The extracts were prepared as described above. Protein concentration was determined as previously described (3).
Western blot analysis.
After electrophoresis, the proteins were transferred to nitrocellulose membranes and the blots were processed for immunodetection as previously described (14). Primary antibodies were diluted 1:1,500 (anti-CypA and anti-α-tubulin; 18 h), 1:2,000 (anti-Hsp70 and anti-VV; 1 h), 1:800 (anti-F11L; 18 h), 1:5,000 (anti-A18R; 1 h), 1:500 (anti-D12L, anti-H3L, anti-A12L, and anti-D1R; 1 h), and 1:100 (anti-D8L; 16 h). The bound antibodies were detected with horseradish peroxidase-conjugated goat anti-rabbit (Amersham/Pharmacia or Santa Cruz Biotechnology) or goat anti-mouse (Amersham/Pharmacia) immunoglobulin G (IgG), followed by incubation with the ECL Western blotting detection reagents (Amersham/Pharmacia), as suggested by the manufacturer.
Affinity chromatography.
Anhydrous coupling of 8′A-Cs to Affi-Gel 10 (Bio-Rad) resin was carried out essentially as previously described (16). Control columns were prepared by the same procedure, except that CsA replaced 8′A-Cs. Postnuclear cell extracts (200 μg) were loaded onto affinity minicolumns, and unbound proteins were recovered by washing the columns with TBAz buffer (20 mM Tris-HCl [pH 7.5], 5 mM β-mercaptoethanol, 0.033% sodium azide). Proteins specifically bound to the matrix were eluted with 2 mg of CsA per ml in 50% TBAz-50% methanol (16). After precipitation with cold acetone, proteins were resolved by SDS-13.5% PAGE. The gels were dried and exposed to X-ray films. In pulse-labeling assays, quantitation of 35S-labeled CypA was performed by densitometric analysis of the Coomassie blue-stained gels and the respective autoradiograms with the image processing and analysis program Scion Image (Beta Release 4; Scion Corporation). For each time point, the value obtained by analysis of the autoradiogram was corrected by the corresponding amount of CypA recovered in the Coomassie blue-stained gel. The ratios obtained for the chase lanes were then divided by the ratio obtained for the pulse lane. The resulting values are presented as percentages of the baseline with reference to the amount of CypA synthesized at 2 h (pulse).
Virus purification.
VV particles were purified from cytoplasmic extracts of infected BSC-40 cells by high-speed centrifugation through a 36% sucrose cushion followed by sedimentation in 25 to 40% sucrose gradient, as previously described (14). For further purification, 2.4 × 1011 purified particles obtained in the first gradient sedimentation were banded on two successive 25 to 40% sucrose gradients. After centrifugation, 450-μl fractions were collected from the bottom of the tube and washed with 10 mM Tris-HCl (pH 7.5). VV particles were recovered by centrifugation at 12,000 × g for 30 min at 4°C and resuspended in 10 mM Tris-HCl (pH 7.5). The number of virus particles was determined from the optical density at 260 nm (OD260) with the formula 1 U of OD260 = 1.2 × 1010 particles/ml. The fractions were analyzed by SDS-PAGE and Western blotting as already described.
Detergent extraction and protease treatment of virions.
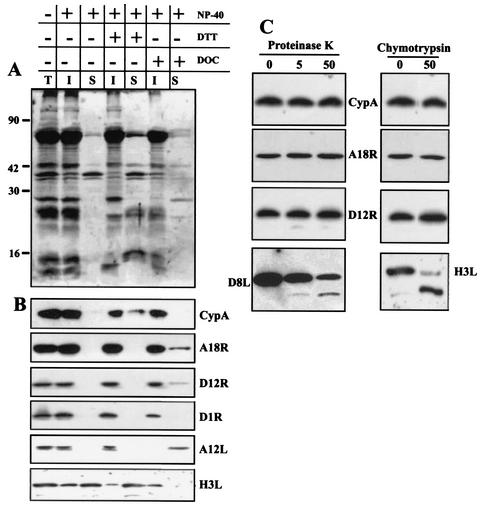
Purified intracellular mature virus (IMV; 7.5 × 109 particles) was incubated for 60 min at 37°C in a reaction mixture containing 0.5% NP-40 and 10 mM Tris-HCl (pH 7.5) in the presence or absence of 50 mM dithiothreitol (DTT). The soluble envelope proteins and insoluble core protein fractions were collected by centrifugation at 12,000 × g for 30 min at 4°C. Viral cores obtained after 0.5% NP-40 treatment were resuspended in DOC buffer (0.2% deoxycholate, 100 mM Tris-HCl [pH 7.5], 250 mM NaCl, 10 mM DTT) and incubated at 4°C for 30 min. The insoluble protein fraction was separated from the solubilized core fraction by centrifugation at 12,000 × g for 10 min at 4°C. The proteins in each fraction were resolved by SDS-PAGE, followed by immunoblotting as described above, and visualized by silver staining.
Purified IMV (7.5 × 109 particles) was digested with either 5 or 50 μg of proteinase K (Gibco-BRL) per ml in 10 mM Tris-HCl (pH 7.5) for 10 min on ice. Alternatively, purified IMV was also digested with 50 μg of chymotrypsin (Sigma) per ml for 30 min at 37°C. The reactions were stopped by addition of 5 mM phenylmethylsulfonyl fluoride (Sigma), and the reaction mixtures were quick-frozen in liquid nitrogen immediately. SDS-PAGE loading buffer was added to the samples still frozen. Proteins were analyzed by electrophoresis, followed by Western blotting, as described above.
Conventional immunofluorescence and confocal laser scanning microscopy.
BSC-40 cells, grown on 13-mm-diameter round glass coverslips in 24-well plates, were infected with VV at an MOI of 10 and incubated at 37°C as described above. In some experiments, the cells were infected with wild-type (WT) or ts53 mutant VV (MOI of 5) and incubated at 31 or 40°C. When indicated, the cells received fresh medium containing 35 μM CsA, 40 μg of cytosine arabinoside (AraC) per ml, 10 mM hydroxyurea (HU), or 100 μg of cycloheximide (CHX) per ml. Procedures for immunofluorescence microscopy were similar to those described previously (7). At the indicated times p.i., the cells were fixed in 4% paraformaldehyde in PHEM buffer (60 mM PIPES, 20 mM HEPES, 10 mM EGTA, 50 mM MgCl2, 70 mM KCl [pH 7.2]) for 10 min at room temperature and 50 min at 4°C and washed in PBS. The cells were then permeabilized in 0.5% Triton X-100 for 3 to 5 min and rinsed in PBS. The coverslips were incubated for 1 h in P-BSA (PBS-1% bovine serum albumin [BSA]) and then for 30 min in 50 mM NH4Cl. The coverslips were washed in P-BSA and incubated for 1.5 h with the anti-CypA (1:100), anti-D8L (1:50), anti-α-tubulin (1:50) primary antibodies. The cells were then rinsed with P-BSA and incubated for 2 h with Cy3-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) diluted 1:800 and/or Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes) diluted 1:500. Cellular and viral DNAs were stained with 1 μg of 4′,6′-diamidino-2-phenylindole (DAPI) per ml for 1 min. The coverslips were washed in P-BSA and then in PBS, mounted in 0.2 M n-propyl gallate, and analyzed with a Zeiss conventional fluorescence microscope (Axiophot). Confocal microscopy was also performed with Zeiss confocal laser scanning microscopes (CLSM 310 and CLSM 510). Each channel was collected separately, and then the channels were merged.
Electron microscopy. (i) Immunogold labeling of thin sections of VV-infected cells.
BSC-40 cells were infected with VV (MOI = 10), and at 20 h p.i., the monolayers were processed for immunolocalization as previously described (40). Briefly, the cells were washed in PBS and fixed with a solution containing 0.4% glutaraldehyde type I, 4% paraformaldehyde, 1% picric acid, 5 mM CaCl2, and 0.5% sucrose in PHEM buffer for 10 min at room temperature and for 50 min at 4°C. The cells were scraped, washed in PBS, and incubated with 50 mM NH4Cl for 30 min on ice. After washing in PBS, the cell pellets were dehydrated in a graded ice-cold ethanol series (30 to 100%) and infiltrated with Unicryl (British Biocell) at −20°C under UV illumination. Ultrathin sections on 400-mesh nickel grids were immunolabeled as follows. The grids were treated with 200 mM NH4Cl in PBS for 20 min, rinsed in PBS, and after incubation with blocking solution (1.5% BSA, 0.5% fish skin gelatin, 0.01% Tween 20 in PBS [pH 7.2]) for 1 h, anti-CypA antibody diluted in blocking solution was added and the mixture was incubated overnight at 4°C. The grids were washed in blocking solution and then incubated for 1 h with 10- or 15-nm colloidal gold-conjugated goat anti-rabbit IgG (BBInternational). The material was rinsed in deionized water, stained with uranyl acetate and lead citrate, and observed in a Zeiss EM 900 transmission electron microscope.
