Amajor step in understanding the molecular interactions that lead to entry of enveloped viruses into their target cells is obtaining structural information on the viral surface glycoproteins at the atomic level. In addition to carrying the main antigenic determinants of the virus, these proteins are responsible for the major steps involved in the entry process, which involve receptor recognition and fusion between viral and cellular membranes. A huge step forward in the field was made >20 years ago, when the laboratories of Don Wiley and John Skehel obtained the crystal structure of the influenza virus hemagglutinin (HA) (1), which carries both receptor-binding and membrane fusion functions. Since then, the x-ray structures of relatively few soluble ectodomains of viral glycoproteins have been determined, compared to the exponential increase of protein structures deposited in the Protein Data Bank in the same period. In particular, structural information on viral envelope proteins combining both receptor-binding and membrane fusion functions is very scarce. Over the years, only the structures of the tick-borne encephalitis (TBE) flavivirus major envelope (E) protein (2) and of the influenza C virus HA-esterase glycoprotein (3) were added to the database. In this issue of PNAS, Modis et al. (4) report the crystal structure of the E protein from dengue virus, a mosquito-borne flavivirus responsible for the highest rate of disease and mortality among members of this viral genus. This structure is therefore a very important addition to the current repertoire.
Dengue virus is the most important arthropod-borne human pathogen. The incidence of dengue fever epidemics has increased dramatically over the last few decades (5), and it is estimated that up to 100 million cases occur annually. In addition, a severe form of the disease, dengue hemorrhagic fever (DHF), has emerged in the same period causing ≈ 500,000 cases worldwide each year (6). There are four different serotypes of dengue virus, and it is believed that DHF may result from sequential infection by different virus serotypes, in a process known as antibody-mediated disease enhancement (ADE) (7). This feature makes finding a vaccine against dengue virus a nontrivial issue, because a vaccine that would not protect effectively against all four dengue serotypes could contribute to ADE. In this context, knowledge of the 3D structure of the envelope glycoprotein can be of major help in designing potent immunogens that protect safely against disease. In addition, the reported structure highlights possible regions of the E protein that can be targeted to block viral entry. The design of small molecules capable of inhibiting the process of entry of pathogenic microbes is appealing, because such drugs do not need to gain access to the interior of cells and therefore are expected to display increased bioavailability. Because different flaviviruses (i.e., TBE, dengue, yellow fever, West Nile, Japanese encephalitis viruses, and others) display ≈ 40% amino acid conservation in their envelope protein (6), the overall fold and the arrangement of domains of these proteins is expected to be similar. This prediction has been further substantiated by the finding that the fusion glycoprotein E1 of Semliki Forest virus (SFV), an alphavirus belonging to the togaviridae family of viruses, displays structural homology, with the same fold and domain arrangement despite the absence of any noticeable sequence similarity (8). In contrast to flaviviruses, the membrane fusion and the receptor-binding functions of alphaviruses are carried out by two different glycoproteins (9). The distinctive three-domain arrangement shared by the fusion proteins from these two viral families is likely to have been conserved because of a fundamental structural requirement in the membrane fusion process, introducing the concept of a distinct class (class II) of membrane fusion proteins. Although the structure of dengue E confirms this prediction (see Fig. 1), it also highlights the importance of knowing the precise conformation of the 60% of amino acids that are different between E proteins from TBE and dengue viruses. Indeed, knowing the conformational details is of paramount importance if specific information is to be used for drug design purposes. In fact, the dengue E molecule was different enough from that of TBE E that standard molecular replacement methods could not be used to determine its structure. The reason is that different domains adopt somewhat different rotation angles with respect to each other, revealing a flexibility that is thought to be necessary for function.
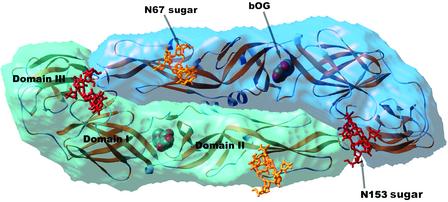
Fig. 1.
Semitransparent surface representation of the dengue virus E protein. The two subunits in the dimer are shown in different shades of blue. The backbone of the molecule is superposed in a yellow ribbon representation. The two glycosylation sites are indicated. To illustrate the location and size of the sugar moiety on a molecule produced in insect cells, the complete high-mannose glycans have been modeled, superposed to the core N-acetyl glucosamine residues that were visible in the crystal structure. The sugars linked to Asn-67 and -153 are shown as yellow and red sticks, respectively. The β-octyl glucoside molecule (indicated by bOG) is displayed as spheres colored according to atom type (red, oxygen; gray, carbon). It lies in a pocket at the hinge region between domains I and II. Figure prepared with the program ribbons (19).