(ii) Immunogold labeling and negative staining of purified IMV particles.
Purified IMV particles (1.5 × 108) were adsorbed to glow-discharged, Formvar-coated 400-mesh nickel grids for 15 min. Subsequently, the grids were incubated or not with 0.5% NP-40-50 mM DTT in 10 mM Tris-HCl (pH 7.5) for 45 min at 37°C. Alternatively, the virus-covered grids were fixed with 4% paraformaldehyde in PHEM buffer for 45 min and then treated or not with 0.2% Triton X-100 in PBS for 15 min at room temperature. After both detergent treatments, the grids were incubated with 20 mM glycine in PBS for 20 min and blocked and immunolabeled as described above, except for the primary antibodies, which were added for 1 h of incubation. After a final wash in PBS, samples were fixed with 2.5% glutaraldehyde in PBS for 15 min and rinsed in deionized water. Virus particles were negatively stained with 5% uranyl acetate for 5 min and analyzed in a Zeiss EM 900 transmission electron microscope.
RESULTS
CypA accumulation and synthesis in VV-infected BSC-40 cells.
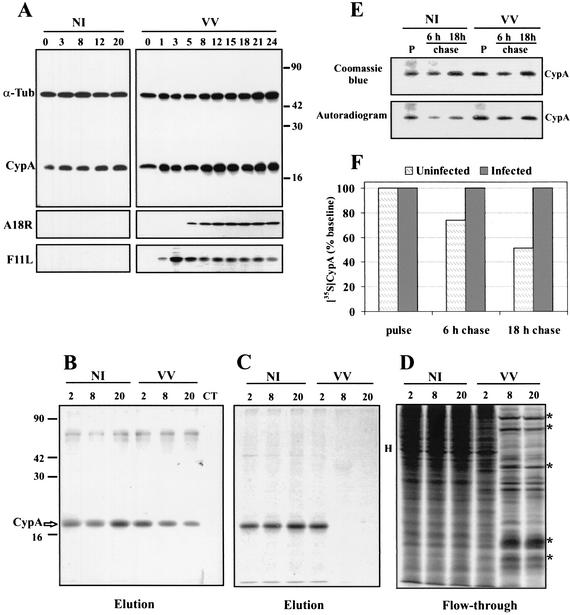
Previous work indicated the involvement of CypA in the antiviral effect of CsA on VV replication. This suggestion was based on the analysis of a panel of CsA analogs with different levels of affinity to CypA. Analogs with strong CypA-binding activity also exhibited a strong antiviral effect. These data led to the proposition that CypA could be required for progression of the VV replicative cycle (16). To gain insight into this issue, we first evaluated the expression of CypA throughout VV infection. BSC-40 cells were infected with VV (MOI = 10) and collected at different times p.i. for preparation of postnuclear extracts. Equal amounts of protein were analyzed by Western blotting. As shown in Fig. 1A, the accumulation of CypA remained practically invariable from early to late times of infection, with levels comparable to those of mock-infected cells. Minor variations in CypA expression detected at some time points were also noticed in α-tubulin expression, which was used as an endogenous control in double-labeled blots. A parallel immunoblot with anti-F11L (early viral protein) and anti-A18R (early/late viral protein) antibodies was performed as a positive control for the progression of VV infection (Fig. 1A).
FIG. 1.
Temporal synthesis and accumulation of CypA in VV-infected cells. BSC-40 cells were mock infected (NI) or infected with VV at 10 PFU/ml, and at the indicated times p.i., postnuclear cell extracts were prepared. (A) Proteins were resolved by SDS-13.5% PAGE, followed by Western blot analysis with anti-α-tubulin, anti-CypA, anti-F11L, and anti-A18R antibodies. (B, C, and D) At the indicated times p.i., the monolayers were pulse-labeled with [35S]methionine for 1 h, extracts were prepared, and 200-μg samples were loaded onto 8′A-Cs-Affi-Gel columns. The proteins retained by the resin were eluted with 2 mg of CsA per ml. Both the unbound (flowthrough) and eluted proteins were analyzed by SDS-13.5% PAGE and staining with Coomassie blue (B), followed by autoradiography (C and D). Asterisks indicate late VV proteins; H indicates a major host protein. CT refers to the control column unable to retain CypA. (E) At 2 h p.i., the monolayers were labeled with [35S]methionine (pulse; P) as described above and chased with medium containing unlabeled methionine for an additional 6 and 18 h. The extracts were prepared and loaded onto 8′A-Cs-Affi-Gel 10 columns as already described. The eluted proteins were resolved by SDS-PAGE and staining with Coomassie blue, followed by autoradiography. (F) The values obtained by densitometric analysis of the autoradiograms in panel E were corrected by the amount of eluted CypA obtained for the corresponding lane in the Coomassie blue-stained gels. The resulting ratio was calculated as described in Materials and Methods, and the final values are expressed as percentages of the 35S-labeled CypA synthesized at the time of the pulse (baseline). The values to the right of panel A and the left of panel B are molecular sizes in kilodaltons.
To investigate CypA synthesis, mock- and VV-infected BSC-40 monolayers were metabolically pulse-labeled at various times p.i. and postnuclear extracts were prepared. The 35S-labeled extracts were loaded onto 8′A-Cs affinity columns that specifically retain CsA-binding proteins, in particular, the major Cyp isoform CypA (16). Proteins specifically bound to 8′A-Cs columns were eluted with an excess of CsA and analyzed by SDS-PAGE and autoradiography. Coomassie blue staining of the gel confirmed the specificity of 8′A-Cs column binding for CypA, which was not retained in control columns (Fig. 1B) as previously reported (16). Consistent with the results shown in Fig. 1A, VV-infected cells accumulated CypA to levels of mock-infected cells (Fig. 1B). However, the autoradiogram analysis revealed a drastic inhibition of CypA synthesis at late times of infection (Fig. 1C). At 2 h p.i., CypA was synthesized to control levels, but after 8 h of infection, only a faint band could be seen (Fig. 1C). This result was not totally unexpected because infection of cells with VV is known to induce a severe shutoff of host protein synthesis (17, 41). This effect is observed in Fig. 1D, which shows the diminished background of host proteins synthesized during late times of infection and eluted from the columns in the flowthrough fractions.
The data in Fig. 1A and C indicate that VV infection may alter CypA stability. To investigate this possibility, we measured CypA turnover in mock- and VV-infected cells by metabolic labeling of the cells at 2 h p.i. (pulse) and chasing for the indicated times with unlabeled medium. The extracts were loaded onto 8′A-Cs affinity columns, and the CypA eluted with CsA was analyzed by SDS-PAGE and autoradiography (Fig. 1E). To quantify the 35S-labeled CypA at each time point, we performed densitometric analyses of the gels and autoradiograms (Fig. 1F). To properly express the amount of 35S-labeled CypA, the values obtained for each lane of the autoradiogram were first corrected by the total amount of CypA in the corresponding lane of the Coomassie blue-stained gel (Fig. 1E). The values presented in Fig. 1F were calculated as described in Materials and Methods and are expressed with reference to the pulse time point (baseline). Figure 1F shows that in mock-infected cells, the [35S]methionine-labeled CypA was progressively degraded, reaching 50% of the baseline value after an 18-h chase. In contrast, when the cells were infected with VV, the half-life of pulse-labeled CypA was markedly extended, with 100% of the labeled protein recovered after an 18-h chase.
Subcellular localization of CypA in VV-infected cells.
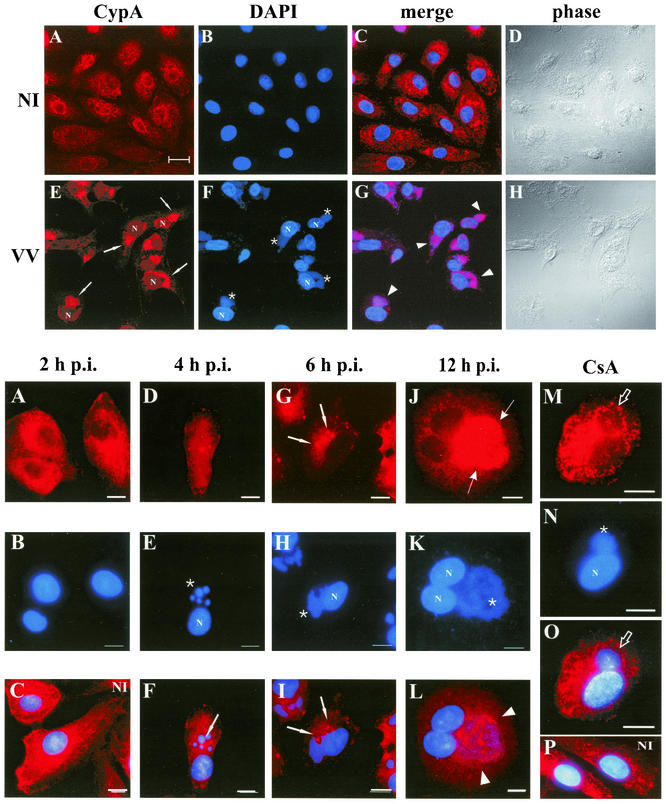
To further investigate the possible involvement of CypA in the VV life cycle, we next examined the distribution pattern of CypA in mock- and VV-infected BSC-40 cells by confocal microscopy (Fig. 2). Mock-infected cells presented a slight punctate staining superimposed upon a fainter diffuse background dispersed throughout the cytoplasm, in agreement with the cytosolic localization of CypA (28, 37). Notably, a weaker CypA immunoreactivity was also found in the nucleus (Fig. 2A). Similar nuclear labeling has also been described by others (37, 47). Infection of BSC-40 cells with VV for 20 h induced a striking reorganization of the distribution pattern of CypA in the cytoplasm. CypA was mostly relocated to regions juxtaposed to the nuclear periphery (Fig. 2E, arrows), and this strong anti-CypA labeling fully colocalized with viral factories, as defined by DNA staining (Fig. 2F, asterisks). Colocalization was clearly evident when the two images were merged (Fig. 2G). It is important to note that the complete coalignment of CypA with VV factories was consistently detected in distinct experiments, corresponding to 96.3% of the cells counted (n = 600) in random fields.
FIG. 2.
(Top panels) Intracellular localization of CypA in VV-infected cells. BSC-40 cells were mock infected (NI) or infected with VV at 10 PFU/ml. At 20 h p.i., the cells were processed for confocal immunofluorescence microscopy with anti-CypA detected with a Cy3-conjugated secondary antibody (red channel; A and E). Viral and cellular DNAs were stained with DAPI (B and F). The merged images are shown in panels C and G. Phase-contrast images are shown in panels D and H. Arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Arrowheads point to colocalization of CypA with viral factories in the merged images. Representative fields are shown. N, nucleus; bar, 20 μm.
To gain additional understanding of the redistribution of CypA during VV infection, a time course experiment was performed. BSC-40 cells were infected with VV, and at the indicated times p.i., the monolayers were processed for indirect immunofluorescence microscopy as described in Materials and Methods. As shown in Fig. 3A, no changes in the CypA distribution pattern were visible at 2 h p.i. compared to that in mock-infected cells (Fig. 3C). Also, cytoplasmic viral factories were not revealed by DAPI staining (Fig. 3B). As the infection proceeded, CypA distribution shifted to a mainly punctate, granular pattern, with the appearance of intensely labeled spots (Fig. 3D and G, arrows). Virosomes were clearly distinguished by DAPI staining, initially as round structures at 4 h p.i. (Fig. 3E, asterisk), evolving to irregularly shaped bodies later in infection (Fig. 3H, asterisk). The granular structures intensely decorated with CypA were dispersed within the cytoplasm and did not colocalize with viral factories at 4 and 6 h p.i. (Fig. 3F and I, arrows), although most of these spotty structures were in close proximity to the virosomes. At 12 h p.i., full colocalization of CypA and viral factories was clearly evident (Fig. 3J to L), as observed at 20 h p.i. (shown in Fig. 2).
FIG. 3.
(Bottom panels) Time course of CypA relocalization. BSC-40 cells were mock infected or infected with VV as described in Materials and Methods and harvested for immunofluorescence assay at the indicated times (A to L). In panels M to P, the cells were mock infected (P) or infected with VV (M, N, and O) and treated with 35 μM CsA at time zero. The infection proceeded for 12 h p.i., when the cells were processed for immunofluorescence assay. Anti-CypA labeling (red channel) is visualized in panels A, D, G, J, and M. Viral and cellular DNAs were stained with DAPI (blue channel), as shown in panels B, E, H, K, and N. Merged images are visualized in panels C, F, I, L, O, and P. White arrows indicate granular CypA structures in the cytoplasm. Thin arrows indicate the redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Arrowheads point to colocalization of CypA with viral factories in the merged images. Open arrows indicate CypA labeling outlining viral factories. Representative fields are shown. NI, mock-infected cells; N, nucleus; bars, 10 μm.
The immunosuppressant CsA was reported to severely inhibit VV progeny production (15), and this effect was correlated with its ability to bind CypA (16). Therefore, we were most interested in investigating the effect of CsA on CypA rearrangement during infection. As shown in Fig. 3M, in the presence of 35 μM CsA, CypA no longer colocalized with viral factories after 12 h of infection. In contrast, staining was dispersed throughout the cytoplasm, except for the virosome region, which appeared to be excluded from CypA labeling. Some labeling outlined viral factories, suggesting a halo-type pattern surrounding the virosomes in the presence of CsA (Fig. 3M and O, open arrows). Additionally, we observed that DNA factories were smaller in CsA-treated cells (Fig. 3N, asterisk) than in untreated cells (Fig. 3K), which is consistent with previous reports that viral DNA replication is inhibited in VV-infected cells treated with CsA (15, 16). Under these conditions, virus progeny production was inhibited to a level 87.4% lower than that in untreated VV-infected BSC-40 cells. Interestingly, although CypA was mainly distributed throughout the cytoplasm, granular staining was still detected in VV-infected cells treated with CsA, although less frequently than in untreated cells at early stages of infection. This pattern of CypA was not visible in mock-infected cells treated with CsA (Fig. 3P) or not treated with CsA (Fig. 3C), indicating that CsA inhibited the relocation of CypA to viral factories induced by VV infection but could not restore the regular distribution pattern observed in uninfected cells.
Specificity of CypA redistribution in VV-infected cells.
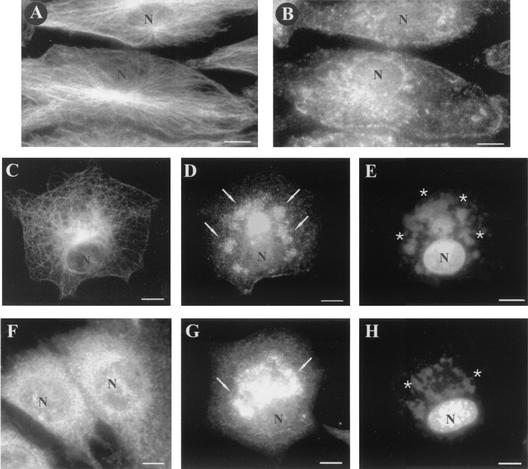
To evaluate the specificity of CypA rearrangement and exclude a general effect of virus infection on cellular proteins under our assay conditions, we analyzed the distribution pattern of another abundant cellular protein, α-tubulin, in VV-infected cells. As previously described (43), the microtubule cytoskeleton was dramatically reorganized during infection (Fig. 4C), but its distribution did not colocalize with the virosomes (Fig. 4E), in contrast to the CypA pattern (Fig. 4D). As a control, the distribution patterns of α-tubulin and CypA in mock-infected cells are shown in Fig. 4A and B, respectively. CypA relocation to viral DNA factories was also detected in mouse embryo fibroblasts (Fig. 4G and H) and HEp-2, RK13, and HeLa cells (data not shown) infected with VV for 20 h.
FIG. 4.
Distribution of CypA and α-tubulin in VV-infected cells. BSC-40 cells (A to E) and mouse embryo fibroblasts (F to H) were mock infected (A, B, and F) or infected with VV (C, D, E, G, and H) as described in Materials and Methods. At 20 h p.i., the monolayers were processed for immunofluorescence microscopy with anti-α-tubulin (A and C) and anti-CypA (B, D, F, and G) antibodies. Viral and cellular DNAs were stained with DAPI (E and H). Arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Representative fields are shown. N, nucleus; bars, 10 μm.
Alteration of the CypA staining pattern requires VV early gene expression.