Like influenza viruses and alphaviruses, flaviviruses enter cells by receptor-mediated endocytosis (10). The acidic environment of the endosomes is responsible for triggering a fusogenic conformational change in the fusion glycoproteins, a change that has been well documented structurally in the case of influenza and other class I enveloped viruses (11). An important finding in the reported work on the dengue virus E protein structure is the identification of a binding pocket that accommodates a hydrophobic ligand, in this case a molecule of the detergent β-N-octylglucoside added during crystallization (4). This pocket lies in a region that has been previously proposed as a possible “hinge” between the extended finger-like domain II and the central domain I (see Fig. 1) (2). This proposal was made on the basis of the clustering in this region of mutations, in different flaviviruses, that affect virulence and alter the pH of the conformational change necessary to trigger membrane fusion (2). Now, Modis et al. (4) find that most of the side chains identified by these mutations line the pocket, which opens as a result of the transfer of a β-hairpin from one β-sheet to a neighboring, apposed β-sheet. Flexibility in the hinge region of the fusion protein is likely to be a hallmark of class II enveloped viruses, because different conformations about this area were also identified in the case of the SFV E1 glycoprotein (8). This feature is very likely to be involved in the conformational change that is necessary to trigger membrane fusion. Binding of a hydrophobic ligand is also known to be necessary for alphavirus membrane fusion, which specifically requires cholesterol and sphingolipids in the target membrane (12, 13). No particular lipid requirement has been identified so far for flaviviruses, but a very recent report on West Nile flavivirus suggests that the conformational change can be triggered by interactions with liposomes even at neutral pH, depending on the particular lipid composition (14). The mechanism that triggers the fusion-promoting conformational change is not known, but the fact that mutations altering the pH of the transition map to this pocket is intriguing. The first step in the conformational change is dimer dissociation, and, in this process, the β-hairpin in question might need to move out of its original position in the dimer. The detergent may have thus stabilized an early intermediate state in which the hinge region has flexed but the dimer has not yet dissociated. The relation between the hydrophobic pocket and the pH of the conformational change is clearly worth further study, because it seems to hold the key for understanding the molecular trigger that leads to membrane fusion.
Knowledge of the 3D structure of the envelope protein will bring antidengue virus research to a new phase.
Concerning the receptor-binding function of protein E, it has been pointed out that the presence of mannose residues is important for viral entry (15). Furthermore, two very recent reports show that mosquito-grown dengue virus relies on interactions with the C-type lectin DC-SIGN to efficiently enter cells (16, 17). Like all flaviviruses, dengue virus requires a replication step in the arthropod vector before its transmission to humans. In a natural infection, the virus is deposited by an infected mosquito into the skin during a blood meal. Dendritic cells are normal residents of the skin and express DC-SIGN at their surface. This lectin is specific for the high-mannose-type carbohydrates that are present on virions produced in insect cells. Fig. 2 shows the distribution of glycosylation sites on the surface of a dengue virion, based on the reported dengue E crystal structure and on an electron microscopy reconstruction of dengue virus into which the TBE E protein structure had been placed (18). There are two putative N-linked glycosylation sites in the dengue virus E-protein, and the structure shows that they are both used. The site corresponding to Asn-153 is conserved among many flaviviruses, whereas the Asn-67 site is unique to dengue viruses. The presence of both N-linked carbohydrates is required for recognition by DC-SIGN (17), suggesting that this tetrameric lectin may need to interact with more than one sugar at the same time for efficient internalization of the virus.
Fig. 2.
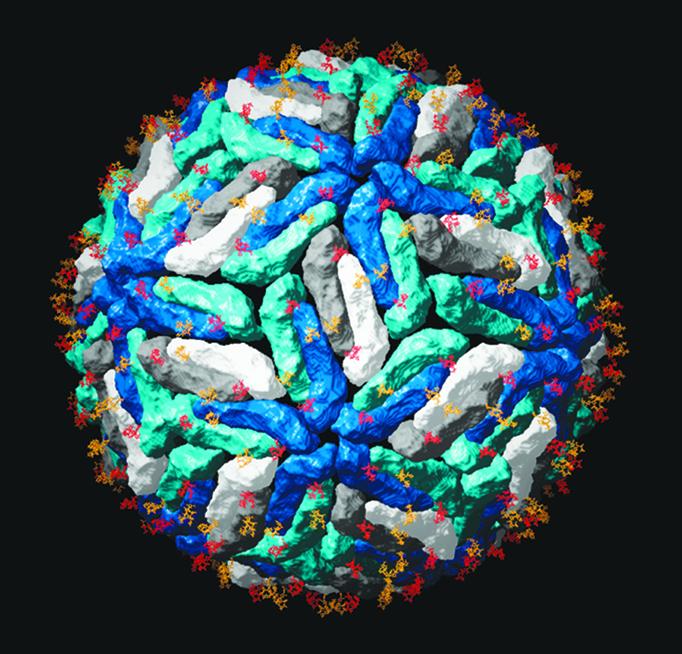
Carbohydrate distribution on the viral surface. The dengue virus E protein was superposed on the Protein Data Bank ID code 1K4R coordinates, which correspond to the tick-borne encephalitis E protein ectodomain placed in the reconstruction of dengue virus obtained by cryoelectron microscopy (18). Each dengue E dimer is represented as in Fig. 1, with the dimers that lie at the icosahedral twofold axis in dark and light gray, and the dimers lying on local twofold axes in two shades of blue. The dark- and light-blue subunits form the five- and threefold contacts, respectively. The sugars are as indicated in Fig. 1, with the same color coding. The high-mannose glycans present at the surface of mosquito-grown virions have been identified as important for the interactions with DC-SIGN in the initial steps leading to entry. Figure prepared with the program ribbons (19).