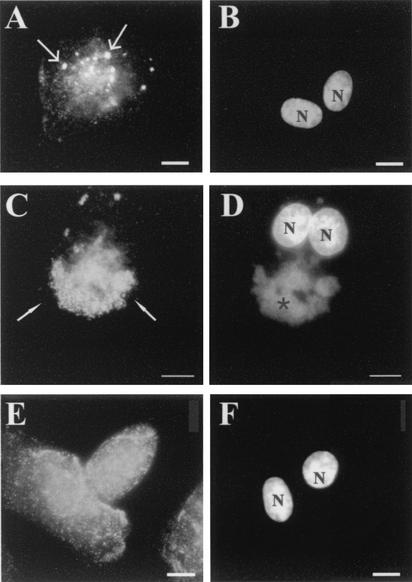
In an attempt to relate CypA redistribution to the stages of VV gene expression, we next examined whether the replication of viral DNA and all subsequent postreplicative events are required for CypA rearrangement. BSC-40 cells were infected with VV for 10 h in the presence of 40 μg of AraC per ml to block viral DNA synthesis. As expected, under this condition, the formation of viral DNA factories was completely abolished (Fig. 5B). Nevertheless, CypA changed its labeling pattern, displaying punctate staining dispersed all over the cytoplasm with brightly labeled granular structures. No redistribution of CypA was observed (Fig. 5A). This pattern was already observed at 6 h posttreatment (data not shown) and was quite similar to that observed in untreated cells at early times p.i., as shown in Fig. 3D and G. Infection of the monolayers for 6 h in the presence of 100 μg of CHX per ml did not alter the subcellular localization or staining pattern of CypA (Fig. 5E) compared to that in mock-infected cells (Fig. 3C), indicating the requirement of de novo protein synthesis to launch CypA rearrangement. Together, these data suggest that the occurrence of the first discernible changes in the CypA distribution pattern during VV infection requires only the early stage of the cycle. The subsequent redistribution of CypA to the virosomes appears to depend on the next steps of infection.
FIG. 5.
Effects of metabolic inhibitors on the distribution of CypA. BSC-40 cells infected with VV were treated with 40 μg of AraC per ml for 10 h (A and B) or with 100 μg of CHX per ml for 6 h (E and F). Cells were processed for immunofluorescence microscopy with anti-CypA (A, C, and E) antibody. Viral and cellular DNAs were stained with DAPI (B, D, and F). (C and D) Untreated cells infected for 10 h. Thin arrows indicate granular CypA structures in the cytoplasm. White arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Representative fields are shown. N, nucleus; bars, 10 μm.
Redistribution of CypA to the virosomes requires viral DNA replication and postreplicative protein synthesis.
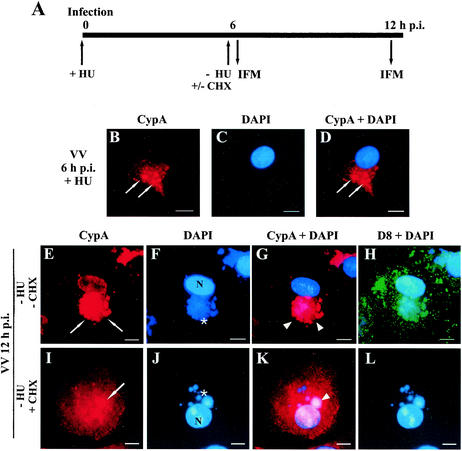
To further examine whether DNA replication and/or postreplicative protein synthesis are required for the subsequent colocalization of CypA with viral factories, we used two experimental approaches. Initially, we performed the assay outlined in Fig. 6A. BSC-40 cells were infected with VV in the presence of 10 mM HU to block DNA replication, allowing the early phase of infection to occur. After 6 h of infection, the HU-containing medium was removed to release viral DNA synthesis at the same time that postreplicative protein synthesis was inhibited by the addition of 100 μg of CHX per ml to the monolayers and incubation for another 6 h. After this period, CypA localization was analyzed by indirect immunofluorescence microscopy. Confirming previous data obtained with AraC, virus DNA replication was not required for the initial changes in the staining pattern of CypA represented by the appearance of intense punctate labeling with dense granular structures at 6 h p.i. in the presence of HU (Fig. 6B). As expected, removal of HU released the replication of viral DNA (Fig. 6F and J) in the presence or absence of postreplicative protein synthesis, which was monitored by expression of the IMV late protein D8L (Fig. 6H and L). In agreement with previous observations made by others, the size of DNA factories did not increase over time when DNA replication occurred in the presence of CHX (Fig. 6J) (2, 20). When virus postreplicative protein synthesis occurred, CypA was recruited to DNA factories and colocalized completely (Fig. 6E and G). Conversely, in the absence of postreplicative protein synthesis, CypA remained mostly scattered throughout the cytoplasm with bright spots dispersed randomly. On limited occasions, we detected CypA colocalization with a few virosomes per cell, corresponding to 14.9% of the cells (n = 464) counted in random fields (Fig. 6I and K). This indicates that virus postreplicative protein synthesis is necessary for full redistribution of CypA to viral factories. Nevertheless, these observations should be interpreted with caution since the use of metabolic inhibitors affects both viral and cellular functions.
FIG. 6.
Localization of CypA in VV-infected cells in the absence of postreplicative protein synthesis. (A) Diagram of the assay. BSC-40 cells were infected with VV in the presence of 10 mM HU. At 6 h p.i. (B to D), the medium was replaced with HU-free medium in the absence (E to H) or presence (I to L) of 100 μg of CHX per ml. The cells were incubated for an additional 6 h and processed for immunofluorescence (IFM) with anti-CypA (B, E, and I) and anti-D8L (green channel; H and L) antibodies. Viral and cellular DNAs were stained with DAPI (C, F, and J). Merged images are shown in panels D, G, K, H, and L. Thin arrows indicate granular CypA structures in the cytoplasm. White arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Arrowheads point to colocalization of CypA with viral factories. Representative fields are shown. N, nucleus; bars, 10 μm.
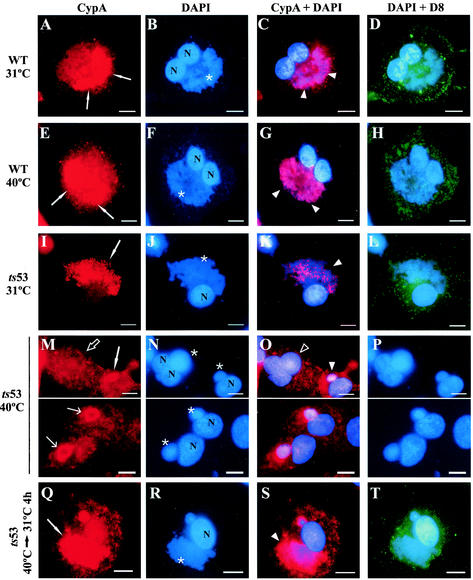
As a second experimental strategy to investigate the involvement of postreplicative protein synthesis in CypA relocation to the virosomes, we performed experiments with the temperature-sensitive VV mutant ts53. This mutant has been previously characterized by Condit and coworkers as defective in late protein synthesis but fully competent in DNA replication (30). The mutation was mapped to the 147-kDa subunit of the viral RNA polymerase (54). Infection of BSC-40 cells with either WT or ts53 VV was performed at 31°C (permissive temperature) or 40°C (nonpermissive temperature), and after 12 h of infection, the cells were processed for immunofluorescence assay as described in Materials and Methods. At 31°C, intense colocalization of CypA with the virosomes was observed in ts53-infected cells (Fig. 7I to K) and the pattern of CypA redistribution to viral factories was quite similar to that of WT infections at both 31°C (Fig. 7A to C) and 40°C (Fig. 7E to G). As a control, the D8L protein was used to examine the synthesis of late proteins (Fig. 7D, H, and L). Consistent with previous data (30), cells infected with ts53 at the nonpermissive temperature exhibited large, round virosomes (Fig. 7N) and were unable to synthesize virus late polypeptides (Fig. 7P). Under this condition, CypA remained mostly dispersed all through the cytoplasm (Fig. 7M, open arrow) and its redistribution to the virosomes was scarcely observed (Fig. 7M, white arrow). In some of these few positive cases, only partial colocalization was detected, particularly at the periphery of the virosomes (Fig. 7M, thin arrows). Table 1 shows that the coalignment of CypA with viral factories was distinguished in 19.3% of the ts53-infected cells at 40°C, in contrast to more than 75% of the cells displaying full colocalization at 31°C.
FIG. 7.
Localization of CypA in ts53-infected cells. BSC-40 cells were infected with WT VV (A to H) or the VV mutant ts53 (I to T) at 31 or 40°C. After 12 h, the cells were processed for immunofluorescence microscopy with anti-CypA (A, E, I, M, and Q) and anti-D8L (green channel; D, H, L, P, and T) antibodies. Viral and cellular DNAs were stained with DAPI (B, F, J, N, and R). Merged images are shown in panels C, D, G, H, K, L, O, P, S, and T. In panels Q to T, ts53-infected cells were incubated for 12 h at 40°C and shifted to 31°C for 4 h, after which they were processed for immunofluorescence assay. White arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Open arrows point to infected cells with no redistribution of CypA. Thin arrows point to redistribution of CypA to the periphery of the virosomes. Asterisks indicate viral factories. White arrowheads indicate colocalization of CypA with viral factories. Open arrowheads indicate the absence of colocalization of CypA with virosomes. Representative fields are shown. N, nucleus; bars, 10 μm.
TABLE 1.
Colocalization of CypA with viral factories in cells infected with VV mutant ts53a
| Virus (temp °C) | 12 h p.i.
|
Shiftdown to 31°C
|
||||
|---|---|---|---|---|---|---|
| 2 h
|
4 h
|
|||||
| No. of cells | Colocalization (%) | No. of cells | Colocalization (%) | No. of cells | Colocalization (%) | |
| WT (31) | 396 | 349 (88.1) | ||||
| WT (40) | 224 | 165 (73.7) | ||||
| ts53 (31) | 234 | 176 (75.2) | ||||
| ts53 (40) | 373 | 72 (19.3) | 249 | 120 (48.2) | 305 | 242 (79.3) |
BSC-40 cells were infected with WT VV or ts53 (MOI = 5) at both 31 and 40°C and harvested at 12 h p.i. for immunofluorescence microscopy. A sample of cells infected with ts53 at 40°C for 12 h was shifted to 31°C for an additional 2 or 4 h. The cells in more than 40 random fields were counted.
To evaluate whether the redistribution of CypA to viral factories would recover with the resumption of postreplicative protein synthesis, the cells were infected nonpermissively with ts53 for 12 h and then shifted to 31°C for 2 or 4 h. In agreement with Hooda-Dhingra et al. (30), the temperature downshift allowed the restoration of virus late protein synthesis detected by anti-D8L staining (Fig. 7T). Simultaneously, full colocalization of CypA with the virosomes was progressively evident (Fig. 7Q to S). Quantitation of the coalignment reinforced this observation and revealed that at 2 h after the temperature downshift, 48.2% of the cells displayed a clear redistribution of CypA to viral factories. Control levels were reached within 4 h after incubation at 31°C (Table 1). Together, these results suggest that postreplicative protein synthesis is important for accurate colocalization of CypA with virosomes.
Association of CypA with virosomes and virus particles in infected cells.
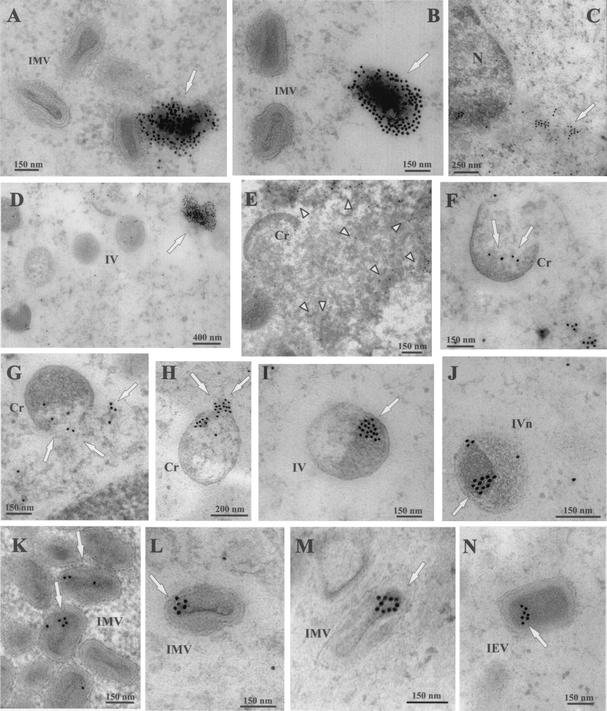
The immunofluorescence assays described above clearly indicated the specific association of CypA with viral factories during VV infection. To investigate this interaction in greater detail, immunoelectron microscopy was performed on ultrathin sections of VV-infected cells that were incubated with anti-CypA antibody, followed by immunogold labeling. As shown in Fig. 8, strong labeling of CypA was detected in association with an amorphous electron-dense material located in the proximity of IMV particles and assembly intermediates (Fig. 8A, B, and D). These dense labeled masses were not observed in areas of the cytoplasm devoid of VV assembly forms or in mock-infected cells (Fig. 8C). Significant labeling was also seen surrounding and within the viroplasm (Fig. 8E). Remarkably, gold particles were consistently observed decorating distinct viral forms of VV morphogenesis. CypA labeling was detected in close association with the viroplasm underlying the concave side of the viral crescents (Fig. 8F to H). Spherical immature particles (IV) and IVs with nucleoid were mainly labeled in the internal region and also surrounding the electron-dense nucleoid (Fig. 8I and J). In mature particles (IMV and intracellular enveloped virus), CypA labeling was primarily found associated with the core and in the space between the core and the IMV envelope (Fig. 8K to N). At all stages of virus maturation, gold grains were rarely detected in association with the particle membranes. We also observed that mature forms were labeled less extensively than IVs, probably because of reduced access of the antibodies to CypA epitopes in the densely packed cores. Similar observations have been reported by others who used immunoelectron microscopy assays to evaluate the localization of distinct VV proteins, including the internal proteins A4L (10) and A30L (52). Also, it is interesting that the CypA labeling associated with the immature and mature forms of the virus was essentially distributed unevenly and in clusters inside the particles. This distribution pattern was detected in 86.7% of the labeled virus particles (n = 117) counted in random fields. The asymmetrical arrangement could also contribute to the weak detection of CypA in some section planes of mature virions.
FIG. 8.
Localization of CypA in VV-infected cells by immunoelectron microscopy. BSC-40 cells mock infected (C) or infected with VV for 20 h were fixed and processed for immunogold labeling with anti-CypA antibody, followed by a colloidal gold-conjugated secondary antibody. Representative fields are shown. CypA labeling is indicated by arrows. In panels A, B, and D, note the intense labeling of amorphous electron-dense material in the proximity of VV particles. Arrowheads in panel E point to gold labeling in the periphery and within the viroplasm. In panels F to H, note the association of gold grains with the viroplasm being encapsidated by the crescent forms. CypA labeling is also found associated with electron-dense material and nucleoid of IVs (I and J). In mature particles, gold labeling is primarily found in the core (K to N). Cr, crescent forms; IVn, IVs with nucleoid; IEV, intracellular enveloped virus; N, nucleus.
CypA is incorporated in purified virions and packaged in viral cores.
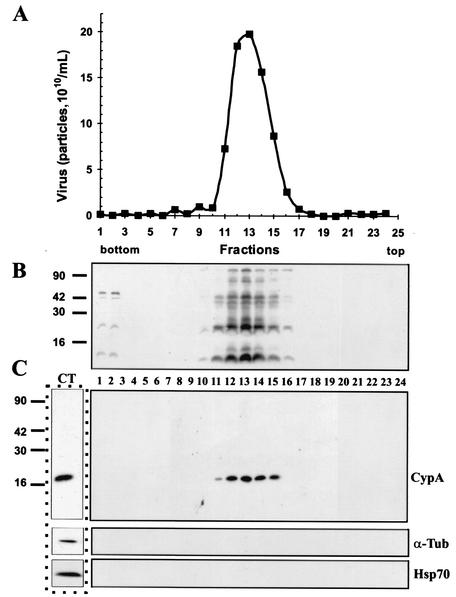
To further examine the association of CypA with VV particles, intracellular virions were purified by three successive sucrose gradient centrifugations and fractions from the third gradient were collected and subjected to SDS-PAGE, followed by Western blotting as already described. Figure 9C shows that CypA detection coincided with the virus-containing fractions, as determined by OD measurement (Fig. 9A) and immunoblotting of VV proteins with antiserum raised against virion structural proteins (Fig. 9B). The copurification of CypA with VV particles was specific since we did not detect other cellular proteins, such as α-tubulin or Hsp70, in association with the virion-containing fractions. To estimate the amount of CypA present in the virus particles, we analyzed the virion proteins by SDS-PAGE, followed by Western blotting. Parallel lanes of the blot contained increasing amounts (10 to 150 μg) of purified CypA that were used as reference samples in the evaluation. Densitometric analysis of the CypA bands was performed, and taking into account the CypA molecular mass, we determined that approximately 156 ± 0.9 CypA molecules were present per VV particle (two separate assays run in duplicate).
FIG. 9.