In conclusion, the determination of the 3D structure of the dengue virus envelope protein will bring research efforts to fight dengue disease to a new phase. With this knowledge, both preventive and therapeutic approaches to counteract the spread of this pathogen can be tackled in a more rational way. In addition to its obvious applications in public health, the information obtained from the structure also sheds light on the fundamental processes of many other enveloped viruses, such as attachment of envelope proteins to receptors and their interactions with lipids to bring about membrane fusion.
Acknowledgments
I thank S. C. Harrison for making the coordinates of the dengue E protein available before publication for illustration purposes.
See companion article on page 6986.
References
- 1.Wilson, I. A., Skehel, J. J. & Wiley, D. C. (1981) Nature 289, 366–378. [DOI] [PubMed] [Google Scholar]
- 2.Rey, F. A., Heinz, F. X., Mandl, C., Kunz, C. & Harrison, S. C. (1995) Nature 375, 291–298. [DOI] [PubMed] [Google Scholar]
- 3.Rosenthal, P. B., Zhang, X., Formanowski, F., Fitz, W., Wong, C. H., Meier-Ewert, H., Skehel, J. J. & Wiley, D. C. (1998) Nature 396, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modis, Y., Ogata, S., Clements, D. & Harrison, S. C. (2003) Proc. Natl. Acad. Sci. USA 100, 6986–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monath, T. P. (1994) Proc. Natl. Acad. Sci. USA 91, 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke, D. S. & Monath, T. P. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 1043–1125.
- 7.Halstead, S. B. (1997) in Dengue and Dengue Hemorragic Fever, eds. Gubler, D. J. & Kuno, G. (Cab International, London), pp. 23–44.
- 8.Lescar, J., Roussel, A., Wien, M. W., Navaza, J., Fuller, S. D., Wengler, G. & Rey, F. A. (2001) Cell 105, 137–148. [DOI] [PubMed] [Google Scholar]
- 9.Schlesinger, S. & Schlesinger, M. J. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 895–916.
- 10.Lindenbach, B. D. & Rice, C. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 941–1042.
- 11.Weissenhorn, W., Dessen, A., Calder, L. J., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1999) Mol. Membr. Biol. 16, 3–9. [DOI] [PubMed] [Google Scholar]
- 12.Wilschut, J., Corver, J., Nieva, J. L., Bron, R., Moesby, L., Reddy, K. C. & Bittman, R. (1995) Mol. Membr. Biol. 12, 143–149. [DOI] [PubMed] [Google Scholar]
- 13.Kielian, M., Chatterjee, P. K., Gibbons, D. L. & Lu, Y. E. (2000) in Fusion of Biological Membranes and Related Problems, eds. Hilderson, H. & Fuller, S. D. (Plenum, New York), Vol. 34, pp. 409–455. [Google Scholar]
- 14.Koschinski, A., Wengler, G., Wengler, G. & Repp, H. (2003) J. Gen. Virol. 84, 1711–1721. [DOI] [PubMed] [Google Scholar]
- 15.Hung, S. L., Lee, P. L., Chen, H. W., Chen, L. K., Kao, C. L. & King, C. C. (1999) Virology 257, 156–167. [DOI] [PubMed] [Google Scholar]
- 16.Tassaneetrithep, B., Burgess, T. H., Granelli-Piperno, A., Trumpfheller, C., Finke, J., Sun, W., Eller, M. A., Pattanapanyasat, K., Sarasombath, S., Birx, D. L., et al. (2003) J. Exp. Med. 197, 823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro-Sanchez, E., Altmeyer, R., Mara, A., Schwartz, O., Fieschi, F., Virelizier, J. L., Arenzana-Seisdedos, F. & Desprès, P. (2003) EMBO Rep., in press. [DOI] [PMC free article] [PubMed]
- 18.Kuhn, R. J., Zhang, W., Rossman, M. G., Pletnev, S. V., Corver, J., Lenches, E., Jones, C. T., Mukhopadhyay, S., Chipman, P. R., Strauss, E. G., et al. (2002) Cell 108, 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carson, M. (1987) J. Mol. Graphics 5, 103–106. [Google Scholar]