Association of CypA with purified VV. Intracellular virus was sedimented on a sucrose cushion and banded on three successive sucrose gradients. Fractions were collected, and the number of particles was determined by OD measurement (A). Aliquots of each fraction were analyzed by SDS-PAGE, followed by a Western blot assay with antiserum raised against total VV proteins (B), anti-CypA (C), anti-α-tubulin, and anti-Hsp70 (C) antibodies. In lane CT, uninfected BSC-40 cell proteins were analyzed by Western blotting as a positive control for the anti-CypA, anti-α-tubulin, and anti-Hsp70 antibodies. Molecular size markers (kilodaltons) are shown to the left of panels B and C.
To determine the localization of CypA within the virion, we used three different approaches. First, we treated the purified VV particles with 0.5% NP-40 in the presence or absence of 50 mM DTT and then separated the core and envelope fractions by centrifugation as described in Materials and Methods. The proteins from each fraction were analyzed by SDS-PAGE, followed by silver staining (Fig. 10A) or immunoblotting. As shown in Fig. 10B, CypA was not extracted by the NP-40 treatment, indicating a core localization, and was only partially released from the virions when treated with the detergent plus the reducing agent. As a control, the immunoblots were stripped and reprobed with antibodies to the viral core proteins A18R, D12L, D1R, and A12L. Consistent with their core location, these polypeptides remained associated with the insoluble fraction after NP-40 treatment. H3L was used as a control for IMV envelope protein and was considerably extracted from VV particles treated with NP-40, as reported before (12). The core fraction was further treated with deoxycholate-containing buffer to separate soluble and insoluble core proteins. Similarly to A18R, D12L, and D1R, CypA was also found entirely associated with the insoluble core fraction after treatment of cores with deoxycholate. In contrast, the core protein A12L was completely extracted to the soluble core fraction.
FIG. 10.
Association of CypA with VV cores. (A and B) Purified VV (7.5 × 109 particles) was incubated with 0.5% NP-40 in the presence or absence of 50 mM DTT. The insoluble core fraction was recovered by centrifugation and treated with 0.2% deoxycholate (DOC). Insoluble and soluble fractions were obtained by centrifugation. The proteins were resolved by SDS-PAGE, followed by silver staining (A). The samples were also analyzed by Western blotting (B) with anti-CypA, anti-A18R, anti-D12R, anti-D1R, anti-A12L, and anti-H3L antibodies. T refers to untreated VV. I and S refer to insoluble and soluble fractions, respectively. (C) Purified VV (7.5 × 109 particles) was digested with different concentrations of proteinase K or with 50 μg of chymotrypsin per ml. The proteins in each sample were resolved by SDS-PAGE, followed by Western blot analysis with anti-CypA, anti-A18R, anti-D12R, anti-D8L, and anti-H3L antibodies. The values to the left of panel A are molecular sizes in kilodaltons.
We also investigated the subviral location of CypA after treatment of purified virions with proteinase K and chymotrypsin, followed by protein analysis by Western blotting. As shown in Fig. 10C, even the highest concentration of proteinase K or chymotrypsin treatment did not produce digestion of CypA, consistent with its association to the viral core. A similar pattern was observed with other core proteins, such as A18R and D12L. As a control for membrane proteins susceptible to protease digestion, the blots were stripped and reprobed with anti-H3L (proteinase K treatment) and anti-D8L (chymotrypsin treatment). As expected, both proteins were digested into smaller products.
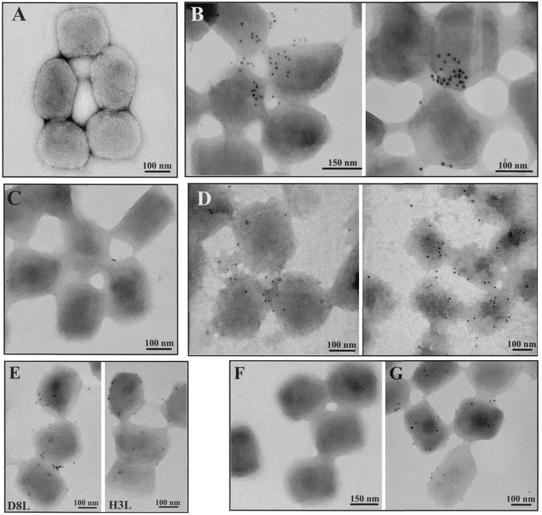
As a third strategy to verify the internal location of CypA in virions, we performed immunolabeling experiments with purified VV particles. After adsorption to nickel grids, the IMV particles were permeabilized with 0.5% Triton X-100 or not permeabilized and then incubated with anti-CypA, followed by a secondary antibody conjugated to 10-nm colloidal gold. Alternatively, the virus-covered grids were incubated with 0.5% NP-40-50 mM DTT to remove the viral membranes, as described in Materials and Methods. Figure 11 shows that intact untreated virus particles were not labeled significantly with anti-CypA (Fig. 11A and C). As expected, untreated IMV particles were labeled with antibodies to H3L and D8L, both envelope proteins (Fig. 11E). After the detergent treatments, immunogold labeling of IMV particles with anti-CypA was detected, implying that CypA is not exposed to the virion surface (Fig. 11B and D). As a control, permeabilized virus was also labeled with anti-D12L (Fig. 11G), a core protein, whereas the intact virus showed no significant labeling (Fig. 11F). Together, these data indicate that CypA is packaged within the virions and is specifically located in the viral core.
FIG. 11.
Detection of CypA in purified VV by immunogold labeling and negative staining. Purified VV (1.5 × 108 particles) was adsorbed to Formvar-coated nickel grids and left unpermeabilized (A, C, E, and F) or permeabilized with Triton X-100 (B and G) or NP-40-DTT (D). The grids were incubated separately with anti-CypA (A to D), anti-D8L, anti-H3L (E), or anti-D12R (F and G) antibody, followed by a colloidal gold-conjugated secondary antibody. Representative fields are shown.
DISCUSSION
It was previously reported that the immunosuppressive drug CsA inhibits VV replication in BSC-40 cells (15). The major CsA-binding protein CypA was suggested to play a role in the ability of CsA to inhibit VV replication, indicating that CypA could be involved in the VV replicative cycle (16). In this study, we have focused on this issue and evaluated the expression pattern and intracellular distribution of CypA during VV infection. CypA synthesis is completely blocked at late times of the VV replicative cycle, which is consistent with the overall shutoff of host translation induced by VV infection, as described by others (17, 41). Despite that, CypA accumulation is sustained throughout infection. Pulse-chase experiments confirmed our assumption that CypA stability is altered, showing an extended half-life in VV-infected cells. This may reflect a broader mechanism induced by VV infection, and it is unlike to occur only with CypA. It has been shown that steady-state levels of other cellular proteins involved in VV replication, such as actin (32) and α-tubulin (this paper), are sustained during the course of infection despite the general shutoff of host translation induced in VV-infected cells. Nevertheless, the turnover rate of actin and α-tubulin during VV infection has never been directly addressed. Our findings may suggest the possible existence of a mechanism triggered by virus infection to control the turnover of specific cellular proteins that are required at optimal levels during VV replication. A similar mechanism was described in human cytomegalovirus-infected cells, where the steady-state level of p53 is increased during infection because of a slower rate of degradation than in uninfected cells. Likewise, p53 distribution is altered and sequestered to the nuclear foci of human cytomegalovirus replication (23).
In mock-infected cells, CypA is distributed uniformly throughout the cell cytoplasm, in accordance with previous reports (28, 37, 47). Late in infection, CypA markedly changes its intracellular organization, with full relocation to the virosomes. Interestingly, CypA rearrangement appears to occur as a two-stage process. Initially, the punctate pattern becomes more intense with the emergence of bright granular structures. At late times p.i., when viral factories are fully developed, CypA relocates to the virosomes completely. The first step requires only VV early protein synthesis, since it occurs in the presence of DNA synthesis inhibitors and is abolished by CHX treatment. The second stage occurs after the onset of virus DNA replication and requires late protein synthesis for full colocalization of CypA with DNA factories.
The presence of CsA during infection prevents CypA redistribution to viral DNA factories. CypA staining remains scattered throughout the cytoplasm and mostly excluded from the virosome region. This suggests that the CsA-binding pocket of the CypA molecule plays an important role in CypA rearrangement during VV infection. We envision two possibilities for the inhibitory effect of CsA on CypA redistribution. It is well documented in the literature that CsA binding to CypA inhibits its PPIase activity (22, 49) and disrupts its association with CypA-binding proteins, such as HIV Gag polyprotein (24, 39, 53). Therefore, we postulate that upon binding to CsA, CypA would be unavailable to interact with other proteins involved in CypA rearrangement. Alternatively, the formation of the CsA-CypA complex could promote the binding of a third partner to the complex, preventing CypA movement to virosomes. This gain-of-function model is well studied for the mechanism of immunosuppression induced by CsA. In T lymphocytes, CsA associates with Cyps, and the complex, but not Cyp alone, binds and inhibits another cytosolic protein, calcineurin, a Ca2+- and calmodulin-dependent phosphatase, leading to a transcriptional block of many genes involved in the launch of the immune response. Binding of calcineurin to the CsA-Cyp complex has been shown to play a critical role in the immunosuppression pathway (19, 38). Previous data from our group, however, suggest that the antiviral mechanism of CsA is not correlated with its immunosuppressive activity, since some nonimmunosuppressive CsA analogs still retain a strong CypA-binding activity and exert a severe antiviral effect although they bind calcineurin weakly (16).
Recently, Hung et al. (32) reported that the chaperone Hsp90 is redistributed transiently to viral factories during VV infection, leaving the virosomes when they start producing new virions. In contrast, CypA colocalizes entirely with the virosomes until late time p.i., even when the factories are fully active in releasing VV progeny. Immunoelectron micrographs convincingly show CypA in the viroplasm in close association with VV particles in distinct stages of maturation. CypA is apparently distributed unevenly in the virus particles, where small clusters are observed. Recently, Griffiths et al. (27) reported that VV has a highly asymmetrical topography that is also reflected in the structure of the inner core. This peculiar aspect of VV particle assembly could account for the unequal distribution of CypA inside virions.
During virus morphogenesis, CypA is encapsidated into mature virions and localizes specifically in the core, with some labeling in the space between the core and the IMV envelope. Immunolabeling assays and biochemical evidence confirm the location of CypA within the core fraction. This internal location suggests that CypA incorporation is specific and that the coelution of CypA with VV particles does not result from cellular contamination during virus purification. Moreover, other abundant cellular proteins, such as α-tubulin and Hsp70, are not found in association with VV particles, as well as Hsp90, which Hung et al. (32) were unable to detect in purified IMV particles, despite its intense accumulation in viral factories. CypA packaging into mature virions has also been described in HIV particles (24, 53). The incorporation is mediated by CypA binding to a proline-rich region of the capsid domain of Gag during assembly, and the inhibition of CypA encapsidation leads to production of noninfectious HIV particles (8, 24, 53). Many studies have attempted to indicate a role for CypA in HIV capsid assembly (4, 51), virus entry into host cells (44, 48), and particle uncoating during capsid disassembly following penetration in a subsequent infection (26, 57). However, the mechanism of CypA activity in HIV replication is not understood.
Attempts to detect a VV protein that interacts with CypA have been unsuccessful. Coimmunoprecipitation assays with anti-CypA IgG failed to identify specific proteins in virus-infected cell extracts (data not shown). Likewise, we could not detect specific interaction of CypA with a virus protein by the yeast two-hybrid system after screening two VV genomic libraries and analyzing more than 400,000 transformants under different selection conditions. Yeast strain Y474 CHY1 (kindly provided by M. Cardenas), in which the cpr1 gene is disrupted, was also used to avoid competition with the endogenous yeast CypA homolog (data not shown). Efforts to pursue this investigation with different experimental strategies are in progress.
It is interesting to speculate what roles CypA could develop in the VV life cycle. Cyps, together with the FK-506-binding proteins and parvulins, are phylogenetically conserved families of folding helper enzymes with PPIase activity (19, 49). It is generally accepted that Cyps may be involved in important processes of cell physiology. CypA, for example, reduces aggregation of incompletely folded molecules of carbonic anhydrase (25) and catalyzes the isomerization of the hormone calcitonin (35). CypA has also been implicated in the cytoplasmic trafficking of proteins and other molecules. It mediates the import of fructose-1,6-bisphosphatase into intermediate transport vesicles in Saccharomyces cerevisiae. The transport of this key regulatory enzyme in yeast gluconeogenesis apparently does not involve a direct protein-protein interaction with CypA (5). Interestingly, in mouse cells, CypA forms a lipoprotein-chaperone complex with caveolin, Cyp40, and Hsp56 that mediates the transport of cholesterol between the endoplasmic reticulum and the membrane caveolae in a vesicle-independent mechanism (55). It is tempting to hypothesize that CypA could be recruited during VV infection to mediate the transport of virus proteins to virosomes. Alternatively, CypA could be required to accumulate in DNA factories to catalyze conformational changes in virus proteins important to the particle assembly process. Indeed, CypA may be involved in the proper folding of VV proteins during the formation of the internal core structure, and therefore, its packaging within the virus particle would be essential to carry out some steps of VV maturation from spherical IV to IMV. Another attractive role for CypA in the VV replicative cycle could be in the uncoating of viral cores. In order to replicate viral DNA, VV cores must disassemble during the early stage of infection. This putative role could be correlated with our previous observations (15), confirmed in this study, that a reduced amount of VV DNA is synthesized in cells treated with CsA, although we cannot exclude the possibility that other CsA-sensitive isoforms of Cyps are involved in this process. In conclusion, our data strengthen the suggestion that CypA could be involved in the VV replicative cycle. Further experiments addressing the potential roles of CypA during VV infection are currently under investigation in our laboratory.
Acknowledgments
We are grateful to Narcisa L. Cunha-e-Silva for many stimulating discussions during the course of this work, Richard Condit for carefully reviewing the manuscript, and Novartis Pharma AG (Basel, Switzerland) for CsA and 8′A-Cs. We also thank Loraine Araujo for help with confocal microscopy, Juliana Dutra and Hilda Petrs for advice on immunofluorescence assays, and Noemia Rodrigues for technical assistance with electron microscopic processing.
This work was supported by grants to C.R.A.D. from CNPq, FUJB and Faperj. A.P.V.C. and N.M. are recipients of a fellowship from CNPq.
REFERENCES
- 1.Arevalo-Rodriguez, M., M. E. Cardenas, X. Wu, S. D. Hanes, and J. Heitman. 2000. Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. 19:3739-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaud, G., and R. Beaud. 1997. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J. Gen. Virol. 78:3297-3302. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bristow, R., J. Byrne, J. Squirell, H. Trencher, T. Carter, B. Rodgers, E. Saman, and J. Duncan. 1999. Human cyclophilin has a significantly higher affinity for HIV-1 recombinant p55 than p24. J. Acquir. Immune Defic. Syndr. Hum. Retrovir. 20:334-336. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. R., D. Y. Cui, G. G. Hung, and H. L. Chiang. 2001. Cyclophilin A mediates Vid22p function in the import of fructose-1,6-bisphosphatase into Vid vesicles. J. Biol. Chem. 276:48017-48026. [DOI] [PubMed] [Google Scholar]
- 6.Broyles, S. S., X. Liu, M. Zhu, and M. Kremer. 1999. Transcription factor YY1 is a vaccinia virus late promoter activator. J. Biol. Chem. 274:35662-35667. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho, T. M., A. G. Ferreira, E. S. Coimbra, C. T. Rosestolato, and W. De Souza. 1999. Distribution of cytoskeletal structures and organelles of the host cell during evolution of the intracellular parasitism by Trypanosoma cruzi. J. Submicrosc. Cytol. Pathol. 31:325-333. [PubMed] [Google Scholar]
- 8.Colgan, J., H. E. Yuan, E. K. Franke, and J. Luban. 1996. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. J. Virol. 70:4299-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colley, N. J., E. K. Baker, M. A. Stamnes, and C. S. Zuker. 1991. The cyclophilin homolog ninaA is required in the secretory pathway. Cell 67:255-263. [DOI] [PubMed] [Google Scholar]
- 10.Cudmore, S., R. Blasco, R. Vincentelli, M. Esteban, B. Sodeik, G. Griffiths, and J. Krijnse Locker. 1996. A vaccinia virus core protein, p39, is membrane associated. J. Virol. 70:6909-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 12.da Fonseca, F. G., E. J. Wolffe, A. Weisberg, and B. Moss. 2000. Characterization of the vaccinia virus H3L envelope protein: topology and posttranslational membrane insertion via the C-terminal hydrophobic tail. J. Virol. 74:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dales, S., and L. Siminovitch. 1961. The development of vaccinia virus in Earle's strain L cells as examined by electron microscopy. J. Biophys. Biochem. Cytol. 10:475-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damaso, C. R., J. J. Esposito, R. C. Condit, and N. Moussatché. 2000. An emergent poxvirus from humans and cattle in Rio de Janeiro State: Cantagalo virus may derive from Brazilian smallpox vaccine. Virology 277:439-449. [DOI] [PubMed] [Google Scholar]
- 15.Damaso, C. R., and S. J. Keller. 1994. Cyclosporin A inhibits vaccinia virus replication in vitro. Arch. Virol. 134:303-319. [DOI] [PubMed] [Google Scholar]
- 16.Damaso, C. R., and N. Moussatché. 1998. Inhibition of vaccinia virus replication by cyclosporin A analogues correlates with their affinity for cellular cyclophilins. J. Gen. Virol. 79:339-346. [DOI] [PubMed] [Google Scholar]
- 17.Damaso, C. R., and N. Moussatché. 1992. Protein synthesis in vaccinia virus-infected cells. I. Effect of hypertonic shock recovery. Arch. Virol. 123:295-308. [DOI] [PubMed] [Google Scholar]
- 18.Damaso, C. R., M. F. Oliveira, S. M. Massarani, and N. Moussatché. 2002. Azathioprine inhibits vaccinia virus replication in both BSC-40 and RAG cell lines acting on different stages of virus cycle. Virology 300:79-91. [DOI] [PubMed] [Google Scholar]
- 19.Dolinski, K., and J. Heitman. 1997. Peptidyl-prolyl isomerases—an overview of the cyclophilin, FKBP and parvulin families, p. 359-369. In M.-J. Gething (ed.), Guidebook to molecular chaperones and protein-folding catalysts. Oxford University Press, Oxford, England.
- 20.Esteban, M., and J. A. Holowczak. 1978. Replication of vaccinia DNA in mouse L cells. IV. Protein synthesis and viral DNA replication. Virology 86:376-390. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira, L. R., N. Moussatché, and V. Moura Neto. 1994. Rearrangement of intermediate filament network of BHK-21 cells infected with vaccinia virus. Arch. Virol. 138:273-285. [DOI] [PubMed] [Google Scholar]
- 22.Fischer, G., B. Wittmann-Liebold, K. Lang, T. Kiefhaber, and F. X. Schmid. 1989. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337:476-478. [DOI] [PubMed] [Google Scholar]
- 23.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 25.Freskgard, P. O., N. Bergenhem, B. H. Jonsson, M. Svensson, and U. Carlsson. 1992. Isomerase and chaperone activity of prolyl isomerase in the folding of carbonic anhydrase. Science 258:466-468. [DOI] [PubMed] [Google Scholar]
- 26.Grättinger, M., H. Hohenberg, D. Thomas, T. Wilk, B. Müller, and H. G. Kräusslich. 1999. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology 257:247-260. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths, G., R. Wepf, T. Wendt, J. K. Locker, M. Cyrklaff, and N. Roos. 2001. Structure and assembly of intracellular mature vaccinia virus: isolated-particle analysis. J. Virol. 75:11034-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handschumacher, R. E., M. W. Harding, J. Rice, R. J. Drugge, and D. W. Speicher. 1984. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226:544-547. [DOI] [PubMed] [Google Scholar]
- 29.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooda-Dhingra, U., C. L. Thompson, and R. C. Condit. 1989. Detailed phenotypic characterization of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J. Virol. 63:714-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horowitz, D. S., E. J. Lee, S. A. Mabon, and T. Misteli. 2002. A cyclophilin functions in pre-mRNA splicing. EMBO J. 21:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung, J. J., C. S. Chung, and W. Chang. 2002. Molecular chaperone Hsp90 is important for vaccinia virus growth in cells. J. Virol. 76:1379-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jindal, S., and R. A. Young. 1992. Vaccinia virus infection induces a stress response that leads to association of Hsp70 with viral proteins. J. Virol. 66:5357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joklik, W. K., and Y. Becker. 1964. The replication and coating of vaccinia DNA. J. Mol. Biol. 10:452-474. [DOI] [PubMed] [Google Scholar]
- 35.Kern, D., T. Drakenberg, M. Wikstrom, S. Forsen, H. Bang, and G. Fischer. 1993. The cis/trans interconversion of the calcium regulating hormone calcitonin is catalyzed by cyclophilin. FEBS Lett. 323:198-202. [DOI] [PubMed] [Google Scholar]
- 36.Klappa, P., R. B. Freedman, and R. Zimmermann. 1995. Protein disulphide isomerase and a lumenal cyclophilin-type peptidyl prolyl cis-trans isomerase are in transient contact with secretory proteins during late stages of translocation. Eur. J. Biochem. 232:755-764. [PubMed] [Google Scholar]
- 37.Le Hir, M., Q. Su, L. Weber, G. Woerly, A. Granelli-Piperno, and B. Ryffel. 1995. In situ detection of cyclosporin A: evidence for nuclear localization of cyclosporine and cyclophilins. Lab. Investig. 73:727-733. [PubMed] [Google Scholar]
- 38.Liu, J., M. W. Albers, T. J. Wandless, S. Luan, D. G. Alberg, P. J. Belshaw, P. Cohen, C. MacKintosh, C. B. Klee, and S. L. Schreiber. 1992. Inhibition of T cell signaling by immunophilin-ligand complexes correlates with loss of calcineurin phosphatase activity. Biochemistry 31:3896-3901. [DOI] [PubMed] [Google Scholar]
- 39.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 40.Melo, A. C., D. Valle, E. A. Machado, A. P. Salerno, G. O. Paiva-Silva, E. S. N. L. Cunha, W. de Souza, and H. Masuda. 2000. Synthesis of vitellogenin by the follicle cells of Rhodnius prolixus. Insect Biochem. Mol. Biol. 30:549-557. [DOI] [PubMed] [Google Scholar]
- 41.Moss, B. 1968. Inhibition of HeLa cell protein synthesis by the vaccinia virion. J. Virol. 2:1028-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2885-2921. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 43.Ploubidou, A., V. Moreau, K. Ashman, I. Reckmann, C. Gonzalez, and M. Way. 2000. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 19:3932-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pushkarsky, T., G. Zybarth, L. Dubrovsky, V. Yurchenko, H. Tang, H. Guo, B. Toole, B. Sherry, and M. Bukrinsky. 2001. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA 98:6360-6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rassow, J., K. Mohrs, S. Koidl, I. B. Barthelmess, N. Pfanner, and M. Tropschug. 1995. Cyclophilin 20 is involved in mitochondrial protein folding in cooperation with molecular chaperones Hsp70 and Hsp60. Mol. Cell. Biol. 15:2654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosales, R., G. Sutter, and B. Moss. 1994. A cellular factor is required for transcription of vaccinia viral intermediate-stage genes. Proc. Natl. Acad. Sci. USA 91:3794-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryffel, B., G. Woerly, B. Greiner, B. Haendler, M. J. Mihatsch, and B. M. Foxwell. 1991. Distribution of the cyclosporine binding protein cyclophilin in human tissues. Immunology 72:399-404. [PMC free article] [PubMed] [Google Scholar]
- 48.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schiene-Fischer, C., and C. Yu. 2001. Receptor accessory folding helper enzymes: the functional role of peptidyl prolyl cis/trans isomerases. FEBS Lett. 495:1-6. [DOI] [PubMed] [Google Scholar]
- 50.Smith, T., L. R. Ferreira, C. Hebert, K. Norris, and J. J. Sauk. 1995. Hsp47 and cyclophilin B traverse the endoplasmic reticulum with procollagen into pre-Golgi intermediate vesicles. A role for Hsp47 and cyclophilin B in the export of procollagen from the endoplasmic reticulum. J. Biol. Chem. 270:18323-18328. [DOI] [PubMed] [Google Scholar]
- 51.Streblow, D. N., M. Kitabwalla, and C. D. Pauza. 1998. Gag protein from human immunodeficiency virus type 1 assembles in the absence of cyclophilin A. Virology 252:228-234. [DOI] [PubMed] [Google Scholar]
- 52.Szajner, P., A. S. Weisberg, E. J. Wolffe, and B. Moss. 2001. Vaccinia virus A30L protein is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 75:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Göttlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, C. L., U. Hooda-Dhingra, and R. C. Condit. 1989. Fine structure mapping of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J. Virol. 63:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uittenbogaard, A., Y. Ying, and E. J. Smart. 1998. Characterization of a cytosolic heat-shock protein-caveolin chaperone complex: involvement in cholesterol trafficking. J. Biol. Chem. 273:6525-6532. [DOI] [PubMed] [Google Scholar]
- 56.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiegers, K., G. Rutter, U. Schubert, M. Grättinger, and H. G. Kräusslich. 1999. Cyclophilin A incorporation is not required for human immunodeficiency virus type 1 particle maturation and does not destabilize the mature capsid. Virology 257:261-274. [DOI] [PubMed] [Google Scholar]
- 58.Wright, C. F., B. W. Oswald, and S. Dellis. 2001. Vaccinia virus late transcription is activated in vitro by cellular heterogeneous nuclear ribonucleoproteins. J. Biol. Chem. 276:40680-40686. [DOI] [PubMed] [Google Scholar]